Abstract
In outcrossing populations, “Haldane’s sieve” states that recessive beneficial alleles are less likely to fix than dominant ones, because they are less exposed to selection when rare. In contrast, selfing organisms are not subject to Haldane’s sieve and are more likely to fix recessive types than outcrossers, as selfing rapidly creates homozygotes, increasing overall selection acting on mutations. However, longer homozygous tracts in selfers also reduce the ability of recombination to create new genotypes. It is unclear how these two effects influence overall adaptation rates in partially selfing organisms. Here, we calculate the fixation probability of beneficial alleles if there is an existing selective sweep in the population. We consider both the potential loss of the second beneficial mutation if it has a weaker advantage than the first one, and the possible replacement of the initial allele if the second mutant is fitter. Overall, loss of weaker adaptive alleles during a first selective sweep has a larger impact on preventing fixation of both mutations in highly selfing organisms. Furthermore, the presence of linked mutations has two opposing effects on Haldane’s sieve. First, recessive mutants are disproportionally likely to be lost in outcrossers, so it is likelier that dominant mutations will fix. Second, with elevated rates of adaptive mutation, selective interference annuls the advantage in selfing organisms of not suffering from Haldane’s sieve; outcrossing organisms are more able to fix weak beneficial mutations of any dominance value. Overall, weakened recombination effects can greatly limit adaptation in selfing organisms.
Keywords: adaptation, selection interference, dominance, self-fertilization, recombination
SELF-FERTILIZATION—reproduction where both gametes arise from the same parent—frequently evolves from outcrossing species in nature. Self-fertilization is widespread in angiosperms (Igic and Kohn 2006), some groups of animals (Jarne and Auld 2006), and fungi (Billiard et al. 2011; Gioti et al. 2012). It confers an initial benefit to an individual’s fecundity, including up to a 50% transmission advantage (Fisher 1941) and reproductive assurance under mate limitation (Baker 1955, 1967; Pannell et al. 2015). Both factors should allow selfing organisms to rapidly spread upon invasion of new habitats, unless countered by high levels of inbreeding depression (Lande and Schemske 1985). However, empirical studies usually find that selfing lineages are a “dead end,” since back transitions to outcrossing are rare, and high extinction rates have been inferred from comparative studies of related selfing–outcrossing taxa (Igic et al. 2008; Goldberg et al. 2010; Wright and Barrett 2010; Wright et al. 2013).
Self-fertilization has therefore been posited to be detrimental in the long term. For an organism with selfing rate σ, the population has an inbreeding rate equivalent to Wright’s (1951) statistic. The effective population size is reduced by a factor of at least (Pollak 1987; Caballero and Hill 1992; Charlesworth 1992). Furthermore, the effective recombination rate is reduced in proportion to the inbreeding rate (Golding and Strobeck 1980; Nordborg 2000; Roze 2009). This joint reduction in both effective population size and recombination can lead to a decrease in the efficacy of selection. Deleterious mutations can therefore accumulate more rapidly in selfing organisms, leading to population extinction (Heller and Maynard Smith 1978; Lynch et al. 1995).
Whether this mechanism is a major cause of extinction of self-fertilizing species is still under debate (reviewed in Glémin and Galtier 2012; Igic and Busch 2013; Hartfield 2016). Some sister-species comparisons of selfing and outcrossing taxa reveal evidence of increased mutation accumulation in selfers, as demonstrated with either increased nonsynonymous-to-synonymous polymorphism ratio () or weaker codon usage bias. Conversely, analyses of divergence rates generally do not show evidence for relaxed selection. Part of the reason for this lack of evidence could arise due to recent transitions to selfing in most of these species, as explicitly demonstrated in Capsella rubella by Brandvain et al. (2013), thus leaving little time for mutation accumulation to act.
Less investigated is the idea that selfing reduces the ability of a species to adapt, especially to new environmental conditions, although it was the one initially formulated by Stebbins (1957). For adaptation at a single locus, selfing organisms are more likely than outcrossers to fix new recessive adaptive mutations (Haldane 1927; Charlesworth 1992) but are generally less efficient in adapting from standing variation (Glémin and Ronfort 2013). Yet the effect of adaptation at multiple loci in partially selfing organisms has received much less attention. Of particular interest is how the reduction in recombination efficacy in highly selfing organisms impedes the overall adaptation rate. A well-established phenomenon in low-recombining genomes is the “Hill–Robertson effect,” where the efficacy of selection acting on a focal mutation is reduced, due to simultaneous selection acting on linked loci (Hill and Robertson 1966; Charlesworth et al. 2009). Outcrossing can therefore break down these effects and unite beneficial mutations from different individuals into the same genome, greatly increasing the adaptation rate (Fisher 1930; Muller 1932; Felsenstein 1974; Otto and Barton 1997).
Historically, the effect of advantageous mutations on mating system evolution has been neglected, since most observable spontaneous mutations are deleterious in partial selfers (Slotte 2014), and the inbreeding depression they cause plays a central role in mating system evolution. Analyses using divergence data from the Arabidopsis genome show low numbers of genes exhibiting signatures of positive selection (Barrier et al. 2003; Clark et al. 2007; Slotte et al. 2010, 2011), and only ∼1% of genes have signatures of positive selection in Medicago truncatula (Paape et al. 2013). These analyses reflect broader findings that the proportion of adaptive substitutions in the coding regions of selfing plants is not significantly different from zero (Gossmann et al. 2010; Hough et al. 2013). However, widespread local adaptation to climate in Arabidopsis is observed (Fournier-Level et al. 2011; Hancock et al. 2011; Ågren et al. 2013), which is expected to leave a weaker signature on the genome at the species scale (Slotte 2014). However, the power to detect local selection can increase once demography and population structure are accounted for (Huber et al. 2014).
Finally, both outcrossing and selfing domesticated plant and crop species can also be used to demonstrate recent adaptation. Ronfort and Glémin (2013) showed how adaptive traits obtained from quantitative trait loci tended to be dominant in outcrossers and recessive in selfers, in line with “Haldane’s sieve” (where dominant mutants are more likely to fix than recessive types in outcrossers). Hence, while beneficial mutations may not be as frequent as deleterious alleles, evidence exists that they arise frequently enough to affect local adaptation and domestication in self-fertilizing species. Furthermore, due to the reduced effective recombination rate in selfers, adaptive alleles should interfere with a greater region of the genome than in outcrossing organisms.
Recently, Hartfield and Glémin (2014) investigated the effect of a linked deleterious mutation on a selective sweep. In contrast to single-locus results, Hartfield and Glémin (2014) demonstrated how weakly recessive beneficial alleles (i.e., those with h just ) can contribute more to fitness increases in outcrossers than in selfers, as they are less likely to fix deleterious alleles via hitchhiking. This model indicated how breaking apart selection interference at linked sites could provide an advantage to some degree of outcrossing, leading to mixed mating being the optimal evolutionary state. A multilocus simulation study by Kamran-Disfani and Agrawal (2014) verified that background selection impedes genome-wide adaptation rates in selfing organisms. These studies clearly showed how linkage to deleterious mutations can limit adaptation in selfers, yet it remains an open question to what extent multiple beneficial mutations interfere in highly selfing species.
This article extends previous analyses to consider how interference between beneficial mutations at linked sites affects their emergence in partially selfing species. Haploid two-locus analytical models of the Hill–Robertson effect are altered to take dominance and selfing into account, and then examined to quantify how adaptation is affected.
Outline of the Problem
General modeling approach
We wish to determine how the effect of existing beneficial mutations at linked loci impedes the fixation of novel adaptive alleles in partially selfing organisms. We consider two-locus models to ensure tractability. Notation used in this study is outlined in Table 1.
Table 1. Glossary of Notation.
| Symbol | Usage |
|---|---|
| Number of haplotypes in the population (of size N) | |
| Locus at which first, second beneficial allele arises | |
| r | Recombination rate between two loci |
| Fitness coefficients of original and new advantageous alleles | |
| Dominance coefficients of original and new advantageous alleles | |
| p | Frequency of first advantageous allele at time point |
| q | Frequency of second advantageous allele at time point (if ) |
| σ | Proportion of matings that are self-fertilizing |
| F | Wright’s (1951) inbreeding coefficient, |
| Probability of identity by descent at two loci (Roze 2009) | |
| Fixation probability of original and new allele if unaffected by linkage (Equation 1) | |
| Fixation probability of both mutants in absence of interference | |
| Actual fixation probability of both mutants, after accounting for interference | |
| Average over one or several selected substitutions at locus A | |
| Ratio of actual to noninterference double-allele fixation probability, | |
| Average R over one or several selected substitutions at locus A | |
| Fixation probability of second allele with wild-type background | |
| Fixation probability of haplotype carrying both sweeps | |
| τ | Time taken for first sweep to reach frequency p |
| T | Scaled time, |
| Average fixation probability of second allele if it does not replace the first sweep | |
| Probability that second sweep replaces first if | |
| Fixation probability of novel allele if appearing on neutral or already beneficial genetic background | |
| Relative selective advantage of second allele if residing on either beneficial or neutral background | |
| Relative selective advantage of recombinant haplotype carrying both alleles | |
| Difference in fixation probability between different backgrounds, | |
| φ | Scaled advantage of new beneficial allele, |
| ρ | Scaled recombination rate, |
| Rate of selected substitution at locus A | |
| Population rate of beneficial mutation at single locus, |
We assume a diploid population of fixed size N, so there are haplotypes present. Each haplotype consists of two liked loci with recombination occurring at a rate r between them. At a first locus A, consider a beneficial mutation with selective coefficient so the fitness of individuals carrying it is in heterozygote form and in homozygote form. Similarly at a linked locus B, the fitness of individuals carrying the beneficial mutation is in heterozygotes and in homozygotes. The wild-type alleles are denoted the four haplotypes are and We further assume that selection acts on genotypes at the diploid stage and that fitness between loci is additive. As an example, an individual composed of the genotype will have fitness
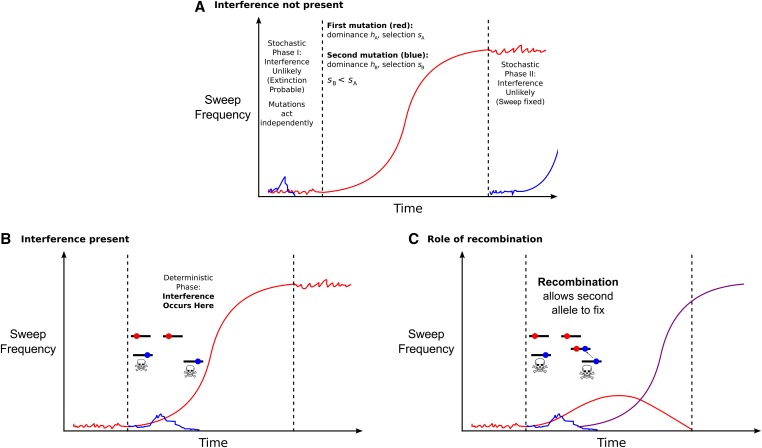
For strongly selected mutants (assuming ), the trajectory of a beneficial mutation can be decomposed into three parts. A schematic is shown in Figure 1. The stages are (i) an initial stochastic phase at low frequency where extinction by drift is likely; (ii) conditioned on escaping initial extinction (i.e., emergence), the allele increases in frequency on a quasi-deterministic trajectory; and (iii) a second stochastic phase at very high frequency where fixation is almost certain (Kaplan et al. 1989). If two mutations segregate simultaneously at low frequency in the stochastic zone, they do not influence each other and their fates can be assumed to be independent. However, as soon as one mutant has emerged and started to sweep quasi-deterministically it affects the fate of the other mutation. When considering a single unlinked mutation, once it has emerged its ultimate fixation is almost certain (which corresponds to the branching process approximation). The probability of fixation is thus equal to the probability of emergence. However, when two (or more) mutations interfere, a mutation that has emerged can be replaced by a linked competitor and ultimately lost, which is well known in asexual species as the “clonal interference” effect (Gerrish and Lenski 1998). If so, the probability of fixation can be lower than the probability of emergence, which can also be lowered by interference. Under tight linkage, or with a high selfing rate (as the loss of heterozygosity will reduce the effective recombination rate), this process has to be taken into account.
Figure 1.
Schematic of the model for the case i.e., the second mutant is weaker than the first. We assume here for simplicity. (A) The red line denotes the frequency of the initial beneficial mutation over time. When it is at low () and high [] frequencies, short-term changes in frequency are determined stochastically. Any linked beneficial alleles that appear during these phases will act independently as one genetic background dominates that the second mutant can appear on. (B) Once the first allele is sufficiently prevalent, it will increase in frequency over time in a regular manner (the “deterministic” phase). Any secondary alleles that appear during this phase will be affected by selection interference, so will be less likely to fix even if they do emerge. These alleles are represented by blue solid circles, with the trajectory shown by a blue line. (C) Interference can be broken down if recombination moves the blue allele onto the fitter background containing the red allele, and this new haplotype (whose frequency is shown by a purple line) emerges in the population.
We assume that mutation is the first to reach a high copy number and escape extinction by drift (although it could have been the second to arise), so is segregating in the population. Its trajectory can be modeled using deterministic equations. Without interference, the probability of fixation of the two mutations, is simply equal to the single-locus probability of fixation of the second mutation:
| (1) |
(Caballero and Hill 1992; Charlesworth 1992). can also be defined as the fixation probability of both alleles given that has already fixed in the population, and hence does not appear. Equation 1 leads to the classical result that the probability of fixation is higher under outcrossing than under selfing when More generally, the emergence of mutation depends on the genetic background it appears on and the switching rate between backgrounds through recombination, which is the cause of the Hill–Robertson effect we wish to model (Hill and Robertson 1966). Denoting the actual fixation probability of both mutants as we then define the degree of interference as the ratio measures to what extent fixation probability is reduced due to the presence of linked mutation. For example, means that the fixation probability of a linked mutant is 1/10th what it would be if unlinked, given the first allele is at frequency p. Hence it is a measure of how severe selection interference is at impeding fixation of subsequent beneficial mutations.
In the simplifying case the dynamics of how the second mutation emerges will differ depending on whether or vice versa. Hence a key parameter in the full model is the ratio If (), the fixation probability of the first mutation is not influenced by the second one and cannot be replaced. We thus need only to compute the emergence probability of the second mutation, which is certain to go extinct unless it appears on or recombines onto the background carrying the first beneficial mutation. Barton (1995) outlined a general model to calculate this effect for a haploid organism. We demonstrate how diploidy and selfing can be accounted for in that model and subsequently compute which is the contribution to arising from selection interference alone.
If (), then the second mutation can replace the first one if it arises on the wild-type background, and no successful recombinant occurs. We can calculate the probability of this effect by adjusting the analysis of Hartfield and Glémin (2014) to consider two beneficial mutations. We thus need to subtract from the probability that mutation replaces mutation once it has emerged, denoted by In the general case, the degree of interference is given by
| (2) |
In practice, instead of calculating we instead measure This is the R quantity for mutation arising when mutation is at a given frequency p, averaged over the whole possible origin times (or allele frequencies p) of the second mutation. Formal definitions of these conditions are presented below.
A simple first analysis: complete selfing vs. outcrossing with free recombination
Before deriving the full model, we can compare the two most extreme cases that can be investigated. Under outcrossing and free recombination, the fates of the two mutations are essentially independent. Hence the fixation probability of the second mutation, conditioned on the first one having emerged, is simply the single-locus probability of fixation given by Equation 1 with (Haldane 1927). At the other extreme with complete selfing (), recombination is suppressed so interference is maximized. Here, a second mutation can fix only if it appears on the same genetic background as the original beneficial allele, which is present at frequency p. Previous theory (Hartfield and Otto 2011; Hartfield and Glémin 2014) on emergence in this scenario gives the fixation probability of the double mutant as
| (3) |
See equation 7 of Hartfield and Glémin (2014) with and The mean probability of fixation of both alleles thus involves integrating Equation 3 over the entire sweep, assuming that the second mutation arises at a time that is uniformly distributed during the first sweep,
| (4) |
where τ is the duration of the first sweep. We can also solve Equation 4 over p from to if so we need to rescale the integral to remove time dependence,
| (5) |
where Solving in the limit of large population size [i.e., ] leads to (see Supplemental Material, File S2): full linkage reduces the emergence probability by one-half (). Intuitively, this can be explained by the fact that as population size increases, the deterministic phase of the first sweep becomes shorter compared to the initial and final stochastic phases [ vs. ] (Ewing et al. 2011). The second mutation thus occurs roughly half of the time during the initial stochastic phase where its probability of arising on the beneficial background, and hence of emerging, is very low ( in Equation 4). Alternatively, it can appear half of the time during the last stochastic phase where it almost always originates in the beneficial background and its probability of emerging is approximately ( in Equation 4).
By comparing this result to that with outcrossing and free recombination (), outcrossing is more able to fix both mutants if instead of without interference. However, the advantage to outcrossing may not be as high, since the true degree of inference depends on the strength of both mutations and the recombination rate. In addition, the degree of stochastic interference also depends on the flow of beneficial mutations, which depends on the mating system. We now turn to the full model to exactly quantify this effect.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Additional information on derivations are provided in File S1, and Mathematica File S2. Simulation code is available as File S3 and at http://github.com/MattHartfield/TwoAdvSelfSims.
Modeling Framework
Deriving the reduction in emergence probability due to interference
We first need to determine the reduction in the emergence probability of the second mutation when it arises, given the first one is at frequency p. We use branching process methods for calculating mutation emergence if acting over multiple genetic backgrounds. In a seminal article, Barton (1995) outlined how to calculate the emergence probability of a focal beneficial allele that changes between different backgrounds in a haploid population. If the probability of switching between backgrounds is of the same order as selection coefficients, s, and difference in emergence probability over backgrounds is of order Barton (1995) showed that the emergence probability of a novel beneficial allele in background i at time t, is a solution to the following differential equation:
| (6) |
The right-hand side of Equation 6 can be decomposed into three terms. The first one is a direct selection term: the fixation probability increases if the beneficial allele has a higher fitness advantage in background i, is the probability that offspring in background i move to background j per generation; in this case, this effect arises from recombination changing the genetic background of the focal allele. Finally, there is a negative term, denoting how genetic drift can cause the allele to go extinct. Note that the differential equation term is negative, since this system of equations is considered going back in time. Barton (1995) subsequently used this framework to investigate the fixation probability of a second beneficial allele, given that it arises in close linkage to an existing sweep.
Our goal is to derive equivalent equations for a diploid, self-fertilizing population. In File S1 and File S2, we show how similar equations can be derived for outcrossing species. However, when selfing is included, then the full system of equations becomes unwieldy. To obtain analytical solutions, we proceed by using a separation-of-timescales argument. When recombination is low, as assumed in Equation 6, haplotypes carrying either the first beneficial allele or the wild type quickly reach their genotypic equilibria with inbreeding (Wright 1951); specifically, we assume that equilibrium is reached between recombination events. If selection acts on genotypes, we can use the relative fitness advantage of each genotype to determine that of the second beneficial allele, depending on the haplotype it appears on. We can then create a variant of Equation 6 with these steady-state values. Using this argument, we obtain a tractable form for the transition probabilities between backgrounds (File S1).
With high selfing rates, Equation 6 remains valid for any recombination rate as the probability of moving from one background to another can be low. But as sweep effects can span large genetic map distances under high selfing, it is important to analyze this special case properly. Roze (2009, 2015) derived the equilibrium genotype frequencies at two loci under partial selfing as a function of the probability of identity by descent at a single locus, F, and at two loci, This approach takes into account correlations of homozygosity between linked loci. In File S1 and File S2, we show that Φ equilibrates as quickly as F, so the separation-of-timescales argument can be used for any recombination level, assuming that two-locus equilibrium genotype frequencies for given selfing and recombination rates are instantaneously reached, compared to change in allele frequencies. This approximation should work well at equilibrium, but not necessarily in nonequilibrium conditions such as during selective sweeps. However, because equilibrium values are quickly reached, these calculations are also accurate under more general conditions (see simulation comparisons in the Results section).
To complete the model, we rederive each component of Equation 6 in turn to account for diploidy and selfing.
Direct selection of the second allele:
Let denote the probability that the new allele fixes, given that it arises in linkage with the existing mutant (which is at frequency p). is the fixation probability if the second allele appears on the wild-type (neutral) background. We further denote the relative selective advantage of each haplotype (containing either both advantageous alleles or the second allele only) by and These are derived by calculating the frequency of each potential haplotype background that the second allele appears on and the relative proportions of each. Calculations are outlined in File S1:
| (7) |
| (8) |
Note that Equations 7 and 8 are the same for both the general and the low recombination cases. As in Barton (1995) the trajectory of the first mutation is assumed to be deterministic, and hence described by the following differential equation:
| (9) |
Furthermore we can rescale time by selection, i.e., setting (Barton 1995).
Rescaling recombination:
Nordborg (2000) used a coalescent argument to show that in selfing populations, when recombination is of order the recombination rate is scaled by a factor (see also Golding and Strobeck 1980). This arises as a proportion F of recombination events are instantly “repaired” due to selfing. We show in File S2 that the same result can be obtained by considering the decay of linkage disequilibrium in an infinite population with the less restrictive condition of r being small () but not necessarily However, this scaling breaks down for high recombination and selfing rates, as it becomes likely that recombination occurs between genotypes within individual lineages (Padhukasahasram et al. 2008). Relaxing the assumption of low recombination, we can derive a more exact rescaling term following Roze (2009, 2015): which reduces to the coalescent rescaling with low recombination. The equilibrium value for Φ can then be used (see Equation A8 in File S1 and Roze and Lenormand 2005; Roze 2009). Furthermore, in File S2 we show that the error will be at most a factor with high recombination. Given that the effective recombination rate will remain small with high selfing rates irrespective of the scaling term, using the coalescent rescaling should capture the average fixation probability, but more accurate expression can be obtained using instead.
Creating the system of equations:
Hence the system of equations in Barton (1995) is modified to
| (10) |
| (11) |
Equations 10 and 11 account for the relative selective advantage of the second allele, θ (with p verifying Equation 9), and the decrease in effective recombination rate [or for greater accuracy]; and the terms are scaled by due to an increase in genetic drift (Pollak 1987; Caballero and Hill 1992; Charlesworth 1992).
Calculating
We next follow the approach of Barton (1995) and investigate the average fixation probability over haplotypes given the first beneficial allele is at a certain frequency, defined as and the difference in emergence probability between the backgrounds, These terms are scaled by the probability of fixation of the second allele if unlinked, so Π lies between 0 and 1. We also introduce the rescaled parameters and to determine how the relative selective strengths and recombination rates affect interference. We thus obtain
| (12) |
| (13) |
where and Note that if we use the more exact recombination rescaling term it is not possible to fully factor out from Equations 10 and 11 as Φ is a function of r.
For a given time of origin of the second mutation, t, the joint solution of this system and Equations 9, 12, and 13 gives These equations must be solved numerically by, e.g., using the “NDSolve” function in Mathematica (Wolfram Research 2014). Alternatively, to remove the time dependence and directly obtain we can divide both Equations 12 and 13 by (Equation 9). Boundary conditions can be found by looking at the behavior of the system as or In this case, we observe that reflective of the fact that as the first mutation fixes, the second allele is certain to arise alongside it. Hence the second allele’s fixation probability is not reduced. Boundary conditions for Δ can be calculated by assuming (as used in Barton 1995) and as In this case Δ tends to which reflects the probability that the second allele can recombine onto the fitter background if appearing on a wild-type chromosome; otherwise it is guaranteed to be lost (Barton 1995). Although this condition assumes small φ, the system of equations appears to work well even with larger φ when compared to simulations.
Integration over the sweep trajectory
To obtain the average effect of interference we need to consider all possible origins of the second mutation. The average R for mutation arising uniformly in a long time interval spanning the sojourn of mutation is given by
| (14) |
As previously shown by Barton (1995), can be approximated by
| (15) |
Integration from very ancient time (equivalently, a frequency ) reflects the fact that mutation affects the fate of mutation even if it appears afterward, when mutation is still at a low frequency in the stochastic zone. Hence it is not obvious if a natural choice for is the sojourn time of the first mutation. Moreover, because selfing and dominance affect the fixation time of alleles (Glémin 2012), averaging over this time would not allow direct comparison between different selfing rates and dominance levels. For example, the effect of linked mutation is expected to be stronger under selfing than under outcrossing but the time interval when interference can occur is shorter. Finally, interference also depends on the substitution rate at locus A, which is also affected by selfing and dominance. All these effects can be taken into account by assuming a steady state of substitutions at a low rate at locus A (i.e., no multiple substitutions). The rate of selected substitution at locus A is given by
| (16) |
where time is measured in generations. Following Barton (1995) we use
| (17) |
The justification is as follows. The waiting time between two sweeps is exponentially distributed with mean If interference between mutations and thus occurs for a proportion of time On average, the effect of on is thus
| (18) |
leading to Equation 17.
Deriving the probability of sweep replacement,
The previous analysis focused primarily on the case where the second mutant is weaker than the first, so However, if selection acting on is sufficiently strong, then it is possible that replaces so only fixes. We need to calculate the probability of this replacement occurring and subtract it from the baseline reduction This probability can be calculated by altering the model of Hartfield and Glémin (2014), which investigated a deleterious allele hitchhiking with a sweep. In our case, the “deleterious” allele is the wild-type allele at the first locus and the “advantageous” allele is the second fitter sweep Hartfield and Otto (2011) implemented a similar rescaling for a haploid model, while Yu and Etheridge (2010) provided a general stochastic algorithm for investigating this behavior.
A fuller derivation is included in File S1. By calculating from first principles, we can infer that
| (19) |
where and are given by Equations 12 and 13. Equation 6 of Hartfield and Glémin (2014) is then used to calculate
| (20) |
for where q is the frequency of allele As above, can be used instead of to create more precise expressions. is the emergence probability of the haplotype carrying both beneficial alleles () if it formed by recombination. It is the solution of the following equation, where is the relative fitness of this haplotype:
| (21) |
The mean interference effect is similar to Equation 17. However, contrary to emergence, the replacement of mutation by mutation can occur only if mutation arises when mutation has already emerged, that is, for This condition is a bit too restrictive because we should also consider the case when mutation arises after but emerges before mutation Moreover, the distribution of should be used instead of the average value. However, these complications have only minor quantitative effects (not shown). If considering replacement, Equation 17 is written as
| (22) |
Simulations
We tested the accuracy of analytical solutions by comparing them to stochastic simulations written in C; code is available in File S3 and online from http://github.com/MattHartfield/TwoAdvSelfSims. When measuring the first allele was seeded at initial frequency p; given this frequency and selfing rate σ, the proportions of mutant homozygotes, heterozygotes, and wild-type homozygotes were calculated based on standard equations with inbreeding (Wright 1951). The second allele was subsequently introduced onto a random background with frequency (i.e., as a single copy). Frequencies of each genotype were altered deterministically by a factor due to selection, where is the fitness of each genotype and is the population mean fitness. Recursion equations derived by Hedrick (1980, equation 3) then calculated how genotype frequencies changed due to partial selfing. A life cycle was completed by resampling N genotypes from a multinomial distribution to implement random drift. The second allele was tracked until one haplotype fixed, with the simulation repeated until 5000 fixations of the second beneficial allele occurred. It was noted how often each haplotype fixed; from these data we subsequently calculated the second allele fixation probability, relative to the expected result without interference. When measuring we instead measured how often the haplotype carrying solely the fitter mutant fixed. Confidence intervals were calculated using the method of Agresti and Coull (1998).
Results
Validating the analytical approach
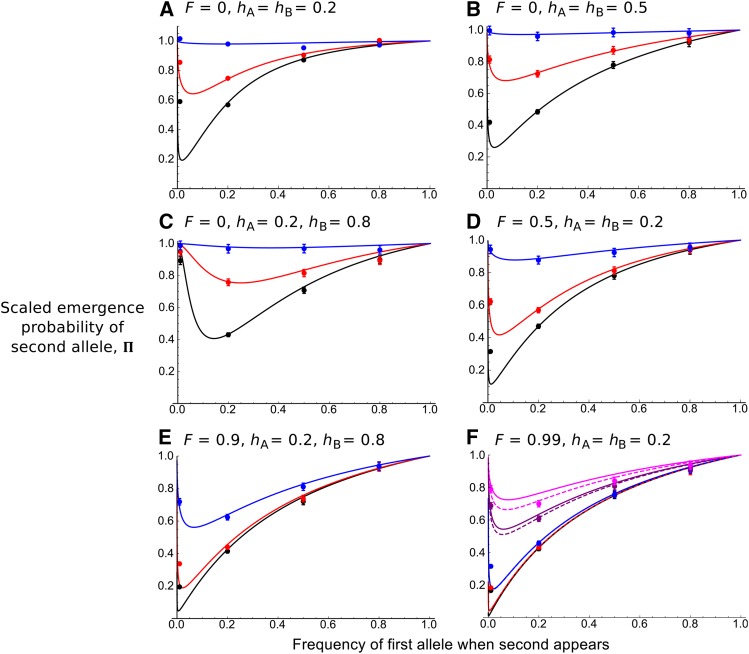
Testing
We first tested the accuracy of as given by Equation 12, with stochastic simulations. A subset of comparisons is shown in Figure 2; fuller comparisons are given in File S2. We see that on the whole, the analytical solutions provide an accurate match with simulations for a wide variety of selfing and dominance values. These include cases of high selfing and recombination, despite our model assuming low recombination. Nevertheless, simulation results slightly underestimate the original analytical solutions with very high values (e.g., see Figure 2F). Using the more exact rescaled recombination rate, improves the fit. Some inaccuracies are also apparent if the first allele is at a low frequency when the second one appears (). This discrepancy likely arises due to the trajectory of the first beneficial allele not being completely deterministic when starting at low frequency.
Figure 2.
Probability of fixation of the second allele relative to the unlinked case, as a function of the first allele frequency, p. = 0.01, = 0.005 (so ), and from bottom to top in A–E, 0.001, 0.01 (corresponding to 0.1, 1). In F, and 0.25 results are also added (corresponding to and 25). Other parameters used are listed above each subplot. Curves correspond to solutions provided by the analytical system of differential equations (Equation 12), rescaled so they are a function of p. In F, dashed lines are analytical results using the more exact recombination rate, Points correspond to 5000 stochastic simulations for which the second beneficial allele has fixed. Bars represent 95% confidence intervals; if the intervals cannot be seen, they lie within the plotted points. Note that in A, simulation results are presented after rescaling fixation probability by the diffusion equation solution, to account for recessive alleles (see main text for details).
In addition, if the second allele is highly recessive where there is outcrossing (), the simulated scaled allele fixation probability can be higher than in single-locus models. This is simply because the fixation probability of recessive beneficial mutants is underestimated using the branching-process solution without considering homozygote genotypes (Equation 1, which holds for highly recessive alleles only in very large population sizes, i.e., at least ). For smaller population sizes a diffusion-equation solution, offers the correct baseline emergence probability (Caballero and Hill 1992). Hence rescaling the simulations by this solution, (instead of where is given by Equation 1) causes simulations to match with analytical solutions.
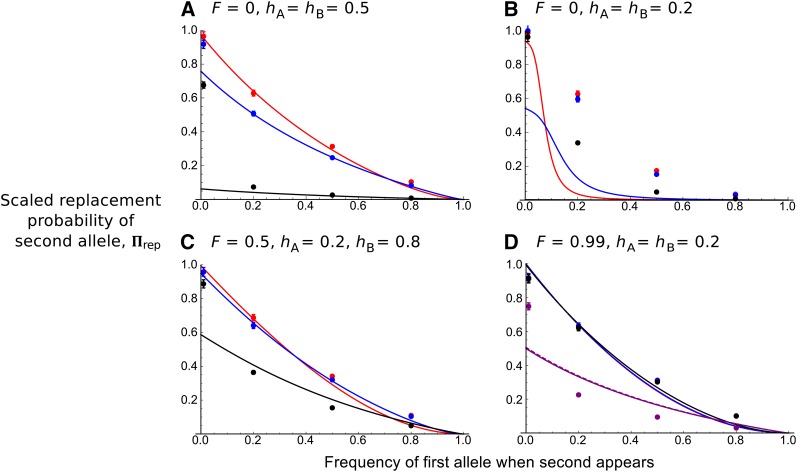
Testing
Figure 3 shows the estimate of compared to simulation data if and the first sweep is at frequency p. Generally, if the first mutation is not recessive, recombination is low (and/or selfing high), and p is not too low (), then the analytical solution matches well with simulations. The fit is not improved using the recombination term, since the unscaled recombination rate remains low (). However, if recombination increases ( approaches 1) and mutations are recessive, then the actual replacement probability can be underestimated (for example, with Figure 3B). By tracking the frequencies of individual haplotypes over time, we can determine that in cases where the model fails, it is because two key assumptions are violated (File S2). In particular, we assumed that recombination occurs only between haplotypes carrying one of the beneficial alleles. But in this case, the wild-type haplotype is not rapidly eliminated. Hence only a fraction of recombination events occur between haplotypes carrying beneficial alleles, and Equation 20 would overestimate the effect of recombination. This error would not be large if net recombination is low. Furthermore, the first beneficial allele does not increase in frequency at the start of the process. This behavior violates the assumption that it will compete with the second allele. These modeling violations are also observed if both alleles are dominant in outcrossing populations ( see File S2).
Figure 3.
(A–D) Probability that a second beneficial allele with advantage replaces an existing sweep with selective advantage where as a function of the first sweep frequency p when the second sweep appears. and (red), 0.1 (blue), 1 (black), or 100 (purple, D only). In D, dashed lines are analytical results using the more exact recombination rate, Other parameters are indicated above each panel. Points correspond to 5000 stochastic simulations for which the second beneficial allele has fixed. If confidence intervals cannot be seen, they lie within the plotted points.
To calculate a more accurate replacement probability in this case, it would be necessary to explicitly account for the frequency of the neutral haplotype (carrying no beneficial alleles). In addition, it would be desirable to explicitly track drift effects as recessive beneficial mutations are present at a low frequency. Unfortunately it will probably be unfeasible to produce tractable analytical solutions in either scenario. Hence in subsequent analyses when we will focus on codominant or dominant mutations ().
Codominant case
Under codominance (), selfing has no effect on the single-locus fixation probabilities. Here, we can therefore analyze the effect of selfing on recombination only. Moreover, for this specific case, results can be directly obtained by rescaling haploid models. Equations 12 and 13 become
| (23) |
| (24) |
where Equations 23 and 24 are similar to Barton’s (1995) equations 6a and 6b for haploids, except (i) with terms in the denominator since our equations are as a function of the first sweep frequency and (ii) that the recombination rate is decreased by The latter scaling reflects how the population size is increased by a factor of 2 in diploids compared to haploids; how inbreeding magnifies drift by a factor increasing the speed at which the first sweep fixes and reducing the potential for recombination to act; and how the effective recombination rate is reduced by (Caballero and Hill 1992). Here, we can use the approximations given by equations 8 and 9a of Barton (1995) with the appropriate rescaling to find simplified forms for the total reduction in the relative fixation probability:
| (25) |
| (26) |
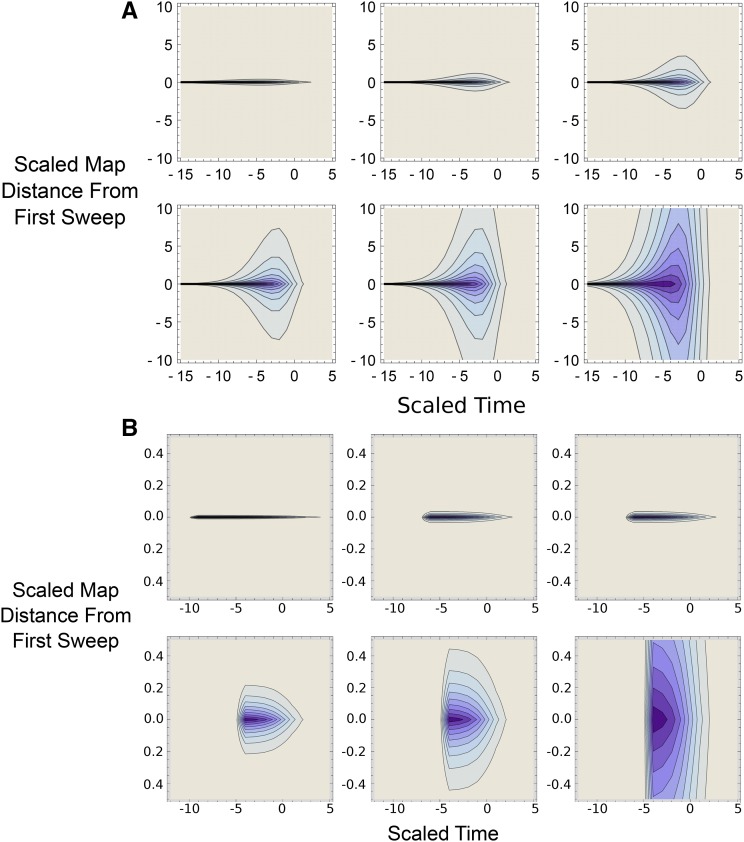
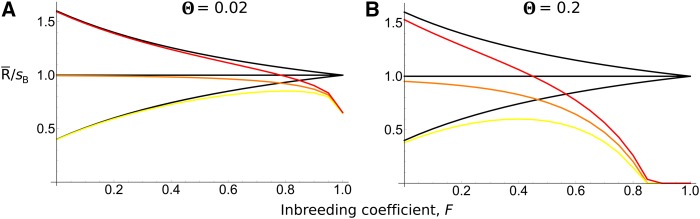
Approximations for the replacement probability can also be obtained (see details in File S1). Quantitative inspection of previous equations shows that the emergence effect (or “stochastic interference” effect) is more important than the replacement effect (Figure 4). The emergence effect is higher for low φ values and can be very high; Equation 25 tends to infinity when φ or tends toward 0. On the contrary, for the replacement case tends toward a small, finite value as φ tends toward infinity. This difference appears because (i) mutations are more sensitive to interference in the stochastic zone than once they have emerged and (ii) selection interference is more pronounced when than when (and the second allele can replace the first one). Consequently, the effect of selfing is more important for low φ values when emergence is the most important process than for high φ values when replacement predominates, as illustrated in Figure 4. Figure 4 also illustrates how the effect of a sweep can extend across long chromosome tracts with high selfing rates.
Figure 4.
(A and B) Contour plots showing degree of interference, measured by Equation 2 with Π defined by Equation 23 (for ) and defined by Equation 15 in File S1 (for ). Both beneficial mutations are additive (). In both A and B, darker colors indicate higher degree of interference (with the darkest representing R approaching 0). x-axis denotes time of the sweep (with the sweep reaching 50% frequency at ); y-axis is the map distance from the first sweep (scaled to ). Top rows of plots are for F values of 0, 0.5, and 0.8, respectively; bottom rows are F values of 0.9, 0.95, and 0.99. Other parameters are and (A) so or (B) so Note different y-axis scaling for A and B.
In previous equations, the scaling factor arises because the length of the sweep is in but we also scaled time by to conserve the same scaling for any selfing rate. Equations 25 and 26 demonstrate the two opposite effects of selfing: the reduction in effective recombination reduces the probability of emergence and also increases the replacement probability but over a shorter period of time as alleles fix more quickly (Glémin 2012). For loose linkage, the effect of selfing on recombination is stronger so that selfing globally decreases the probability of fixation. However, for tight linkage interference occurs for any selfing rate, such that the dominant effect of selfing is the reduction in sweep length (Figure A2 in File S1). Boundary conditions can be found for the two extreme cases when φ is either small or large. When replacement is more likely under outcrossing than under complete selfing if (see File S2). When emergence of the second beneficial allele is more likely under selfing than under outcrossing only for very tight linkage, that is for where ϵ is the residual outcrossing rate under selfing (see File S2).
Effect of dominance on interference
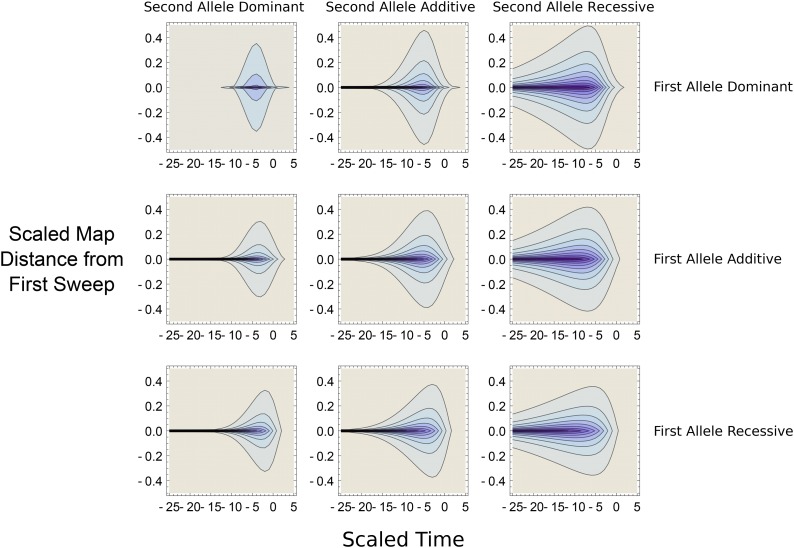
For high selfing rates, the interference process is well approximated by the additive case. However, to get a complete picture of the effect of selfing we need to analyze how dominance affects the process. We first consider outcrossing populations before turning to the effect of mating system on the adaptation rate. When investigating dominance, two questions arise. Which kind of mutations cause the strongest interference? And which ones are the most sensitive to interference and are hence more likely to be lost?
The effects of interference for different combinations of dominance levels are presented in Figure 5 for The main difference in sweep dynamics arises from the length of the two stochastic phases. Because a mutation causes interference mainly during its deterministic trajectory, which is similar for any dominance level for any (Ewing et al. 2011), the dominance level of mutation has thus only a weak effect on the emergence probability of mutation However, the sensitivity of mutation to interference strongly depends on its dominance level, as it depends on the length of its initial stochastic phase, which is (Ewing et al. 2011). Recessive mutations are thus more sensitive to interference than additive and dominant ones. Interference thus reinforces Haldane’s sieve, in the sense that recessive mutations are even less likely to emerge in outcrossing populations if tightly linked to the initial mutation.
Figure 5.
Contour plots showing degree of interference, as measured by Equation 2 with Π defined by Equation 23 and (as ), for different dominance values. In all panels, darker colors indicate higher degree of interference (R approaching 0); x-axis denotes time of the sweep (with the sweep reaching 50% frequency at ); y-axis is the map distance from the first sweep (scaled to ). Labels denote the dominance value of the first and the second mutation, with recessive mutants having additive mutations dominant mutations Other parameters are and so
In the case of strong interference, this effect can be substantial as illustrated in Figure 5. Interestingly, this result is not symmetrical since dominant mutations exhibit only slightly less interference than additive mutations. As far as we know this outcome has not been described before and leads to the prediction that the dominance spectrum of fixed beneficial mutations (i.e., the expected density of dominance values observed in fixed alleles) should vary with recombination rates (Figure A3 in File S1). Recessive mutations seldom fix in low-recombination regions, as they are present at low frequencies for longer periods of time than dominant alleles.
Conditions under which selection is more efficient under outcrossing than under selfing
We now have all the ingredients to study the range of conditions under which selfing reduces the adaptation rate. Without interference and other factors increasing drift in selfers, selfing reduces (respectively increases) adaptation from new dominant (respectively recessive) mutations (Caballero and Hill 1992; Charlesworth 1992). How does interference affect this behavior? This question can be explored by considering a steady flow of mutations and analyzing where is given by Equation 17. As shown in File S2, the total effect of interference on replacement will be no more than of the order of (which is always lower than a few tens) while the effect on emergence can be much more important. In what follows we therefore focus on the case where
Figure 6 illustrates how selfing can affect the probability of fixation of the second mutation compared to the single-locus case. Under a low adaptation regime ( for the per-locus beneficial mutation rate) interference is weak and the probability of fixation is reduced only in highly selfing species. This reduction is moderate and selfing species are still better than outcrossers at fixing recessive mutations. Under a stronger adaptation regime (), interference can be substantial even in mixed-mating species and adaptation can be fully impeded in highly selfing species if
| (27) |
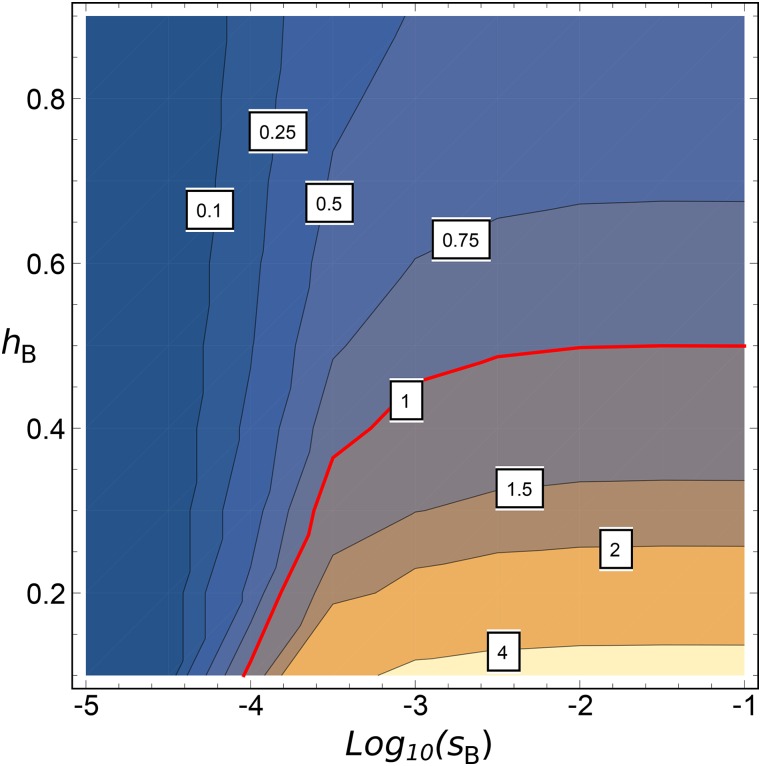
(see Barton 1995). This threshold depends on φ, which means that, even under a low adaptation regime, weakly beneficial mutations can be affected by interference in highly selfing species. Figure 7 shows the joint dominance and selection spectrum for which selection is more efficient in outcrossing than in highly selfing () species. Strongly beneficial mutations are very weakly affected by interference, so only dominant mutations are more efficiently selected in outcrossing than in selfing species. However, (very) weak beneficial mutations are better fixed in outcrossing populations, whatever their dominance level.
Figure 6.
(A and B) Plots of the total effect of interference, as defined using Equation 17, as a function of F. The y-axis is the probability of emergence scaled to the expected emergence probability with There is a continual rate of mutation (A) or 0.2 (B). and () and (yellow line), 0.5 (orange line), or 0.8 (red). Black lines show expected fixation probability in the absence of interference.
Figure 7.
Contour plot of the ratio of (Equation 17) for and as a function of (on a scale) and Values less than 1 indicate that outcrossers have the higher fixation probability, and values greater than 1 indicate that populations have the higher probability. Other parameters are and
Discussion
Interference between beneficial mutations with partial selfing and dominance
Multilocus models of adaptation in partial self-fertilizing species can inform on how the interplay between homozygote creation and reduction in recombination jointly affects selection acting on multiple sites. It is already known that the presence of linked deleterious variation means that mildly recessive beneficial mutations (h just less than 1/2) are more able to fix in outcrossers than in selfing organisms by recombining away from the deleterious allele, in contrast to single-locus theory (Hartfield and Glémin 2014). More generally, genome-wide background selection can substantially reduce adaptation in highly selfing species (Kamran-Disfani and Agrawal 2014). Yet the extent that linkage between beneficial mutations affects mating-system evolution remains poorly known.
Here we extended several previous models of selection interference to consider how adaptation is impeded in partially selfing organisms. We considered two possibilities. First, given that an existing sweep is progressing through the population, subsequent mutations confer a lesser selective advantage and can fix only if recombining onto the fitter genetic background (the “emergence” effect). Alternatively, a second mutant could be fitter and replace the existing sweep, unless recombination unites the two alleles (the “replacement” effect). We found that the emergence effect is generally stronger than the replacement effect and is more likely to lead to loss of beneficial mutations (Figure 4).
Furthermore, selection interference has two opposite effects on Haldane’s sieve. In mainly outcrossing populations (where it operates), Haldane’s sieve is reinforced because recessive mutations are even more likely to be lost when rare compared to dominant ones, compared to single-locus results. However, when comparing different mating systems, interference reduces or nullifies the advantage of selfing of not being affected by Haldane’s sieve. Consequently, weakly beneficial mutations are more likely to be fixed in outcrossers, irrespective of their dominance level (Figure 7). These findings thus contribute to a body of literature as to when the predictions of Haldane’s sieve should break down or otherwise be altered. Other examples include the fixation probability of mutations being independent from dominance if arising from previously deleterious variation (Orr and Betancourt 2001); more generally, outcrossers are more able to fix mutations with any dominance level compared to selfers if arising from standing variation and when multiple linked deleterious variants are present (Glémin and Ronfort 2013). Conversely, dominant mutations can be lost in metapopulations due to strong drift effects (Pannell et al. 2005).
In our model we assumed that no more than two beneficial mutations simultaneously interfere in the population. However, even if mutation does not occur frequently enough to lead to multiple mutations interfering under outcrossing, the presence of a few sweeping mutations throughout a genome can jointly interfere in highly selfing species. Obtaining a general model of multiple substitutions in a diploid partially selfing population is a difficult task, but it is likely that the rate of adaptation would be further reduced compared to the two-locus predictions (as found in haploid populations by Weissman and Barton 2012).
It is also of interest to ask whether our calculations hold with different types of inbreeding (such as sib mating). For a single unlinked mutant, Caballero and Hill (1992) showed how various inbreeding regimes determine the value of F used in calculating fixation probabilities (Equation 1). However, it is unclear how effective recombination rates will be affected. For example, Nordborg’s (2000) rescaling argument relies on the proportion of recombination events that are instantly repaired by direct self-fertilization; these dynamics would surely be different under alternative inbreeding scenarios. Further work would be necessary to determine how other types of inbreeding affect net recombination rates and thus the ability for selection interference to be broken down.
Causes of limits to adaptation in selfing species
We have already shown in a previous article how adaptation can be impeded in low-recombining selfing species due to the hitchhiking of linked deleterious mutations (Hartfield and Glémin 2014), with Kamran-Disfani and Agrawal (2014) demonstrating that background selection can also greatly limit adaptation. Hence the question arises of whether deleterious mutations or multiple sweeps are more likely to impede overall adaptation rates in selfing species.
Background selection due to strongly selected deleterious alleles causes a general reduction in variation across the genome by reducing (Nordborg et al. 1996); here the overall reduction in emergence probability is proportional to where is mediated by the strength and rate of deleterious mutations (Barton 1995; Johnson and Barton 2002) and thus affects all mutations in the same way [note that this process becomes more complicated with weaker deleterious mutations (McVean and Charlesworth 2000)]. Because of background selection, selfing is thus expected to globally reduce adaptation without affecting the spectrum of fixed mutations. Similarly, adaptation from standing variation, which depends on polymorphism level, is expected to be affected by the same proportion (Glémin and Ronfort 2013). Alternatively, interference between beneficial mutations is mediated by φ, the ratio of the selection coefficients of the sweeps. For a given selective effect at locus A, weak mutations at locus B are thus more affected by interference than stronger ones, and the net effect of interference cannot be summarized by a single change in (Barton 1995; Weissman and Barton 2012). Because of selective interference, selfing is also expected to shift the spectrum of fixed mutations toward those of strong effects. Interestingly, Weissman and Barton (2012) showed that neutral polymorphism can be significantly reduced by multiple sweeps, even if they do not interfere among themselves. This suggests that in selfing species, adaptation from standing variation should be more limited than predicted by single-locus theory (Glémin and Ronfort 2013). Selective interference could thus affect both the number and the type of adaptations observed in selfing species.
Reflecting on this logic, both processes should interact and we therefore predict that background selection will have a diminishing-returns effect. As background selection lowers then the substitution rate of beneficial mutations will be reduced (since it is proportional to for μ the per-site mutation rate), and hence interference between beneficial mutations will subsequently be alleviated. No such respite will be available with a higher adaptive mutation rate; on the contrary, interference will increase (Figure 6). Impediment of adaptive alleles should play a strong role in reducing the fitness of selfing species, causing them to be an evolutionary dead end. Further theoretical work teasing apart these effects would be desirable. Given the complexity of such analyses, simulation studies similar to those of Kamran-Disfani and Agrawal (2014) would be a useful approach to answering this question.
In a recent study, Lande and Porcher (2015) demonstrated that once the selfing rate became critically high, selfing organisms then purged a large amount of quantitative trait variation, limiting their ability to respond to selection in a changing environment. This mechanism provides an alternative basis as to how selfing organisms are an evolutionary dead end. However, they consider only populations at equilibrium; our results suggest that directional selection should further reduce quantitative genetic variation due to selective interference among mutations. Subsequent theoretical work is needed to determine the impact of interference via sweeps on the loss of quantitative variation. Furthermore, complex organisms (i.e., those where many loci underlie phenotypic selection) are less likely to adapt to a moving optimum compared to when only a few traits are under selection (Matuszewski et al. 2014) and can also purge genetic variance for lower selfing rates (Lande and Porcher 2015). Complex selfing organisms should thus be less able to adapt to environmental changes.
Empirical implications
The models derived here lead to several testable predictions for the rate of adaptation between selfing and outcrossing sister species. These include an overall reduction in the adaptive substitution rate in selfing populations; a shift in the distribution of fitness effects in selfing organisms to include only strongly selected mutations that escape interference; and a difference in the dominance spectrum of adaptive mutations in outcrossers compared to selfers, as already predicted by single-locus theory (Charlesworth 1992) and observed with quantitative trait loci (QTL) for domesticated crops (Ronfort and Glémin 2013).
Few studies currently exist that directly compare adaptation rates and potential between related selfing and outcrossing species, but they are in agreement with the predictions of the model. In plants, the self-incompatible C. grandiflora exhibited much higher adaptation rates [where of nonsynonymous substations were estimated to be driven by positive selection, using the McDonald–Kreitman statistic (Slotte et al. (2010)] than in the related selfing species Arabidopsis thaliana (where α is not significantly different from zero). Similarly, the outcrossing snail Physa acuta exhibited significant adaptation rates (), while no evidence for adaptation in the selfing snail was obtained (Burgarella et al. 2015); in fact, evidence suggests that deleterious mutations segregate due to drift (). In agreement with the predicted inefficacy of selection on weak mutations, Qiu et al. (2011) also observed significantly lower selection on codon usage in the Capsella and Arabidopsis selfers than in their outcrossing sister species.
In addition, as only strong advantageous mutations are expected to escape loss through selection interference, this result can explain why selective sweeps covering large tracts of a genome are commonly observed, as with A. thaliana (Long et al. 2013) and Caenorhabditis elegans (Andersen et al. 2012). Extended sweep signatures can also be explained by reduced effective recombination rates in selfing genomes. Finally, selective interference between beneficial mutations could explain why maladaptive QTL are observed as underlying fitness components, as detected in A. thaliana (Ågren et al. 2013). Direct QTL comparisons between selfing and outcrossing sister species would therefore be desirable to determine to what extent selection interference leads to maladaptation in selfing species.
Acknowledgments
We thank Joachim Hermisson, Denis Roze, and an anonymous reviewer for their very helpful comments on the manuscript. M.H. was funded by an Action Thématique et Incitative sur Programme (ATIP)-Avenir grant from the Centre National de la Recherche Scientifique (CNRS) and Institut National de la Santé et de la Recherche Médicale (INSERM) to Samuel Alizon and a Marie Curie International Outgoing Fellowship grant MC-IOF-622936 (project SEXSEL) and also acknowledges additional support from the CNRS and the Institut de Recherche pour le Développement (IRD). S.G. is supported by the French CNRS and a Marie Curie Intra-European Fellowship, grant IEF-623486 (project SELFADAPT). This work was also supported by two grants from the Agence Nationale de la Recherche (TRANS, ANR-11-BSV7-013-03; and SEAD, ANR-13-ADAP-0011).
Footnotes
Communicating editor: J. Hermisson
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188821/-/DC1.
Literature Cited
- Ågren J., Oakley C. G., McKay J. K., Lovell J. T., Schemske D. W., 2013. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 110: 21077–21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti A., Coull B. A., 1998. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52: 119–126. [Google Scholar]
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker H. G., 1955. Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution 9: 347–349. [Google Scholar]
- Baker H. G., 1967. Support for Baker’s law–as a rule. Evolution 21: 853–856. [DOI] [PubMed] [Google Scholar]
- Barrier M., Bustamante C. D., Yu J., Purugganan M. D., 2003. Selection on rapidly evolving proteins in the Arabidopsis genome. Genetics 163: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1995. Linkage and the limits to natural selection. Genetics 140: 821–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard S., López-Villavicencio M., Devier B., Hood M. E., Fairhead C., et al. , 2011. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol. Rev. Camb. Philos. Soc. 86: 421–442. [DOI] [PubMed] [Google Scholar]
- Brandvain Y., Slotte T., Hazzouri K. M., Wright S. I., Coop G., 2013. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9: e1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgarella C., Gayral P., Ballenghien M., Bernard A., David P., et al. , 2015. Molecular evolution of freshwater snails with contrasting mating systems. Mol. Biol. Evol. 32: 2403–2416. [DOI] [PubMed] [Google Scholar]
- Caballero A., Hill W. G., 1992. Effects of partial inbreeding on fixation rates and variation of mutant genes. Genetics 131: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1992. Evolutionary rates in partially self-fertilizing species. Am. Nat. 140: 126–148. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Betancourt A. J., Kaiser V. B., Gordo I., 2009. Genetic recombination and molecular evolution. Cold Spring Harb. Symp. Quant. Biol. 74: 177–186. [DOI] [PubMed] [Google Scholar]
- Clark R. M., Schweikert G., Toomajian C., Ossowski S., Zeller G., et al. , 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342. [DOI] [PubMed] [Google Scholar]
- Ewing G., Hermisson J., Pfaffelhuber P., Rudolf J., 2011. Selective sweeps for recessive alleles and for other modes of dominance. J. Math. Biol. 63: 399–431. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Fisher R. A., 1941. Average excess and average effect of a gene substitution. Ann. Eugen. 11: 53–63. [Google Scholar]
- Fournier-Level A., Korte A., Cooper M. D., Nordborg M., Schmitt J., et al. , 2011. A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89. [DOI] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103: 127–144. [PubMed] [Google Scholar]
- Gioti A., Mushegian A. A., Strandberg R., Stajich J. E., Johannesson H., 2012. Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Mol. Biol. Evol. 29: 3215–3226. [DOI] [PubMed] [Google Scholar]
- Glémin S., 2012. Extinction and fixation times with dominance and inbreeding. Theor. Popul. Biol. 81: 310–316. [DOI] [PubMed] [Google Scholar]
- Glémin, S., and N. Galtier, 2012 Genome evolution in outcrossing vs. selfing vs. asexual species. Methods Mol. Biol. 855: 311–335. [DOI] [PubMed] [Google Scholar]
- Glémin S., Ronfort J., 2013. Adaptation and maladaptation in selfing and outcrossing species: new mutations vs. standing variation. Evolution 67: 225–240. [DOI] [PubMed] [Google Scholar]
- Goldberg E. E., Kohn J. R., Lande R., Robertson K. A., Smith S. A., et al. , 2010. Species selection maintains self-incompatibility. Science 330: 493–495. [DOI] [PubMed] [Google Scholar]
- Golding G. B., Strobeck C., 1980. Linkage disequilibrium in a finite population that is partially selfing. Genetics 94: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T. I., Song B.-H., Windsor A. J., Mitchell-Olds T., Dixon C. J., et al. , 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection, part V: selection and mutation. Math. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Hancock A. M., Brachi B., Faure N., Horton M. W., Jarymowycz L. B., et al. , 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86. [DOI] [PubMed] [Google Scholar]
- Hartfield M., 2016. Evolutionary genetic consequences of facultative sex and outcrossing. J. Evol. Biol. 29: 5–22. [DOI] [PubMed] [Google Scholar]
- Hartfield M., Glémin S., 2014. Hitchhiking of deleterious alleles and the cost of adaptation in partially selfing species. Genetics 196: 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfield M., Otto S. P., 2011. Recombination and hitchhiking of deleterious alleles. Evolution 65: 2421–2434. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., 1980. Hitchhiking: a comparison of linkage and partial selection. Genetics 94: 791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R., Maynard Smith J., 1978. Does Muller’s ratchet work with selfing? Genet. Res. 32: 289–293. [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Hough J., Williamson R. J., Wright S. I., 2013. Patterns of selection in plant genomes. Annu. Rev. Ecol. Evol. Syst. 44: 31–49. [Google Scholar]
- Huber C. D., Nordborg M., Hermisson J., Hellmann I., 2014. Keeping it local: evidence for positive selection in Swedish Arabidopsis thaliana. Mol. Biol. Evol. 31: 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B., Busch J. W., 2013. Is self-fertilization an evolutionary dead end? New Phytol. 198: 386–397. [DOI] [PubMed] [Google Scholar]
- Igic B., Kohn J. R., 2006. The distribution of plant mating systems: study bias against obligately outcrossing species. Evolution 60: 1098–1103. [PubMed] [Google Scholar]
- Igic B., Lande R., Kohn J. R., 2008. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 169: 93–104. [Google Scholar]
- Jarne P., Auld J. R., 2006. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution 60: 1816–1824. [DOI] [PubMed] [Google Scholar]
- Johnson T., Barton N. H., 2002. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamran-Disfani A., Agrawal A. F., 2014. Selfing, adaptation and background selection in finite populations. J. Evol. Biol. 27: 1360–1371. [DOI] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., Porcher E., 2015. Maintenance of quantitative genetic variance under partial self-fertilization, with implications for evolution of selfing. Genetics 200: 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., Schemske D. W., 1985. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39: 24–40. [DOI] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A., et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J., Burger R., 1995. Mutation accumulation and the extinction of small populations. Am. Nat. 146: 489–518. [Google Scholar]
- Matuszewski S., Hermisson J., Kopp M., 2014. Fisher’s geometric model with a moving optimum. Evolution 68: 2571–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B., 2000. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1932. Some genetic aspects of sex. Am. Nat. 66: 118–138. [Google Scholar]
- Nordborg M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 67: 159–174. [DOI] [PubMed] [Google Scholar]
- Orr H. A., Betancourt A. J., 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Barton N. H., 1997. The evolution of recombination: removing the limits to natural selection. Genetics 147: 879–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paape T., Bataillon T., Zhou P., Kono T. J. Y., Briskine R., et al. , 2013. Selection, genome-wide fitness effects and evolutionary rates in the model legume Medicago truncatula. Mol. Ecol. 22: 3525–3538. [DOI] [PubMed] [Google Scholar]
- Padhukasahasram B., Marjoram P., Wall J. D., Bustamante C. D., Nordborg M., 2008. Exploring population genetic models with recombination using efficient forward-time simulations. Genetics 178: 2417–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell J. R., Dorken M. E., Eppley S. M., 2005. ‘Haldane’s Sieve’ in a metapopulation: sifting through plant reproductive polymorphisms. Trends Ecol. Evol. 20: 374–379. [DOI] [PubMed] [Google Scholar]
- Pannell J. R., Auld J. R., Brandvain Y., Burd M., Busch J. W., et al. , 2015. The scope of Baker’s law. New Phytol. 208: 656–667. [DOI] [PubMed] [Google Scholar]
- Pollak E., 1987. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics 117: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S., Zeng K., Slotte T., Wright S., Charlesworth D., 2011. Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species. Genome Biol. Evol. 3: 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronfort J., Glémin S., 2013. Mating system, Haldane’s sieve, and the domestication process. Evolution 67: 1518–1526. [DOI] [PubMed] [Google Scholar]
- Roze D., 2009. Diploidy, population structure, and the evolution of recombination. Am. Nat. 174: S79–S94. [DOI] [PubMed] [Google Scholar]
- Roze D., 2015. Effects of interference between selected loci on the mutation load, inbreeding depression, and heterosis. Genetics 201: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze D., Lenormand T., 2005. Self-fertilization and the evolution of recombination. Genetics 170: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., 2014. The impact of linked selection on plant genomic variation. Brief. Funct. Genomics 13: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J. P., Hazzouri K. M., Wright S. I., 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821. [DOI] [PubMed] [Google Scholar]
- Slotte T., Bataillon T., Hansen T. T., St. Onge K., Wright S. I., et al. , 2011. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol. Evol. 3: 1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G. L., 1957. Self fertilization and population variability in the higher plants. Am. Nat. 91: 337–354. [Google Scholar]
- Weissman D. B., Barton N. H., 2012. Limits to the rate of adaptive substitution in sexual populations. PLoS Genet. 8: e1002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research , 2014. Mathematica Edition: Version 10. Wolfram Research, Champaign, IL. [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Barrett S. C. H., 2010. The long-term benefits of self-rejection. Science 330: 459–460. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Kalisz S., Slotte T., 2013. Evolutionary consequences of self-fertilization in plants. Proc. Biol. Sci. 280: 20130133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Etheridge A. M., 2010. The fixation probability of two competing beneficial mutations. Theor. Popul. Biol. 78: 36–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Additional information on derivations are provided in File S1, and Mathematica File S2. Simulation code is available as File S3 and at http://github.com/MattHartfield/TwoAdvSelfSims.