Abstract
As a highly conserved protein, the translationally controlled tumor protein (TCTP) carries out vital roles in various life processes. In rubber tree, two TCTP genes, HbTCTP and HbTCTP1, were cloned, but only HbTCTP1 was studied in details. In this study, cis-acting regulatory elements, expression patterns, subcellular localization, interacting proteins, and antioxidant activity of HbTCTP were systematically analyzed. Besides the common cis-acting regulatory elements, HbTCTP promoter also harbored various known cis-elements that respond to hormone/stresses. Being consistent with the aforementioned results, HbTCTP was regulated by drought, low temperature, high salt, ethylene (ET), wounding, H2O2, and methyl jasmonate (MeJA) treatments. HbTCTP was expressed throughout different tissues and developmental stages of leaves. In addition, HbTCTP was associated with tapping panel dryness (TPD). HbTCTP was localized in the membrane, cytoplasm and the nucleus, and interacted with four proteins rubber elongation factor (REF), 17.5 kDa heat shock family protein, annexin, and REF-like stress related protein 1. Being similar to HbTCTP1, HbTCTP also indicated antioxidant activity in metal-catalyzed oxidation (MCO) system. Our results are useful for further understanding the molecular characterization and expression profiles of HbTCTP, but also lay a solid foundation for elucidating the function of HbTCTP in rubber tree.
Keywords: expression analyses, Hevea brasiliensis, stress/hormone response, DNA protection activity, TCTP, TPD
Introduction
The translationally controlled tumor protein (TCTP) is a highly conserved protein, and it is widely expressed in all eukaryotic organisms (Bommer and Thiele, 2004). Since Yenofsky et al. (1983) reported the first TCTP in mouse, the TCTPs have been cloned and characterized in different organisms. In animals, the TCTPs were reported to be associated with several biological processes such as cell growth, cell cycle progression, differentiation, malignant transformation, protection against various stress conditions and apoptosis (Amson et al., 2013).
In contrast to animals, the studies on plant TCTPs are very limited. The first plant TCTP gene was cloned in Medicago sativa (Pay et al., 1992). Currently, TCTP genes have been isolated from different plants species. Besides being related to plant growth and development (Lu et al., 2007; Brioudes et al., 2010; Nakkaew et al., 2010; Qin et al., 2011), TCTPs were also involved in a wide range of stimulus response, such as darkness (Sage-Ono et al., 1998), aluminum (Ermolayev et al., 2003), pathogens (Jones et al., 2006; Li et al., 2010b), NaCl (Vincent et al., 2007; Cao et al., 2010), mercury (Wang et al., 2012), heat, cold, and drought (Kim et al., 2012; Li et al., 2013), as well as growth regulators such as auxins, ABA (Berkowitz et al., 2008; Cao et al., 2010; Kim et al., 2012), and methyl jasmonate (MeJA) (Li et al., 2013). Interestingly, the TCTPs have been detected in castor and winter squash phloem saps, suggesting that the TCTPs might be involved in regulating the destination-selective long-distance movement of phloem proteins by interacting with phloem proteins (Barnes et al., 2004; Aoki et al., 2005; Hinojosa-Moya et al., 2006). Recently, lupin and pumpkin TCTP mRNA have also been found in the phloem sap transcriptome and phloem sap exudates, respectively (Rodriguez-Medina et al., 2011; Hinojosa-Moya et al., 2013).
TCTP varies in its gene number among different organisms. Mammals have more than two TCTP-like sequences, while plants and fungi usually contain one or two TCTP genes (Hinojosa-Moya et al., 2008). Based on this wide range of functions and the different TCTP versions, Gutiérrez-Galeano et al. (2014) suggested that TCTPs might take place function specialization in plants with two members. They further proposed that there were two groups of TCTP genes, AtTCTP1-like and CmTCTP-like clades, and their respective functions might be inferred from their corresponding groups.
There are two TCTP genes identified in rubber tree (Li et al., 2013). Liang et al. (2009) firstly reported that HbTCTP (GenBank accession: FJ156098) was constitutively expressed in latex, leaves and barks and induced by ethylene (ET) treatment. Deng et al. (2012) further analyzed the gene structure and developed molecular markers of HbTCTP. Li et al. (2013) cloned another TCTP gene (HbTCTP1) in rubber tree. HbTCTP1 was expressed throughout different tissues and developmental stages of leaves. Besides being related to tapping panel dryness (TPD), HbTCTP1 was regulated by drought, low temperature, high salt, ET, wounding, H2O2, and MeJA. HbTCTP1 possessed supercoiled DNA protection activity in metal-catalyzed oxidation (MCO) system. The aforementioned results suggested that HbTCTP1 was a multifunctional gene and associated with hormone/stress response and TPD. However, the other TCTP gene, HbTCTP, was not studied and reported in details until now. Here, we further analyzed expression patterns, cis-regulatory elements of HbTCTP, as well as interacting proteins, subcellular location, and antioxidant activity of HbTCTP protein. Our results not only are useful for further understanding the molecular characterization and expression profiles of HbTCTP, but also lay a solid foundation for elucidating HbTCTP function in rubber tree.
Materials and methods
Plant materials
Reyan 8-79, a high-yielding rubber tree clone, was planted at the experimental farm of Chinese Academy of Tropical Agricultural Sciences. The plant tissues, latex, male and female flowers, anthers, leaves and barks were separately collected from three 17-year-old rubber trees regularly tapped on the s/2 d/3 system with 1% ET stimulation for the past 11 years, and each rubber tree was referred as one biological replicate. According to Hao and Wu (2000), 6-year-old virgin trees were applied with ET, H2O2 and MeJA treatments prior to the first tapping. The wounding treatment was performed according to the method provide by Tang et al. (2010). One-year old seedlings of Reyan 7-33-97 were separately treated with drought (16% PEG), low temperature 8(8°C), and high salt (1M NaCl), and the leaves from five treated and five control (1-year old seedlings of Reyan 7-33-97 were grown at normal condition without any treatments including drought, low temperature, or high salt) materials were separately collected and equivalently pooled at different times for total RNA extraction. The leaves from each five seedlings were referred as one biological replicate. The leaves of different developmental stages were separately harvested from three 6-year-old Reyan 8-79, and each rubber tree was referred as one biological replicate. The trees with a partial or complete stoppage of latex flow are defined as the initial and advanced stages of TPD trees, respectively. In this study, the latex and barks were collected from the TPD trees at initial stage. The latex and barks from five healthy and five TPD rubber trees were separately collected and equivalently pooled for total RNA exaction, and the samples from each five trees were referred as one biological replicate. All samples for real-time RT-PCR experiments were harvested with three biological replicates.
RNA and DNA extraction
Total RNAs from different tissues were prepared according to Xu's method (Xu et al., 2010), and then treated with RNase-free RQ1 DNase (Promega, USA). The genomic DNA was isolated from the young leaves with the CTAB method (Porebski et al., 1997). The quantity and quality of RNA and DNA were determined by agarose gel electrophoresis and the spectrophotometer.
Cloning and analyses of HbTCTP promoter
To obtain the promoter sequence of HbTCTP, the PCR primers were designed according to the sequences of contig338249 (GenBank accession: AJJZ010271614.1) (Rahman et al., 2013). The promoter sequence of HbTCTP was isolated by PCR amplification using GenomeWalker universal kit (Clontech, USA). The primary PCR was performed with the outer adaptor primer (AP1) and the outer gene-specific primer 5′-CAGAATCACTCACCCAAACG-3′ using Advantage2 polymerase mix (Clontech, USA). The secondary PCR was performed with the nested adaptor primer (AP2) and the nested gene-specific primer 5′-CAGATCCCCATGTACACAAC-3′. The PCR product was cloned into the pMD18-T vector and sequenced. The cis-acting regulatory elements within the promoter of HbTCTP were predicted using PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace).
Real-time RT-PCR
First-strand cDNA was obtained using ReverAid™ first strand cDNA synthesis kit (Thermo Scientific, USA). In each real-time RT-PCR reaction, the gene-specific primers were used, and 18S rRNA gene (GenBank accession: AB268099) was used as the internal reference according to Tang et al. (2010). The forward (F) and reverse (R) primers are the following: for 18S rRNA (F 5′-GCTCGAAGACGATCAGATACC-3′ and R 5′-TTCAGCCTTGCGACCATAC-3′); for HbTCTP (F 5′-TTGTGGATCGATCCGAGAGA-3′ and R 5′-CAACCC ACTTCCCCTCAACT-3′). Real-time RT-PCR was performed with the LightCycler 2.0 system (Roche Diagnostics, Germany) and SYBR premix Ex Taq™ II (Takara, Japan). The reactions were carried out as follows: 30 s at 94°C for denaturation, followed by 45 cycles of 94°C for 5 s, 60°C for 20 s, and 72°C for 20 s. The relative abundance was calculated by the LightCycler Relative Quantification Software 4.05. The specificity of each primer pairs was verified by the melting curves and sequencing the PCR products. All real-time RT-PCR experiments described here were reproduced with three biological replicates, and the values were presented as mean ± S.D. The statistical analysis was performed using SPSS software version 19.0 (IBM, Armonk, New York) and one-way ANOVA with the Student-Newman-Keuls test was used for comparison of HbTCTP expressions across different tissues and development stage. The t-test was performed for comparison of HbTCTP expressions under the treatments of stresses and hormones, or healthy and TPD rubber trees.
Subcellular localization of HbTCTP
To determine its subcellular localization, the ORF of HbTCTP was cloned into the vector CD3-1103 (TAIR Accession: 5016231514). In this construct, the gene was in-frame fused at its 5′ end to an enhanced Yellow Fluorescent Protien (eYFP) gene. The forward and reverse primers were 5′-CTCAAGCTTCGAATTCTATGTTGGTCTATCAGGATTTGCT-3′ and 5′-CTAGATCAGGTGGATCCGCATTTGACCTCCT TCAAAGCA-3′, respectively. The PCR-amplified fragment was ligated into the vector basing on the method of homologous recombination. The resultant construct was transformed into rice protoplasts for transient expression, following the methods described by Bart et al. (2006). The fluorescent signals were examined with a laser confocal microscope (Leica TCS SP5), stimulated by light of 512 nm wavelength [To indicate cell nucleus, the nuclear mCherry expression vector CD3-1106 (TAIR Accession: 5016231517) was optionally cotransformed into rice protoplasts together with the construct. mCherry fluorescence was stimulated by light of 585 nm wavelength].
Yeast two-hybrid screen and assays
HybriZAP®-2.1 two-hybrid predigested vector/gigapack® cloning kit (Stratagene, CA) was used to screen the proteins interacting with HbTCTP in the study. The coding sequence of HbTCTP was cloned into the bait vector pBD-GAL4 Cam. Saccharomyces cerevisiae strain YRG-2 were first transformed with the bait pBD-HbTCTP and subsequently transformed with latex cDNA library plasmid DNA by the lithium acetate method. The transformants were plated onto SD medium lacking leucine, tryptophan and histidine (SD/-Leu/-Trp/-His). The colonies growing on selection media were restreaked onto SD/-Leu/-Trp/-His, transferred to the filter paper, and assayed for β-galactosidase (LacZ) activity by the filter lift assay. The LacZ-positive colonies growing on SD/-Leu/-Trp/-His medium were considered putative positive clones. The plasmids were isolated from these yeast colonies, transformed, and amplified in the E. coli XL1-Blue strain. The isolated plasmids were then transformed back into YRG-2 cells either alone or in combination with control plasmids. The retransformation assays were able to elicit expression of the two reporter genes only in the presence of pBD-HbTCTP bait, but not alone or in combination with control plasmids, were considered true positives. The validated positive clones were selected to be sequenced.
Purification and MCO assay of HbTCTP fusion protein
The coding sequences of HbTCTP with the introduced EcoR I and BamH I sites were ligated into pET28a plasmid. The correct pET28a-HbTCTP was confirmed by sequencing and transferred into E. coli strain BL21 (DE3). In LB medium with 100 mg/L kanamycin, the E. coli BL21 cells containing pET28a-HbTCTP or pET28a plasmids were separately grown at 37°C overnight. One mL transformants cultured overnight were inoculated into 200 mL fresh LB medium with 100 mg/L kanamycin. After the OD600 value of the transformants reached 0.4, the transformants were further incubated at 37°C for ~3 h with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) induction. To obtain HbTCTP protein, the pET28a-HbTCTP was purified and substituted by Zeba™ Desalt Spin Columns (Thermo Scientific Pierce, USA). The MCO assay was employed to determine whether HbTCTP could protect supercoiled DNA according to the method described by Li et al. (2004). Fifty microliters reaction mixtures contain 33 μM FeCl3, 3.3 mM DTT and 200 μg/mL purified HbTCTP or HbTCTP1 proteins. The pUC19 supercoiled DNA (300 ng) were added to each reaction and incubated further at 37°C for 2 h. The DNA protection was evaluated with 10 μL reaction samples by 1% agarose gel electrophoresis. In addition, bovine serum albumin (BSA) was also assayed in similar conditions as controls.
Degradome data
According to German et al. (2009), a library with all degraded mRNA was constructed in rubber tree. After sequencing, analysis with the Cleaveland pipeline, slicing sites of mRNA by small RNA are detected. Degradome data were provided by the DEBA Hevea project funded by Institut Français du caoutchouc. Degradome data were obtained according to an adapted protocol from German et al. (2009) and analyzed by the CLEAVELAND pipeline developed by Addo-Quaye et al. (2009).
Results
Hormone/stresses respond-related cis-elements identified in HbTCTP promoter
Using the PCR-based Genome Walking method, we obtained a 1201-bp promoter sequence of HbTCTP (GenBank accession: KX179468). The cis-acting regulatory elements within HbTCTP promoter were predicted by PLACE software. Besides the common cis-acting regulatory elements including TATA boxes and CAAT boxes, the HbTCTP promoter also harbored various known cis-elements associated with hormone response [ethylene, abscisic acid (ABA), gibberellin (GA), salicylic acid (SA), and auxin responses], stress response (anaerobically induced, disease resistance, copper, CO2, pathogenesis and salt-induced, light, wounding, UV-B irradiation, heat shock, water, and pro- or hypo-osmolarity responses), as well as stress and hormone responses (drought and ABA, dehydration, ABA and cold, GA and sugar). In addition, some special cis-acting regulatory elements were also found in HbTCTP promoter, such as phytochrome regulated element, and sugar response element, etc…(Supplementary Table S1). The results indicated that HbTCTP expression might be involved in stress/hormone response.
Expression analysis of HbTCTP
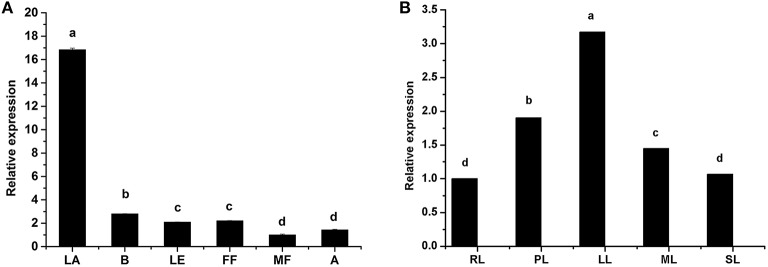
We systematically analyzed HbTCTP expression by real time RT-PCR. As shown in Figure 1A, HbTCTP was highly expressed in latex, followed by barks, female flowers, leaves, anthers and male flowers. Gébelin et al. (2012) predicted that eleven contigs encoding rubber tree TCTP genes could be targeted by miRNA. Therefore, we set out to detect whether this occurs to HbTCTP. As shown in Table 1, three contigs (CL1994Contig3, CL18Contig4, and CL18Contig13) being identified to HbTCTP are experimentally validated by degradome analysis in at least two tissues. In addition, HbTCTP expression indicated a significant change in different leaf development stages, with the highest level in light young leaves, the lowest in red leaves (Figure 1B).
Figure 1.
Real time RT-PCR analyses of HbTCTP expression patterns in different tissues and developmental stages of leaves. Panels (A,B) represent HbTCTP expression profiles in different tissues and developmental stages of leaves, respectively; B, LA, LE, FF, MF, and A represent barks, latex, leaves, female flowers, male flowers, and anthers, respectively. RL, PL, LL, ML, and SL separately are red leaves, pale-green young, light young, mature, and senescence leaves. 18S rRNA is used as the internal control for real time RT-PCR analyses. The standard bars were obtained from at least three independent replications (different RNA preparations from different biological replicates) and the results were given as mean ± S.D. The different letters indicate significant difference at P < 0.05.
Table 1.
The information of HbTCTP experimentally validated by degradome analysis.
| HbTCTP | |||||||
|---|---|---|---|---|---|---|---|
| SiteID | CL1994Contig3:25 | CL1994Contig3:25 | CL18Contig4:316 | CL18Contig4:316 | CL18Contig13:280 | CL18Contig13:280 | CL18Contig13:280 |
| Query | acc_635132_22 | acc_635132_22 | acc_298609_20 | acc_81739_20 | acc_298609_20 | acc_298609_20 | acc_298609_20 |
| Query GenBank accession | KX230767 | KX230767 | KX230768 | KX230769 | KX230768 | KX230768 | KX230768 |
| miRNA homology | ppt-miR1023b-3p | ppt-miR1023b-3p | ptc-miRf12236-akr | ptc-miRf12236-akr | ptc-miRf12236-akr | ptc-miRf12236-akr | ptc-miRf12236-akr |
| Transcript | CL1994Contig3 | CL1994Contig3 | CL18Contig4 | CL18Contig4 | CL18Contig13 | CL18Contig13 | CL18Contig13 |
| Transcript GenBank accession | KX230764 | KX230764 | KX230765 | KX230765 | KX230766 | KX230766 | KX230766 |
| TStart | 12.0 | 12.0 | 303.0 | 303.0 | 267.0 | 267.0 | 267.0 |
| TStop | 34.0 | 34.0 | 325.0 | 325.0 | 289.0 | 289.0 | 289.0 |
| TSlice | 25.0 | 25.0 | 316.0 | 316.0 | 280.0 | 280.0 | 280.0 |
| MFEperfect | −34.6 | −34.6 | −31.5 | −31.5 | −31.5 | −31.5 | −31.5 |
| MFEsite | −22.7 | −22.7 | −20.6 | −20.6 | −20.6 | −20.6 | −20.6 |
| MFEratio | 0.65606936416185 | 0.65606936416185 | 0.65396825396825 | 0.65396825396825 | 0.65396825396825 | 0.65396825396825 | 0.65396825396825 |
| AllenScore | 10.0 | 10.0 | 11.5 | 11.5 | 11.5 | 11.5 | 11.5 |
| Paired | 3-5,32-30;8-18,27-17;19-22,15-12 | 3-5,32-30;8-18,27-17;19-22,15-12 | 4-17,322-309;18-20,305-303 | 4-17,322-309;18-20,305-303 | 4-17,286-273;18-20,269-267 | 4-17,286-273;18-20,269-267 | 4-17,286-273;18-20,269-267 |
| Unpaired | 1-2,34-33[UP5];6-7,29-28[SIL];x-x,16-16[BULt] | 1-2,34-33[UP5];6-7,29-28[SIL];x-x,16-16[BULt] | 1-3,325-323[UP5];x-x,308-306[BULt] | 1-3,325-323[UP5];x-x,308-306[BULt] | 1-3,289-287[UP5];x-x,272-270[BULt] | 1-3,289-287[UP5];x-x,272-270[BULt] | 1-3,289-287[UP5];x-x,272-270[BULt] |
| Structure | ((((.(((((((((((.(((.&.))).)))))))))))-)))) | ((((.(((((((((((.(((.&.))).)))))))))))-)))) | (((…((((((((((((((…&…))))))))))))))—))) | (((…((((((((((((((…&…))))))))))))))—))) | (((…((((((((((((((…&…))))))))))))))—))) | (((…((((((((((((((…&…))))))))))))))—))) | (((…((((((((((((((…&…))))))))))))))—))) |
| Sequence | CUCUCGCGCUCUCUUUUU UUCCC& AAGAAUUGAAGAGAGUGU-AGAG | CUCUCGCGCUCUCUUUUUUUCCC&AAGAAUUGAAGAGAGUGU-AGAG | CCAAAUUUUUGGGGGGGGGAGAG&CAUUCCUCUUCCUAAAA—UGG | CCAAAUUUUUGGGGGGGGGAGAG&CAUUCCUCUUCCUAAAA—UGG | CCAAAUUUUUGGGGGGGGGAGAG&CAUUCCUCUUCCUAAAA—UGG | CCAAAUUUUUGGGGGGGGGAGAG&CAUUCCUCUUCCUAAAA—UGG | CCAAAUUUUUGGGGGGGGGAGAG&CAUUCCUCUUCCUAAAA—UGG |
| DegradomeCategory | 2 | 3 | 4 | 4 | 4 | 4 | 4 |
| DegradomePval | 0.057064826793158 | 0.043840735949404 | 0.043696995831537 | 0.040822193474199 | 0.079977935468352 | 0.085484564218372 | 0.11412318662399 |
| Librairy | bark | leaf | root | latex | latex | root | leaf |
| Length | 22 | 22 | 20 | 20 | 20 | 20 | 20 |
SiteID, a unique name for a putative splicing site in the form of “[transcript]:[slice_site]”; Query, small RNA accession; miRNA homology, column added by the authors with the best hit after blasting the small RNA sequence against PMRD database (Zhang et al., 2010); Tstart, one-based start position of the alignment within the transcript; Tstop, one-based stop position of the alignment within the transcript; Tslice, one-based position of the alignment opposite position 10 of the query; MFEperfect, minimum free energy of a perfectly matched site (approximate); MFEsite, minimum free energy of the aligment in question; MFEratio, MFEsite/MFEperfect; AllenScore, penalty score calculated by Allen (Allen et al., 2005); Paired, string representing paired position in the query and transcript; Unpaired, string representing unpaired positions in the query and transcript; Structure, aligned secondary structure; Sequence, aligned sequences of transcript and query; Degradome category, category 4: just one read at this position; category 3: >1 read, but below or equal to the average depth of coverage on the transcript; category 2: >1 read, equal to the average depth of coverage on the transcript; category 1: >1 read, equal to the maximum of the average depth of coverage on the transcript when there is >1 position at maximum value; category 0: >1 read, equal to the maximum of the average depth of coverage on the transcript when there is just one position at maximum value; DegradomePval, p-value for this degradome hit; Library, column added by the authors, name of the tissue from which degradome library was sequenced; Length, column added by the authors, small RNA length in nucleotide.
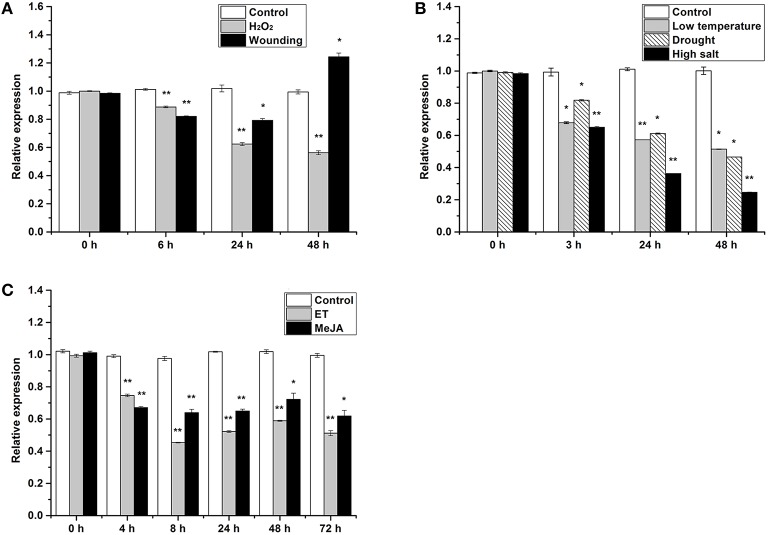
HbTCTP expression was also analyzed under several treatments including drought, low temperature, high salt, ET, wounding, H2O2 and MeJA. As shown in Figure 2, HbTCTP was regulated by all the treatments, but its expression profiles widely varied depending on the different treatments. The statistical analyses showed that the changes of HbTCTP expression indicated significant differences under all treatments. Under drought, low temperature, high salt, and H2O2 treatments, HbTCTP transcripts steadily decreased (Figures 2A,B). Under ET and MeJA treatment, the HbTCTP was totally downregulated, but indicated slight oscillations in different times (Figure 2C). Under wounding treatment, HbTCTP transcripts were decreased at 6 h after treatment, and then upregulated at 48 h (Figure 2A). In contrast, HbTCTP did not significantly change in their corresponding controls (Figure 2). These results suggested that HbTCTP might be associated with ET, MeJA and stress responses in rubber tree.
Figure 2.
Real time RT-PCR analyses of HbTCTP expression patterns under different treatments. Panels (A–C) represent real time RT-PCR analyses of HbTCTP expression profiles under H2O2 and wounding; low temperature, drought, and high salt; ET and MeJA treatments, respectively; 18S rRNA is used as the internal control for real time RT-PCR analyses. The standard bars were obtained from at least three independent replications (different RNA preparations from different biological replicates) and the results were given as mean ± S.D. Compared with the control, one asterisk shows significant difference with a P < 0.05, and two asterisks show very significant difference with a P < 0.01.
Li et al. (2013) reported that HbTCTP1 transcripts were upregulated in healthy rubber tree, suggesting that HbTCTP1 was associated with TPD; therefore, HbTCTP expression was also investigated in healthy and TPD trees. As shown in Figure 3, the HbTCTP transcripts were higher in TPD rubber tree than healthy one. The statistical analyses indicated that the changes of HbTCTP expression had significant differences between healthy and TPD rubber trees. The results suggested that HbTCTP might be associated with TPD processes in rubber tree.
Figure 3.
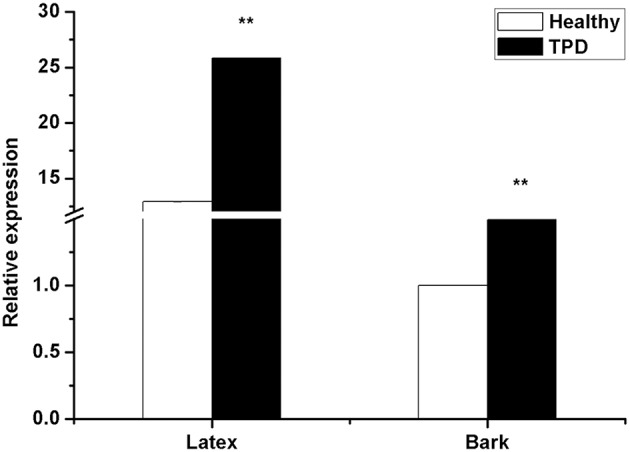
Real time RT-PCR analyses of HbTCTP expression in healthy and TPD rubber trees. 18S rRNA is used as the internal control for real time RT-PCR analyses. The standard bars were obtained from at least three independent replications (different RNA preparations from different biological replicates) and the results were given as mean ± S.D. Compared with the healthy rubber tree, two asterisks show very significant difference with a P < 0.01.
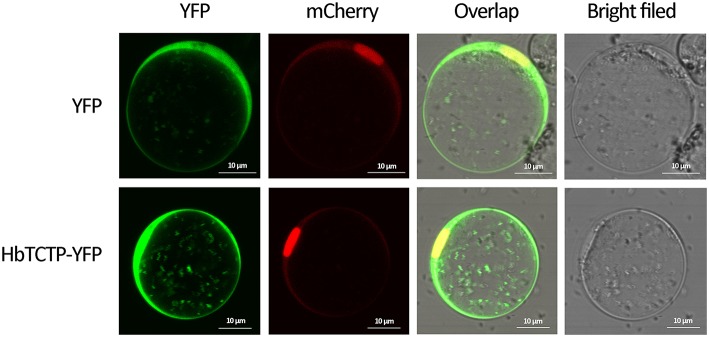
HbTCTP located in whole cell
To investigate the subcellular localization of HbTCTP protein, we transiently expressed HbTCTP: eYFP fusion protein in rice protoplasts and examined with a laser confocal microscope. As shown in Figure 4, the fluorescent signals of the fusion protein were visualized in the membrane, the cytosol and the nucleus, as similar as those of eYFP alone. The results suggested that HbTCTP has no preference in subcellular localization.
Figure 4.
Subcellular localization of HbTCTP-YFP fusion protein in rice protoplasts.
Identification of HbTCTP-interacting proteins
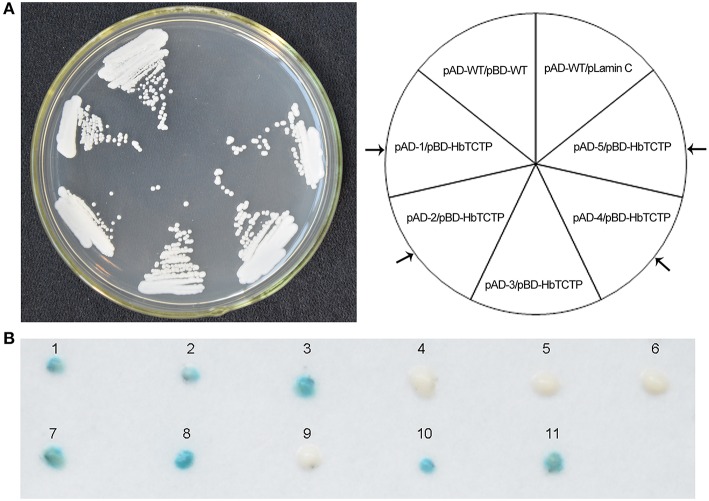
Next, we screened and identified the HbTCTP-interacting proteins by yeast two-hybrid (Y2H) assay. The initial screening identified five colonies, which were subsequently validated for activating the LacZ gene. As shown in Figure 5, only four colonies can activate LacZ gene. The plasmid DNAs were isolated from the four colonies and retransformed into the YRG-2 strain to confirm whether the activation is indeed due to the presence of the fusion protein. We observed that all the four plasmids were able to activate the LacZ gene only in the presence of pBD-HbTCTP. All the four sequences contained complete open reading frames, and the blastx results indicated that the four sequences (GenBank accession: KX179469–KX179472) encoded different proteins including rubber elongation factor protein, 17.5 kDa heat shock family protein, annexin, and REF-like stress related protein 1.
Figure 5.
Identification of the interaction protein of HbTCTP by yeast two-hybird system. (A) Yeast cells transformed with bait and prey vectors were streaked onto SD/-Leu/-Trp/-His. pAD-WT/pBD-WT and pAD-WT/pLamin C were used as positive and negative controls, respectively. (B) β-Galactosidase assays. The numbers 1–11 indicate yeast cells transformed with the following plasmid combinations: 1, pAD-WT/pBD-WT; 2, pAD-MUT/pBD-MUT; 3, pGAL4; 4, pAD-WT/pLamin C; 5, pAD-MUT/pLamin C; 6, pLamin C; 7, pAD-1/pBD-HbTCTP; 8, pAD-2/pBD-HbTCTP; 9, pAD-3/pBD-HbTCTP; 10, pAD-4/pBD-HbTCTP; 11, pAD-5/pBD-HbTCTP. Arrow indicated that the positive clones.
HbTCTP indicating in vitro supercoiled DNA protection activity
To analyze in vitro supercoiled DNA protection activities, HbTCTP was expressed in E. coli BL21 cells and purified. The obtained fusion protein showed an approximately 25.0-kDa band on SDS-PAGE, in agreement with the predicted molecular mass (19.1 kDa for HbTCTP and 6.2 kDa for the His and T7 tags), and was therefore used for DNA protection activity assay. As shown in Figure 6, the MCO components without incubation or separately incubated MCO components did not bring about any damage to supercoiled DNA, whereas the complete nicking of supercoiled DNA occurred in MCO system without HbTCTP protein and with BSA (a negative control). Being consistent with HbTCTP1 identified with supercoiled DNA protection activity (Li et al., 2013), the HbTCTP proteins indicated the protection of supercoiled DNA, suggesting that HbTCTP protein possessed supercoiled DNA protection activity against MCO.
Figure 6.
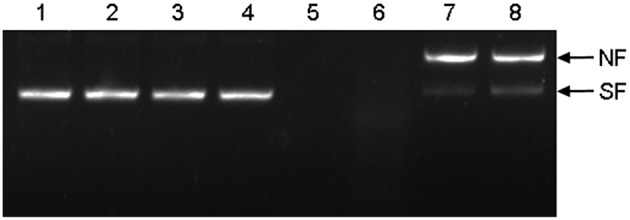
Protection of supercoiled DNA cleavage by recombinant HbTCTP in a MCO system. (1) pUC19 without incubation; (2) pUC19 in water and incubated 37°C for 2 h; (3) pUC19 only with FeCl3; (4) pUC19 only with DTT; (5) pUC19 with MCO system; (6) pUC19 with BSA (0.6 mg/mL) with MCO system as a negative control; (7) pUC19 with HbTCTP protein (200 μg/mL) with MCO system; (8) pUC19 with HbTCTP1 protein (200 μg/mL) with MCO system. NF, nicked form; SF, supercoiled form of the plasmid.
Discussion
TCTP is a highly conserved protein throughout all the eukaryotes, and it is associated with a wide range of cellular functions including cell growth, cell cycle progression, and protection against various stresses, etc. It is reported that there are variable numbers of TCTP genes in different organisms, and plants in general contain one or two TCTP genes (Hinojosa-Moya et al., 2008). Rubber tree contains two TCTP genes, HbTCTP and HbTCTP1, but only HbTCTP1 was analyzed and reported in detail. In this study, HbTCTP was systematically studied. Being different from HbTCTP1, HbTCTP was highly expressed in latex, which differ from the previous results that there was no difference in the expression of HbTCTP among barks, leaves and latex (Liang et al., 2009). With leaf growth and development, HbTCTP expression was initially increased, and then decreased. The highest and lowest expression of HbTCTP was separately in light young and red leaves, whereas the correspondent in mature and light young leaves as far as HbTCTP1 is concerned.
Besides its expression widely varied on different tissues and developmental stage, TCTP was reported to be regulated in various stresses (Ermolayev et al., 2003; Jones et al., 2006; Vincent et al., 2007; Cao et al., 2010; Li et al., 2010b, 2013; Kim et al., 2012; Wang et al., 2012) and growth regulators (Berkowitz et al., 2008; Liang et al., 2009; Cao et al., 2010; Kim et al., 2012; Li et al., 2013). In accordance with the aforementioned results, HbTCTP was regulated by ET, wounding, MeJA, low temperature, high salt, H2O2 and drought treatments. The cis-acting regulatory elements that respond to hormones and stresses were identified in HbTCTP promoter, which might explain its expression profiles under hormone and stresses treatments. Although HbTCTP1 was also regulated by all the aforementioned treatments, the expression patterns of HbTCTP evidently differ from that of HbTCTP1. Moreover, the HbTCTP1 transcripts were obviously higher in healthy rubber tree than TPD tree (Li et al., 2013). On the contrary, HbTCTP was significantly downregulated in healthy rubber tree. According to the expression patterns of two TCTP genes, it is reasonable that there is a functional specialization between HbTCTP and HbTCTP1.
Hoepflinger et al. (2013) reported that AtTCTP-GFP indicated cytosolic distribution. Mammalian TCTP was predominantly found in cytosol and nucleus in vivo, although it was described as a protein functioning in mitochondria (Zhang et al., 2002). In our study, HbTCTP was found to be localized in membrane, cytosol, and nucleus in rice protoplasts, which is consistent with the result that no signal peptide was predicted within HbTCTP protein by using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/). Of course, the subcellular location of HbTCTP need be further validated in rubber tree. TCTP is a multifunctional protein, which is inferred from the fact that it is capable of interacting with diverse targets, including cytoskeletal components, factors involved in cell repair, apoptosis (pro- and antiapoptotic), protein synthesis, and even general metabolism (Amson et al., 2013). In this study, we identified that HbTCTP interacted with HbREF, 17.5 kDa heat shock protein, annexin, and stress-related protein-like. Consisting with our result, Zeng et al. (2011) also reported that HbREF interacted with HbTCTP. REF is necessary for prenyltransferases from a number of sources to add multiple cis-isoprene units to rubber molecules (Dennis and Light, 1989). In addition, HbTCTP was preferential expression in latex. Therefore, HbTCTP might be involved in rubber biosynthesis (RB) by interacting with HbREF.
An uncompensated oxidative stress occurred on the onset of TPD (Yusof et al., 1995). In TPD tree, the NAD(P)H oxidase activities increased, whereas variable peroxidase and SOD levels decreased (Xu and Xiao, 1988). In addition, Peng et al. (2011) reported that TPD rubber tree indicated typical characteristics of programmed cell death (PCD). Li et al. (2010a) proposed that reactive oxygen species (ROS) metabolism, ubiquitin proteasome pathway (UPP), PCD, and RB pathways might play important roles in TPD. Gébelin et al. (2013) found that the target genes of TPD-related microRNAs were associated with RB, ROS-scavenging systems, and PCD. By identifying and analyzing TPD-related genes in rubber tree bark, Li et al. (2016) further reported that the TPD-related genes significantly enriched in eight GO terms and five KEGG pathways were closely associated with ROS metabolism, PCD, and RB. In this study, the HbTCTP transcripts were higher in TPD rubber tree than healthy rubber one. HbTCTP indicated antioxidant activity in vitro. Interestingly, HbTCTP also interacted with 17.5 kDa heat shock family protein, and annexin. sHSPs are molecular chaperones that prevent misfolding and irreversible aggregation of their client proteins; moreover sHSPs have roles in differentiation, proteasomal degradation, anti-apoptotic property and development (Bakthisaran et al., 2015). Plant annexins are involved in abiotic stress responses (Divya et al., 2010; Huh et al., 2010) and oxidative stress responses (Richards et al., 2014). Given the aforementioned results, HbTCTP might participate TPD by regulating ROS metabolism, PCD, and RB.
Conclusion
In summary, HbTCTP was regulated by drought, low temperature, high salt, ET, wounding, H2O2, and MeJA. HbTCTP was localized in the membrane, cytoplasm, and nucleus of rice protoplasts. Being similar to HbTCTP1, HbTCTP indicated antioxidant activity in vitro. Interacting with HbREF, 17.5 kDa heat shock protein, annexin, and REF-like stress related protein 1, HbTCTP might be involved in TPD by regulating ROS metabolism, PCD, and RB pathways in rubber tree.
Author contributions
DL and ZX conceived and designed the experiments. DL wrote the manuscript. ZZ, ZD, and LJ revised the manuscript. ZD and JC performed most of the experiments and data analysis. LJ and PM performed miRNA degradome analysis. ZZ carried out subcellular location analysis. CL, HL, and HY performed experimental samples collection, DNA and RNA extraction. All authors have read and approved the manuscript.
Conflict of interest statement
The reviewer LZ declared a shared affiliation, though no other collaboration, with one of the authors ZZ to the handling Editor, who ensured that the process nevertheless met the standards of a fair and objective review. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the earmarked funds from National Natural Science Foundation of China (31270651, 31200514 and 31570684) and Rubber Research Institute, CATAS (1630022015003 and 1630022014006).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00789
References
- Addo-Quaye C., Miller W., Axtell M. J. (2009). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131. 10.1093/bioinformatics/btn604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A. M., Carrington J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Amson R., Pece S., Marine J. C., Di Fiore P. P., Telerman A. (2013). TPT1/ TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol. 23, 37–46. 10.1016/j.tcb.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Aoki K., Suzui N., Fujimaki S., Dohmae N., Yonekura-Sakakibara K., Fujiwara T., et al. (2005). Destination-selective long-distance movement of phloem proteins. Plant Cell 17, 1801–1814. 10.1105/tpc.105.031419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthisaran R., Tangirala R., Rao C. M. (2015). Small heat shock proteins: role in cellular functions and pathology. Biochim. Biophys. Acta 1854, 291–319. 10.1016/j.bbapap.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Barnes A., Bale J., Constantinidou C., Ashton P., Jones A., Pritchard J. (2004). Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. J. Exp. Bot. 55, 1473–1481. 10.1093/jxb/erh161 [DOI] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C. J., Bartley L., Ronald P. C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2:13. 10.1186/1746-4811-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O., Jost R., Pollmann S., Masle J. (2008). Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20, 3430–3447. 10.1105/tpc.108.061010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer U. A., Thiele B. J. (2004). The translationally controlled tumour protein (TCTP). Int. J. Biochem. Cell Biol. 36, 379–385. 10.1016/S1357-2725(03)00213-9 [DOI] [PubMed] [Google Scholar]
- Brioudes F., Thierry A. M., Chambrier P., Mollereau B., Bendahmane M. (2010). Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc. Natl. Acad. Sci. U.S.A. 107, 16384–16389. 10.1073/pnas.1007926107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Lu Y., Chen G., Lei J. (2010). Functional characterization of the translationally controlled tumor protein (TCTP) gene associated with growth and defense response in cabbage. Plant Cell Tiss. Organ Cult. 103, 217–226. 10.1007/s11240-010-9769-6 [DOI] [Google Scholar]
- Deng Z., Qin B., Feng F., Li D. (2012). Gene structure analysis and molecular marker development of HbTCTP. Chin. J. Trop. Crops 33, 1450–1455. 10.3969/j.issn.1000-2561.2012.08.023 [DOI] [Google Scholar]
- Dennis M. S., Light D. R. (1989). Rubber elongation factor from Hevea brasiliensis. identification, characterization, and role in rubber biosynthesis. J. Biol. Chem. 264, 18608–18617. [PubMed] [Google Scholar]
- Divya K., Jami S. K., Kirti P. B. (2010). Constitutive expression of mustard annexin, AnnBj1 enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant Mol. Biol. 73, 293–308. 10.1007/s11103-010-9615-6 [DOI] [PubMed] [Google Scholar]
- Ermolayev V., Weschke W., Manteuffel R. (2003). Comparison of Al-induced gene expression in sensitive and tolerant soybean cultivars. J. Exp. Bot. 54, 2745–2756. 10.1093/jxb/erg302 [DOI] [PubMed] [Google Scholar]
- Gébelin V., Argout X., Engchuan W., Pitollat B., Duan C., Montoro P., et al. (2012). Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol. 12:18. 10.1186/1471-2229-12-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gébelin V., Leclercq J., Kuswanhadi Argout X., Chaidamsari T., Hu S., et al. (2013). The small RNA profile in latex from Hevea brasiliensis trees is affected by tapping panel dryness. Tree Physiol. 33, 1084–1098. 10.1093/treephys/tpt076 [DOI] [PubMed] [Google Scholar]
- German M. A., Luo S., Schroth G., Meyers B. C., Green P. J. (2009). Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 4, 356–362. 10.1038/nprot.2009.8 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Galeano D. F., Toscano-Morales R., Calderón-Pérez B., Xoconostle-Cázares B., Ruiz-Medrano R. (2014). Structural divergence of plant TCTPs. Front. Plant Sci. 5:361. 10.3389/fpls.2014.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B. Z., Wu J. L. (2000). Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43. 10.1006/anbo.1999.0995 [DOI] [Google Scholar]
- Hinojosa-Moya J. J., Xoconostle-Cázares B., Lucas W. J., Ruiz-Medrano R. (2006). Differential accumulation of a translationally controlled tumor protein mRNA from Cucurbita maxima in response to CMV infection, in Biology of Plant Microbe Interactions, Vol. 5, eds Sánchez F., Quinto C., Lopez-Lara I. M., Geiger O. (St. Paul, MN: International Society for Molecular Plant-Microbe Interactions; ), 242–246. [Google Scholar]
- Hinojosa-Moya J. J., Xoconostle-Cázares B., Piedra-Ibarra E., Méndez-Tenorio A., Lucas W. J., Ruiz-Medrano R. (2008). Phylogenetic and structural analysis of translationally controlled tumor proteins. J. Mol. Evol. 66, 472–483. 10.1007/s00239-008-9099-z [DOI] [PubMed] [Google Scholar]
- Hinojosa-Moya J. J., Xoconostle-Cázares B., Toscano-Morales R., Ramírez-Ortega F., Cabrera-Ponce J. L., Ruiz-Medrano R. (2013). Characterization of the pumpkin translationally-controlled tumor protein CmTCTP. Plant Signal. Behav. 8:e26477. 10.4161/psb.26477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepflinger M. C., Reitsamer J., Geretschlaeger A. M., Mehlmer N., Tenhaken R. (2013). The effect of translationally controlled tumour protein (TCTP) on programmed cell death in plants. BMC Plant Biol. 13:135. 10.1186/1471-2229-13-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S. M., Noh E. K., Kim H. G., Jeon B. W., Bae K., Hu H. C., et al. (2010). Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses. Plant Cell Physiol. 51, 1499–1514. 10.1093/pcp/pcq111 [DOI] [PubMed] [Google Scholar]
- Jones A. M. E., Thomas V., Bennett M. H., Mansfield J., Grant M. (2006). Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 142, 1603–1620. 10.1104/pp.106.086231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. M., Han Y. J., Hwang O. J., Lee S. S., Shin A. Y., Kim S. Y., et al. (2012). Overexpression of Arabidopsis translationally controlled tumor protein gene AtTCTP enhances drought tolerance with rapid ABA-induced stomatal closure. Mol. Cells 33, 617–626. 10.1007/s10059-012-0080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Deng Z., Chen C., Xia Z., Wu M., He P., et al. (2010a). Identification and characterization of genes associated with tapping panel dryness from Hevea brasiliensis latex using suppression subtractive hybridization. BMC Plant Biol. 10:140. 10.1186/1471-2229-10-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Deng Z., Liu X., Qin B. (2013). Molecular cloning, expression profiles and characterization of a novel translationally controlled tumor protein in rubber tree (Hevea brasiliensis). J. Plant Physiol. 170, 497–504. 10.1016/j.jplph.2012.11.014 [DOI] [PubMed] [Google Scholar]
- Li D., Wang X., Deng Z., Liu H., Yang H., He G. (2016). Transcriptome analyses reveal molecular mechanism underlying tapping panel dryness of rubber tree (Hevea brasiliensis). Sci. Rep. 6:23540. 10.1038/srep23540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu X. Y., Li X. P., Wang Z. Y. (2010b). Cloning of a TCTP gene in wheat and its expression induced by Erysiphe graminis. Bull. Bot. Res. 30, 441–447. 10.7525/j.issn.1673-5102.2010.04.011 [DOI] [Google Scholar]
- Li J., Zhang W. B., Loukas A., Lin R. Y., Ito A., Zhang L. H., et al. (2004). Functional expression and characterization of Echinococcus granulosus thioredoxin peroxidase suggests a role in protection against oxidative damage. Gene 326, 157–165. 10.1016/j.gene.2003.10.027 [DOI] [PubMed] [Google Scholar]
- Liang X. L., Li H. L., Peng S. Q. (2009). Cloning and expression of HbTCTP from Hevea brasiliensis. Mol. Plant Breeding 7, 188–193. 10.3969/j.issn.1672-416X.2009.01.031 [DOI] [Google Scholar]
- Lu C., Wallis J. G., Browse J. (2007). An analysis of expressed sequence tags of developing castor endosperm using a full-length cDNA library. BMC Plant Biol. 7, 42. 10.1186/1471-2229-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakkaew A., Chotigeat W., Phongdara A. (2010). Molecular cloning and expression of EgTCTP, encoding a calcium binding protein, enhances the growth of callus in oil palm (Elaeis guineensis, Jacq). Songklanakarin J. Sci. Technol. 32, 561–569. [Google Scholar]
- Pay A., Heberle-Bors E., Hirt H. (1992). An alfalfa cDNA encodes a protein with homology to translationally controlled human tumor protein. Plant Mol. Biol. 19, 501–503. [DOI] [PubMed] [Google Scholar]
- Peng S. Q., Wu K. X., Huang G. X., Chen S. C. (2011). HbMyb1, a Myb transcription factor from Hevea brasiliensis, suppresses stress induced cell death in transgenic tobacco. Plant Physiol. Biochem. 49, 1429–1435. 10.1016/j.plaphy.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Porebski S., Bailey L. G., Baum B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. 10.1007/BF02772108 [DOI] [Google Scholar]
- Qin X., Gao F., Zhang J., Gao J., Lin S., Wang Y., et al. (2011). Molecular cloning, characterization and expression of cDNA encoding translationally controlled tumor protein (TCTP) from Jatropha curcas L. Mol. Biol. Rep. 38, 3107–3112. 10.1007/s11033-010-9980-x [DOI] [PubMed] [Google Scholar]
- Rahman A. Y. A., Usharraj A. O., Misra B. B., Thottathil G. P., Jayasekaran K., Feng Y., et al. (2013). Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics 14:75. 10.1186/1471-2164-14-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S. L., Laohavisit A., Mortimer J. C., Shabala L., Swarbreck S. M., Shabala S., et al. (2014). Annexin 1 regulates the H2O2-induced calcium signature in Arabidopsis thaliana roots. Plant J. 77, 136–145. 10.1111/tpj.12372 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Medina C., Atkins C. A., Mann A. J., Jordan M. E., Smith P. M. C. (2011). Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.). BMC Plant Biol. 11:36. 10.1186/1471-2229-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage-Ono K., Ono M., Harada H., Kamada H. (1998). Dark-induced accumulation of mRNA for a homolog of translationally controlled tumor protein (TCTP) in Pharbitis. Plant Cell Physiol. 39, 357–360. 10.1093/oxfordjournals.pcp.a029377 [DOI] [PubMed] [Google Scholar]
- Tang C. R., Huang D. B., Yang J. H., Liu S. J., Sark S., Li H. P., et al. (2010). The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cell Environ. 33, 1708–1720. 10.1111/j.1365-3040.2010.02175.x [DOI] [PubMed] [Google Scholar]
- Vincent D., Ergül A., Bohlman M. C., Tattersall E. A., Tillett R. L., Wheatley M. D., et al. (2007). Proteomic analysis reveals differences between Vitis vinifera L. cv. chardonnay and cv. cabernet sauvignon and their responses to water deficit and salinity. J. Exp. Bot. 58, 1873–1892. 10.1093/jxb/erm012 [DOI] [PubMed] [Google Scholar]
- Wang F., Shang Y., Yang L., Zhu C. (2012). Comparative proteomic study and functional analysis of translationally controlled tumor protein in rice roots under Hg2+ stress. J. Environ. Sci. 24, 2149–2158. 10.1016/S1001-0742(11)61062-0 [DOI] [PubMed] [Google Scholar]
- Xu J., Aileni M., Abbagani S., Zhang P. (2010). A reliable and efficient method for total RNA isolation from various members of spurge family (Euphorbiaceae). Phytochem. Anal. 21, 395–398. 10.1002/pca.1205 [DOI] [PubMed] [Google Scholar]
- Xu W. X., Xiao X. Z. (1988). Studies on peroxidase and superoxidase dismutase isozymes in dry rubber trees. Chinese J. Trop. Crops 9, 31–36. [Google Scholar]
- Yenofsky R., Cereghini S., Krowczynska A., Brawerman G. (1983). Regulation of mRNA utilization in mouse erythroleukemia cells induced to differentiate by exposure to dimethyl sulfoxide. Mol. Cell Biol. 3, 1197–1203. 10.1128/MCB.3.7.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof F., Siti Arija M. A., Ghandimathi H., Hamzah Z., Sivakumaran S., Yeang H. Y. (1995). Changes in some physiological latex parameters in relation to over exploitation and onset of induced tapping panel dryness. J. Nat. Rubb. Res. 10, 182–198. [Google Scholar]
- Zeng R. Z., Li X. K., Nie Z. Y., Dai L. J., Li Y., Duan C. F. (2011). Screening interaction proteins of rubber elongation factor (REF) by yeast two-hybrid system. J. Trop. Subtrop. Bot. 19, 400–406. 10.3969/j.issn.1005-3395.2011.05.002 [DOI] [Google Scholar]
- Zhang D., Li F., Weidner D., Mnjoyan Z. H., Fujise K. (2002). Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and fortilin. the potential role of MCL1 as a fortilin chaperone. J. Biol. Chem. 277, 37430–37438. 10.1074/jbc.M207413200 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yu J., Li D., Zhang Z., Liu F., Zhou X., et al. (2010). PMRD: plant microRNA database. Nucleic Acids Res. 38 (Suppl. 1), 806–813. 10.1093/nar/gkp818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.