Abstract
Wolff–Parkinson–White (WPW) syndrome is a common cause of supraventricular tachycardia that carries a risk of sudden cardiac death. To date, mutations in only one gene, PRKAG2, which encodes the 5’ -AMP-activated protein kinase subunit γ-2, have been identified as causative for WPW. DNA samples from five members of a family with WPW were analyzed by exome sequencing. We applied recently designed prioritization strategies (VAAST/pedigree VAAST) coupled with an ontology-based algorithm (Phevor) that reduced the number of potentially damaging variants to 10: a variant in KCNE2 previously associated with Long QT syndrome was also identified. Of these 11 variants, only MYH6 p.E1885K segregated with the WPW phenotype in all affected individuals and was absent in 10 unaffected family members. This variant was predicted to be damaging by in silico methods and is not present in the 1,000 genome and NHLBI exome sequencing project databases. Screening of a replication cohort of 47 unrelated WPW patients did not identify other likely causative variants in PRKAG2 or MYH6. MYH6 variants have been identified in patients with atrial septal defects, cardiomyopathies, and sick sinus syndrome. Our data highlight the pleiotropic nature of phenotypes associated with defects in this gene.
Keywords: Wolff–Parkinson–White, whole exome sequencing, MYH6
INTRODUCTION
Wolff Parkinson, and White first identified a series of patients with the following features: (i) a bundle branch block; (ii) a short PR interval; and (iii) paroxysms of tachycardia. In 1940, this combination of pre-excitation and paroxysmal tachycardia was termed Wolff–Parkinson–White (WPW) syndrome [Wolff et al., 2006]. WPW is commonly associated with paroxysmal supraventricular tachycardia (1.5–3.1 per 1,000 persons) [Guize et al., 1985] that is maintained by accessory pathway(s) secondary to a developmental cardiac defect in atrioventricular electrical insulation [Kent, 1893]. Although electrophysiological studies remain the gold standard for differentiating various phenotypes, the existence of identical electrophysiological characteristics in a single cohort argues for a common genetic cause. The familial occurrence of the WPW syndrome is well documented, is typically inherited in an autosomal dominant pattern, and is sometimes associated with familial cardiomyopathy [Harnischfeger, 1959;Massumi, 1967; Schneider, 1969]. The molecular genetics of WPW have been investigated but to date only a single gene, PRKAG2, has been identified and mutations in this gene explain only a small fraction (<5%) of WPW (Gollob et al., 2001 a,b).
Advances in next generation sequencing modalities, such as whole-exome sequencing (WES), have revolutionized the study of genetic susceptibility to disease. However, in light of the large number of variants identified in these datasets, variant prioritization and identification of disease-causing variants remain a significant challenge. Variant prioritization tools, such as SIFT [Kumar et al., 2009] and PolyPhen2 [Adzhubei et al., 2010] rely on phylogenetic conservation to identify damaging variants and thus are unable to score a large number of variants that reside in non-conserved regions of unknown structure. Moreover, these tools often have low specificity and sensitivity [Flanagan et al., 2010]. The variant annotation, analysis, and search tool (VAAST) utilizes a more accurate and comprehensive approach to variant prioritization, by using the global, genome-wide frequency of observing an amino acid substitution in any gene [Yandell et al., 2011; Hu et al., 2013]. This approach allows VAAST to score any coding change regardless of conservation [Rope et al., 2011; Hu et al., 2013; McElroy et al., 2013; Shirley et al., 2013]. Pedigree-VAAST (pVAAST) [Hu et al., 2014] performs linkage analysis by calculating a novel gene-based LOD score specifically designed for sequence data. The LOD score at each locus is incorporated directly into VAAST to increase the accuracy and greatly decrease the bioinformatic complexity of family-based disease-gene identification efforts. Finally, the phenotype driven variant ontological re-ranking (Phevor) [Singleton et al., 2014] tool integrates knowledge from multiple biomedical ontologies with VAAST output to reprioritize candidate disease-causing variants. In this report, we combine these novels variant prioritization strategies to identify disease-causing variants in whole exomes from a multi-generation family with WPW.
MATERIALS AND METHODS
Patient Enrollment and Phenotyping
A multigenerational Caucasian family with WPW (K32326) was identified in the Heart Center at Primary Children’s Hospital. Clinical evaluation of family members was performed by an electrophysiologist, a medical geneticist, and genetic counselor, who excluded confounding alternatives such as the mitochondrial MELAS syndrome [Aggarwal et al., 2001]. WPW patients were diagnosed using strict electrocardiographic criteria: (i) short PR interval and (ii) presence of a ventricular pre-excitation or delta wave, as determined by an electrophysiologist (SPE, EVS, MT-F).
With University of Utah Institutional Review Board approval, members of the family were enrolled in the University of Utah Pediatric Cardiology Genotype-Phenotype Core after obtaining written informed consent. All human subjects’ research were performed in accordance with relevant guidelines and regulations. DNA was isolated from peripheral blood samples using a Centra Autopure LS (Qiagen, Valencia, CA) in the University of Utah Center for Clinical and Translational Science. DNA samples were analyzed by agarose gel electrophoresis to confirm the DNA integrity and quantitated using a Nanodrop.
Analysis of PRKAG2
PCR primers were designed to amplify the coding exons, as well as at least 50 nucleotides of the surrounding introns, of PRKAG2 using the ExonPrimer utility in the UCSC genome browser (http://genome.ucsc.edu/: PCR Primers in Supplemental Table SII) and used to amplify DNA from two of the patients. An aliquot of DNA was analyzed by agarose gel electrophoresis and then the PCR product was purified by treating with 4 µl of Exo-SAP-IT (Affymetrix) at 37°C for 2 hr and 80°C for 15 min. The PCR product was then submitted to the University of Utah DNA sequencing core for analysis and results compared to the published PRKAG2 sequences using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Exome Sequencing
DNA from five members of K32326 (Fig. 1: I:2, II:2, II:5, III:6, and III:7) were sent to the Baylor Hopkins Center for Mendelian Genomics for WES. In brief, 1 µg of DNA was used to construct an Illumina paired-end pre-capture library according to the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_-Guide_1005361_D). The complete protocol and oligonucleotide sequences are accessible from the Baylor Human Genome Sequencing Center (HGSC) website (https://hgsc.bcm.edu/sites/default/files/documents/Illumina_Barcoded_Paired-End_Capture_Library_Preparation.pdf). Four pre-captured libraries were pooled and then hybridized in solution to the HGSC CORE design [Bainbridge et al., 2011] (52Mb, NimbleGen) according to the manufacturer’s protocol NimbleGen SeqCap EZ Exome Library SR User’s Guide (Version 2.2) with minor revisions. The sequencing run was performed in paired-end mode using an Illumina HiSeq 2000 platform, with sequencing-by-synthesis reactions extended for 101 cycles from each end and an additional 7 cycles for the index read. With a sequencing yield of 12 Gb, coverage depth of 20X or greater was achieved for 92% of the targeted exome bases. Illumina sequence analysis was performed using the HGSC Mercury analysis pipeline (https://www.hgsc.bcm.edu/software/mercury) that moves data through various analysis tools from the initial sequence generation on the instrument to annotated variant calls (SNPs and intra-read in/dels). Reads were mapped to the GRCh37 Human reference genome (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/) using the Burrows-Wheeler aligner[Li and Durbin, 2009] (BWA, http://bio-bwa.sourceforge.net/) to produce BAM [Li et al., 2009] (bi-] (binary alignment/map) files. Quality recalibration was performed using GATK [DePristo et al., 2011] (http://www.broadinstitute.org/gatk/), and where necessary separate sequence-event BAMs were merged into a single sample-level BAM. Using the software package SAMtools [Li et al., 2009], the aligned sequencing reads were converted and merged into sorted and indexed BAM files. The SAMtools utilities mpileup and bcftools were implemented to call sequence variants. To reduce the number of false positives in the call-set, the five individuals in the family were called together with 139 individuals from the 1,000 Genomes project. ANNOVAR was used to identify variants not previously reported in the 1,000 Genome Project (Phase 1 All-Sites (2011_05)), dbSNP databases [Sherry et al., 2001] (dbSNP build 132), or present with a Minor Allele Frequency (MAF) <0.1% in Caucasians. To predict deleterious effects of non-synonymous amino acid changes, ANNOVAR utilizes various functional annotation algorithms such as SIFT [Kumar et al., 2009], PolyPhen2 [Adzhubei et al., 2010], and MutationTaster [Schwarz et al., 2010]. AlignGVD [Tavtigian et al., 2006] predictions were also made using Alamut software (v2.3: Interactive Biosoftware, Rouen, France).
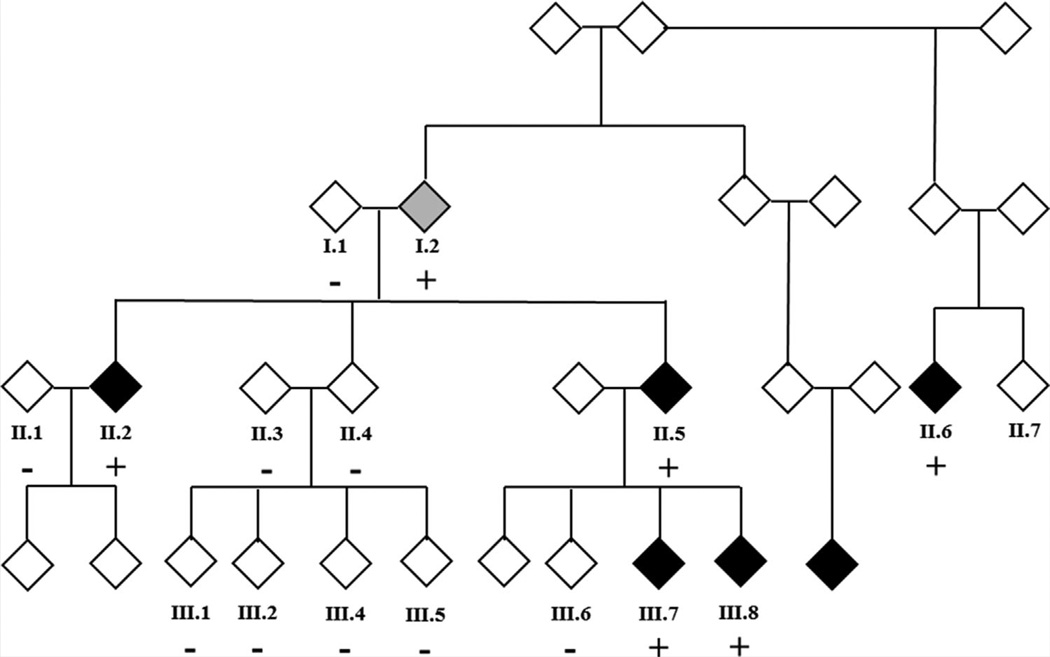
FIG. 1.
Family K32326 pedigree. The gender of family members is masked for confidentiality. Black symbols represent patients with WPW and a gray symbol represents the patient with a diagnosis of SVT (Patient 1:2): autosomal dominant inheritance, with incomplete penetrance, is the most likely genetic model. Only participants in the study for whom DNA is available for analysis are numbered. +: Positive for MYH6 c.5653G>A, p.Glu1885Lys; −: Negative for MYH6 c.5653G>A, p.Glu1885Lys.
Further variant prioritization was accomplished through the use of VAAST [Yandell et al., 2011], which combines variant frequency data, mutation severity, and conservation into a single score that is compared genome wide. The analysis was performed following best practices as described in the publication by Kennedy et al. [2014]. Because all of the sequenced individuals are related, pedigree-VAAST (pVAAST) was chosen over standard VAAST analysis [Hu et al., 2014]. pVAAST further empowers the standard VAAST algorithm by probabilistically calculating the degree to which variants follow a specified inheritance pattern. In this case, the disease follows a dominant mode of inheritance with reasonably high penetrance, so the pVAAST analysis was parameterized accordingly. pVAAST results were then re-ranked using Phevor [Singleton et al., 2014]. The Phevor tool takes rankings from gene prioritization tools and re-ranks them based on phenotype information though terms in biomedical ontologies such as GO [Ashburner et al., 2000] and HPO [Kohler et al., 2014]. For this analysis, the Phevor was run using the following HPO terms: prolonged QRS complex (HP:0006677), shortened PR interval (HP:0005165), paroxysmal supraventricular tachycardia (HP:0004763), sudden cardiac death (HP:0001645), ventricular pre-excitation with multiple accessory pathways (HP:0006684), paroxysmal atrial fibrillation (HP:0004757), stroke (HP:0001297), cardiomyopathy (HP:0001638), palpitations (HP:0001962) which describe the WPW phenotype. Cardiac expression of candidate genes was assessed from GenAtlas data accessed by searches of the BioGPS portal (biogps.org).
Validation of Variants Identified by Exome Sequencing
PCR primers were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) to amplify and validate variants detected by Exome Sequencing (PCR primers described in Supplemental Table SIII). The PCR product was purified and sequenced as described above. Once a variant was confirmed all available family members, affected and unaffected, were screened in the same way to assess variant segregation.
Analysis of the Replication Cohort
Forty-seven unrelated patients diagnosed with WPW by ECG and enrolled into the Genotype-Phenotype Core served as a comparison cohort: eight of these patients reported a family history of WPW. Primers were designed to amplify coding exons and exon-intron boundaries of candidate genes using the ExonPrimer utility, as described above (Supplemental Table SII). All patient DNA samples (10 ng per sample) were amplified by PCR in duplicate, along with negative (water) controls, using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) and analyzed on a Lightscanner (Biofire, Salt Lake City, UT), as described previously [Arrington et al., 2008, 2012]. Samples giving abnormal profiles were analyzed by DNA sequencing, as described above.
RESULTS
Clinical Characteristics of Family K32326
The pedigree for K32326 is shown in Figure 1. All the enrolled family members were previously diagnosed with WPW or diagnosed by ECG at the time of enrollment.
Patient I.2 presented to a local emergency department at age 61 years with chest pain and palpitations. The patient’s heart rate was 216 bpm and his rhythm was supraventricular tachycardia (SVT) with a probable rate-related bundle branch block. With adenosine administration, the tachycardia terminated transiently but returned within minutes. Ultimately, the patient converted to sinus rhythm with intravenous diltiazem administration. While no delta wave was present when in sinus rhythm, the response to adenosine and family history suggests the presence of a concealed accessory pathway. Subsequently, the patient was diagnosed with aortic stenosis and later, with atrial fibrillation (AF): we were unable to obtain surgical record to determine whether the aortic stenosis was calcific.
Patient II.2 was diagnosed with WPW and a subaortic membrane as a child. At age 19, the patient underwent resection of the subaortic membrane and a septal myectomy. With regard to the subaortic membrane, there was not a muscular component to the membrane itself, rather there was additional septal hypertrophy below the membrane that was felt to be a secondary consequence of the obstructive membrane. An intraoperative electrophysiology study was also performed with ablation of multiple accessory atrioventricular pathways. In the immediate post-operative period, the patient was hemodynamically unstable with ventricular tachycardia and underwent a catheter-based electrophysiology study/ ablative procedure. This procedure was complicated by complete heart block and possible incomplete ablation of ventricular tachycardia necessitating placement of an ICD. The patient subsequently developed recurrent left-ventricular outflow tract obstruction at age 29 years and underwent repeat subaortic membrane resection and a valve sparing Konno procedure.
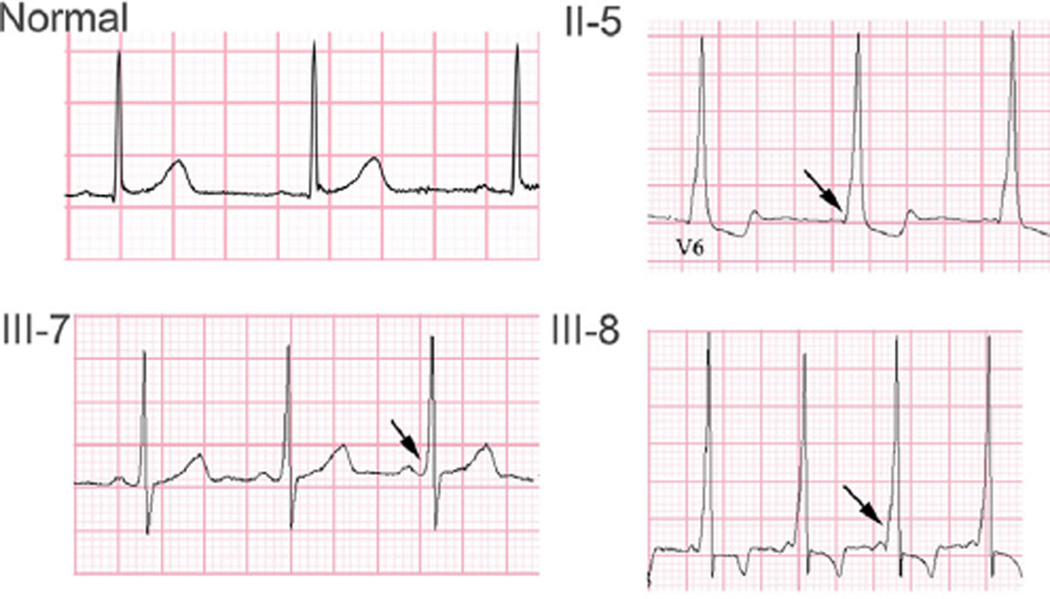
Patient II.5 was diagnosed with WPW at age 28 years after presenting with SVT following a routine surgical procedure. The patient’s heart was structurally normal with normal ventricular function. A radiofrequency (RF) ablation procedure was performed that identified two left-sided accessory pathways and a right-sided fasciculoventricular pathway, that was not a participant in SVT circuit. The two left-sided accessory pathways were ablated. Shortly thereafter, the patient experienced a syncopal episode and was noted to be in a wide QRS complex tachycardia, consistent with SVT with a rate-related right bundle branch pattern (Fig. 2). Over the next year, the patient underwent two additional RF ablation procedures to ablate the recurrence of the manifest left-sided pathways.
FIG. 2.
Representative ECGs from selected members of K32326. Top row: example of a normal resting ECG (left panel). Right panel, resting ECG from patient II-5 in sinus rhythm, showing classic short PR interval and ventricular pre-excitation or delta wave (arrow). Bottom row: ECG from patient III-7 shows short PR interval and subtle ventricular pre-excitation (left panel). ECG from patient III-8 reveals markedly short PR interval and prominent ventricular pre-excitation (right panel).
Patient II.6 was diagnosed with WPW as at 5 years of age. This patient experienced multiple episodes of SVT, often with a rate-related right bundle branch pattern. After experiencing three episodes of syncope despite beta-blocker therapy, the patient underwent RF ablation of a left lateral accessory pathway at age 14. The ablation procedure was transiently effective, but subsequent ECGs demonstrated persistent ventricular pre-excitation. The patient’s heart is structurally normal with normal ventricular function.
Patient III.7 was found to have ventricular pre-excitation at the time of enrollment in this research project by a screening ECG (Fig. 2). The patient has no history of tachyarrythmia or other cardiac symptoms.
Patient III.8 presented with neonatal SVT and was diagnosed with WPW syndrome (Fig. 2). The rhythm was initially controlled with propranolol, which was discontinued at around 1 year of age. The patient is now 10 years old, still has evidence of ventricular preexcitation on ECG but has not had any further episodes of SVT. The patient’s heart is structurally and functionally normal.
Screening members of this family by PCR and DNA sequencing failed to identify any disease-causing variants in PRKAG2. Therefore, this kindred was considered an excellent candidate family for whole exome sequencing for the identification of novel disease associated loci.
Exome Sequence Analysis and Segregation of Variants
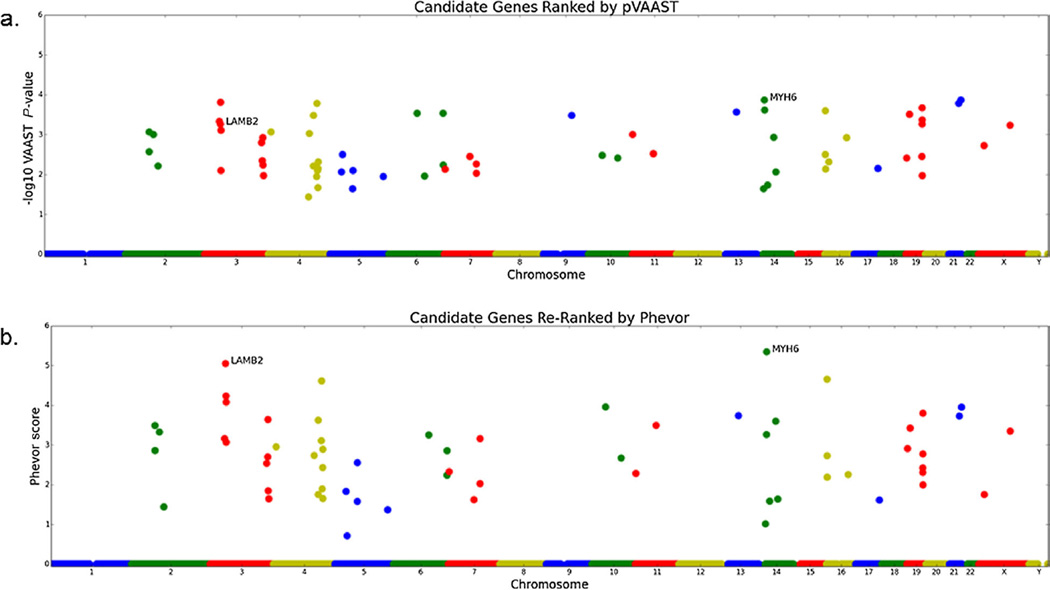
DNA from five family members I:2, II:2, II:5, III:6, and III:7 were analyzed by whole exome sequencing. Analysis of shared variants identified several hundred that were novel (not in ESP or 1,000 g databases) or rare (MAF <0.1%), predicted to be damaging by at least one in silico algorithm (SIFT, PolyPhen2, MutationTaster, and AlignGVD) and found in genes with a potential role in cardiac development or function. To determine which of the shared variants were most likely causative, they were prioritized using VAAST and pVAAST and then Phevor. This analysis found two variants of interest with higher scores than any other (laminin (β-2 chain (LAMB2) c.1750C>T: p.Arg584Cys, and myosin heavy chain 6 (MYH6) c.5653G>A, p.Glu1885Lys), with the MYH6 variant having the highest rank (Fig. 3 and Table I). Because none of the variants reached genome wide significance threshold for pVAAST (2.54 × 10−6), and Phevor is a non-parametric test, we had to consider many of the top ranked genes. This was done through additional genetic screening of un-sequenced family members (see below). An additional eight variants ranked by pVAAST or Phevor could not be eliminated as WPW candidates because of their reported function and/or cardiac expression (Table I). In addition, a variant in KCNE2 (p.Ile57Thr), previously associated with Long QT syndrome [Abbott et al., 1999] and present in ClinVar as a disease-causing variant (http://www.ncbi.nlm.nih.gov/clinvar/RCV000006426/), was identified and considered a candidate because of this association, even though it was not ranked (Table I]. All available family members (both affected and unaffected) were screened to determine segregation of these variants. The MYH6 variant was the only variant present in all affected individuals in the extended pedigree (including 1.2) and absent in all unaffected family members available for screening (Fig. 1). Of note, the KCNE2 variant was not detected in two of the affected individuals and was present in four of the unaffected family members.
FIG. 3.
Manhattan plots of the pVAAST and Phevor results. A: Manhattan plot of pVAAST scores for all protein-coding genes in Human Genome release hg19; each dot is a single gene. B: Manhattan plot of Phevor scores obtained by using the pVAAST results in conjunction with the phenotype terms describing symptoms of Wolff–Parkinson–White. The x-axis shows the genomic location of each gene arranged by position along the chromosomes. The y-axis is the Phevor or pVAAST score. MYH6 and LAMB2 are highlighted in each plot as they are the highest scoring genes by the combination of analyses.
TABLE I.
Summary of Variants Ranked by pVAAST/Phevor Showing Pathogenicity Predictions of Four In Silico Algorithms
| Gene | Phevor rank |
pVAAST rank |
Variant (transcript level) |
Variant (protein level) |
AlignGVD score |
SIFT score | Polyphen2 score |
MutationTaster score |
|---|---|---|---|---|---|---|---|---|
| MYH6 | 1 | 2 |
NM_002471.3: c.5653G>A |
p. Glu1885Lys |
C55 | Deleterious (0.00) |
Possibly damaging (0.716) |
Disease causing (1.0) |
| LAMB2 | 2 | 17 |
NM_002292.3: c.1750C>T |
p. Arg584Cys |
C65 | Deleterious (0.00) |
Probably damaging (0.997) |
Disease causing (1.0) |
| CCDC154 | 3 | 8 |
NM_001143980.1: c.959T>A |
p. Leu320Gln |
C65 | Deleterious (0.00) |
Probably damaging (0.999) |
Polymorphism (0.940) |
| IFRD2 | 5 | 3 |
NM_006764.4: c.1175G>A |
p. Arg392His |
C0 | Tolerated (0.10) |
Probably damaging (0.945) |
Disease causing (0.983) |
| DIP2A | 8 | 1 |
NM_015151.3: c.3319C>T |
p. Arg1107Trp |
C0 | Deleterious (0.01) |
Probably damaging (0.948) |
Disease causing (0.995) |
| SIGLEC11 | 9 | 18 |
NM_052884.2: c.342C>G |
p. Cys114Trp |
C15 | Deleterious (0.00) |
Probably damaging (1.000) |
Disease causing (1.0) |
| LTF | 22 | 16 |
NM_002343.3: c.293C>T |
p.Ala98Val | C0 | Tolerated (0.07) |
Possibly damaging (0.689) |
Polymorphism (0.554) |
| FUZ | 32 | 6 |
NM_025129.4: c.272C>T |
p.Ser91Phe | C0 | Deleterious (0.02) |
Probably damaging (0.946) |
Disease causing (0.998) |
| INTU | 33 | 23 |
NM_015693.3: c.2512T>C: rs144025772 |
p. Cys838Arg |
C0 | Deleterious (0.01) |
Probably damaging (0.974) |
Disease causing (1.000) |
| MYH14 | 42 | 15 |
NM_001145809.1: c.1919G>A: rs199696801 |
p. Arg640Gln |
C0 | Deleterious (0.04) |
Benign (0.055) |
Polymorphism (0.934) |
| KCNE2 | - | - |
NM_172201.1: c.170T>C: rs74315448 |
p.lle57Thr | C65 | Deleterious (0.00) |
Probably damaging (0.918) |
Disease causing (0.999) |
Note: AlignGVD scores variants on a scale C0, C15, C25, C35, C45, C55, C65, with CG5 representing the highest risk allele. The KCNE2 variant was not ranked by Phevor but was considered a candidate variant because of its association with Long OT syndrome [Abbott et al., 1999] and is present in ClinVar as a disease-causing variant (http://www.ncbi.nlm.nih.gov/clinvar/RCV000006426/).
To further confirm the results of the VAAST analysis, results from the screens of family members who were not analyzed by WES were placed into a second pVAAST/Phevor analysis (Supplemental Fig. S1). This analysis confirmed that MYH6 is by far the most likely causative gene among the top scoring candidates. When considered with the additional screened family members, the pVAAST P-value is very close to genome wide significance (P= 9.77 × 10−6).
In addition to having the highest score in Phevor, the MYH6 variant is predicted to be likely deleterious/damaging by four commonly used in silico algorithms: Align GVD, SIFT, Polyphen2, and MutationTaster (Table I). This missense mutation (p.Glu1885Lys) alters an amino acid residing in the myosin tail domain and is perfectly conserved between species from zebrafish to humans (Fig. 4).
FIG. 4.
Conservation analysis of MYH6. Alignments of MYH6 across several species of the amino acids surrounding Glutamic acid 1885 generated using the PRALINE multiple sequence alignment tool (http://www.ibi.vu.nl/programs/pralinewww/). [Heringa, 1999] The scoring scheme is from 0 for the least conserved alignment position, up to 10 for the most conserved alignment position (represented by *) and the color assignments for each score are shown at the bottom. Note the complete conservation of this amino acid and the high degree of conservation across the domain.
The distinct cohort of 47 unrelated WPW patients was screened for the mutations in PRKAG2 and MYH6. Two novel synonymous variants were identified in PRKAG2 (c.231C>T: p.Phe77 and c.1485C>A. p.Thr495 (NM_016203.3)): neither are predicted to alter splicing. One novel non-synonymous variant was identified in MYH6 (c.635C>T; p.Ala212Val), which is predicted to be benign/ not damaging by AlignGVD, SIFT, MutatioTaster, and Polyphen2. Therefore, neither PRKAG2 nor MYH6 are commonly mutated in our cohort of patients with WPW, confirming a high degree of genetic heterogeneity in this disease.
DISCUSSION
This study represents the first reported exome sequence analysis of a family with WPW. This family was initially screened for mutations in PRKAG2, the only gene robustly associated with WPW, and was negative for candidate mutations. We, therefore, performed WES on five family members (three affected, one unaffected, and an obligate carrier) in order to identify the disease-causing gene in this highly penetrant kindred. We employed recently designed prioritization strategies (VAAST/ pedigree VAAST) coupled with the ontology-based algorithm Phevor to generate a limited number of potential candidate alleles. This strategy was previously successful in identifying disease-causing alleles in small, family-based WES analyses [Singleton et al., 2014]. Of the highly ranked alleles identified in our WPW family, only one segregated with the phenotype: MYH6 c.5653G>A; p.Glu1885Lys. This variant affects a highly conserved glutamic acid in the myosin tail, is predicted to be pathogenic/deleterious by four separate in silico algorithms and is ranked as the strongest candidate variant by Phevor [Singleton et al., 2014]. This evidence supports the conclusion that the MYH6 variant is the most likely causative mutation responsible for the WPW phenotype in this family, although there was variable/incomplete penetrance in this family. The KCNE2 variant (I57T) is listed in ClinVar as a pathogenic variant. However, given the lack of segregation with phenotype in this WPW family, it is likely that this variant is not pathogenic or a modifier, at best.
These data expand the cardiac phenotypes associated with mutations in MYH6. The original association of MYH6 mutations with cardiac disease was in a family with hypertrophic cardiomyopathy (HCM) [Niimura et al., 2002]. No members of the family studied here had signs of HCM: one patient (II.2) had mild septal hypertrophy but this was probably due to the elevated pressure within the left ventricle as a result of a subaortic membrane, rather than HCM. Subsequently, there have been other reports of mutations associated with HCM and dilated cardiomyopathy [Carniel et al., 2005]. In 2005, Ching et al. (2005) reported the identification of a mutation in MYH6 in a family with atrial septal defects (ASD): other MYH6 mutations have also been linked to ASD [Granados-Riveron et al., 2010; Arrington et al., 2012].
In addition to the role of Myh6 in structural heart disease, variations in MYH6 have previously been linked to cardiac arrhythmias and heart rate, but not pre-excitation. In a prior study to identify variants that modulate heart rate, PR interval and QRS duration in individuals of European descent, a large genome-wide association study (GWAS) identified MYH6 as a locus linked to heart rate [Holm et al., 2010], and this was confirmed in a recent replication study [den Hoed et al., 2013]. In a GWAS study of Icelandic patients with sick sinus syndrome (SSS), Holm et al. (2011) identified an association with SNPs at chromosome 14q11. Whole genome sequencing analysis of four patients carrying one of the associated SNPs identified a missense variant in MYH6 (c.2161C>T, p.Arg721Trp). Analysis of additional Icelandic patients and controls identified this variant in 2.1% of SSS patients but only 0.21% of controls. The authors calculated that the lifetime risk of being diagnosed with SSS is around 6% for non-carriers of MYH6 c.2161C>T, but approximately 50% for carriers of the variant. In addition, there was a residual association, after exclusion of SSS cases, with other arrhythmias, including atrial fibrillation.
There is also an indirect link between MYH6 and arrhythmias. The microRNA, miR-208a, is encoded within an intron of MYH6. MiR-208a Tg mice develop cardiac arrhythmias, have abnormal cardiac conduction, and altered expression of the cardiac transcription factors, including GATA4, and connexin 40 [Callis et al., 2009]. Further, the expression of this microRNA directly correlates with the expression of MYH6 [Diniz et al., 2013]. Therefore, mutations that alter MYH6 mRNA stability could impact the expression of MiR-208a, and consequently downstream target genes.
In summary, we utilized whole exome sequencing in a high-risk pedigree to identify a novel candidate locus for WPW, MYH6. The variant identified in this family (p.Glu1885Lys) is novel, predicted to be deleterious, and potentially alters the structure of a highly conserved functional domain. This coiled-coil myosin heavy chain tail region is responsible for interactions between myosin molecules and provides the structural backbone of the thick filament [Strehler et al., 1986]. The major limitations of this study are (i) that while WES provides data for the coding regions of most genes in the human genome there are still other regions that are not well covered (Supplemental Table SI). Therefore, disease causing variants in these regions would be missed, (ii) No functional data are provided to support the conclusion, that the MYH6 variant is causative. However, we believe the combination of bioinformatics approaches and the known role for this protein in cardiac disease makes this a very strong candidate. We did not identify variants in MYH6 or PRKAG2 in other individuals from our cohort of 47 unrelated WPW patients, underscoring the high degree of genetic heterogeneity in this condition and suggesting that additional investigation in large WPW cohorts will be needed to replicate this finding. It is unclear why mutations in MYH6 cause such pleiotropic effects on cardiac structure and function, ranging from atrial septal defects, to cardiomyopathies, to cardiac arrhythmias. From the locations of the variants associated with these phenotypes, genotype-phenotype correlations are not obvious (Supplemental Fig. S2) and will require more detailed modeling.
Supplementary Material
Acknowledgments
This work was supported by funds from the American Heart Association (12GRNT11090001: Bowles, P.I. and 10FTF3920017: Jou, P.I), National Institutes of Health (1X01HL115006: Bowles and Leppert, P.I.), and the Primary Children’s Foundation (Gruber, P.I.). Portions of this work were supported by NIGMS R01GM104390 and NHGRI R44HG006579 (Yandell, P.I.). Brett Kennedy was supported by a training award from the National Center for Advancing Translational Sciences, National Institutes of Health (1ULTR001067). DNA extractions were performed in the University of Utah Center for Clinical and Translational Science, which is funded by Public Health Services research grant no. M01-RR00064 from the National Center for Research Resources, the Children’s Health Research Center at the University of Utah, and the Clinical Genetics Research Program at the University of Utah. Exome sequencing was performed by the Baylor-Hopkins Center for Mendelian Genomics, which is funded by National Human Genome Research Institute (NHGRI) grant U54HG006542 to Dr. Jim Lupski.
Footnotes
Conflicts of interests: none.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal P, Gill-Randall R, Wheatley T, Buchalter MB, Metcalfe J, Alcolado JC. Identification of mtDNA mutation in a pedigree with gestational diabetes, deafness, Wolff–Parkinson–White syndrome and placenta accreta. Hum Hered. 2001;51:114–116. doi: 10.1159/000022950. [DOI] [PubMed] [Google Scholar]
- Arrington CB, Bleyl SB, Matsunami N, Bonnell GD, Otterud BE, Nielsen DC, Stevens J, Levy S, Leppert MF, Bowles NE. Exome analysis of a family with pleiotropic congenital heart disease. Circ Cardiovasc Genet. 2012;5:175–182. doi: 10.1161/CIRCGENETICS.111.961797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrington CB, Sower CT, Chuckwuk N, Stevens J, Leppert MF, Yetman AT, Bowles NE. Absence of TGFBR1 and TGFBR2 mutations in patients with bicuspid aortic valve and aortic dilation. Am J Cardiol. 2008;102:629–631. doi: 10.1016/j.amjcard.2008.04.044. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Wu Y, Newsham I, Muzny DM, Jefferies JL, Albert TJ, Burgess DL, Gibbs RA. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel E, Taylor MR, Sinagra G, Di Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov D, Graw SL, Feiger J, Zhu XZ, Dao D, Ferguson DA, Bristow MR, Mestroni L. Alpha-myosin heavy chain: A sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54–59. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
- Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37:423–428. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, Nolte IM, Segre AV, Holm H, Handsaker RE, Westra HJ, Johnson T, Isaacs A, Yang J, Lundby A, Zhao JH, Kim YJ, Go MJ, Almgren P, Bochud M, Boucher G, Cornelis MC, Gudbjartsson D, Hadley D, van der Harst P, Hayward C, den Heijer M, Igl W, Jackson AU, Kutalik Z, Luan J, Kemp JP, Kristiansson K, Ladenvall C, Lorentzon M, Montasser ME, Njajou OT, O’Reilly PF, Padmanabhan S, St Pourcain B, Rankinen T, Salo P, Tanaka T, Timpson NJ, Vitart V, Waite L, Wheeler W, Zhang W, Draisma HH, Feitosa MF, Kerr KF, Lind PA, Mihailov E, Onland-Moret NC, Song C, Weedon MN, Xie W, Yengo L, Absher D, Albert CM, Alonso A, Arking DE, de Bakker PI, Balkau B, Barlassina C, Benaglio P, Bis JC, Bouatia-Naji N, Brage S, Chanock SJ, Chines PS, Chung M, Darbar D, Dina C, Dorr M, Elliott P, Felix SB, Fischer K, Fuchsberger C, de Geus EJ, Goyette P, Gudnason V, Harris TB, Hartikainen AL, Havulinna AS, Heckbert SR, Hicks AA, Hofman A, Holewijn S, Hoogstra-Berends F, Hottenga JJ, Jensen MK, Johansson A, Junttila J, Kaab S, Kanon B, Ketkar S, Khaw KT, Knowles JW, Kooner AS, Kors JA, Kumari M, Milani L, Laiho P, Lakatta EG, Langenberg C, Leusink M, Liu Y, Luben RN, Lunetta KL, Lynch SN, Markus MR, Marques-Vidal P, Mateo Leach I, McArdle WL, McCarroll SA, Medland SE, Miller KA, Montgomery GW, Morrison AC, Muller-Nurasyid M, Navarro P, Nelis M, O’Connell JR, O’Donnell CJ, Ong KK, Newman AB, Peters A, Polasek O, Pouta A, Pramstaller PP, Psaty BM, Rao DC, Ring SM, Rossin EJ, Rudan D, Sanna S, Scott RA, Sehmi JS, Sharp S, Shin JT, Singleton AB, Smith AV, Soranzo N, Spector TD, Stewart C, Stringham HM, Tarasov KV, Uitterlinden AG, Vandenput L, Hwang SJ, Whitfield JB, Wijmenga C, Wild SH, Willemsen G, Wilson JF, Witteman JC, Wong A, Wong Q, Jamshidi Y, Zitting P, Boer JM, Boomsma DI, Borecki IB, van Duijn CM, Ekelund U, Forouhi NG, Froguel P, Hingorani A, Ingelsson E, Kivimaki M, Kronmal RA, Kuh D, Lind L, Martin NG, Oostra BA, Pedersen NL, Quertermous T, Rotter JI, van der Schouw YT, Verschuren WM, Walker M, Albanes D, Arnar DO, Assimes TL, Bandinelli S, Boehnke M, de Boer RA, Bouchard C, Caulfield WL, Chambers JC, Curhan G, Cusi D, Eriksson J, Ferrucci L, van Gilst WH, Glorioso N, de Graaf J, Groop L, Gyllensten U, Hsueh WC, Hu FB, Huikuri HV, Hunter DJ, Iribarren C, Isomaa B, Jarvelin MR, Jula A, Kahonen M, Kiemeney LA, van der Klauw MM, Kooner JS, Kraft P, Iacoviello L, Lehtimaki T, Lokki ML, Mitchell BD, Navis G, Nieminen MS, Ohlsson C, Poulter NR, Qi L, Raitakari OT, Rimm EB, Rioux JD, Rizzi F, Rudan I, Salomaa V, Sever PS, Shields DC, Shuldiner AR, Sinisalo J, Stanton AV, Stolk RP, Strachan DP, Tardif JC, Thorsteinsdottir U, Tuomilehto J, van Veldhuisen DJ, Virtamo J, Viikari J, Vollenweider P, Waeber G, Widen E, Cho YS, Olsen JV, Visscher PM, Willer C, Franke L, Erdmann J, Thompson JR, Pfeufer A, Sotoodehnia N, Newton-Cheh C, Ellinor PT, Stricker BH, Metspalu A, Perola M, Beckmann JS, Smith GD, Stefansson K, Wareham NJ, Munroe PB, Sibon OC, Milan DJ, Snieder H, Samani NJ, Loos RJ. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz GP, Takano AP, Barreto-Chaves ML. MiRNA-208a and miRNA-208b are triggered in thyroid hormone-induced cardiac hypertrophy—role of type 1 Angiotensin II receptor (AT1R) on miRNA-208a/ alpha-MHC modulation. Mol Cell Endocrinol. 2013;374:117–124. doi: 10.1016/j.mce.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010;14:533–537. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, Ahmad F, Lozado R, Shah G, Fananapazir L, Bachinski LL, Roberts R. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001a;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- Gollob MH, Seger JJ, Gollob TN, Tapscott T, Gonzales O, Bachinski L, Roberts R. Novel PRKAG2 mutation responsible for the genetic syndrome of ventricular preexcitation and conduction system disease with childhood onset and absence of cardiac hypertrophy. Circulation. 2001b;104:3030–3033. doi: 10.1161/hc5001.102111. [DOI] [PubMed] [Google Scholar]
- Granados-Riveron JT, Ghosh TK, Pope M, Bu’Lock F, Thornborough C, Eason J, Kirk EP, Fatkin D, Feneley MP, Harvey RP, Armour JA, David Brook J. Alpha-cardiac myosin heavy chain (MYH6) mutations affecting myofibril formation are associated with congenital heart defects. Hum Mol Genet. 2010;19:4007–4016. doi: 10.1093/hmg/ddq315. [DOI] [PubMed] [Google Scholar]
- Guize L, Soria R, Chaouat JC, Chretien JM, Houe D, Le Heuzey JY. Prevalence and course of Wolf-Parkinson-White syndrome in a population of 138,048 subjects. Ann Med Interne (Paris) 1985;136:474–478. [PubMed] [Google Scholar]
- Harnischfeger WW. Hereditary occurrence of the pre-excitation (Wolff-Parkinson-White) syndrome with re-entry mechanism and concealed conduction. Circulation. 1959;19:28–40. doi: 10.1161/01.cir.19.1.28. [DOI] [PubMed] [Google Scholar]
- Heringa J. Two strategies for sequence comparison: Profile-preprocessed and secondary structure-induced multiple alignment. Comput Chem. 1999;23:341–364. doi: 10.1016/s0097-8485(99)00012-1. [DOI] [PubMed] [Google Scholar]
- Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U, Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, Magnusson OT, Helgason A, Saemundsdottir J, Gylfason A, Stefans-dottir H, Gretarsdottir S, Matthiasson SE, Thorgeirsson GM, Jonasdottir A, Sigurdsson A, Stefansson H, Werge T, Rafnar T, Kiemeney LA, Parvez B, Muhammad R, Roden DM, Darbar D, Thorleifsson G, Walters GB, Kong A, Thorsteinsdottir U, Arnar DO, Stefansson K. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M. VAAST 2.0: Improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;37:622–634. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, Durtschi JD, Tavtigian SV, Shankaracharya W, Wu W, Scheet P, Wang S, Xing J, Glusman G, Hubley R, Li H, Garg V, Moore B, Hood L, Galas DJ, Srivastava D, Reese MG, Jorde LB, Yandell M, Huff CD. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotech. 2014;32:663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B, Kronenberg Z, Hu H, Moore B, Flygare S, Reese MG, Jorde LB, Yandell M, Huff C. Using VAAST to identify disease-associated variants in next-generation sequencing data. Curr Protoc Hum Genet. 2014;81:6. doi: 10.1002/0471142905.hg0614s81. 14 11–16 14 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent AF. Researches on the structure and function of the mammalian heart. J Physiol. 1893;14:i2–i254. [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jahn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park SM, Riggs ER, Scott RH, Sisodiya S, Van Vooren S, Wapner RJ, Wilkie AO, Wright CF, Vulto-van Silfhout AT, de Leeuw N, de Vries BB, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN. The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massumi RA. Familial Wolff-Parkinson-White syndrome with cardiomyopathy. Am J Med. 1967;43:951–955. doi: 10.1016/0002-9343(67)90254-9. [DOI] [PubMed] [Google Scholar]
- McElroy JJ, Gutman CE, Shaffer CM, Busch TD, Puttonen H, Teramo K, Murray JC, Hallman M, Muglia LJ. Maternal coding variants in complement receptor 1 and spontaneous idiopathic preterm birth. Hum Genet. 2013;132:935–942. doi: 10.1007/s00439-013-1304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millat G, Chevalier P, Restier-Miron L, Da Costa A, Bouvagnet P, Kugener B, Fayol L, Gonzalez Armengod C, Oddou B, Chanavat V, Froidefond E, Perraudin R, Rousson R, Rodriguez-Lafrasse C. Spectrum of pathogenic mutations and associated polymorphisms in a cohort of 44 unrelated patients with long QT syndrome. Clin Genet. 2006;70:214–227. doi: 10.1111/j.1399-0004.2006.00671.x. [DOI] [PubMed] [Google Scholar]
- Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, Swensen JJ, Johnson WE, Moore B, Huff CD, Bird LM, Carey JC, Opitz JM, Stevens CA, Jiang T, Schank C, Fain HD, Robison R, Dalley B, Chin S, South ST, Pysher TJ, Jorde LB, Hakonarson H, Lillehaug JR, Biesecker LG, Yandell M, Arnesen T, Lyon GJ. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RG. Familial occurrence of Wolff-Parkinson-White syndrome. Am Heart J. 1969;78:34–37. doi: 10.1016/0002-8703(69)90255-5. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucl Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, Cohen B, North PE, Marchuk DA, Comi AM, Pevsner J. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MV, Guthery SL, Voelkerding KV, Chen K, Kennedy B, Margraf RL, Durtschi J, Eilbeck K, Reese MG, Jorde LB, Huff CD, Yandell M. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet. 2014;94:599–610. doi: 10.1016/j.ajhg.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehler EE, Strehler-Page MA, Perriard JC, Periasamy M, Nadal-Ginard B. Complete nucleotide and encoded amino acid sequence of a mammalian myosin heavy chain gene. Evidence against intron-dependent evolution of the rod. J Mol Biol. 1986;190:291–317. doi: 10.1016/0022-2836(86)90003-3. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff L, Parkinson J, White PD. Bundle-branch block with short P-R interval in healthy young people prone to paroxysmal tachycardia. 1930. Ann Noninvasive Electrocardiol. 2006;11:340–353. doi: 10.1111/j.1542-474X.2006.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, Huff C, Hu H, Singleton M, Moore B, Xing J, Jorde LB, Reese MG. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.