Abstract
The Illumina Infinium HumanMethylation450 BeadChip (450k) is widely used for the evaluation of DNA methylation levels in large-scale datasets, particularly in cancer. The 450k design allows copy number variant (CNV) calling using existing bioinformatics tools. However, in cancer samples, numerous large-scale aberrations cause shifting in the probe intensities and thereby may result in erroneous CNV calling. Therefore, a baseline correction process is needed. We suggest the maximum peak of probe segment density to correct the shift in the intensities in cancer samples.
Availability and implementation: CopyNumber450kCancer is implemented as an R package. The package with examples can be downloaded at http://cran.r-project.org.
Contact: nour.marzouka@medsci.uu.se
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
DNA methylation is the most studied epigenetic modification in cancer and many other diseases. Many technologies and platforms have been developed to facilitate DNA methylation analysis, such as bisulphite conversion followed by analysis using DNA methylation arrays. The Illumina Infinium HumanMethylation450 BeadChip (450k) provides detection of methylation levels of approximately 485000 CpG loci across the human genome (Sandoval et al., 2011).
The 450k platform is based on similar biochemical reaction principle and technology as the Infinium SNP arrays, making it possible to use the 450k platform to detect copy number variants (CNVs) as a zero-cost byproduct of methylation studies (Feber et al., 2014). Recently, bioinformatics tools were developed for copy number (CN) calling from methylation data (e.g. ChAMP (Morris et al., 2014), CopyNumber450k and conumee (www.bioconductor.org)).
CN calling consists of the following principal steps: normalization of probe intensities, calculation of the Log R Ratios (LRRs), segmentation and determination of copy number status for each segment based on a chosen cutoff level or a P-value threshold. In each step different algorithms, methods and parameters can be applied. However, in cancer samples the large and numerous chromosomal duplications and deletions cause a shift in the LRRs away from the hypothetical baseline level (i.e. 2 n/diploid level) resulting in erroneous CNV calling. The conventional normalization methods (e.g. Quantile and Functional normalization (Fortin et al., 2014)) as well as the median/average sample centering cannot overcome this problem. A similar centering problem was observed in copy number calling from array-based Comparative Genomic Hybridization (aCGH) data for cancer samples. The density of the probes was suggested to help determine the center of the samples and avoid erroneous CNV calls (Lipson et al., 2007). However, to date there is no tool available to resolve this problem in CN data derived from the 450k array.
2 Description
To resolve the cancer-specific problem of erroneous CN calling in data derived from the 450k array, we provide a freely available R package denoted CopyNumber450kCancer that can run on all operating systems with installed R (version > 3.0) and provides a novel functionality to correct the center in segmentation data obtained from CN calling tools such as CopyNumber450k and ChAMP.
3 Baseline estimation
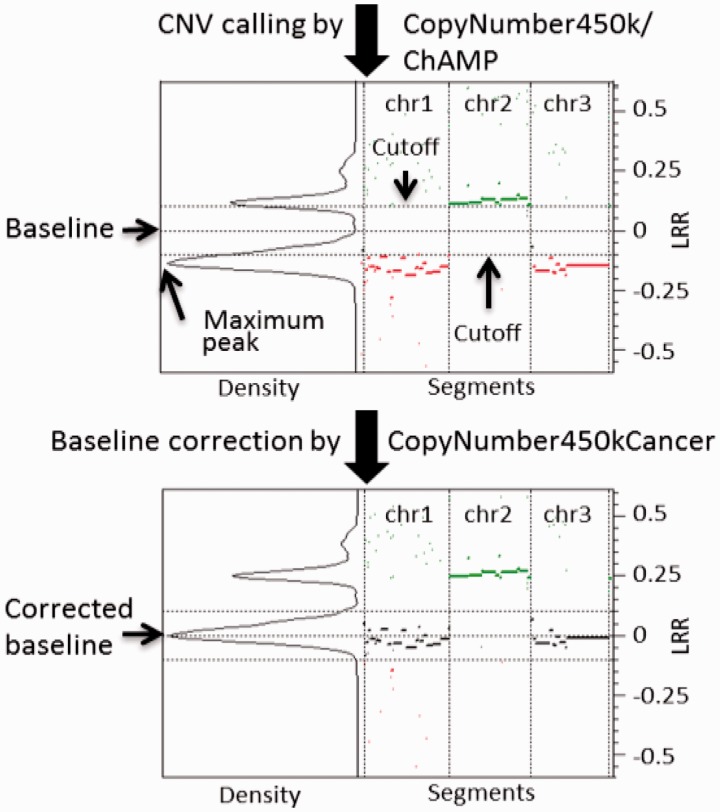
For the baseline estimation we assumed that the majority of the probes are located close to the correct baseline. Therefore a density function based on segments (weighted on the number of probes) should show maximum peak at the correct baseline. For the correction, the sample log values are shifted in an amount equal to the difference between the sample baseline and the maximum peak level (Fig. 1). We call this method Maximum Density Peak Estimation (MDPE). MDPE avoids the challenges and limitations facing the CN calling from 450k data, such as the absence of B-allele frequency that is available from SNP genotyping arrays. CopyNumber450kCancer has a function for manual revision where samples with inaccurate automatic baseline determination can be interactively corrected.
Fig. 1.
The upper plot shows the CN calling from a cancer sample by CopyNumber450k (first 3 chromosomes shown). The segments above the cutoff represent amplifications, and the segments under the cutoff represent deletions. The horizontal lines represent the cutoffs and the zero level (i.e. baseline). The lower panel shows the segments after the baseline correction by CopyNumber450kCancer
To assess the accuracy of the correction method, we performed the auto-correction for 764 public acute lymphoblastic leukemia samples (Nordlund et al., 2013). All the generated plots (Supplementary 1 and 2) were manually reviewed in comparison to the karyotyping data.
The auto-correction changed the baseline for 760 samples, the rest 4 samples (0.5%) had correct baseline without any changes. For 740 samples (96.9%) the auto-correction was correct. Manual modification was needed for 20 samples (2.6%). The difference between the corrected and uncorrected CN data was significant in all the cytogenetic subtypes (Supp. 3).
4 Input and output
CopyNumber450kCancer uses a simple input data structure that can be generated by a wide range of segmentation tools. The package requires two input files. The first file contains the genomic regions for all samples with log values and number of the probes in each segment. The second file contains the samples names. To facilitate the analysis, the tool can directly read the segmentation output file from the CopyNumber450k package.
CopyNumber450kCancer generates a corrected segmentation file, corrected plots, a QC file and a baseline shifting file.
5 Quality control
There is no well-defined quality control (QC) standard for 450k data segmentation. Therefore we selected QC standards developed for SNP array data. A QC file is generated with the following SNP QC standards for each sample; InterQuartile Range (IQR), Median Absolute Pairwise Difference (MAPD), number of segments and standard deviation (SD). An additional QC measurement called Maximum Density Peak Sharpness (MDPS) is also reported. The QC values are calculated based on the log values of the segments instead of the individual probes. CopyNumber450kCancer does not provide any fixed QC thresholds as they may differ from one experiment to another and are also dependent on the type of analysis. The user can use the QC file to exclude samples that have low quality QC values. However, we strongly recommend the visual reviewing option in order to recognize low-quality samples.
6 Interactive revision
CopyNumber450kCancer provides graphical interactive plots as an option to supervise/review the baseline estimation. The user can select a log ratio interval wherein the baseline should be located. For the reviewing step, we strongly recommend to use any external sample information (e.g. karyotyping) that can help the user decide the correct baseline.
The package comes with example files that can be run directly for auto-correction and interactive revision:
regions <- system.file(“extdata”, “regions.csv”, package = “CopyNumber450kCancer”)
sample_list <- system.file(“extdata”,
“sample_list.csv”,package= “CopyNumber450kCancer”)
object <- ReadData(regions, sample_list)
object <- AutoCorrectPeak(object)
object <- ReviewPlot(object)
7 Conclusion
CN calling from the 450k data is possible, but faces some difficulties in some cancer samples due to incorrect sample centering and baseline shifting. Without solving this issue the CN calling will be inaccurate. We successfully tested the MDPE method on 450k cancer segmentation data. CopyNumber450kCancer package implements the MDPE method together with interactive reviewing to efficiently correct the baseline in cancer samples. The main advantages for CopyNumber450kCancer are: fast auto-correction (few seconds per sample), high accuracy rate, in-sample correction, no input parameters needed, low computer resources required and adaptable to 450k-similar technologies.
Supplementary Material
Funding
This work was supported by The Swedish Cancer Society [CAN2010/592]; The Swedish Research Council for Science and Technology [90559401] and The Swedish Foundation for Strategic Research [RBc08-008].
Conflict of Interest: none declared.
References
- Feber A., et al. (2014) Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol., 15, R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P., et al. (2014) Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol., 15, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson D. (2007) Determining the center of array-CGH data. In: Computational Aspects of DNA Copy Number Measurement. Technion – Israel Institute of Technology, Computer Science Department, pp. 105–110. [Google Scholar]
- Morris T.J., et al. (2014) ChAMP: 450k chip analysis methylation pipeline. Bioinformatics, 30, 428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund J., et al. (2013) Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol., 14, r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J., et al. (2011) Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics Off. J. DNA Methylation Soc., 6, 692–702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.