Abstract
Motivation: Metagenomics characterizes the taxonomic diversity of microbial communities by sequencing DNA directly from an environmental sample. One of the main challenges in metagenomics data analysis is the binning step, where each sequenced read is assigned to a taxonomic clade. Because of the large volume of metagenomics datasets, binning methods need fast and accurate algorithms that can operate with reasonable computing requirements. While standard alignment-based methods provide state-of-the-art performance, compositional approaches that assign a taxonomic class to a DNA read based on the k-mers it contains have the potential to provide faster solutions.
Results: We propose a new rank-flexible machine learning-based compositional approach for taxonomic assignment of metagenomics reads and show that it benefits from increasing the number of fragments sampled from reference genome to tune its parameters, up to a coverage of about 10, and from increasing the k-mer size to about 12. Tuning the method involves training machine learning models on about 108 samples in 107 dimensions, which is out of reach of standard softwares but can be done efficiently with modern implementations for large-scale machine learning. The resulting method is competitive in terms of accuracy with well-established alignment and composition-based tools for problems involving a small to moderate number of candidate species and for reasonable amounts of sequencing errors. We show, however, that machine learning-based compositional approaches are still limited in their ability to deal with problems involving a greater number of species and more sensitive to sequencing errors. We finally show that the new method outperforms the state-of-the-art in its ability to classify reads from species of lineage absent from the reference database and confirm that compositional approaches achieve faster prediction times, with a gain of 2–17 times with respect to the BWA-MEM short read mapper, depending on the number of candidate species and the level of sequencing noise.
Availability and implementation: Data and codes are available at http://cbio.ensmp.fr/largescalemetagenomics.
Contact: pierre.mahe@biomerieux.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Recent progress in next-generation sequencing technologies allow to access large amounts of genomic data within a few hours at a reasonable cost (Soon et al., 2013). In metagenomics, next-generation sequencing is used to analyze the genomic content of microbial communities by sequencing all DNA present in an environmental sample (Riesenfeld et al., 2004). It gives access to all organisms present in the sample even if they do not grow on culture media (Hugenholtz et al., 2002) and allows us to characterize with an unprecedented level of resolution the diversity of the microbial realm (Peterson et al., 2009).
The raw output of a metagenomics experiment is a large set of short DNA sequences (reads) obtained by high-throughput sequencing of the DNA present in the sample. There exist two main approaches to analyze these data, corresponding to slightly different goals. On the one hand, taxonomic profiling aims to estimate the relative abundance of the members of the microbial community, without necessarily assigning each read to a taxonomic class. Recent works like WGSQuikr (Koslicki et al., 2014) or GASIC (Lindner and Renard, 2012) proved to be very efficient for this purpose. Taxonomic binning methods, on the other hand, explicitly assign each read to a taxonomic clade. This process can be unsupervised, relying on clustering methods to assign reads to several bins (i.e. clusters), or supervised, in which case reads are individually assigned to nodes of the taxonomy (Mande et al., 2012). While binning is arguably more challenging than profiling, it is a necessary step for downstream applications which require draft-genome reconstruction. This may notably be the case in a diagnostics context, where further analyses could aim to detect pathogen microorganisms (Miller et al., 2013) or antibiotic resistance mechanisms (Schmieder and Edwards, 2012).
In this article, we focus on the problem of supervised taxonomic binning, where we wish to assign each read in a metagenomics sample to a node of a pre-defined taxonomy. Two main computational strategies have been proposed for that purpose: (i) alignment-based approaches, where the read is searched against a reference sequence database with sequence alignment tools like BLAST (Huson et al., 2007) or short read mapping tools (e.g. BWA, Li and Durbin, 2009) and (ii) compositional approaches, where a machine learning model such as a naive Bayes (NB) classifier (Parks et al., 2011; Wang et al., 2007) or a support vector machine (SVM, McHardy et al., 2007; Patil et al., 2012) is trained to label the read based on the set of k-mers it contains. Recently, a very fast compositional approach using long k-mers and not based on machine learning models, called Kraken (Wood and Salzberg, 2014), has also been proposed. Since the taxonomic classification of a sequence by compositional approaches is only based on the set of k-mers it contains, they can offer significant gain in terms of classification time over similarity-based approaches. Training a machine learning model for taxonomic binning can, however, be computationally challenging. Indeed, compositional approaches must be trained on a set of sequences with known taxonomic labels, typically obtained by sampling error-free fragments from reference genomes. In the case of NB classifiers, explicit sampling of fragments from reference genomes is not needed to train the model: instead, a global profile of k-mer abundance from each reference genome is sufficient to estimate the parameters of the NB model, leading to simple and fast implementations (Parks et al., 2011; Rosen et al., 2011; Wang et al., 2007). On the other hand, in the case of SVM and related discriminative methods, an explicit sampling of fragments from reference genomes to train the model based on the k-mer content of each fragment is needed, which can be a limitation for standard SVM implementations. For example, Patil et al. (2012) sampled approximately 10 000 fragments from 1768 genomes to train a structured SVM (based on a k-mer representation with k = 4, 5, 6) and reported an accuracy competitive with similarity-based approaches. Increasing the number of fragments sampled to train a SVM may improve its accuracy and allow us to investigate larger values of k. However, it also raises computational challenges, as it involves machine learning problems where a model must be trained from potentially millions or billions of training examples, each represented by a vector in 107 dimensions for, e.g. k = 12.
In this work, we investigate the potential of compositional approaches for taxonomic label assignment using modern, large-scale machine learning algorithms. We propose a new, rank-flexible compositional approach trained with large-scale machine learning methods and assess its performance in different situations. We show that it provides an interesting trade-off in speed and accuracy compared to the state-of-the-art, particularly when confronted to species absent from the reference database, and for a moderate number of candidate species.
2 Methods
2.1 Linear models for read classification
In most of compositional metagenomics applications, a sequence is represented by its k-mer profile, namely, a vector counting the number of occurrences of any possible word of k letters in the sequence. Only the nucleotides are usually considered to define k-mer profiles, that are therefore -dimensional vectors. Although the size of the k-mer profile of a sequence of length l increases exponentially with k, it contains at most non-zero elements since a sequence of length k contains different k-mers.
Given a sequence represented by its k-mer profile , we consider linear models to assign it to one of K chosen taxonomic classes. A linear model is a set of weight vectors that assign x to the class
| (1) |
where is the standard inner product between vectors. To train the linear model, we start from a training set of sequences with known taxonomic labels . An NB classifier, e.g. is a linear model where the weights are estimated from the k-mer count distributions on each class. Another class of linear models popular in machine learning, which include SVM, are the discriminative approaches that learn the weights by solving an optimization problem which aims to separate the training data of each class from each other. More precisely, to optimize the weight wj of the jth class, one typically assigns a binary label yi to each training example (yi = 1 if ci = j or otherwise) and solves an optimization problem of the form
| (2) |
where is a loss function quantifying how ‘good’ the prediction t is if the true label is y, and is a regularization parameter to tune, helpful to prevent overfitting in high dimension. An SVM solves (2) with the hinge loss , but other losses such as the logistic loss or the squared loss are also possible and often lead to models with similar accuracies. These models have met significant success in numerous real-world learning tasks, including compositional metagenomics (Patil et al., 2012). In this work, we use the squared loss function and choose λ = 0, a setting that seemed appropriate from preliminary experiments.
2.2 Large-scale learning of linear models
Although learning linear models by solving (2) is now a mature technology implemented in numerous softwares, metagenomics applications raise computational challenges for most standard implementations, due to the large values that n (number of reads in the training set), (dimension of the models) and K (number of taxonomic classes) can take.
The training set is typically obtained by sampling fragments from reference genomes with known taxonomic class. For example, Patil et al. (2012) sampled approximately n = 10 000 fragments from 1768 genomes to train SVM models based on k-mer profiles of size k = 4, 5, 6. However, the number of distinct fragments that may be drawn from a genome sequence is approximately equal to its length (by sampling a fragment starting at each position in the genome), hence can reach several millions for each microbial genome, leading to potentially billions of training sequences when thousands of reference genomes are used. While considering every possible fragment from every possible genome may not be the best choice because of the possible redundancy between the reads, it may still be useful to consider a significant number of fragments to properly account for the intra and inter species genomic variability. Similarly, exploring models with k larger than 6, say 10 or 15, may be interesting but requires (i) the capacity to manipulate the corresponding -dimensional vectors () and (ii) large training sets since many examples are needed to learn a model in high dimension. Finally, real-life applications involving actual environmental samples may contain several hundreds microbial species, casting the problem into a relatively massive multiclass scenario out of reach of most standard implementations of SVM.
To solve (2) efficiently when n, k and K take large values, we use a dedicated implementation of stochastic gradient descent (SGD Bottou, 1998) available in the Vowpal Wabbit software (VW, Agarwal et al., 2014; Langford et al., 2007). In short, SGD exploits the fact that the objective function in (2) is an average of n terms, one for each training example, to approximate the gradient at each step using a single, randomly chosen term. Although SGD requires more steps to converge to the solution than standard gradient descent, each step is n times faster and the method is overall faster and more scalable. In addition, although the dimension of the data is large, VW exploits the fact that each training example is sparse, leading to efficient memory storage and fast updates at each SGD step. We refer the interested reader to Bottou (2010) for more discussion about the relevance of SGD in large-scale learning. In practice, VW can train a model with virtually no limit on n as long as the data can be stored on a disk (they are not loaded in memory). As for k, VW can handle up to 232 distinct features, and the count of each k-mer is randomly mapped to one feature by a hash table. This means that we have virtually no limit on k, except that when k approaches or exceeds the limit (such that , i.e. k = 16), collisions will appear in the hash table and different k-mers will be counted together, which may impact the performance of the model.
2.3 Rank-specific and rank-flexible read classification
The classification approach described in Section 2.1 can be readily applied by labelling sampled fragments according to a given taxonomic rank and learning read classification models tailored to this level of resolution, which is sometimes referred to as rank-specific approaches (Parks et al., 2011). We build such rank-specific classifiers at the species, genus and family levels.
In addition, we implement a rank-flexible classifier to automatically choose the most adequate level where a read should be classified in the taxonomy or leave it unclassified if it looks too different from the reads used to train the model. For that purpose, we assess the reliability of a rank-specific prediction at any level by means of two criteria: the maximum score returned by the linear model (1) and the difference between the two largest scores. According to the terminology proposed by Gammerman and Vovk (2007), the former criterion accounts for the credibility of the prediction: if the sequence is not granted a sufficient score for any class, it may be considered unusual with respect to the training dataset. The latter criterion accounts for the confidence of the prediction: if the scores of the two top-scoring classes are comparable, both classes may be considered plausible. By combining both criteria we can reject predictions that are unlikely or ambiguous. To combine rank-specific classifiers into a rank-flexible one, we start from the model built at the lowest rank—species in our case—and iteratively allow a rejected read to be classified at the upper rank. If a read is rejected by all rank-specific models considered, it is left unclassified.
The reject option mechanism underlying this rank-flexible procedure heavily depends on thresholds on the maximum score of the linear model and on the difference between the two largest scores, which can be set globally or on a taxon-by-taxon basis. A strategy to optimize these thresholds is described in Supplementary Materials (Section 2), together with an illustration of the trade-offs that can be achieved in terms of recall and precision.
3 Data
We assemble three databases of genomes, which we refer to below as the small, the medium and the large databases. Each comprises a set of reference genomes to train the models and a set of validation genomes from which reads are generated to evaluate the performance of the different classification methods.
The small database contains as references 356 complete genome sequences covering 51 bacterial species, listed in Supplementary Table S1. For validation, we choose 52 genomes not present in the reference database but originating from one of the 51 species (two genomes are indeed available for the Francisella tularensis species, one of which originating from the novicida subspecies). This small database is of limited biological interest but is convenient to extensively test the different methods and vary their parameters.
The medium and large databases are meant to represent more realistic situations, involving a larger number of candidate bacterial species and a larger number of reference genomes. We download the 5201 complete bacterial and archeal genomes available in the NCBI RefSeq database as of July 2014 (Pruitt et al., 2012), by means of a functionality embedded in the Fragment Classification Package (Parks et al., 2011). We then filter these sequences according to a criterion proposed in Parks et al. (2011), only keeping genomes that belong to genera represented by at least three species. We also remove genomes represented by less than 106 nucleotides to filter draft genome sequences, plasmids, phages, contigs and other short sequences. The 2961 remaining genomes originate from 774 species, among which 193 are represented by at least two strains and 110 by at least three strains. To build the medium database, we randomly pick one strain within each of the 110 species with at least three strains as a validation set and combine all other sequences in the 193 species with at least two strains as reference database. The rationale of this split is to ensure that each species in the reference database is represented by at least two strains, which allows to optimize the values of the thresholds involved in the classification procedure on a taxon-by-taxon basis, by means of an internal validation process. To define the large database, we randomly pick one strain within each of the 193 species represented by at least two strains to define the validation database and keep the remaining sequences of the 774 species as reference database. This ensures that each strain in the validation set comes from a species that is present in the reference database.
In addition, we create two additional validation sets to assess the performance of models trained on the large reference database in more challenging situations. First, we assemble a novel lineage validation set composed of genomes in the NCBI RefSeq database from species not represented in the large reference database. Details about the number of strains in the novel lineage validation set are provided in Supplementary Table S2. Second, we consider a real dataset composed of actual reads, the HMP Microbial Mock Community dataset (Even, Low Concentration, 454 GS FLX Titanium, SRA accession SRX030841). This dataset contains 1 386 198 reads coming from a mock sample made of a genomic DNA mixture obtained from 20 bacterial and 1 archaeal strains [see Martin et al. (2012) for further details about the considered strains]. Reads with quality score below 20 are trimmed, and reads shorter than 25 bp are filtered out.
4 Results
4.1 Proof of concept on the small database
In this section, we present a study on the small dataset, aiming to evaluate the impact of increasing the number of fragments used to train the model as well as the length of the k-mers considered. For that purpose, we consider a rank-specific setting defined at the species level, without any reject option mechanism, and learn several classification models based on fragments of length L = 200 or L = 400 sampled from the 356 reference genomes in the small reference database, represented by k-mers of size in . The number of fragments used to learn the models is gradually increased by drawing several ‘batches’ of fragments to cover, on average, each nucleotide of the reference genomes a pre-defined number of times c. We vary the coverage c between 0.1 to its maximal value, equal to the length of the fragments considered. This leads to learning models from around , for c = 0.1 and L = 400, up to around fragments, when c reaches its maximal value. This is way beyond the configurations considered for instance in (Patil et al., 2012), where SVM models were learned from approximately 104 fragments drawn from 1768 genomes.
To assess the performance of these models, we consider two sets of 134 319 fragments, of respective length 200 and 400, drawn from the 52 complete genomes that are not in the reference database used to train the models. Performance is measured in terms of species-level recall. We first compute the prediction recall within each species, i.e. the proportion of fragments originating from this species that are correctly classified and consider the average recall observed across species. In a multiclass setting, this indicator is indeed less biased toward over-represented classes than the global rate of correct classification.
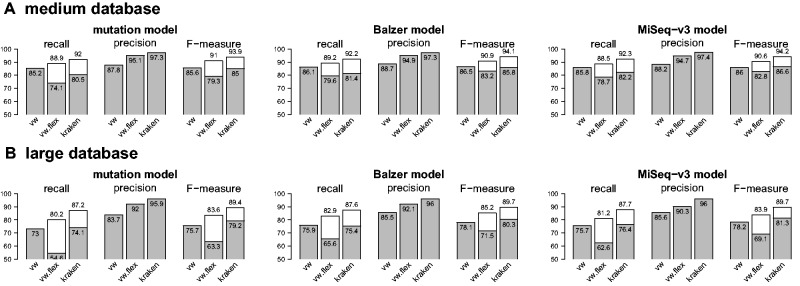
Figure 1 shows the performance reached by models based on fragments of length 200 (left) or 400 (right), for different values of k (horizontal axis) and different coverages (different colors). We first note that for c = 0.1, i.e. for a limited number of fragments, the classification performance starts by increasing with the size of the k-mers (up to k = 8 and k = 10 for fragments of length 200 and 400, respectively) and subsequently decreases. This suggests that the number of fragments considered in this setting is not sufficient to efficiently learn when the dimensionality of the feature space becomes too large. Note that twice as many fragments of length 200 as fragments of size 400 are drawn for a given coverage value, which may explain why performance still increases beyond k = 8 with smaller fragments. Increasing the number of fragments confirms this hypothesis: performance systematically increases or remains steady with k for and for , the performance is significantly higher than that obtained at c = 0.1, for both length of fragments. Increasing the coverage from c = 1 to c = 10 has a positive impact in both cases, although marginally for fragments of length 400. Further increasing the number of fragments does not bring any noticeable improvement.
Fig. 1.
Increasing the number of fragments and k-mer size on the small datasets. Left: L = 200 bp fragments. Right: L = 400 bp fragments. These figures show the average species-level recall obtained by linear predictors trained with Vowpal Wabbit from fragments covering each reference genome with a mean coverage c from 0.1 to L. Performances are reported as a function of k-mer sizes
Altogether, the optimal configuration on this small dataset involves k-mers of size 12 and drawing fragments at a coverage for the two lengths of fragments considered. Further increasing the size of the k-mers did not bring improvements and actually proved to be challenging. Indeed, as mentioned above, VW proceeds by hashing the input features into a vector offering at most 232 entries. This hashing operation can induce collisions between features, which can be detrimental to the model if the number of features becomes too high with respect to the size of the hash table. This issue is even more stressed in a multiclass setting, where the number of hash table entries available per model is divided by the number of classes considered. On this dataset, 51 models have to be stored in the hash table, which reduces the number of entries available per model to . We have empirically observed that performance could not increase for k greater than 12 and actually decreased for k-mers greater than 15.
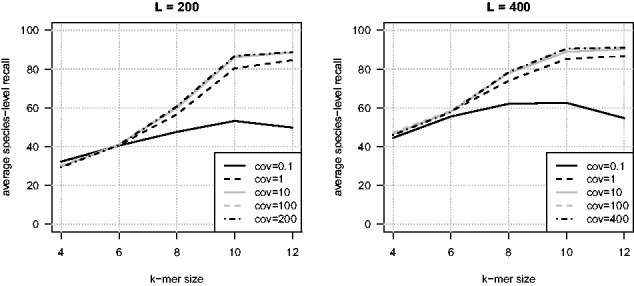
We now compare these results to two well-established approaches: a comparative approach based on the BWA-MEM sequence aligner (Li, 2013) and a compositional approach based on the generative NB classifier (Rosen et al., 2011). The NB experiments rely on the Fragment Classification Package implementation (Parks et al., 2011) and are carried out in the same setting as VW: we compute profiles of k-mers abundance for the 356 genomes of the reference database and use them to assign test fragments to their most likely genome. BWA-MEM is configured to solely return hits with maximal score (option -T 0). Unmapped fragments are counted as misclassifications, and a single hit is randomly picked in case of multiple hits, to obtain a species-level prediction. This latter random hit selection process is repeated 20 times, and the performance indicator reported below corresponds to its median value obtained across repetitions. Results are shown in Figure 2. We first note that k-mer-based approaches, either generative or discriminative, never outperform the alignment-based approach. Comparable results are nevertheless obtained for k = 12 with VW, while the NB shows slightly lesser performance (around 4 points in both cases). Performances obtained for shorter k-mers are markedly lower than that obtained by BWA-MEM. We note finally that VW generally outperforms the NB classifier, except for small k-mers ().
Fig. 2.
Comparison between Vowpal Wabbit and reference methods on the small datasets. Left: L = 200 bp fragments. Right: L = 400 bp fragments. These figures show the average species-level recall obtained by linear predictors trained with Vowpal Wabbit from fragments covering each reference genome with a mean coverage equal to 10 (solid line). Performances are reported as afunction of k-mer sizes. This approach is compared to the standard compositional NB approach (dashed line) and an alignment-based approach based on BWA (dash-dotted line)
In summary, these experiments demonstrate the relevance and feasibility of large-scale machine learning for taxonomic binning: we obtain a performance comparable to that of the well-established alignment-based approach, provided a sufficient number of fragments and long enough k-mers are considered to learn the k-mers-based predictive models.
4.2 Evaluation on the medium and large reference databases
We now proceed to a more realistic evaluation involving a larger number of candidate microbial species and a larger number of reference genomes, using the medium and large reference databases. We learn classification models based on results obtained on the small database: we consider k-mers of size 12 and a number of fragments allowing to cover each base of the reference genomes 10 times in average. We limit our analysis to fragments of length 200, which leads to models learned from around and fragments for the medium and large reference databases, respectively. Note that due to the larger number of species involved, around ≃ 412 and ≃ 411 entries of the VW hash table are available per model for each of these reference databases. We evaluate the performance of the models on fragments extracted from the genomes of the corresponding validation databases and draw a number of fragments necessary to cover each base of each genome once in average, which represents around and sequences for the medium and large databases, respectively.
We consider the rank-specific (at the species level) and rank-flexible VW strategies, as explained in Section 2.3, and compare them to BWA-MEM and NB, also set to work in a species-level rank-specific setting, as well as to the rank-flexible Kraken method trained on the same reference database (Wood and Salzberg, 2014). We assess the classification performance in terms of precision and recall. For a given species, recall (or sensitivity) is defined as the proportion of test sequences originating from this species that are classified as such. Precision (or positive predictive value) corresponds to the proportion of test sequences actually originating from this species among sequences classified as such. To compare the classification performance of the various systems, we compute the precision and recall observed for each species of the validation dataset and report their average value. We also report the average species-level F-measure, defined as the harmonic mean of precision and recall. For rank-flexible strategies, we consider a second definition of recall, referred to as the upper recall, and the corresponding upper F-measure, in which a sequence is considered to be correctly predicted if it is classified into a ancestor taxon of its species.
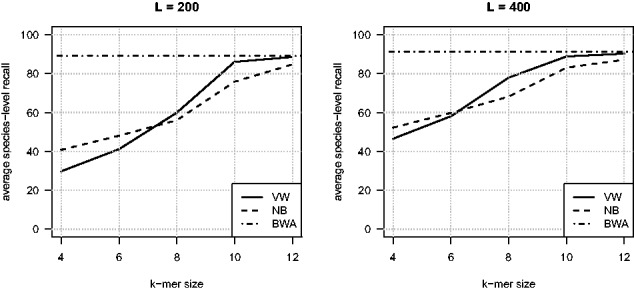
Results are shown in Figure 3. We first note that for the medium reference database, rank-specific VW and BWA-MEM performances are very similar in terms of species-level recall (mean value of 86.2% and 86.8%, respectively). The NB classifier, on the other hand, has a lower species-level recall, with 6 points less than the alignment-based approach. Rank-flexible strategies also exhibit a lower species-level recall but offer a higher level of upper recall, with Kraken providing slightly better performance than VW (1 point in terms of species-level recall and 3 points in upper recall). On the other hand, rank-flexible strategies offer a significantly higher precision than the three rank-specific approaches, which reach comparable values. This is due to the rank-flexible ability to reject predictions and to classify reads at upper ranks, which positively impacts the precision, measured at the species-level. Interestingly, we note that while both rank-flexible approaches reject the same amount of predictions (around 5% per species in average) and classify the same amount of fragments at the species-level (around 84% per species in average), Kraken shows a higher precision than VW (97.5% vs. 95%). These results therefore indicate that trade-offs can be met in terms of precision and recall. Rank-specific approaches based on BWA and VW can indeed offer a higher species-level recall, at the price of classification errors hence of precision. Rank-flexible strategies, on the other hand, manage to maintain a high level of precision, at the price of a lower species-level recall. This drop is, however, counterbalanced by the flexibility of classifying sequences at upper ranks, leading to a higher upper-recall than the species-level recall of rank-specific approaches.
Fig. 3.
Performance on the medium and large reference databases. This figure shows the classification performance measured on genomic fragments in terms of average species-level recall, precision and F-measure, for the various classification strategies considered. For rank-flexible approaches, the average upper recall and upper F-measure are shown as white bars on top of the gray ones, representing species-level indicators
Considering a larger number of candidate species in the large reference database proves to be challenging for methods based on short k-mers. Indeed, a drop of 10 points in terms of species-level recall is observed for the VW and NB rank-specific strategies, while BWA suffers of a lesser drop of around 6 points. Kraken turns out to be the most competitive approach compared to BWA. Indeed, Kraken species-level recall becomes comparable to that of rank-specific VW but still offers a greater precision and the flexibility of retrieving predictions at upper ranks. The rank-flexible strategy based on VW is less competitive on the large database than it was on the medium one.
4.3 Robustness to sequencing errors
The evaluation performed in the previous section is based on taxonomic classification of DNA fragments drawn from reference genomes without errors. In real life, sequencing errors may alter the read sequences and make the classification problem more challenging. To evaluate the robustness of the classifiers to sequencing errors, we generate new reads simulating three realistic types of sequencing errors: homopolymeric stretches, which are commonly encountered in pyrosequencing (e.g. Roche 454) and ion sequencing (e.g. IonTorrent) technologies, general mutations (substitutions and insertions/deletions) and an error model tailored to the Illumina MiSeq technology. For the two former models, we rely on the Grinder read simulation software (Angly et al., 2012). More precisely, we consider the Balzer homopolymeric error model meant to reproduce the Roche 454 technology (Balzer et al., 2010) and the 4th degree polynomial proposed by Korbel et al. (2009) to study general mutations. In this latter case, however, we modify the parameters of the error model to reach a median error rate of 2%, in agreement with the results of the original publication (Korbel et al., 2009). The Illumina MiSeq error model was defined internally, according to a procedure described in Supplementary Materials (Section 3). To compare the results of the fragment- and read-based evaluations, each dataset simulated from the medium and large validation databases includes as well around or sequences, respectively.
Figure 4 presents the performance obtained by the rank-specific and rank-flexible VW strategies and those obtained by Kraken. On the medium database, we note that the impact of these sequencing errors is usually limited, where a drop of at most one point is observed with respect to the results obtained on fragments, essentially due do a decrease of the recall. This is, however, not the case for the general mutation error model with the VW rank-flexible strategy, where a drop of 6 points in terms of species-level recall is observed. The upper recall, on the other hand, remains comparable, indicating that a larger number of predictions are made (correctly) at an upper rank in this case. Considering a larger number of species in the large database also has a limited impact for the VW rank-specific strategy and for Kraken, for the Balzer and Miseq error models (drop of <0.5 point with respect to the fragments results). The mutation error model is, however, more challenging, leading to a drop of species-level recall of 3 points for VW and 1.5 point for Kraken (reduced to a 0.7 point in terms of upper recall). The rank-flexible VW strategy, on the other hand, is more impacted. We note indeed a higher drop in terms of species-level recall, even for the Balzer and MiSeq error model (2 and 5 points, respectively). The situation is even worse for the mutation error model, where a drop of almost 12 points is observed. This is due, at least in part, to an increase of the rejection rate with this error model (13% per species in average vs. 8–10% for the same strategy on other error models and 7.5% with Kraken on the same error model). In any case, the drop in terms of upper recall is less severe, indicating that a larger fraction of predictions are made above the species level.
Fig. 4.
Robustness to sequencing errors. Top: medium reference database; Bottom: large reference database. This figure shows the classification performance measured on simulated reads in terms of average species-level recall, precision and F-measure, for the various classification strategies considered. For rank-flexible approaches, the average upper recall and upper F-measure are shown as white bars on top of the gray ones, representing species level
In summary, these results suggest that k-mer-based read classification models are robust to realistic sequencing noise. We note, however, that VW is globally more impacted than Kraken, especially in its rank-flexible setting and the general mutation error model. Additional experiments involving higher levels of noise, described in Supplementary Materials (Section 4), confirm this observation.
4.4 Evaluation on novel lineages
Metagenomic samples encountered in real-life applications may include microorganisms not represented in the reference database used to carry out the taxonomic binning. This may be due to the presence of a previously unknown microorganism, which is common in environmental studies, or of a microorganism for which no reference genome is available. In this section, we thus evaluate the ability of taxonomic binning methods to classify reads coming from novel bacterial lineages at their appropriate taxonomic rank. For this purpose, we extract from the NCBI RefSeq database genomes of species not represented in the large reference database and qualify them according to the rank of their closest relative. For instance, a strain is said to be ‘reachable’ at the genus level when its species is not part of the reference database but when other species of the same genus are represented. To estimate the performance of classifiers for such novel lineages, we extract strains reachable at the genus, family, order, class and phylum levels, draw genomic fragments from their genomes and classify them using the rank-flexible VW strategy and Kraken, on the large reference database. We assess the performance in terms of the proportion of reads that are (i) assigned to the appropriate taxon, (ii) assigned to an ancestor taxon, (iii) assigned to a descendant taxon, (iv) rejected and (v) misclassified. In the two former cases, the prediction is considered to be correct, which allows to define criteria of recall by considering case (i) and upper recall by considering cases (i) and (ii). Predictions assigned to a descendant taxon are said to be too specific.
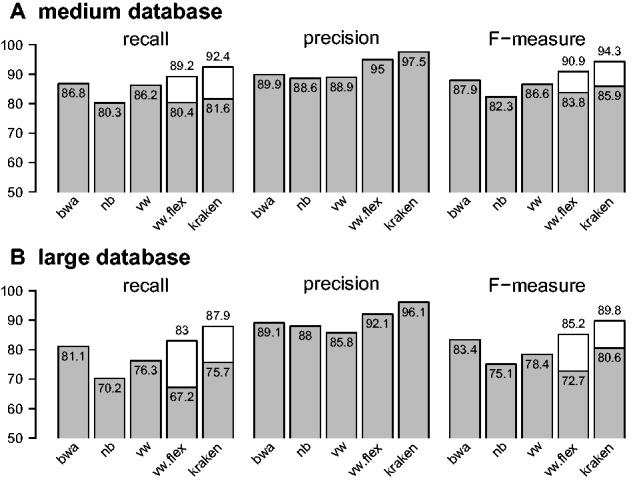
Figure 5 shows the results obtained for novel lineages reachable at the genus, family and order levels. Results obtained at other ranks are provided in Supplementary Materials (Section 5). We can first note from panels A and B that both VW and Kraken reject a large proportion of the predictions (between 56% and more than 93% in average across reachable taxa). The reject rate increases with the taxonomic distance between the novel lineage and the reference database in both cases and is larger with Kraken, especially at the family level and above. The high reject rate of Kraken comes with a very moderate error rate (around 3% in average), while VW suffers from a much higher error rate (around 15%). We note moreover that the recall is very low at the family level and above, suggesting that it is difficult to effectively detect genomic proximity from k-mers at such taxonomic distance. A closer look at novel lineages reachable at the genus level, i.e. strains coming from species for which species of the same genus are available, which probably constitutes the most realistic scenario, highlights important differences between VW and Kraken. We note indeed that the recall is much higher with VW than Kraken: 15.3% vs. 4.2% on average across reachable genera and 19.4% vs. 4.6% in terms of upper recall. Conversely, Kraken classifies a greater proportion of fragments too specifically, assigning 26.3% of the fragments to a sibling species, instead of 10.3% with VW. This is illustrated in Figure 5C, which compares the upper recall on a genus-by-genus basis, as well as in Supplementary Figure S9, which further highlights the trade-off observed in terms of recall and proportion of too specific prediction.
Fig. 5.
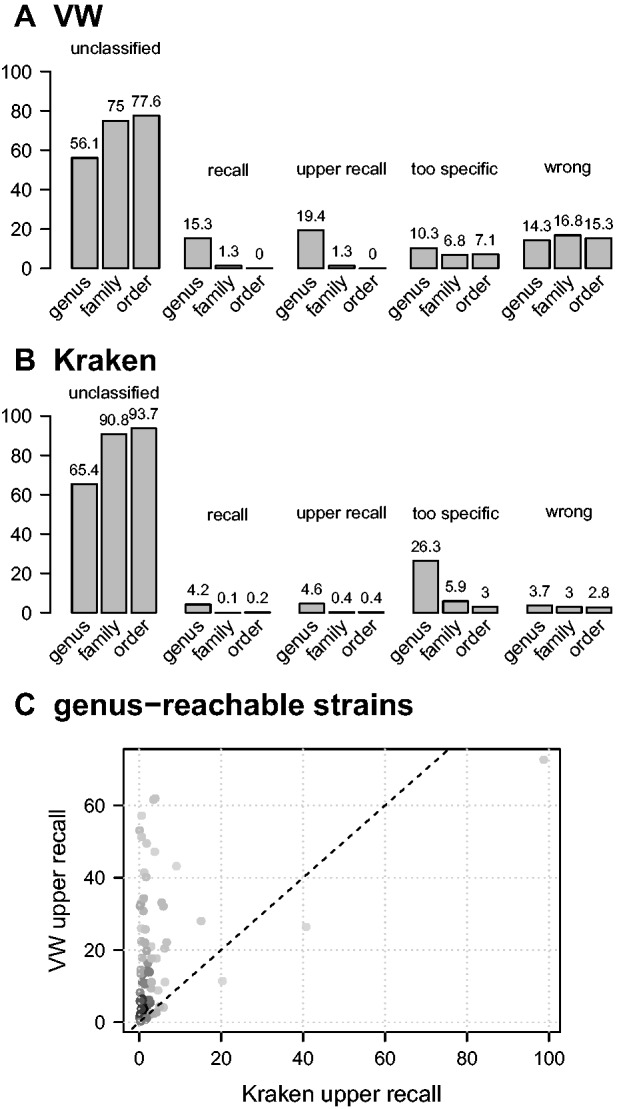
VW and Kraken performance on novel lineages. Panels (A) and (B) present the proportions of unclassified fragments, the recall (fragments assigned to their correct reachable taxon), the upper recall (fragments assigned to their correct reachable taxon or one of its ancestors), the proportion of too specific predictions (fragments assigned to a descendant of their reachable taxon) and the error rate (fragments assigned to a taxon not part of the branch of their reachable taxon). These results are shown for three reachable ranks (genus, family and order) and correspond to average values across reachable taxa of a given rank. Results obtained at higher ranks are provided in Supplementary Figure S7. Panel (C) compares the VW and Kraken upper recall estimations for the 69 genera used to evaluate the performance on the new strains reachable at the genus level
In summary, these results indicate first that the faculty of k-mers-based methods to effectively recognize novel genera and other more taxonomically distant lineages is limited. As for novel species, the rank-flexible VW strategy is more efficient than Kraken, with a greater ability to assign reads to the appropriate genus (or above) where Kraken essentially classify reads at the species level, that is, as a sibling species.
4.5 Evaluation on a real dataset
We finally compare the predictions obtained by VW and Kraken on the real, HMP Microbial Mock Community dataset, using the models learned on the large reference database. As we have no certainty about the correct classification of each individual read, we compare the aggregated predictions over all reads.
Figure 6 shows the abundance profiles obtained by the two approaches, restricted to taxa accounting for at least 0.2% of the sample. We note that these abundance profiles are highly comparable. In particular, we note that both approaches reject approximately the same amount of reads, while VW classifies slightly more reads at the genus-level. Eighteen out of the 21 spiked strains are retrieved by both methods (Streptococcus agalactiae and Lactobacillus gasseri are not shown in Figure 6 because their abundance is below 0.2%) and the three remaining ones (Escherichia coli, Rhodobacter sphaeroides and Actinomyces odontolyticus) are not, because they are not represented in the reference database. Interestingly, both methods classify reads coming from the ‘novel’ E. coli strain into Shigella species because of their close relatedness (Lukjancenko et al., 2010), with the advantage for VW to take into account this uncertainty by classifying more reads at the Shigella genus level.
Fig. 6.
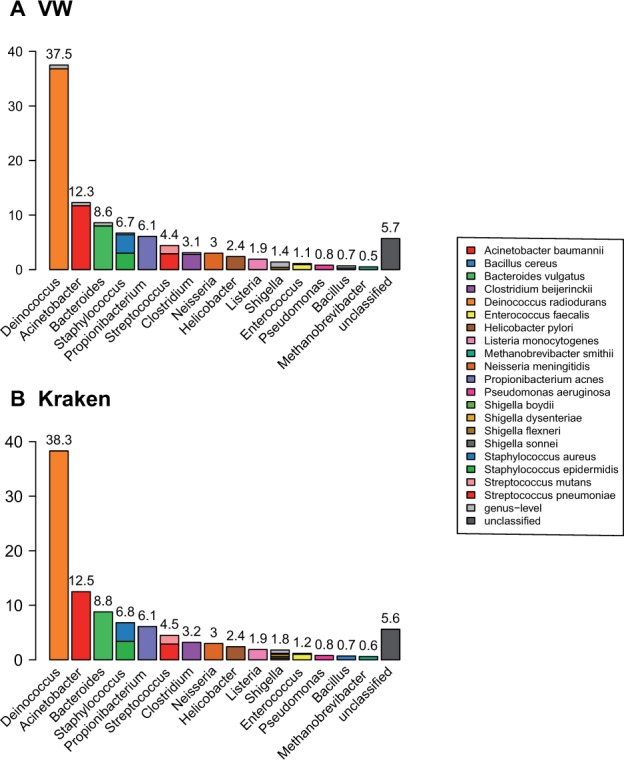
Abundance profiles obtained with VW (A) and Kraken (B) on the HMP spiked dataset. Abundance profiles are presented at the genus level, with colors corresponding to the different predicted species, restricted to species accounting for at least 0.2%) of the samples. Light-gray fractions of the bars correspond to genus-level predictions
4.6 Classification speed
Last but not least, we now turn to the comparison of the comparative and compositional approaches in terms of prediction time. This aspect is indeed of critical importance for the analysis of the large volumes of sequence data provided by next-generation sequencing technologies and constitutes the main motivation of resorting to k-mer-based approaches. To perform this evaluation, we measure the time taken by BWA-MEM and the k-mer-based approaches to process the four test datasets involved in the previous experiments (one fragment dataset, three reads datasets), with the models learned from the medium and large databases, to investigate the impact of the number of species involved in the reference database. We do not make a distinction between the VW and NB compositional approaches: both involve computing a score for each candidate species, defined as a dot product between the k-mer profile of the sequence to classify and a vector of weights obtained by training the model. To compute this dot-product efficiently, we implemented a procedure described in Sonnenburg et al. (2006). With this procedure, each A, T, G, C nucleotide is encoded by two bits, which allows to directly convert a k-mer as in integer between 0 and . Provided that the weight vector is loaded into memory, the score can be computed ‘on the fly’ while evaluating the k-mer profile of the sequence to be classified, by adding the contribution of the current k-mer to the score. The drawback of this procedure lies in the fact that the vectors of weights defining the classification models need to be loaded into memory, which can be cumbersome in a multiclass setting. For 193 and 774 species and k-mers of size 12, this amounted to 12 and 48 gigabytes, respectively.
Computation times are measured on a single CPU (Intel XEON-2.8 Ghz) equipped with 250 GB of memory and are detailed in Supplementary Table S4. The time needed to classify each read or fragment dataset by VW shows little variation, for a given reference database. Its median value obtained across test datasets reaches 4.4 and 8.8 min using the medium and large reference databases, respectively, hence about a 2-fold difference. The time taken by Kraken is smaller and more stable across datasets: 2.9 and 3.3 min using the medium and large reference databases, respectively, hence about 1.5 and 2.7 times faster than VW. On the large database, this therefore amounts to classifying around and 200 bp reads per minute with VW and Kraken, respectively. BWA-MEM shows a different behavior. We indeed observe that the time is longer with reads than fragments, while the size of the reference database has a lesser impact. Compositional approaches systematically offer shorter prediction times than the alignment-based approach, with an improvement of 2.4–12.4 for VW and 7–17.3 times with Kraken, depending on the configuration.
Finally, Figure 7 further investigates the relationship between the number of species involved in the reference database and VW prediction time. For this purpose, we generate random models involving 1 to 1000 classes and measure the time needed to process each test dataset. We note that for problems involving fewer than 100 candidate species, VW can achieve faster prediction times than Kraken. This situation may in particular arise in diagnostic applications involving specific types of specimens or, at the extreme, regarding the issue of filtering reads coming from the human host in microbiome samples. In this latter case, a binary VW model trained to discriminate bacterial from human reads takes around 20 s to process these test datasets, while the dedicated Kraken model takes around 3 min.
Fig. 7.
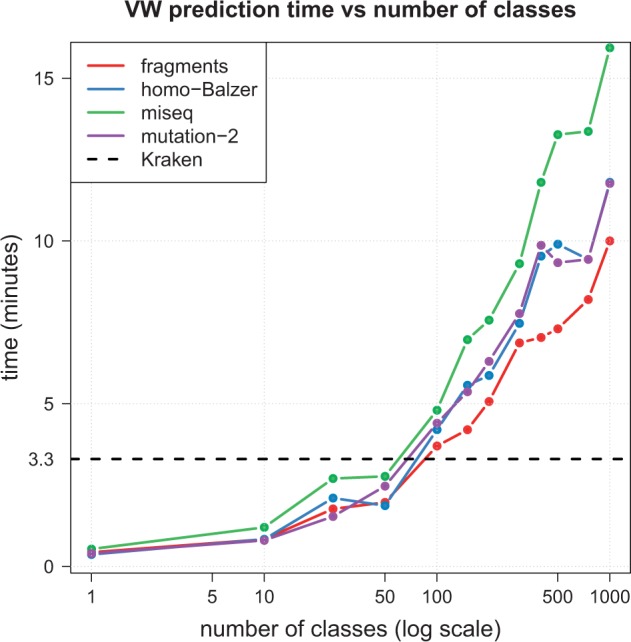
Impact of the number of classes on VW prediction time. Time needed to process each test dataset by VW as the number of classes increases from 1 to 1000. The dashed horizontal line represents the median time measured by Kraken on the large database
5 Discussion
In this work, we investigate the potential of modern, large-scale machine learning approaches for taxonomic binning of metagenomics data and propose a new rank-flexible classification strategy. We extensively evaluate its performance when the scale of the problem increases regarding (i) the length of the k-mers considered to represent a sequence, (ii) the number of fragments used to learn the model and (iii) the number of candidate species involved in the reference database. We also investigate in details its robustness to sequencing errors using simulated reads. We consider three baselines for this evaluation: a comparative approach based on the BWA-MEM sequence aligner and two compositional approaches based on the generative NB classifier and based on Kraken. We demonstrate in particular that increasing the number of fragments used to train the model has a significant impact on the accuracy of the model and allows to estimate models based on longer k-mers. While this could be expected and was already highlighted by previous studies, the resulting configurations are out of reach of standard SVM implementations. We also show that discriminatively trained compositional models usually offer significantly higher performances than generative NB classifiers. The resulting models are competitive with well-established alignment tools and with Kraken for problems involving a small to moderate number of candidate species and for realistic amounts of sequencing errors. Our results suggest, however, that machine learning-based compositional approaches, both discriminative and generative, are still limited in their ability to deal with problems involving more than a few hundreds species. In this case, indeed, compositional approaches exhibit lower performance than alignment-based approaches and are much more negatively impacted by sequencing errors. When confronted with species absent from the training set, we show that our model is more accurate than Kraken, which has a larger level of rejection due to its use of longer k-mers, and affects more reads too specifically to species of the reference dataset. Finally, we confirm that compositional approaches achieve faster prediction times. This is indeed systematically the case in the various configurations listed above, with predictions obtained 2–17 times faster by compositional approaches, and, interestingly, depends on the number of candidate species. We note in particular, that for problems involving fewer than 100 candidate species, which may arise in diagnostic applications involving specific types of specimens, VW can achieve faster prediction times than Kraken. At the extreme, for the binary problem aiming to separate bacterial from human reads, which is commonly used while analyzing a microbiome sample, VW can offer a 9-fold increase in terms of prediction time with respect to Kraken. We emphasize, however, that fast predictions can only be obtained provided that the classification models are loaded in memory, hence for a memory footprint that scales linearly with the number of candidate species and exponentially with the size of the k-mers, which can become important for large reference databases and long k-mers.
At least three simple extensions could be envisioned to make compositional approaches more competitive in accuracy with the alignment-based approach, faster and to limit their memory footprint. First, the robustness to sequencing errors may be improved by learning models from simulated reads instead of fragments. This could indeed allow to tune the model to the sequencing technology producing the reads to be analyzed, provided its error model is properly known and characterized. Second, introducing a sparsity-inducing penalty while learning the model would have the effect of reducing the number of features entering the model, hence to reduce the memory footprint required to load the model into memory. Finally, alternative strategies, known as error correcting tournaments (Beygelzimer et al., 2009), could be straightforwardly considered to reduce the number of models to learn, hence to store into memory during prediction, to address a multiclass problem. Our results indeed suggest that addressing these issues is critical to build state-of-the-art compositional classifiers to analyze metagenomics samples that may involve a broad spectrum of species.
Supplementary Material
Funding
This work was supported by the European Research Council (SMAC-ERC-280032 to J-P.V.) and the French National Research Agency (ANR-11-BINF-0001 to J-P.V.).
Conflict of Interest: none declared.
References
- Agarwal A., et al. (2014) A reliable effective terascale linear learning system. J. Mach. Learn. Res., 15, 1111–1133. [Google Scholar]
- Angly F., et al. (2012) Grinder: a versatile amplicon and shotgun sequence simulator. Nucleic Acids Res., 40, 94–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer S., et al. (2010) Characteristics of 454 pyrosequencing data enabling realistic simulation with flowsim. Bioinformatics, 26, 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beygelzimer A., et al. (2009) Error-correcting tournaments. Algorithmic Learn. Theory, 5809, 247–262. [Google Scholar]
- Bottou L. (1998) Online learning and stochastic approximations. Online Learn. Neural Netw., 17, 9–42. [Google Scholar]
- Bottou L. (2010) Large-scale machine learning with stochastic gradient descent. In: Lechevallier,Y. and Saporta,G. (eds), Proceedings of COMPSTAT’2010, Physica-Verlag, pp. 177–186. [Google Scholar]
- Gammerman A., Vovk V. (2007) Eedging predictions in machine learning. Comput. J., 50, 151–163. [Google Scholar]
- Hugenholtz P., et al. (2002) Exploring prokaryotic diversity in the genomic era. Genome Biol., 3, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D., et al. (2007) MEGAN analysis of metagenomic data. Genome Res., 17, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J., et al. (2009) PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol., 10, 23–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslicki D., et al. (2014) WGSQuikr: fast whole-genome shotgun metagenomic classification. PLoS One, 9, e91784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford J., et al. (2007) Vowpal Wabbit open source project. Technical report. Yahoo. [Google Scholar]
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. [Google Scholar]
- Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner M.S., Renard B.Y. (2012) Metagenomic abundance estimation and diagnostic testing on species level. Nucleic Acids Res., 41, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjancenko O., et al. (2010) Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol., 60, 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mande S., et al. (2012) Classification of metagenomic sequences: methods and challenges. Brief Bioinform., 13, 669–681. [DOI] [PubMed] [Google Scholar]
- Martin J., et al. (2012) Optimizing read mapping to reference genomes to determine composition and species prevalence in microbial communities. PLoS One, 7, e36427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy A.C., et al. (2007) Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods, 4, 63–72. [DOI] [PubMed] [Google Scholar]
- Miller R., et al. (2013) Metagenomics for pathogen detection in public health. Genome Med., 5, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D., et al. (2011) Classifying short genomic fragments from novel lineages using composition and homology. BMC Bioinformatics, 12, 328–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K., et al. (2012) The PhyloPythiaS web server for taxonomic assignment of metagenome sequences. PLoS One, 7, e38581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J., et al. (2009) The NIH human microbiome project. Genome Res., 19, 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K., et al. (2012) NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res., 40, 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenfeld C., et al. (2004) Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet., 38, 525–552. [DOI] [PubMed] [Google Scholar]
- Rosen G., et al. (2011) NBC: the Naive Bayes classification tool webserver for taxonomic classification of metagenomic reads. Bioinformatics, 27, 127–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. (2012) Insights into antibiotic resistance through metagenomic approaches. Future Microbiol., 7, 73–89. [DOI] [PubMed] [Google Scholar]
- Sonnenburg S., et al. (2006) Large scale learning with string kernels. J. Mach. Learn. Res., 7, 1531–1565. [Google Scholar]
- Soon W., et al. (2013) High-throughput sequencing for biology and medicine. Mol. Syst. Biol., 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol., 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol., 15, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.