Summary
KRAB-containing zinc finger proteins (KRAB-ZFPs) are early embryonic controllers of transposable elements (TEs), which they repress with their cofactor KAP1 through histone and DNA methylation, a process thought to result in irreversible silencing. Using a target-centered functional screen, we matched murine TEs with their cognate KRAB-ZFP. We found the paralogs ZFP932 and Gm15446 to bind overlapping but distinguishable subsets of ERVK (endogenous retrovirus K), to repress these elements in embryonic stem cells, and to regulate secondarily the expression of neighboring genes. Most importantly, we uncovered that these KRAB-ZFPs and KAP1 control TEs in adult tissues, in cell culture and in vivo, where they partner up to modulate cellular genes. Therefore, TEs and KRAB-ZFPs establish transcriptional networks that regulate not only development but probably many physiological events. Given the high degree of species-specificity of TEs and KRAB-ZFPs, these results have important implications for understanding the biology of higher vertebrates, including humans.
Introduction
Transposable elements (TEs) account for more than half of the human and murine genomes (Lander et al., 2001; Waterston et al., 2002). Long considered as purely parasitic, they are now recognized as important motors of evolution, yet they also represent genomic threats requiring control from the earliest stages of development. Whether they are DNA transposons or retrotransposons – endogenous retroviruses (ERVS), LINEs SINEs and SVAs, reviewed in Friedli and Trono, 2015 –, TEs can disrupt genes, alter their transcription or serve as ground for recombination, and have been implicated in diseases such as cancer and diabetes (Hancks and Kazazian, 2012; Jern and Coffin, 2008). However, growing evidence indicates that TEs can be coopted for the benefit of the host, with for instance expression of zygotic activation genes driven from the LTR (long terminal repeat) of MERVL (murine endogenous retrovirus L) in the mouse, and many binding sites for pluripotency factors residing within mobile DNA elements in the human genome (Bourque et al., 2008; Chuong, 2013; Dupressoir et al., 2012; Fort et al., 2014; Macfarlan et al., 2012).
TEs are repressed through RNA- and protein-based epigenetic mechanisms instated during the first days of embryogenesis. KRAB-containing zinc finger proteins (KRAB-ZFPs) constitute a large family of transcription factors implicated in this process. KRAB-ZFPs bind to specific DNA sequences through an array of zinc fingers, and recruit their cofactor KAP1, which serves as a scaffold for a heterochromatin-inducing complex encompassing histone methyltransferase, histone deacetylase, nucleosome remodelling and DNA methyltransferase activities (reviewed in Rowe and Trono, 2011). Depletion of KAP1 or its partner histone methyltransferase SETDB1 in murine or human embryonic stem (ES) cells activates the expression of endogenous retroelements (EREs) (Matsui et al., 2010; Rowe et al., 2010; Turelli et al., 2014). This impacts expression of nearby genes, as KAP1 and associated effectors control TE-originating promoter or enhancer effects (Rebollo et al., 2012; Rowe et al., 2013b; Wolf et al., 2015). Furthermore, a few individual KRAB-ZFPs have been confirmed to repress retroelements in pluripotent cells, such as ZFP809 for murine leukemia virus (MLV) and its endogenous relatives (Wolf and Goff, 2007; Wolf and Goff, 2009; Wolf et al., 2015), or Gm6871 and ZNF93 for mouse and human LINEs, respectively (Castro-Diaz et al., 2014; Jacobs et al., 2014). Although recent findings indicate that many KRAB-ZFPs have EREs as their preferential genomic targets (Najafabadi et al., 2015) (and our unpublished results), detailed functional data are missing about most members of the family.
Encoded in the hundreds by the genomes of higher vertebrates, KRAB-ZFPs first emerged in early tetrapods some 350 million years ago, and the expansion of this gene family subsequently mirrored waves of retroviral invasion into the genomes of these species (Thomas and Schneider, 2011). During this process, Krab-zfp genes underwent strong positive selection at positions encoding for amino acids predicted to determine DNA binding specificity, consistent with a role in countering rapidly mutating genetic invaders (Emerson and Thomas, 2009). Furthermore the study of a couple of KRAB-ZFP/TE target pairs suggests the parallel evolution of restriction factors and TE mutants escaping their inhibition (Jacobs et al., 2014). This led to the suggestion of a host-invader arms race, where KRAB-ZFPs were primarily selected to silence TEs.
In ES cells, the KRAB-ZFP-mediated docking of KAP1 and associated epigenetic modifiers at TEs triggers the deposition of repressive chromatin marks such as trimethylation of histone 3 on lysine 9 (H3K9me3), and methylation of CpG dinucleotides by de novo DNA methyltransferases (Rowe and Trono, 2011). Once established, DNA methylation is perpetuated across cell divisions, and its establishment at TEs during early embryogenesis is thought to result in permanent silencing, without need for persistent expression of their cognate KRAB-ZFP repressors (Quenneville et al., 2012; Wolf et al., 2015). Cumulated evidence indicates that DNA methylation is indeed an important mechanism of TE control in somatic tissues (Hutnick et al., 2010; Jackson-Grusby et al., 2001; Rowe et al., 2013a; Walsh et al., 1998). However, we previously observed that a significant fraction of TEs bound by KAP1 in human ES cells still carries the corepressor in mature T lymphocytes (Turelli et al., 2014), and that KAP1 deletion in neuronal progenitors activates some EREs (Fasching et al., 2015). A few ERVs are similarly induced in murine B-lymphocytes deleted for SETDB1, the histone methyltransferase associated with KAP1 (Collins et al., 2015). Moreover, many KRAB-ZFPs are expressed not only in ES cells but also in a variety of tissues (Barde et al., 2013; Corsinotti et al., 2013; Lizio et al., 2015).
In order to investigate KRAB-ZFPs/TE interactions, we developed a functional screen to identify KRAB-ZFPs responsible for recognizing specific DNA sequences. This led us to characterize two members of the family, ZFP932 and its paralog Gm15446, which we found to regulate distinct subsets of endogenous retrovirus-K (ERVK) in murine ES cells. Invalidating current models (Maksakova et al., 2008; Mikkelsen et al., 2007; Rowe and Trono, 2011; Wolf et al., 2015), we further determined that these two KRAB-ZFPs also regulate their TE targets in differentiated tissues, through histone-based mechanisms not always correlated with the DNA methylation status of these loci. Furthermore, the dynamic control of these TEs by their KRAB-ZFP repressors modulated the expression of cellular genes in several adult tissues examined, both in cell culture and in vivo. We conclude that TEs and their KRAB-ZFP controllers are broad regulators of cellular gene expression, likely engaged in influencing multiple aspects of the biology of higher species.
Results
A functional screen identifies KRAB-ZFPs repressing specific DNA targets
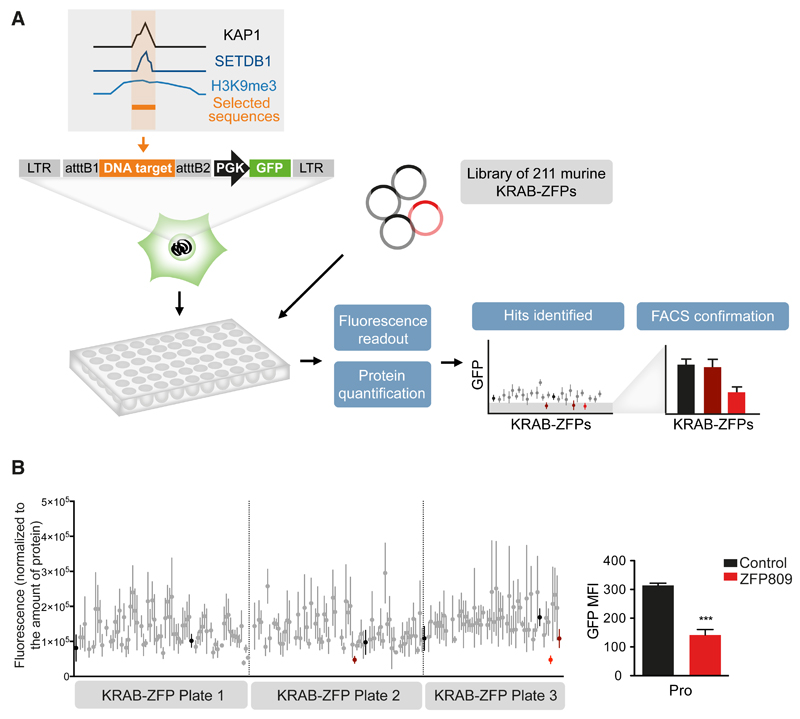
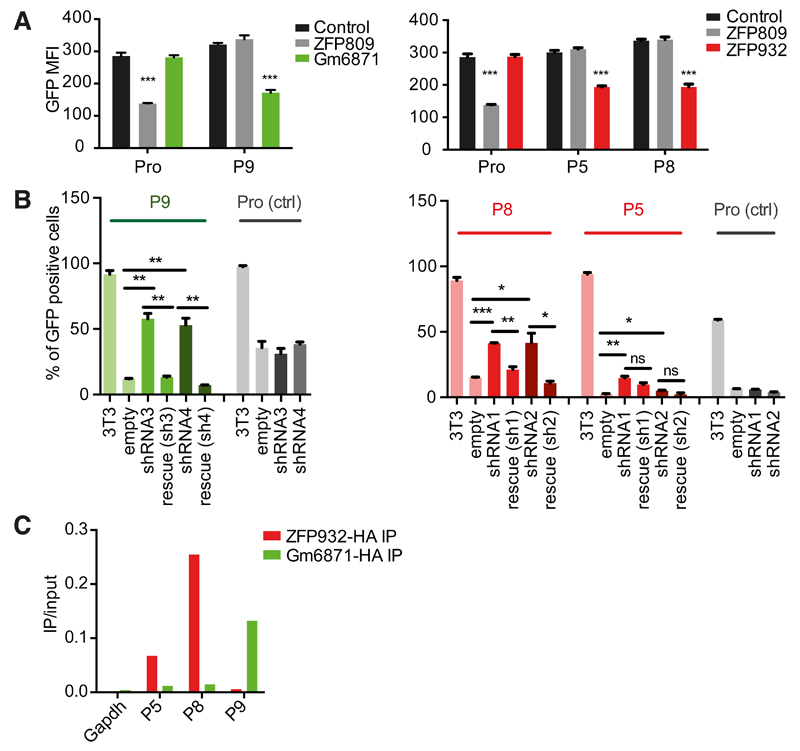
In order to identify KRAB-ZFPs responsible for regulating specific TEs, we developed a large-scale functional screen based on the repression of a reporter cassette through the intermediate of nearby DNA baits (Figure 1A). Putative KRAB-ZFP-binding sequences were selected based on the coincidence of KAP1, SETDB1 and H3K9me3 peaks in chromatin immunoprecipitation / deep sequencing (ChlP-seq) studies of murine ES cells (Bilodeau et al., 2009; Rowe et al., 2013b). Sequences contained within TEs or zinc finger proteins were favored, and imprinting control regions (known for their recruitment of ZFP57) were excluded. We selected 19 such targets (Table S1), and added the PBSLys1,2 sequence – the primer binding site (PBS) sequence of many retroviruses, demonstrated to drive KAP1-mediated repression, but through an unknown KRAB-ZFP intermediate (Wolf et al., 2008). Lentivectors containing these baits upstream of a PGK-GFP cassette were first introduced into murine ES cells, which revealed that 10 out of 20 of these TE-derived fragments induced KAP1-dependent repression of GFP expression (Figure S1). The corresponding vectors were used to engineer stable 293T cell lines, which were transfected in triplicates with individual members of a library of 211 murine KRAB-ZFPs (Table S2) and examined 6 days later for GFP expression with a fluorescence plate reader; the hits were then confirmed by FACS analysis. A control cell line, transduced with a vector containing the MLV PBSPro sequence upstream of the PGK-GFP cassette properly singled out ZFP809 as its cognate repressor, giving us confidence in our approach (Figure 1B). Out of the 10 tested DNA sequences, 3 allowed the identification of a specific KRAB-ZFP ligand (Figures 2A and S2A-S2C). The P9 bait, which comprises a SINE-B2 element, was repressed by Gm6871. The P5 and P8 baits, both derived from ERVKs (RLTR44-int for P5; RLTR9A3, MMERVK10D3-I-int, and RLTR6-int for P8), were repressed by ZFP932. To confirm the matches thereby identified, we introduced the DNA bait-PGK-GFP lentiviral vectors in mouse ES cells depleted for the individual KRAB-ZFPs by RNA interference. For P5, P8 or P9, depleting the corresponding KRAB-ZFP released GFP expression, while it had no effect on a vector containing the PBSPro control sequence (Figure 2B and S2D). Furthermore, repression could be restored by overexpressing an shRNA-resistant form of the specific KRAB-ZFP for P9 and P8 (significantly), and to a lesser extent for P5 (not significantly, possibly due to initial lower values). Finally, we could document the bindings of Gm6871 to the P9 sequence, and of ZFP932 to both of its genomic targets by chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) (Figure 2C and S2E). Interestingly, Gm6871 also binds some LINE-1 elements, which it can repress in murine ES cells (Castro-Diaz et al., 2014). In Gm6871-specific ChIP-seq analyses, not only LINEs, but also the SINE-containing P9 locus was detected (Figure S2B), indicating that a KRAB-ZFP can recognize TEs from different subgroups.
Figure 1. A functional screen to match KRAB-ZFPs repressors with their genomic targets.
(A) Methodological outline. A library of murine KRAB-ZFPs-expressing plasmids was transfected in 293T stable cell lines containing the DNA sequence of interest upstream a PGK-GFP cassette. Fluorescence readout in 96-well plates was normalized for protein content, and hits were tested by FACS for confirmation. (B) Validation of the screen using the previously characterized ZFP809/PBSPro (Pro) pair. Fluorescence readout was performed (left) and identified hits were tested by FACS. The only hit confirmed by FACS was ZFP809 (right panel). Black: transfection control; red: hits identified by plate reader; light red: hits confirmed by FACS. Error bars represent SD, ***p < 0.001, Student’s t test. See also Figure S1, Table S1, and Table S2.
Figure 2. Identification of Gm6871 and ZFP932 as ligands of KAP1-repressed TE sequences.
(A) FACS confirmation of hits identified through screening of selected target sequences. (B) GFP repression assay in control, ZFP932- or Gm6871-knockdown, or ZFP-complemented murine ESC. (C) HA ChIP PCR of Gm6871 and ZFP932 in mES overexpressing the corresponding tagged protein (results representative of 3 independent experiments). Error bars represent SD, * p < 0.05, ** p ≤0.01, *** p ≤ 0.001, ns= not significant, Student’s t test. See also Figure S2.
ZFP932 and its paralog, Gm15446, bind to distinct subsets of ERVK endogenous retroelements
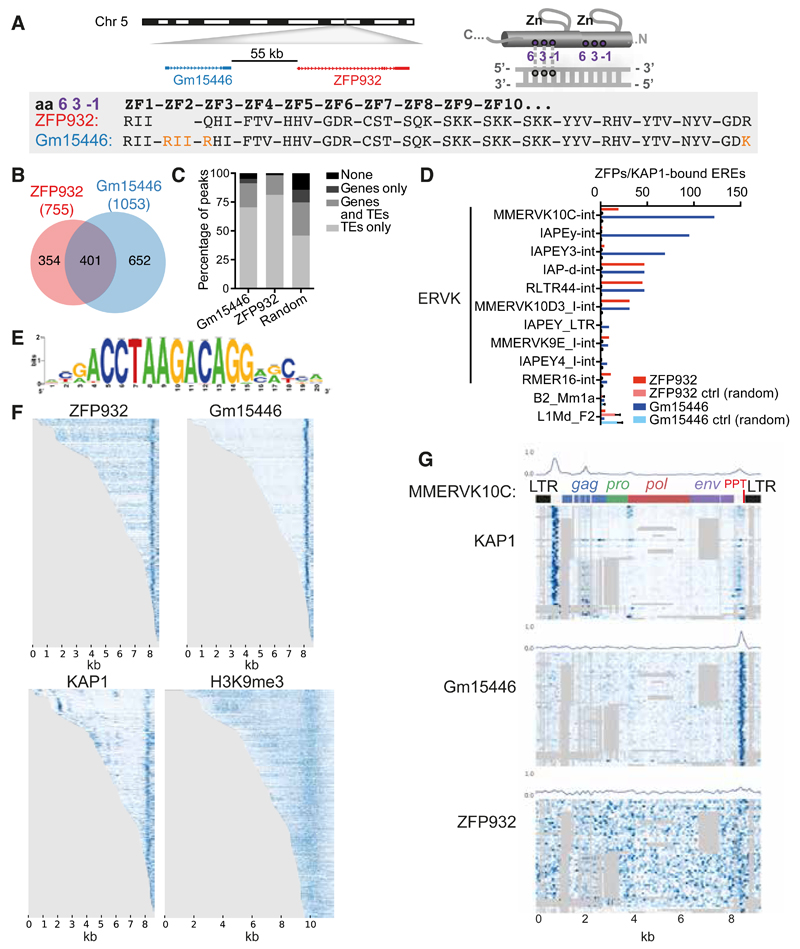
We then turned to the biological characterization of Zfp932. We realized that this mouse-specific KRAB-ZFP gene has a paralog, Gm15446, located in close proximity on chromosome 5 (Figure 3A), and also targeted by our Zfp932-directed siRNAs. When examining the predicted DNA-contacting amino acids within the zinc finger arrays (Liu et al., 2014) of ZFP932 and Gm15446, we saw that the two proteins differed by only two point mutations and the presence of one extra zinc finger in Gm15446. In 293T cells, overexpression of either protein led to repression of the same targets, albeit with some differences in efficiency (Figure S3A).
Figure 3. ZFP932 and its paralog Gm15446 bind with KAP1 to the 3’ end of distinct subsets of ERVK.
(A) Representation of genomic location of Zfp932 and Gm15446 (left), and scheme of the 3 amino acids important for DNA recognition on a zinc finger domain (right). Lower panel depicts the comparison of the 3 amino acids predicted to contact DNA of each zinc finger of ZFP932 and Gm15446, highlighting differences in orange. (B) Venn diagram of ZFP932 and Gm15446 binding sites determined by ChIP-seq in mESC. (C) Percentage of ZFP932, Gm15446, or random control peaks on genic and repeated regions. Random control is based on the mean of 100 random shuffling of the peaks of Gm15446 (KRAB-ZFP ChIP with more peaks). (D) Top 10 ERE families bound by ZFP932 or Gm15446 together with KAP1 in mESC. SINEs and LINEs (bottom) served as negative controls. A control with 100 random shuffling of the binding sites is also shown. (E) Predicted DNA-binding motif for ZFP932 and Gm15446. (F) ChIP-seq coverage plots in mESC on all EREs bound by ZFP932 or Gm15446, ranked by size. Each row is independently normalized, with enrichment proportional to darkness of the blue color. For H3K9me3, representation extends 1 kb up and downstream of the ERE boundaries. (G) Coverage plot on multiple alignment of different ChIP-seqs in ES cells on “full-length” (>5kb) MMERVK10C-int bound by ZFP932 or Gm15446. Repbase MMERVK10C-int consensus is represented on top. Mean of binding coverage is illustrated in each plot. Each row is independently normalized, with enrichment proportional to darkness of the blue color. Gray areas correspond to gaps in multiple alignments. Error bars represent SD. See also Figure S3.
We next investigated by ChIP-seq the genomic targets of ZFP932 and Gm15446 in mouse ES cells overexpressing HA-tagged derivatives of these proteins (Figure S3B). The intersection of duplicate ChIP-seq experiments yielded a total of 755 peaks for ZFP932 and 1053 for Gm15446, amongst which 401 were shared (Figure 3B). Overlapping these data with genic regions and using our updated census of repeats in the mouse genome (see Supplemental Experimental Procedures for details), we observed that most of the KRAB-ZFPs peaks were on TEs (Figure 3C) – either in or outside genes –, the majority of which were ERVKs (68.7% of peaks for ZFP932 and 91.5% for Gm15446). To decrease the possibility of unspecific targets due to overexpression of the proteins, we overlapped the results of these ChIP-seqs with that of similar analysis performed on endogenous KAP1 in murine ES cells. In total, 226 peaks for ZFP932 and 448 for Gm15446 overlapped with KAP1 peaks, mostly at ERVKs, confirming that these KRAB-ZFPs recognize this ERE. Interestingly, some differences were noted between the two paralogs (Figure 3D). For instance, ZFP932 and Gm15446 were similarly enriched at RLTR44-int, IAP-d-int, and MMERVK10D3_I-int elements, but Gm15446 was more frequently found at MMERVK10C-int, IAPEy-int, and IAPEY3-int. Using ChIP-seq data, we identified a motif present in 80% of the ZFP932- and Gm15446-binding sites and 82.7% of ZFPs-enriched ERVKs, but absent from members of the ERV1 family, not targeted by these KRAB-ZFPs (Figure 3E). The underlying sequence is different from a ZFP932-recruiting motif identified the Ptch1 gene promoter in a limb mesenchymal cell line (He et al., 2011). However, we did not detect ZFP932/Gm15446 at this site. It could be that Ptch1 binds ZFP932 only in particular cellular contexts, and via another protein.
We then mapped more precisely the ZFP932/Gm15446/KAP1 binding sites within the genome of their ERV targets. MLV and some IAPs (intracisternal A article, a subtype of ERV) were previously found to recruit KAP1 via the proximal part of their provirus, consistent with the observed repression of their nearby 5’LTR (long terminal repeat) (Rowe et al., 2010; Wolf and Goff, 2009; Wolf et al., 2015). Surprisingly, both ZFP932 and Gm15446 were instead enriched near the 3’end of their target retroelements, together with KAP1 and H3K9me3 (Figure 3F). An analysis using multiple alignments of MMERVK10C-int elements revealed that the two paralogs and KAP1 bound a region situated just after env, upstream of the 3’LTR, partly overlapping with the 3’ polypurine tract (PPT), an element important for reverse transcription (Figure 3G). Interestingly, KAP1 was also enriched at two 5’ sites in these retroelements, where it was likely recruited by other KRAB-ZFPs since neither of the two paralogs was found there. Finally we asked more broadly if KAP1 binds to multiple regions of retroelements other than ZFP932/Gm15446 targets, and we identified several KAP1 peaks on IAPEz, another subtype of ERVs (Figures S3C-S3D). These results demonstrate that KAP1 can bind to multiple locations of the same TE, including its 3’end, via different KRAB-ZFPs.
ZFP932 and Gm15446 regulate ERVK and nearby gene transcription in mouse embryonic stem cells
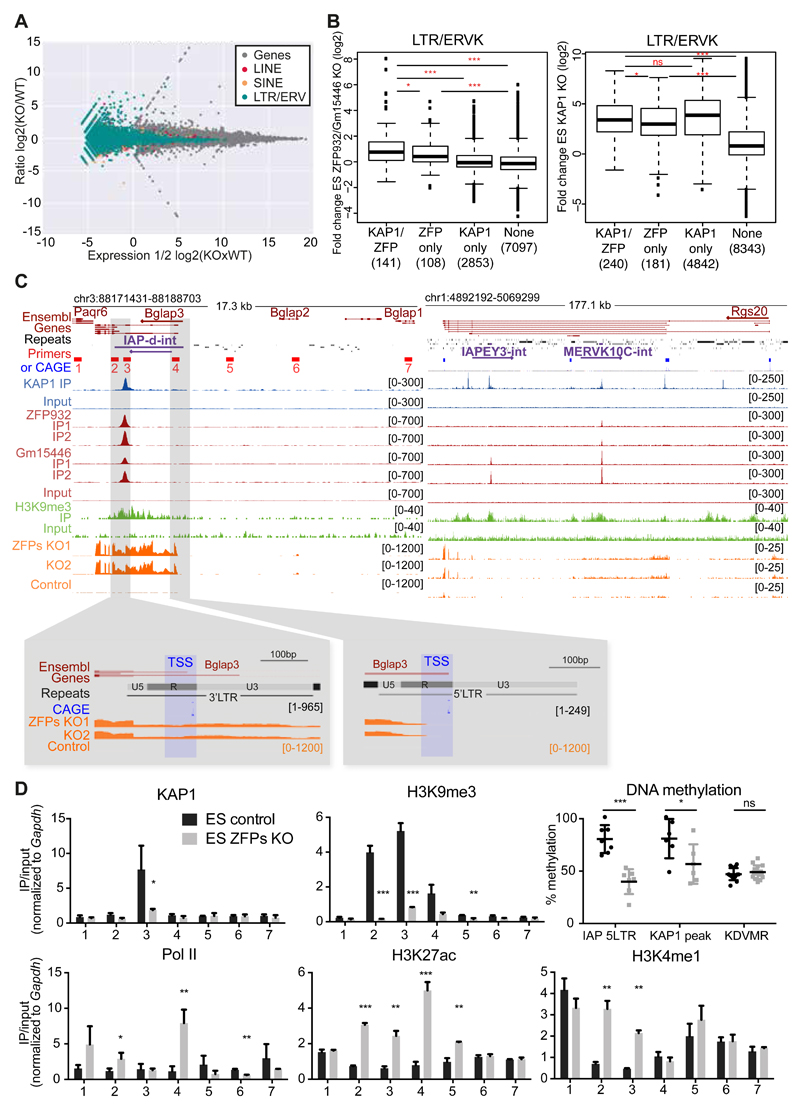
To gain insight on the transcriptional impact of these two KRAB-ZFPs, we next deleted ZFP932 and Gm15446 from murine ES cells by CRISPR/Cas9-mediated editing of the locus and defined the transcriptome of the resulting cells by RNA-seq (Figure 4A). Mainly members of the LTR/ERV class of transposable elements were upregulated, consistent with the ChIP-seq data. Overlapping these results with those of RNA-seq performed in KAP1 knockout (KO) ES cells (Figure S4A) unveiled 141 ERV, 17 LINE and 27 SINE integrants, the expression of which was significantly increased in both settings. ERVs that had been identified as ZFP932/Gm15446 binding targets were more upregulated in cells depleted for either protein or KAP1, compared with ERVs on which these KRAB-ZFPs and KAP1 had not been detected (Figure 4B). Of the bound ERVs with increased expression upon ZFPs KO, many had only the 3’PPT KAP1 peak, or had a 3’ peak markedly more pronounced than its 5’ counterparts.
Figure 4. ZFP932/Gm15446/KAP1 binding regulates expression of ERVs and nearby genes in mESC.
(A) MA-plot of RNA-seq from ZFP932/Gm15446 KO vs. control ES cells. Expression of genes and TEs is shown. (B) Boxplots of ERVK fold expression in ZFP932/Gm15446 KO (left) or KAP1 KO (right) ES cells. (C) Scheme of Bglap3 (left) and Rgs20 (right) genomic loci. ChIP-seq and RNA-seq signal are depicted, along with primers used in panel D (numbered 1-7), and FANTOM CAGE data corresponding to TSSs. Orientation of the genes and TEs is indicated with an arrow. Repeats of interest are highlighted in purple. Lower panels correspond to two regions of zoom in the Bglap3 locus, indicating the TSSs of the gene. CAGE data correspond to mapped TSS peaks and to the max signal of CAGE reads obtained in one of the tissues analysed by the Fantom5 consortium. (D) ChIP-PCR analysis of different epigenetic marks and pyrosequencing determination of DNA methylation at the Bglap3 locus in ZFP932/Gm15446 KO vs. control ES cells. Location of primers is depicted in panel C above. Error bars represent SD, * p < 0.05, ** p ≤0.01, *** p ≤ 0.001, ns= not significant, Student’s t test was used in panel D and Wilcoxon test in panel B. See also Figure S4.
It was previously demonstrated that the KRAB/KAP1-mediated control of endogenous retroelements preserves the transcriptional dynamics of ES cells by preventing TE-originating enhancer and promoter effects on nearby genes (Rowe et al., 2013b; Wolf et al., 2015). Seventy-one genes were significantly upregulated upon ZFP932/Gm15446 KO, and 29 of those were significantly more expressed also in KAP1 KO mES cells, 6 of which were within 50kb of an ERV bound by both the KRAB-ZFPs and their cofactor. Among these genes, Bglap3 stood out as one of the most upregulated genes in the KRAB-ZFPs KO cells (fold-change of 270.9, P value of 0.00000003). Bglap3 has three short and two long transcripts, the latter interestingly containing a ZFP932/Gm15446-bound IAP-d element (Figure 4C). By comparing FANTOM CAGE transcriptional start site (TSS) data with RNA-seq, we observed that both the 5’ and the 3’LTRs of the IAP function as promoters of the Bglap3 gene, with transcripts starting exactly at the beginning of the LTR R region. Both Bglap3 and its IAP were more expressed in murine ES cells deleted for either ZFP932/Gm15446 or KAP1, demonstrating that the retroelement partakes in the controlling unit of this gene (Figures 4C and S4B-S4C). This effect was confirmed through ZFP932/Gm15446 knockdown (KD) experiments, properly controlled by restoration of the repression upon complementation with interfering RNAs-resistant derivatives of the two KRAB-ZFPs (Figure S4D). The Rgs20 locus provided another example of gene-TE pair regulated by ZFP932/Gm15446, in which no TSS overlapped with the TEs, suggesting that this gene is regulated through enhancer effects (Figure 4C and S4B).
We deepened our study of this phenomenon by examining the epigenetic status of the Bglap3 locus (Figure 4D). Upon ZFP932/Gm15446 deletion, ChIP-PCR analyses revealed loss of KAP1 and H3K9me3 and gain of the activation marks H3K27ac and H3K4me1 at the IAP, together with the accumulation of PolII at the predicted TSSs of Bglap3. Furthermore, pyrosequencing unveiled a marked drop in DNA methylation both underneath the KAP1 peak and at the 5’ end of the IAP. These data indicate that, in murine ES cells, ZFP932/Gm15446 epigenetically regulate not only the expression of their target ERVKs but also the transcriptional influence of these TEs on nearby genes, via both promoter and enhancer effects.
The KRAB/KAP1 system regulates TEs in differentiated cells
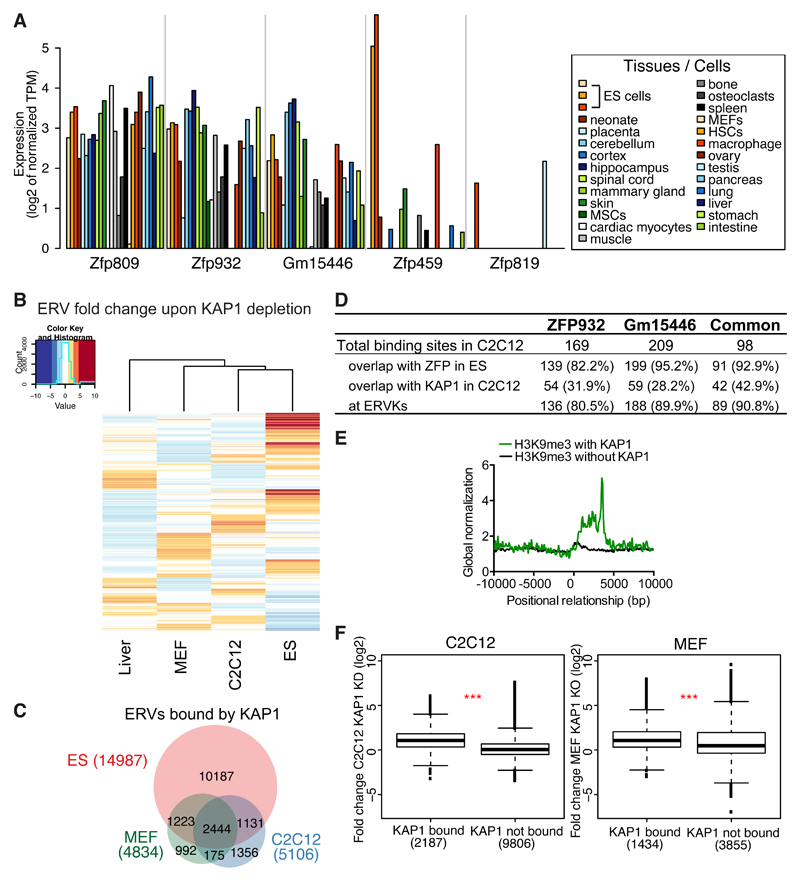
To gain further insight into Zfp932/Gm15446 functions we assessed their expression in representative murine tissues, using CAGE datasets generated by the FANTOM consortium (Lizio et al., 2015) (Figure 5A). Both paralogs were broadly expressed in adult murine cells, as was Zfp809. In contrast, Zfp459 and Zfp819, which we previously found to be highly transcribed in murine pluripotent stem cells (Corsinotti et al., 2013), were largely restricted to undifferentiated cells. Because ZFP932 and Gm15446 displayed a strong preference for ERVK family members as their genomic targets, we suspected that they might control these TEs in adult tissues as well. To probe this hypothesis, we first examined by RNA-seq the transcriptomes of hepatocytes harvested from control and liver-specific Kap1 KO mice, wild type and Kap1-deleted mouse embryonic fibroblasts (MEFs) (Figure S5A), and control and Kap1 KD C2C12 mouse myoblast cells. In all these settings, expression of ERVs was increased upon KAP1 depletion, even though induction was less pronounced than in Kap1 KO ES cells (Figure 5B). We ranked the significantly upregulated families in each dataset and found that MERVK10C-int had the highest number of induced integrants in KAP1-depleted differentiated cells (Figure S5B). General MERVK10C-int primers confirmed by RT-qPCR the increased expression of these elements in liver and C2C12 depleted for KAP1, in spite of the absence of detectable changes in the DNA methylation status of these loci as measured by bisulfite sequencing (Figures S5C-S5D). Interestingly, there were differences in the subsets of ERVs deregulated in the various tissues following KAP1 removal (Figure 5B). For example, particular MMERGLN and ORR1A0 integrants located on chromosome 12 were upregulated in KAP1-depleted liver and ES cells, but not in their C2C12 counterparts (Figure S5C).
Figure 5. The KRAB/KAP1 system controls TE expression in differentiated tissues.
(A) Krab-zfp genes mRNA expression according to FANTOM 5 CAGE data. MSCs, mesenchymal stem cells; HSCs, hematopoietic stem cells. (B) Heatmap of ERVs average fold-change expression (linear scale) upon KAP1 depletion in different tissues. Liver = Kap1 KO versus control, C2C12 = Kap1 KD versus control, MEF = Kap1 KO versus control, ES = Kap1 KO versus control. (C) Venn diagram of ERVs bound by KAP1 in ES, C2C12, or MEF cells. (D) Comparative table of ZFP932, Gm15446 or common ZFP932 and Gm15446 binding sites in C2C12, determined by ChIP-seq. Absolute values and percentage relative to total binding sites are given. (E) Distribution of H3K9me3 ChIP-seq with or without presence of KAP1, in C2C12, relative to the 5’ end of ERVK elements (5’ end corresponds to 0 bp). (F) Boxplot of ERVKs fold-change expression in C2C12 Kap1 KD or MEFs Kap1 KO versus control. *** p ≤ 0.001, Wilcoxon test. See also Figure S5.
Confirming that the KRAB/KAP1 system is involved in the somatic control of TEs, about 24% of ERVs bound by KAP1 in ES cells still bore the corepressor in C2C12 cells and MEFs (Figure 5C). Interestingly, for 30% of the ERV loci bound by KAP1 in C2C12 and 24% in MEFs no significant enrichment was found in ES cells. We also performed ZFP932- and Gm15446-specific ChIP-seq analyses in C2C12 cells expressing HA-tagged forms of these proteins (Figure 5D, Figure S5E). We detected 169 ZFP932 and 209 Gm15446 binding sites, 98 of which were common, and the large majority of which were also bound by either one or both of these KRAB-ZFPs in ES cells. Most resided within TEs, namely ERVKs, and KAP1 was also detected at about a third of them, supporting a role for these ZFPs in regulating such elements in differentiated cells. We then asked if H3K9me3 marks were present at KAP1-bound ERVKs in C2C12. Using H3K9me3 ChIP-seq data in these cells, we observed a higher correlation of H3K9me3 at ERVKs with than without KAP1 (Figure 5E). General ERVK primers revealed upregulation of MERVK10C, IAP-d, and IAPEz in C2C12 cells depleted for SETDB1, suggesting that this histone methyltransferase partners with KAP1 also in differentiated cells to control TEs (Figure S5F). Finally, ERVKs that were bound by KAP1 in C2C12 or MEFs were significantly more expressed than their unbound counterparts when the corepressor was depleted (Figure 5F). Taken together, these results demonstrate a role for the KRAB/KAP1 system in the somatic control of TEs.
TEs and their KRAB-ZFP controllers regulate gene expression in adult tissues
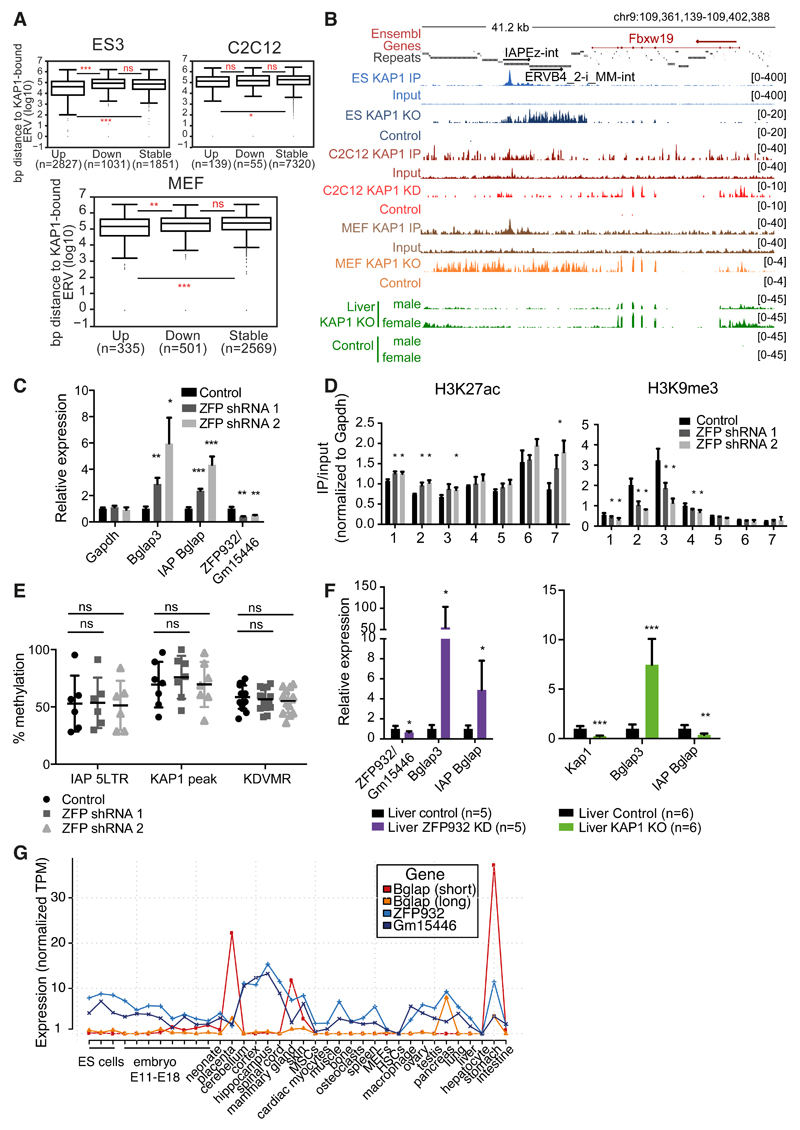
We then asked if the KRAB/KAP1-mediated control of TEs also prevents the illegitimate transcription of nearby genes in adult cells. For this, we first analyzed our RNA-seq of KAP1-depleted cells, separating the genes in stable, up- or downregulated (deregulation criteria was 2-fold change cut-off and P value adjusted ≤ 0.05), and calculated their distance to closest KAP1-bound ERV. As expected, in ES cells upregulated genes were significantly closer to a KAP1-bound ERV than their downregulated or stable counterparts (Figure 6A). In the case of C2C12 KAP1 KD, although the same trend was observed, upregulated genes were only significantly closer to KAP1-bound ERVs when compared to stable but not downregulated genes. Interestingly, for MEFs KAP1 KO we observed the same pattern as in ES cells, suggesting that, even if to a smaller extent, KAP1-regulated TEs can impact neighbouring genes transcription also in differentiated cells.
Figure 6. TEs and their KRAB/KAP1 controllers influence gene expression in adult tissues.
(A) Distance to nearest KAP1-bound ERV of upregulated (fold-change ≥ 2, P adj ≤0.05), stable, or downregulated (fold-change ≤ 2, P adj ≤ 0.05) genes in ES cells Kap1 KO, C2C12 Kap1 KD, or MEFs Kap1 KO. (B) Representative genomic region with ERV and Fbxw19 gene regulated by KAP1 in differentiated cells. KAP1-depleted and control RNA-seq densities of different tissues/cells are depicted along with KAP1 ChIP-seq data. (C) mRNA expression of Bglap3 gene and IAP Bglap in ZFP932/Gm15446 KD in C2C12 cells versus control (normalized to ActinG and Tbp). (D) ChIP-PCR of chromatin marks on Bglap3 locus in ZFP932/Gm15446 KD vs. control C2C12 cells. (E) DNA methylation analysis by pyrosequencing on IAP Bglap in same cells. (F) mRNA expression of Bglap3 gene and IAP Bglap in ZFP932/Gm15446 KD or Kap1 KO mice liver compared to control (normalized to Gapdh and ActinB). (G) Zfp932, Gm15446, and Bglap3 gene expression in different murine tissues/cells according to FANTOM 5 CAGE data. AU, arbitrary units; MSCs, mesenchymal stem cells; HSCs, hematopoietic stem cells. Error bars represent SD, * p < 0.05, ** p ≤0.01, *** p ≤ 0.001, ns= not significant, Wilcoxon test was used in panel A and Student’s t test in panels C-F. See also Figure S6.
We confirmed our hypothesis further by examining in more details a few upregulated genes situated within 100kb of a KAP1-bound ERV. At some loci, such as Fbxw19, KAP1 depletion resulted in activating both the ERV and its nearby gene in all cells examined (Figures 6B and S6A-B). At others, such as Gm13251, removal of the regulator induced expression in liver and in ES cells, but not in C2C12 cells (Figure S6B). We then examined the Bglap3 locus, which we found enriched for ZFP932/Gm15446, KAP1 and H3K9me3 also in C2C12 cells (Figure S6C). Upon RNA interference-mediated KD of these two KRAB-ZFPs, both the IAP and Bglap3 were activated (Figure 6C), coincident with loss of H3K9me3 and KAP1 and mild but significant gain of H3K27ac at the IAP (Figure 6D, and S6D). However, in spite of its transcriptional activation, this ERV remained DNA methylated (Figure 6E).
To expand these findings in vivo, we generated ZFP932/Gm15446 KD mice by transduction of fertilized oocytes with shRNA-expressing lentiviral vectors. We could not assess major phenotypic abnormalities in these animals, but a marked upregulation of Bglap3 gene and the upstream IAP was measured in their liver, albeit again with no detectable loss of DNA methylation at this locus (Figures 6F and S6E). ChIP-PCR experiments in murine wild type liver confirmed the enrichment of KAP1 and H3K9me3 at the IAP (Figure S6F). Interestingly, in the liver of conditional Alb-Cre Kap1 KO mice, in which the regulator is deleted not during early embryogenesis but some three weeks after birth, there was increased expression of Bglap3 but not of the IAP, suggesting an uncoupling of the regulation of the two LTRs of the retroelement (Figure 6F). Also in this setting, we observed that the IAP Bglap remained highly methylated (Figure S6G). Finally, we asked if the Bglap3 gene is expressed in physiological conditions and how does it correlate with the ZFPs transcription. By analysing CAGE expression data we observed that in tissues like placenta, mammary gland, and pancreas Bglap3 transcripts are expressed and correlate with lower Zfp932/Gm15446 levels (the long transcript driven by the IAP 5’LTR, and the short transcript by the 3’LTR) (Figure 6G). Collectively, these data establish that the KRAB/KAP1 system uses TE-residing platforms to regulate gene expression in adult cells.
Discussion
This work establishes that TEs and their KRAB-ZFP controllers can regulate gene expression in adult tissues. Our results thus invalidate a generally accepted model, which assumes that the KRAB/KAP1 system irreversibly silences TEs during early embryonic development (Maksakova et al., 2008; Mikkelsen et al., 2007; Rowe and Trono, 2011; Walsh et al., 1998; Wolf et al., 2015). Furthermore, our study strongly suggests that Krab-Zfp genes are not selected simply to inactivate TEs, but also to domesticate their transcriptional potential for the benefit of the host.
Here, through a target-centric functional screen, we identified several KRAB-ZFPs responsible for the sequence-specific repression of TEs in mouse ES cells, including two KRAB-ZFP paralogs that recognized partly overlapping members of the ERVK family. Not only could we verify that these two KRAB-ZFPs controlled these ERVs and neighboring genes in ES cells, but we further discovered that they were expressed in a broad range of somatic murine cells. We could also demonstrate that, in several differentiated cell types, including in vivo in the liver, KAP1 and KRAB-ZFPs were still bound to and controlled TEs, and modulated the expression of nearby genes via promoter and enhancer effects, even if with a smaller global impact than the observed in ES cells. Remarkably, this process occurred without apparent alteration of the DNA methylation status of these loci, but primarily involved histone-based changes.
Although DNA methylation is traditionally considered as a silencing mechanism, emerging evidence indicates that its impact on gene expression is far more diversified (Jones, 2012; Schubeler, 2012). We had previously observed that docking of the KRAB/KAP1 complex at genomic loci leads to their DNA methylation when it occurs in early embryonic cells but not in their differentiated counterparts, where it induces repression solely by histone-based, hence fully reversible, changes (Groner et al., 2012; Quenneville et al., 2012; Wiznerowicz et al., 2007). The present data further indicate that protein-restricted chromatin modifications can reignite transcription at these sites in adult cells, in spite of persistent DNA methylation. This is consistent with recent observations that some ERVs are activated without changes in their DNA methylation status in B cells depleted for the histone methyltransferase SETDB1 (Collins et al., 2015), and that, in human ES cells, transcription can occur from some highly methylated KAP1-controlled promoters (Turelli et al., 2014).
Whether in the context of murine or human ES cells, we had previously noted that KAP1 depletion does not systematically trigger the activation of all KAP1-bound TEs (Rowe et al., 2010; Turelli et al., 2014). Here, we observed that, in adult cells as well, the range of ERVs activated upon KAP1 or ZFP932/Gm16446 depletion differed depending on the cellular environment. In C2C12, for example, many IAPEz elements bound by KAP1 were not significantly de-repressed upon its removal (not illustrated). At the Gm13251 locus, KAP1 depletion activated two ERVs in liver and in ES cells, but not in MEFs or C2C12 cells. Correspondingly, in Setdb1 KO B-lymphocytes, the upregulation of selected ERVs correlated with the presence within their promoters of binding sites for B cell-specific transcription factors (Collins et al., 2015). It is likely that this type of restriction broadly applies to KRAB-ZFP-controlled loci, the expression of which is conditioned not only by removal or biochemical inactivation of their epigenetic repressors, but also by the presence of a proper set of activators, likely tissue-specific. In addition, specific loci may be subjected to dominant influences imposed by the local chromatin configuration, and other control mechanisms, such as small RNAs-based, may be at play in some environments (Bierhoff et al., 2014; Heras et al., 2013; Marchetto et al., 2013; Pezic et al., 2014). Finally, KAP1 can undergo locus-specific post-translational modifications switching its function from corepressor to coactivator (Singh et al., 2015), and KRAB-ZFPs could also be subjected to this type of regulation, leading to the recruitment of different sets of effector complexes.
Our demonstration that KRAB-ZFPs are involved in controlling TEs not only in embryonic but also differentiated cells is consistent with the widespread expression of these proteins in adult organisms, whether mouse or human (Lizio et al., 2015), and with the finding that a great majority of these transcription factors have TEs as their preferred genomic targets (Najafabadi et al., 2015) (and our unpublished results). It also contributes to explain the broad range of phenotypes induced by the conditional KO of Kap1 in the mouse, even though the master regulator carries out some KRAB-ZFP- and TE-independent functions (Iyengar and Farnham, 2011; Iyengar et al., 2011; McNamara et al., 2016; Singh et al., 2015).
Our study unveils several other unsuspected aspects of the KRAB/KAP1-mediated control of transposable elements. First, we found that a given KRAB-ZFP can recognize different subgroups of TEs, e.g. Gm6871 repressing members of both the LINE and SINE families. Second, we determined that the two ERVK-targeting KRAB-ZFPs identified here tether KAP1 near the 3’ end of their retroviral targets, not at the PBS region or close to their promoter as previously documented for MLV, several other IAPs and most LINEs (Castro-Diaz et al., 2014; Jacobs et al., 2014; Rowe et al., 2010; Wolf and Goff, 2009; Wolf et al., 2015). Interestingly, this 3’end peak overlaps with the 3’PPT, a sequence important for reverse transcription. While it readily explained how ZFP932 and Gm15446 can control the impact of the IAP Bglap 3’LTR on the Bglap3 gene, it was more surprising to observe that this distal location also served to repress the 5’LTR of this element, as demonstrated by the spreading of chromatin marks and its upregulation in ES cells or mice where these two KRAB-ZFPs were depleted. Third, we observed the differential regulation of the two LTRs of the same ERV, and consequently of the gene placed under their influence, via one KRAB/KAP1 -binding site. Depletion of ZFP932/Gm15446 from the earliest times of embryonic development activated the expression of both Bglap3 and its controlling IAP in the liver of adult mice, whereas the deletion of Kap1 in this organ after birth only de-repressed the Bglap3 gene, but not its IAP. It thus seems that the regulation of the 5’ and 3’ LTR of this endogenous retrovirus can be uncoupled. In the absence of obvious differences in the DNA methylation status of these two transcription units, the molecular basis of their distinctive behavior remains to be identified. We could even speculate that, as previously reported (Wolf et al., 2015), the depletion of KRAB/KAP1 regulation at different moments of development could lead to differential TE control due to deposition of other epigenetic marks. Finally, we found that KAP1 can be recruited to several regions of a same ERV, likely via distinct KRAB-ZFPs. It strongly suggests that a TE can be regulated in temporally and functionally differential fashions, hence that the complexity of KRAB/KAP1-mediated regulation of TEs and their gene targets is much greater than envisioned so far.
Our results support a model in which the evolutionary selection of KRAB-ZFP genes is not only an arms race aimed at silencing TEs but also the instrument of their domestication. It is likely that TEs and their KRAB-ZFPs regulators modulate multiple aspects of the biology of tetrapods, superimposing a species-specific layer of control over canonical, conserved regulatory pathways. Indeed, a large fraction of recognizable mobile elements in the genome are unique to the corresponding species and its close relatives, both in sequence and location; accordingly, levels of orthology between KRAB-ZFPs of different organisms are limited. Therefore, KRAB-ZFPs and their targets must play major roles in the speciation of higher vertebrates, including humans.
Experimental Procedures
Cell culture and mouse work
Murine ES cells and MEFs wild type and KO for KAP1 were cultured and generated as previously described (Rowe et al., 2013a; Rowe et al., 2013b). Mouse C2C12 myoblasts were cultured as described (Singh et al., 2015). Two clonal KRAB-ZFPs KO cell lines were generated using an integrase defective lentiviral vector containing the pLentiCRISPR with an sgRNA for Zfp932 and Gm15446 or a control luciferase sgRNA. Knockdown experiments were performed with specific shRNA in pLKO lentiviral vectors. Hepatocyte-specific Kap1 KO mice were generated and genotyped according to (Bojkowska et al., 2012). ZFP932/Gm15446 KD mice were generated by lentiviral transgenesis as described (Rowe et al., 2013a). All shRNAs and sgRNAs used in this study are listed in Supplemental Experimental Procedures. All animal experiments were approved by the local veterinary office and carried out in accordance with the EU Directive (2010/63/EU) for care and use of laboratory animals.
Functional screen
DNA target sequences to be tested were chosen from the overlap of publicly available murine ESC ChIP-seq data for KAP1 (GSE41903), SETDB1 (GSE18371), H3K9me3 (GSE41903), and absence of ZFP57 (GSE31183). PBSLys1,2 sequence plus 19 sequences corresponding to KAP1 peaks were selected based on presence of TEs or of interesting KAP1 targets (such as 3’end of ZFP genes) (see Table S1). These sequences were cloned upstream a PGK-GFP cassette, tested for repression in murine ES, and only repressed sequences were tested in the screen. Lentiviral vectors with these sequences were used to transduce 293T cell lines, which were sorted for GFP presence, generating stable cell lines. The screen was performed by reverse transfection of a library of 211 murine KRAB-ZFPs (Table S2), in 96-well plates, in an automated fashion, in triplicates. Cells were harvested on day 6 after transfection, lysed, and GFP fluorescence was measured by plate reader (excitation at 485 nm and emission at 520 nm). Total protein content was quantified using BCA (BCA Protein Assay Reagent, Thermoscientific), and used to calculate normalized GFP fluorescence. Candidate hits were identified by selecting the 10 KRAB-ZFPs with the lowest normalized fluorescence values per 96-well plate, and only the ones that were present in all 3 replicates were considered. Hits were identified by transfection of the 293T cell line of interest with the candidate KRAB-ZFPs, in 24-well plates, with FACS readout after 6 days. The detailed protocol is given in Supplemental Experimental Procedures.
RT-PCR and RNA-seq
Total RNA was extracted and DNAse-I treated using a spin column-based RNA purification kit (Macherey-Nagel). cDNA was prepared with SuperScript II reverse transcriptase (Invitrogen). Primers (listed in Supplemental Experimental Procedures) were used for SYBR Green qPCR (Applied Biosystems), and specificity was confirmed with dissociation curves. For mRNA sequencing, 100-bp single-end RNA-seq libraries were prepared using the Illumina TruSeq Stranded mRNA reagents (Illumina). Cluster generation was performed with the resulting libraries using the Illumina TruSeq SR Cluster Kit v4 reagents. Sequencing was performed with Illumina HiSeq 2500 in 100-bp reads run. The RNA-seq reads were mapped to the mm9 genome using TopHat (Kim et al., 2013), allowing multimapped reads to be randomly assigned once among the mapped loci. Gene counts were generated with HTseq-count program using default parameters, and TE counts were computed using BEDtools (multicov). Sequencing depth normalization and differential expression analyses were performed using the voom function of Bioconductor package LIMMA (Law et al., 2014).
ChIP-PCR and ChIP-seq
Chromatin immunoprecipitation and library preparation were done according to (Rowe et al., 2013b), with modifications as described in Supplemental Experimental Procedures. Sequencing was performed with Illumina HiSeq 2500 in 100-bp reads run. Reads were mapped to the mouse genome assembly mm9 using Bowtie 2 (Langmead and Salzberg, 2012), using the sensitive-local mode. Peaks were called using MACS (Zhang et al., 2008) or SICER software (for histone modification marks) (Zang et al., 2009), with total input as control.
DNA methylation analysis
Genomic DNA was extracted, converted using an Epitect Bisulfite kit (Qiagen), and used in two rounds of PCR followed by PCR product purification. Pyrosequencing and bisulfite sequencing were performed as previously described (Fasching et al., 2015; Rowe et al., 2013a).
Bioinformatics analyses and statistics
All genome-wide TE analyses were performed using a merged repeats track generated in-house, using RepeatMasker 3.2.8 and merging homonymous ERV-int integrants or attributed LTRs within 400bp or less. Genomic region analyses were done with BEDTools (Quinlan and Hall, 2010). Motif search was performed using with RSAT (Medina-Rivera et al., 2015), using unbound ERVK elements as control. For coverage plots, ChIP-seq signal on each feature of interest were extracted from the bigwig file, scaled between 0 and 1; in some cases, multiple alignment was performed with MAFFT. Statistical difference was assessed by Student’s t test, except for Figures 4B, 5F, and 6A for which Wilcoxon test was applied. Error bars represent ± 1SD. R version 3.1.2 (http://www.R-project.org) or GraphPad Prism version 4.0 (http://www.graphpad.com) were used for statistical analyses. Detailed bioinformatics analyses are provided in Supplemental Experimental Procedures.
Data Access. All next-generation sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database under the accession number GSE74278.
Supplementary Material
Acknowledgments
We thank B. Deplancke and C. Gubelmann for part of the murine KRAB-ZFP library; E. Planet and A. Kapopoulou for help with data analysis; C. Raclot and S. Verp for technical assistance; I. Barde and M. Friedli for advice; the University of Lausanne Genomics Core Facility for sequencing; the EPFL Biomolecular Screeening facility for robotics and advice; A. Reymond (CIG, UNIL, Lausanne) for use of the pyrosequencer; Vital-IT for computing; and the members of the Trono lab for discussions. This work was financed through grants from the Swiss National Foundation, the European Union (FP7/2007-2013/ REA no 290123), and the European Research Council to D.T.
Footnotes
Author Contribuitions
G.E. conceived the study, designed and performed experiments, analysed and interpreted data, and wrote the manuscript. M.C. and A.K. designed, performed and analysed experiments. J.D. analysed data. A.C. performed experiments and made intellectual contributions. S.O. performed experiments. M.I. analysed data and made intellectual contributions. H.M.R. designed experiments and helped conceive the study. P.T. made intellectual contributions and helped supervise the project. D.T. conceived the study, designed experiments, interpreted data, and wrote the manuscript. All authors reviewed the manuscript.
References
- Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science (New York, N.Y.) 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Molecular cell. 2014;54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes & development. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, Santoni de Sio F, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. Liver-specific ablation of Kruppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology (Baltimore, Md.) 2012;56:1279–1290. doi: 10.1002/hep.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome research. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes & development. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB. Retroviruses facilitate the rapid evolution of the mammalian placenta. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:853–861. doi: 10.1002/bies.201300059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8367–8372. doi: 10.1073/pnas.1422187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PloS one. 2013;8:e56721. doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS genetics. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching L, Kapopoulou A, Sachdeva R, Petri R, Jonsson ME, Manne C, Turelli P, Jem P, Cammas F, Trono D, et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell reports. 2015;10:20–28. doi: 10.1016/j.celrep.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nature genetics. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annual Review of Cellular and Developmental Biology. 2015;31:13.11–13.23. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- Groner AC, Tschopp P, Challet L, Dietrich JE, Verp S, Offner S, Barde I, Rodriguez I, Hiiragi T, Trono D. The Kruppel-associated box repressor domain can induce reversible heterochromatization of a mouse locus in vivo. The Journal of biological chemistry. 2012;287:25361–25369. doi: 10.1074/jbc.M112.350884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Current opinion in genetics & development. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Cai J, Lim JW, Kroll K, Ma L. A novel KRAB domain-containing zinc finger transcription factor ZNF431 directly represses Patched1 transcription. The Journal of biological chemistry. 2011;286:7279–7289. doi: 10.1074/jbc.M110.178780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nature structural & molecular biology. 2013;20:1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. The Journal of biological chemistry. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. The Journal of biological chemistry. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, 3rd, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Molecular and cellular biology. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature genetics. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jem P, Coffin JM. Effects of retroviruses on host genome function. Annual review of genetics. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews. Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chang LH, Sun Y, Lu X, Stubbs L. Deep vertebrate roots for mammalian zinc finger transcription factor subfamilies. Genome biology and evolution. 2014;6:510–525. doi: 10.1093/gbe/evu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome biology. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cellular and molecular life sciences : CMLS. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- McNamara RP, Reeder JE, McMillan EA, Bacon CW, McCann JL, D’Orso I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Molecular cell. 2016;61:39–53. doi: 10.1016/j.molcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Rivera A, Defrance M, Sand O, Herrmann C, Castro-Mondragon JA, Delerce J, Jaeger S, Blanchet C, Vincens P, Caron C, et al. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic acids research. 2015;43:W50–56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nature biotechnology. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- Pezic D, Manakov SA, Sachidanandam R, Aravin AA. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes & development. 2014;28:1410–1428. doi: 10.1101/gad.240895.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell reports. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Miceli-Royer K, Zhang Y, Farivar S, Gagnier L, Mager DL. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome biology. 2012;13:R89. doi: 10.1186/gb-2012-13-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development (Cambridge, England) 2013a;140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome research. 2013b;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Schubeler D. Molecular biology. Epigenetic islands in a genetic ocean. Science (New York, N.Y.) 2012;338:756–757. doi: 10.1126/science.1227243. [DOI] [PubMed] [Google Scholar]
- Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, Faralli H, Choi J, Youn HD, Dilworth FJ, et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes & development. 2015;29:513–525. doi: 10.1101/gad.254532.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome research. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome research. 2014;24:1260–1270. doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature genetics. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. The Journal of biological chemistry. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Hug K, Goff SP. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12521–12526. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes & development. 2015;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics (Oxford, England) 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.