Abstract
Objectives
In Klebsiella pneumoniae, overproduction of RamA and RarA leads to increased MICs of various antibiotics; MarA and SoxS are predicted to perform a similar function. We have compared the relative effects of overproducing these four AraC-type regulators on envelope permeability (a combination of outer membrane permeability and efflux), efflux pump and porin production, and antibiotic susceptibility in K. pneumoniae.
Methods
Regulators were overproduced using a pBAD expression vector. Antibiotic susceptibility was measured using disc testing. Envelope permeability was estimated using a fluorescent dye accumulation assay. Porin and efflux pump production was quantified using proteomics and validated using real-time quantitative RT–PCR.
Results
Envelope permeability and antibiotic disc inhibition zone diameters both reduced during overproduction of RamA and to a lesser extent RarA or SoxS, but did not change following overproduction of MarA. These effects were associated with overproduction of the efflux pumps AcrAB (for RamA and SoxS) and OqxAB (for RamA and RarA) and the outer membrane protein TolC (for all regulators). Effects on porin production were strain specific.
Conclusions
RamA is the most potent regulator of antibiotic permeability in K. pneumoniae, followed by RarA then SoxS, with MarA having very little effect. This observed relative potency correlates well with the frequency at which these regulators are reportedly overproduced in clinical isolates.
Introduction
The ability of an antimicrobial drug to cross the Gram-negative bacterial envelope (referred to here as ‘envelope permeability’) is dictated by the rate of drug entry through outer membrane porins and the rate of expulsion by efflux pumps. Global regulatory systems exist to control envelope permeability in enteric bacteria. The paradigm is the Mar system, which has been widely studied in Escherichia coli. Loss-of-function mutations in the transcriptional repressor MarR confer multidrug resistance in E. coli via up-regulation of marA. MarA is an AraC-type transcriptional activator that stimulates transcription of a group of genes, the Mar regulon, including acrAB and tolC, that encode a tripartite efflux system. In addition, MarA causes decreased production of the OmpF porin in E. coli by activating the transcription of micF, an antisense RNA that binds to ompF mRNA, preventing protein synthesis.1,2
Klebsiella pneumoniae is a common cause of serious healthcare-associated infections, particularly in hospitals. Frequently, K. pneumoniae isolates are MDR, and reduced envelope permeability is at least partly responsible.3–5 Overexpression of marA in K. pneumoniae MDR mutants has been seen in one study,6 but overexpression of the marA homologue, ramA, is far more common in clinical isolates and frequently correlates with overexpression of the acrAB efflux pump operon.7–11 Expression of ramA is repressed by RamR in K. pneumoniae in a manner analogous to the control of marA expression by MarR in E. coli.12,13 Multidrug resistance in K. pneumoniae has occasionally also been associated with overexpression of two other marA homologues: rarA, which correlates with overexpression of the oqxAB efflux pump operon,14–16 and soxS.9
The primary aim of the work presented below was to determine the relative effects that RamA, RarA, SoxS and MarA have on antimicrobial drug susceptibility, envelope permeability and efflux pump/porin production when overproduced in K. pneumoniae clinical isolates.
Materials and methods
Bacterial strains and antibiotic susceptibility testing
E. coli TOP10 (Invitrogen, Leek, The Netherlands) and the K. pneumoniae clinical isolates NCTC 5055 (a reference strain) and SM (a gift from Professor Alasdair MacGowan, Department of Microbiology, Southmead Hospital, Bristol, UK) and the otherwise isogenic pair ECL8 and ECL8ΔramR13 were used throughout. Disc susceptibility testing was performed according to CLSI methodology.17
Cloning K. pneumoniae regulator and porin genes and transformation of K. pneumoniae
K. pneumoniae NCTC 5055 genes were amplified by PCR using the primers listed in Table S1 (available as Supplementary data at JAC Online) and the method described previously.18 The PCR amplicons were inserted into the pBAD-TOPO expression vector (Invitrogen) according to the manufacturer's instructions. A religated pBAD-TOPO derivative was used as a plasmid-only no-expression control. The pBAD vector adds a small N-terminal leader peptide to the expressed protein; this inhibited outer membrane targeting of porins (data not shown), so the DNA encoding this leader sequence was entirely deleted using NcoI digestion and re-ligation according to the manufacturer's instructions. Plasmids were used to transform K. pneumoniae NCTC 5055 or SM to ampicillin resistance (50 mg/L) using electroporation as standard for laboratory-strain E. coli.
Fluorescent Hoechst (H) 33342 dye accumulation assay
Each K. pneumoniae pBAD transformant was inoculated into a separate batch of 50 mL of CAMHB (Sigma) containing 50 mg/L ampicillin and arabinose [0.2% (w/v)] in a 250 mL foam-stoppered flask to an initial OD600 of ≈0.05. Cultures were incubated with shaking (160 rpm) until the OD600 had reached 0.5–0.7. Envelope permeability was estimated with an established fluorescent dye accumulation assay19 with black flat-bottomed 96-well plates (Greiner Bio-one, Stonehouse, UK) and a Fluostar Optima (Aylesbury, UK) plate reader. H33342 (Sigma) was used at a final concentration of 2.5 μM (for strain SM) or 25 μM (for strain NCTC 5055).
Measurements of gene expression
Each pBAD transformant was inoculated into a separate batch of 50 mL of CAMHB (Sigma) [for outer membrane protein (OMP) analysis] or Nutrient Broth (Oxoid) (for total envelope proteomics or real-time quantitative RT–PCR) in a 250 mL foam-stoppered flask to an initial OD600 of ≈0.05. Cultures were incubated with shaking (160 rpm) for 3 h, when the OD600 had reached 0.5–0.7. Detailed methods for protein and RNA extraction and analysis are available as Supplementary data at JAC Online.
Results and discussion
RamA, and to a lesser extent RarA or SoxS, overproduction in K. pneumoniae reduces envelope permeability and antibiotic susceptibility, but MarA overproduction does not
In both test K. pneumoniae strains, inhibition zone diameter decreases were seen for 15/20 antimicrobials following RamA overproduction, resulting in intermediate resistance (according to CLSI criteria20) to cefuroxime and cefoxitin in the NCTC 5055 strain and full minocycline resistance in the SM strain. RamA overproduction from pBAD accurately reflected the phenotypic effect of ‘natural’ RamA overproduction from the chromosome because zone diameter changes in the ramR deletion strain ECL8ΔramR compared with its ramR+ parent, ECL8, were similar to those seen when overproducing RamA from pBAD using 0.2% (w/v) arabinose (Table 1 and Table S2).
Table 1.
Disc susceptibility assay for K. pneumoniae NCTC 5055 transformants expressing regulator genes
| Antibiotic (μg in disc) | pBAD (control) zone (mm) | pBAD(marA) zone (difference from control) (mm) | pBAD(soxS) zone (difference from control) (mm) | pBAD(rarA) zone (difference from control) (mm) | pBAD(ramA) zone (difference from control) (mm) | ECL8ΔramR zone (difference from ECL8) (mm) |
|---|---|---|---|---|---|---|
| Amikacin (30) | 24 (S) | NC | NC | NC | NC | NC (S) |
| Gentamicin (10) | 23 (S) | NC | NC | NC | NC | −2 (S) |
| Tobramycin (10) | 22 (S) | −2 (S) | NC | −2 (S) | NC | NC (S) |
| Cefoxitin (30) | 23 (S) | NC | −2 (S) | −2 (S) | −6 (I) | −4 (S) |
| Cefuroxime (30) | 22 (S) | NC | NC | −2 (S) | −5 (I) | −9 (I) |
| Ceftriaxone (30) | 32 (S) | NC | NC | −2 (S) | −3 (S) | −7 (S) |
| Cefotaxime (30) | 35 (S) | NC | −2 (S) | NC | −6 (S) | −4 (S) |
| Ceftazidime (30) | 29 (S) | NC | −2 (S) | −2 (S) | −4 (S) | −5 (S) |
| Cefepime (30) | 30 (S) | NC | NC | NC | −4 (S) | −3 (S) |
| Aztreonam (30) | 34 (S) | −3 (S) | −2 (S) | −4 (S) | −6 (S) | −2 (S) |
| Imipenem (10) | 29 (S) | NC | NC | NC | −2 (S) | NC (S) |
| Meropenem (10) | 31 (S) | NC | NC | NC | NC | NC (S) |
| Doripenem (10) | 30 (S) | NC | NC | NC | NC | NC (S) |
| Ciprofloxacin (5) | 30 (S) | NC | NC | NC | −8 (S) | −3 (S) |
| Norfloxacin (10) | 30 (S) | NC | −2 (S) | −3 (S) | −11 (S) | −9 (S) |
| Ofloxacin (5) | 28 (S) | NC | NC | −3 (S) | −5 (S) | −9 (S) |
| Tigecycline (15) | 21 | NC | NC | −2 | −4 | −5 |
| Minocycline (30) | 21 (S) | NC | NC | NC | −4 (S) | −6 (S) |
| Chloramphenicol (30) | 25 (S) | NC | NC | −3 (S) | −6 (S) | −9 (S) |
| Trimethoprim/sulfamethoxazole (1.25/23.75) | 27 (S) | NC | NC | NC | −4 (S) | NC (S) |
S, susceptible; I, intermediate resistant; NC, no change in zone diameter versus control.
Assays were performed using Mueller–Hinton agar with 0.2% (w/v) arabinose to stimulate expression of the cloned genes and 50 mg/L ampicillin to select for the pBAD plasmids. Otherwise, the assay was performed according to the CLSI protocol.17
Values reported are the means of at least three repetitions rounded to the nearest integer.
Mean changes <2 mm are reported as NC.
Susceptibility breakpoints are as set by the CLSI.20
RarA overproduction caused a smaller number and magnitude of zone diameter decreases than RamA overproduction in both strains. The effect of SoxS overproduction was similar to that of RarA in strain SM, though it had very little effect in strain NCTC 5055. Overproduction of MarA in both strains had almost no effect on inhibition zone diameters for the 20 test antimicrobial discs (Table 1 and Table S2).
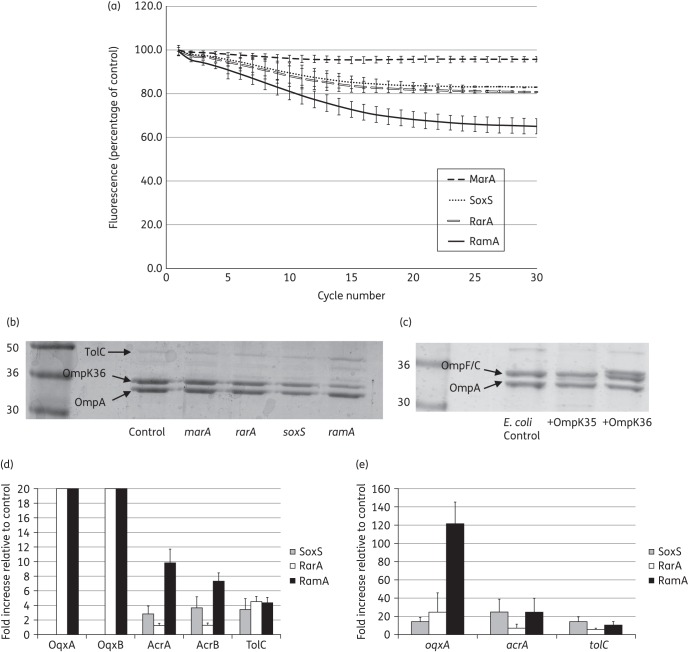
An ∼40% reduction in the accumulation of H33342 (a measure of envelope permeability) by the RamA-overproducing transformant was observed relative to the plasmid-only control (set to 100%) in K. pneumoniae NCTC 5055 (Figure 1a). The reduction was even greater in the SM strain following overproduction of RamA (Figure S1A). In contrast, MarA overproduction had very little effect on envelope permeability and overproduction of RarA or SoxS had an intermediate effect in either strain (Figure 1a and Figure S1A). Thus, the relative effects of the four AraC-type regulators on envelope permeability and antibiotic susceptibility correlate.
Figure 1.
Effect of AraC-type regulator overproduction in K. pneumoniae NCTC 5055 on envelope permeability and gene expression. (a) The accumulation of H33342 dye by K. pneumoniae NCTC 5055 transformants overexpressing ramA, rarA, soxS or marA is represented as a percentage relative to accumulation in the plasmid-only control transformant (set to 100%) over a 30 cycle (45 min) incubation period. Each graph shows the mean data for four biological replicates with eight technical replicates in each, and error bars define the SEM. (b) OMPs were purified from K. pneumoniae NCTC 5055 transformants carrying the control plasmid or those overexpressing ramA, rarA, soxS or marA. The OmpK35 and OmpK36 porins were identified by reference to panel (c), which shows OMPs from E. coli TOP10 and transformants overexpressing ompK35 or ompK36 cloned from K. pneumoniae NCTC 5055. OmpA and TolC are marked by reference to their predicted molecular weight and relatively high abundance. All OMPs were separated using SDS–PAGE and stained as set out in the Materials and methods section. These data are representative of three biological replicates. (d) Fold change in the abundance of selected efflux pump proteins and (e) fold change in the efflux pump gene mRNA levels in K. pneumoniae NCTC 5055 transformants carrying the control plasmid (set to 1) or those overexpressing ramA, rarA or soxS growing in nutrient broth. Data are mean ± SEM; three biological replicates, with four technical replicates for the quantitative RT–PCR.
Real-time quantitative RT–PCR confirmed similar levels of overexpression for each AraC-type regulator gene upon arabinose induction in liquid culture (Figure S2A). Overproduction of one AraC-type regulator did not significantly alter the expression of any of the other regulator genes [P > 0.3 for a t-test comparing transcript levels (CT value) in the control versus overexpressing transformant]. This means that reciprocal effects on regulator gene expression (e.g. down-regulation of ramA expression from the chromosome when marA is overexpressed from pBAD) cannot explain why some regulators had a smaller phenotypic effect than others. A dose–response analysis of arabinose concentration confirmed that the observed differences between the effects of the regulators on antibiotic susceptibility are not due to differential protein production during the assay. Even when using 25 times more arabinose to induce the production of MarA, SoxS and RarA than to induce the production of RamA, this last regulator exerted by far the strongest effect on zone diameter. Furthermore, the effects of MarA, SoxS and RarA overproduction on zone diameter peaked at a lower arabinose concentration than the effect of RamA (Figure S2B).
Effect of RamA, RarA and SoxS overproduction on efflux pump and porin production
OMP profiles revealed that there is evidence for the overabundance of a protein of the expected molecular weight of the efflux pump OMP TolC only in RamA-overproducing NCTC 5055 (Figure 1b); for the SM strain, there is evidence for RamA, RarA and SoxS, but not MarA, inducing TolC overproduction (Figure S1B). Only two porin bands were observed in OMP extracts from K. pneumoniae NCTC 5055. By cloning and overproducing NCTC 5055 porin genes in E. coli, it was possible to confirm that OmpK35 (OmpF) is the non-detectable porin band in NCTC 5055 OMP preparations (Figure 1c). OmpK35 is visible in K. pneumoniae SM (though it is lower in abundance than OmpK36) and there is evidence for reduced production of OmpK35 in transformants overproducing RamA and RarA, but not MarA and SoxS (Figure S1B).
To give more quantitative data on efflux pump and porin protein production/gene expression following overproduction of the regulators, total envelope proteomics and quantitative RT–PCR were performed. Normalized relative protein production/gene expression data in NCTC 5055 transformants overproducing RamA, RarA and SoxS versus the pBAD (control) transformant, set to 1 in each case, are recorded in Figure 1(d and e). MarA was excluded here due to the lack of phenotypic effect seen in previous experiments. In some cases, proteins were found to be below the level of detection in control cells, but they were identifiable in regulator-overproducing cells. Because of this binary result, it is not possible to quantify the fold change in their production; however, this must be considerable and therefore a nominal fold change of 20 is assigned for these proteins (Figure 1d).
As expected given its particularly strong effect on antibiotic susceptibility and envelope permeability, RamA overproduction had the greatest effect on efflux pump protein production (Figure 1d) and this effect is clearly exerted at the level of transcription (Figure 1e). OqxAB, which is particularly associated with quinolone efflux in enteric bacteria21 and AcrAB, which is the most commonly identified efflux pump seen to be overproduced in MDR K. pneumoniae isolates,6–15 were both overproduced >4-fold relative to control cells (Figure 1d). In addition, TolC was also overproduced >4-fold. Even though AcrAB and OqxAB are overproduced in the presence of RamA, the pump primarily responsible for multidrug resistance is likely to be AcrAB. We say this because OqxAB was ∼100 times less abundant than AcrAB [0.7 ± 0.43% (mean ± 95% CI, n = 3)] in RamA-overproducing cells, according to the proteomic data. Proteomic analysis of Ecl8 versus Ecl8ΔramR confirmed that AcrA, AcrB and TolC are overproduced when RamA is overproduced from the chromosome [3.34 ± 0.83-fold, 4.25 ± 0.47-fold and 3.06 ± 0.33-fold, respectively (mean ± SEM, n = 3)].
Overproduction of RarA has previously been shown to control oqxAB and acrAB expression,22 though in strain NCTC 5055 we could only see significant up-regulation of OqxAB (Figure 1d and e). This was, however, sufficient to explain the reduced susceptibility primarily to fluoroquinolones and tigecycline seen in this RarA-overproducing NCTC 5055 recombinant (Table 1). The effect of SoxS overproduction in NCTC 5055 was, as expected from the antibiotic susceptibility data (Table 1), less strong than RamA and RarA (Figure 1d and e). There was evidence for an increase in AcrAB production, though this was about a third of the effect of RamA. We could not detect the low-abundance pump OqxAB in the SoxS-overproducing recombinant, but the quantitative RT–PCR data confirmed that oqxAB is positively controlled by this regulator (Figure 1e).
Porin proteins previously shown to be involved in antibiotic entry were not down-regulated upon overproduction of any of the AraC-type regulators in K. pneumoniae NCTC 5055. For OmpK36, fold changes (mean ± SEM, n = 3) were 1.86 ± 0.62 (SoxS), 0.78 ± 0.11 (RarA) and 1.19 ± 0.54 (RamA). For OmpK35, fold changes were 2.14 ± 0.81 (SoxS), 1.60 ± 0.25 (RarA) and 1.32 ± 0.22 (RamA). In contrast, in the Ecl8 strain, overproduction of RamA caused a repression of OmpK35 porin production (5.24 ± 1.34-fold down-regulated in Ecl8ΔramR versus Ecl8), but no change in OmpK36 production (1.15 ± 0.05-fold). The qualitative OMP analysis (Figure 1b) led us to suggest that that OmpK35 is already down-regulated in NCTC 5055, and the ratio of OmpK36 to OmpK35 was calculated from the raw proteomic abundance data as 6.04 ± 1.28 to 1 in NCTC 5055 and 0.93 ± 0.04 to 1 in Ecl8. Accordingly, the repressive effect of RamA on production of OmpK35, apparent in Ecl8ΔramR [and also in the SM strain (Figure S1B)] is masked in strain NCTC 5055 because OmpK35 production is already reduced by some other mechanism (Figure 1b). Importantly, however, the effect of RamA overproduction in Ecl8ΔramR on antibiotic susceptibility was not significantly more than in NCTC 5055 (Table 1), so this additional effect of reducing OmpK35 levels does not appear to be phenotypically important.
Conclusions
Overall, the strength by which the four AraC-type regulators have been shown here to affect antibiotic susceptibility (RamA > RarA > SoxS > MarA) correlates well with previously published incidences of AraC-type regulator overproduction in clinical K. pneumoniae isolates and laboratory-selected mutants.6–15 The strength of effect of these regulators appears to correlate with their ability to activate efflux pump gene expression; though the involvement of reducing OmpK35 production in some backgrounds cannot be excluded, it does not appear to be of major importance.
RamA overproduction in K. pneumoniae reduces susceptibility to a number of drug classes, including tetracyclines, quinolones and cephalosporins, but excluding aminoglycosides (Table 1 and Table S2). However, resistance based on clinical breakpoints20 was only reached for minocycline, and only in the SM strain. Intermediate resistance to cefuroxime and cefoxitin was also seen, but only in the NCTC 5055 strain; no resistance was seen in Ecl8ΔramR. Clearly, therefore, RamA is not a key antibiotic resistance determinant in K. pneumoniae on its own. It has recently been shown that the RamA regulon includes genes that contribute to other clinically relevant phenotypes, e.g. LPS biosynthesis.23 This, rather than its impact on antibiotic resistance, might therefore explain why RamA overproduction is seen in clinical isolates.6–13 However, it remains to be seen whether RamA-mediated efflux pump overproduction can enhance plasmid-mediated resistance or allow easier selection of MDR mutants with other additional regulatory or antimicrobial drug target site mutations. Indeed, the same must be said for the other regulators studied here and further work is necessary to investigate this possibility.
Funding
This work was supported in part by a grant to M. B. A. from the BSAC and University of Bristol internal funds. J.-C. J.-C. was funded by a postgraduate scholarship from CONACyT, Mexico. W. N. I. W. A. K. was funded by a postgraduate scholarship from the Malaysian Ministry of Education. M. S. T. was funded through the Wellcome Trust Clinical Veterinary Research Training Programme. J. B. was funded by a BSAC vacation scholarship.
Transparency declarations
None to declare.
Supplementary data
Acknowledgements
We are grateful to Lucy Pettman, who generated the rarA and soxS clones used in this study.
References
- 1.Alekshun MN, Levy SB. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol 1999; 7: 410–3. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 2000; 182: 3467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice LB. The clinical consequences of antimicrobial resistance. Curr Opin Microbiol 2009; 12: 476–81. [DOI] [PubMed] [Google Scholar]
- 4.Schultsz C, Geerlings S. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs 2012; 72: 1–16. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009; 9: 228–36. [DOI] [PubMed] [Google Scholar]
- 6.Bratu S, Landman D, George A et al. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother 2009; 64: 278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneiders T, Amyes SG, Levy SB. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother 2003; 47: 2831–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Källman O, Motakefi A, Wretlind B et al. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J Antimicrob Chemother 2008; 62: 986–90. [DOI] [PubMed] [Google Scholar]
- 9.Bialek-Davenet S, Marcon E, Leflon-Guibout V et al. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother 2011; 55: 2795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruzin A, Visalli MA, Keeney D et al. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother 2005; 49: 1017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentschke M, Wolters M, Sobottka I et al. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother 2010; 54: 2720–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenblum R, Khan E, Gonzalez G et al. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents 2011; 38: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Majumdar S, Yu J, Spencer J et al. Molecular basis of non-mutational derepression of ramA in Klebsiella pneumoniae. J Antimicrob Chemother 2014; 69: 2681–9. [DOI] [PubMed] [Google Scholar]
- 14.Veleba M, Higgins PG, Gonzalez G et al. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 2012; 56: 4450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veleba M, Schneiders T. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 2012; 56: 4466–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bialek-Davenet S, Lavigne JP, Guyot K et al. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 2015; 70: 81–8. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests—Ninth Edition: Approved Standard M2-A9. CLSI, Wayne, PA, USA, 2006. [Google Scholar]
- 18.Avison MB, von Heldreich CJ, Higgins CS et al. A TEM-2 β-lactamase encoded on an active Tn1-like transposon in the genome of a clinical isolate of Stenotrophomonas maltophilia. J Antimicrob Chemother 2000; 46: 879–84. [DOI] [PubMed] [Google Scholar]
- 19.Coldham NG, Webber M, Woodward MJ et al. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother 2010; 65: 1655–63. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. CLSI, Wayne, PA, USA, 2014. [Google Scholar]
- 21.Perez F, Rudin SD, Marshall SH et al. OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob Agents Chemother 2013; 57: 4602–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Majumdar S, Veleba M, Finn S et al. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob Agents Chemother 2013; 57: 1603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Majumdar S, Yu J, Fookes M et al. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 2015; 11: e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.