Abstract
Midbrain dopamine (mDA) neurons play critical roles in the regulation of voluntary movement and their dysfunction is associated with Parkinson’s disease. Pitx3 has been implicated in the proper development of mDA neurons in the substantia nigra pars compacta, which are selectively lost in Parkinson’s disease. However, the basic mechanisms underlying its role in mDA neuron development and/or survival are poorly understood. Toward this goal, we sought to identify downstream target genes of Pitx3 by comparing gene expression profiles in mDA neurons of wild-type and Pitx3-deficient aphakia mice. This global gene expression analysis revealed many potential target genes of Pitx3; in particular, the expression of vesicular monoamine transporter 2 and dopamine transporter, responsible for dopamine storage and reuptake, respectively, is greatly reduced in mDA neurons by Pitx3 ablation. In addition, gain-of-function analyses and chromatin immunoprecipitation strongly indicate that Pitx3 may directly activate transcription of vesicular monoamine transporter 2 and dopamine transporter genes, critically contributing to neurotransmission and/or survival of mDA neurons. As the two genes have been known to be regulated by Nurr1, another key dopaminergic transcription factor, we propose that Pitx3 and Nurr1 may coordinately regulate mDA specification and survival, at least in part, through a merging and overlapping downstream pathway.
Keywords: aphakia mice, dopamine neuron, Parkinson’s disease, Pitx3, substantia nigra pars compacta
Among potential transcription factors involved in development and physiological function of midbrain dopamine (mDA) neurons, Nurr1 and Pitx3 may play critical roles in the development of mDA neurons when DA neuronal precursors exit mitotic stage (Riddle and Pollock 2003; Smits and Smidt 2006). Studies with Nurr1 knockout mouse have shown that while it is not essential for the initial formation of DA neuronal precursors, Nurr1 is important for determining the DA neurotransmitter identity and neurotransmission, as well as for the survival and maintenance of DA neurons later on. This conclusion has been derived from, and further supported, by the identification of target genes of Nurr1. So far, the list of known Nurr1 downstream target genes includes tyrosine hydroxylase (TH) (Castillo et al. 1998; Saucedo-Cardenas et al. 1998; Sakurada et al. 1999; Iwawaki et al. 2000; Kim et al. 2003), aromatic amino acid decarboxylase (Hermanson et al. 2003), vesicular monoamine transporter 2 (VMAT2) (Hermanson et al. 2003; Smits et al. 2003), DA transporter (DAT) (Smits et al. 2003), c-Ret (Wallen et al. 2001), p57Kip2 (Joseph et al. 2003), neuropilin (Hermanson et al. 2006) brain-derived neurotrophic factor (Volpicelli et al. 2007) and vasoactive intestinal peptide (Luo et al. 2007).
Pitx3 is another crucial transcription factor involved in the early development of mDA neurons, especially the substantia nigra pars compacta (SNpc) DA neurons (Hwang et al. 2003; van den Munckhof et al. 2003; Nunes et al. 2003; Smidt et al. 2004). Notably, both Nurr1 and Pitx3 are expressed in mDA neurons throughout adulthood, suggesting the interesting possibility that Pitx3 and Nurr1 are important for the maintenance and normal physiology of mature mDA neurons. In line with this, several reports recently showed that mutations of these genes are closely associated with Parkinson’s disease (PD) (Le et al. 2003, 2008; Grimes et al. 2006; Fuchs et al. 2009; Bergman et al. 2008). Therefore, unveiling regulatory cascade of Pitx3 will help us to understand not only basic mechanisms of early development and physiology of mDA neurons but also pathophysiology of PD.
Despite the functional importance of Pitx3, molecular pathways controlled by Pitx3 are only partially understood (Jacobs et al. 2007; Peng et al. 2007). In this report, we attempted to identify downstream target genes of Pitx3 on a genomic scale by comparing gene expression profiles of laser-captured mDA neurons from E12.5 wild-type (wt) and aphakia (ak) mice. Intriguingly, among the list of mostly affected genes by Pitx3 ablation, we identified VMAT2 and DAT genes and found that their in vivo expression is significantly compromised during both early embryonic and adult stages of mDA neurons in ak mice. In addition, our gain-of-function studies using in vitro differentiation of Pitx3-over-expressing mouse embryonic stem cells (mESCs) and chromatin immunoprecipitation (ChIP) analysis further suggested that VMAT2 and DAT are direct target genes of Pitx3. Taken together, we propose that Pitx3, in concert with Nurr1, controls neurotransmission and homeostasis of DA neurons by regulating these two target genes.
Experimental procedures
Animal care
Pitx3-deficient ak mice were maintained as previously described (Hwang et al. 2005). Animal use was in accordance with Institutional Animal Care and Use Committee of McLean Hospital and followed National Institutes of Health guidelines.
In situ hybridization
For in situ hybridization, both wt and ak mouse embryos of the age of E12.5 and E14.5 were fixed in 4% p-formaldehyde/phosphate-buffered saline (PBS) overnight, cryoprotected in 30% sucrose/PBS at 4°C and then frozen in optimal cutting temperature (OCT) compound. Adult brains from 9-week-old mice were snap-frozen on dry ice without OCT. Serial coronal and sagital sections were cut at 14 μm on a cryostat and went through in situ hybridization process as described previously (Hwang et al. 2003). In this study, probes for in situ hybridization were labeled with digoxigenin (DIG) and visualized with an alkaline phosphatase-conjugated anti-DIG antibody using Nitro-Blue Tetrazolium Chloride (NBT)/5-Bromo-4-Chloro-3′-Indolyphosphate p-Toluidine Salt (BCIP) as a substrate. The DNA fragments used for the riboprobe generation were amplified by PCR using the following primer pairs: TH, forward 5′-GTATACGCCACGCTGAGGG-3′, reverse 5′-ATCCTGGACCCCCTCTAAGG-3′; mouse VMAT2, forward 5′-GCAACTTTTCTAGGGGTTTG-3′, reverse 5′-GTTCCAGAACATGAACTGG-3′; mouse DAT, forward 5′-GGCAGATCTTCCAGACACC-3′, reverse 5′-CAGAGAGGTGGAGCTCATC-3′.
Laser capture microdissection
Laser capture microdissection (LCM) of TH-positive neurons was performed using a PixCell II Laser-capture Microscope (Arcturus, Mountain View, CA, USA) and macro LCM caps (CapSure LCM Caps, Arcturus). Briefly, 10-μm cryosections of the brain were fixed in ice-cold acetone for 5 min, followed by air-drying. After rehydration in PBS for 5 s, the sections were stained for 5 min in PBS containing an anti-TH antibody (diluted 1 : 100; Pel-freez, Rogers, AR, USA) and then were rinsed in PBS for 5 s. Subsequently, the slides were stained with an Alexa Fluor 594 anti-rabbit IgG (1 : 100) (Molecular Probes, Eugene, OR, USA) for 5 min, rinsed with PBS for 5 s, dehydrated by sequential exposure to increasing concentrations of ethanol (30 s in 75%, 30 s in 95% and 1 min in 100%) and then treated with xylene for 2 min. Finally, the slides were air-dried for 5 min before LCM. For our experiments, about 1500 TH-positive neurons were isolated from the adult ventral tegmental area (VTA) and the ventral mesencephalon of E12.5 embryos.
RNA preparation and RT-PCR analyses
Total RNA from the laser-captured neurons was isolated using the PicoPure kit (Arcturus) according to the manufacturer’s protocol. Then we used the RiboAmp kit (Arcturus) to perform T7-based linear amplification. The total RNA from the ventral mesencephalic tissue of embryos was prepared using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) followed by treatment with DNase I (Qiagen) and then used for either DNA microarray or RT-PCR experiment. For RT-PCR analysis, 5 μg of RNA was transcribed into cDNA with the SuperScriptTH First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Samples were amplified in Continuous Fluorescence Detector (MJ RESEARCH, Waltham, MA, USA) using DNA Engine Opticon software under the following conditions: denaturing step at 94°C, 30 s; annealing step at 55°C, 30 s; extension step at 72°C, 30 s for 50 cycles and a final extension step at 72°C, 10 min. The cDNA was analyzed in a PCR assay using the following primers; En1, forward 5′-GTGAGTGTGAGTGTGTTTCCTTGTG-3′, reverse 5′-CTCGTAGTCTGTGGGGTTGTATTTC-3′; guanosine triphosphate cyclohydrolase (GTP) cyclohydrolase I (GTPCH), forward 5′-ATCCCTTCTGAACGACCCTG-3′, reverse 5′-AATTCCCCAAATGTGCTCGG-3′; DAT, forward 5′-CATTGCCACATCCTCCATGG-3′, reverse 5′-TAGGCCAGTTTCTCTCGGAA-3′; VMAT2, forward 5′-GTCCACCTGCTAAGGAAGAA-3′, reverse 5′-CAGGAGACACATGTACACAG-3′; TH, forward 5′-TCCAACCTTTCCTGGCCCAG-3′, reverse 5′-GCATGAAGGGCAGGAGGAAT-3′; glyceraldehyde-3-phosphate dehydrogenase, forward 5′-AGGGCATCTTGGGCTACACTG-3′, reverse 5′-TGGGTGGTCCAGGGTTTCTTAC-3′; cRet, forward 5′-TCCCAGAGTGAGTTACGAGACCTG-3′, reverse 5′-GACAGCCCAAAGTCGGAAAT-3′.
DNA microarray analyses
Two micrograms of each linearly amplified aRNA sample derived from LCM-captured TH-positive neurons was subjected to DNA microarray analyses using the Affymetrix GeneChip Mouse Expression Set 430 2.0 (Affymetrix, Santa Clara, CA, USA). These DNA microarray analyses were performed three times per sample (n = 3) in Transcription Analysis Laboratory at California Institute of Technology.
Cultivation and dopaminergic differentiation of mouse embryonic stem cells
The mESC line D3 was obtained from ATCC (Rockland, MD, USA) and cultured on gelatin-coated dishes in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 1× non-essential amino acids (Invitogen), 2 mM glutamine (Invitrogen), 0.001% b-mercaptoethanol (Invitrogen), 10% donor horse serum (Sigma, St Louis, MO) and 2000 U/mL human recombinant leukemia inhibitory factor (R&D, Minneapolis, MN, USA). Dopaminergic differentiation of mESCs was carried out using the 5-stage protocol described previously (Lee et al. 2000).
Retroviral vector, viral production and titration
The Pitx3-expressing recombinant retroviral vector used in this study was generated by inserting Pitx3 cDNA into the pMS retroviral plasmid. As a control, enhanced green fluorescence protein (EGFP)-expressing retroviral vector was also constructed by inserting EGFP into the same retroviral plasmid. To generate retroviral particles, the recombinant retroviral plasmids were transfected into retroviral packaging cells, 293GPG cells, using Lipofectamine 2000 (Invitrogen). Medium containing secreted retroviruses was collected everyday from day 2 to 5, followed by concentration of viral particles by ultracentrifugation (SW28, 50,000 × g, 90 min). Retroviral titration was carried by Retro-X qRT-PCR Titration Kit (Clontech, Mountain View, CA, USA) following manufacturer’s instructions.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay was performed according to manufacturer’s protocol (Upstate, Lake Placid, NY, USA). Briefly, 1 × 106 cells of neural precursor cells derived from mESCs were plated in 60 mm plates and cultured for 2 days and then transduced by hemagglutinin (HA)-tagged Pitx3 in retroviral vector. Cells were cultured 2 days more, differentiated into DA neurons for 7 or 14 days, cross-linked with 1% formaldehyde for 10 min and then harvested in the presence of protease inhibitor (EDTA-free complete; Roche). These cells were then lysed and sonicated to generate 200–500 bp DNA fragments. One-tenth of the lysates was used for input control. The remaining lysates were divided by half and treated with 1 μg of polyclonal anti-HA antibody (Upstate) or normal rabbit IgG as a negative control overnight at 4°C. After treating Salmon sperm DNA/Protein A agarose slurry to immunoprecipitated complexes, the precipitates were extensively washed and then eluted in elution buffer (1% sodium dodecyl sulfate, 0.1 M NaHCO3). The cross-linked protein–DNA complexes were reversed by NaCl treatment. The DNA was recovered by phenol extraction and resuspended in 50 μL of H2O. PCR was performed to detect specifically bound DNA using MasterAmp 2× PCR Premix IN buffer (Epicentre, Madison, WI, USA) using 1 μL of the resuspended sample as a template at 94°C 30 s, 55°C 30 s, 72°C 30 s for 30 cycles with primer sets in 25 μL reaction volume. Primers for VMAT2 are 5′-CATCACGCAGACTTGAAAGAC-3′ and 5′-CGCCTCGCCTTGCTTATCC-3′. Primers for DAT are 5′-GTTAACATTTACCCAACTGG-3′ and 5′-GGACCTCTAAATCAACATG-3′.
Results
Identification of potential downstream target genes of Pitx3 on a genomic scale
In an effort to unveil potential role of Pitx3 in the development and function of mDA neurons, we attempted to identify its downstream target genes on a genomic scale. Toward this goal, we compared gene expression profile of mDA neurons between ak (Pitx3-deficient) and wt (Pitx3-present) mice. As the outcome could be easily biased by the secondary effects resulting from the absence or death of a specific population of mDA neurons, we performed the laser capture experiments at the development stage soon after Pitx3 started to be expressed (E11.5) and before no obvious differences in the mDA neuronal pattern were detected between ak and wt mice. In regards to this issue, no apparent differences in the number and distribution of mDA neurons have been detected at E12.5 between wt and ak mice (Fig. S1), which is in consistent with the previous report from other group (16). On the contrary, a clear defect became evident in SNpc DA neurons from E14.5 (Fig. S2).
About 1500 TH-positive neurons were captured throughout the whole ventral midbrain of both wt and ak E12.5 embryos by LCM. Total RNAs were isolated, linearly amplified and subjected to DNA microarray analyses using the Affymetrix GeneChip Mouse Expression Set 430 2.0 which includes 45 000 probe sets to analyze the expression level of over 39 000 transcripts and variants from well characterized mouse genes. Our microarray analysis revealed that 243 independent genes were down-regulated twofold or more in ak mDA neurons compared with wt mice (Table S1). This gene list included many genes that are known to regulate cell cycle/growth, cell migration and transcriptional regulation. Thus, it is possible that Pitx3 may regulate neurogenesis (DA neuron/precursor generation), neuronal migration and other important functions of DA neurons. Interestingly, among highly affected genes (Fig. 1a, left), included were the DAT and the VMAT2 genes which are primarily responsible for DA reuptake into the nerve terminals and DA storage into synaptic vesicles, respectively. When RT-PCR analyses were performed using total RNAs isolated from independently laser captured tissues, we confirmed that mRNA expression levels of DAT, VMAT2, as well as five randomly chosen additional genes were dramatically reduced by the absence of Pitx3 (Fig. 1a, right).
Fig. 1.
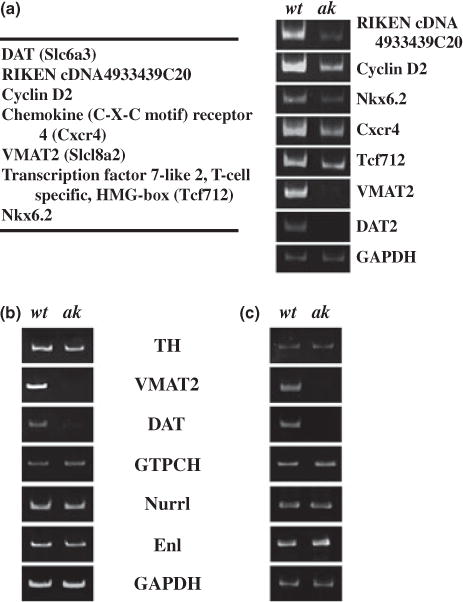
Examination of the expression change of dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2) and several other genes by the absence of Pitx3. (a, left) Among genes significantly down-regulated in laser capture microdissection-captured midbrain dopamine (mDA) neurons in aphakia (ak) mice as examined by DNA microarray experiment, five genes were randomly chosen in addition to DAT and VMAT2 for RT-PCR analysis. (a, right) RT-PCR analysis confirms that all these seven genes are affected in mDA neurons of ak mice compared with those of wild-type (wt) mice and thus validates our microarray experiment. In particular, expression of VMAT2 and DAT was most strikingly affected in ak mice. (b) The level of several mDA neuronal markers was examined in total RNAs from laser-captured mDA neurons of wt and ak embryos at E12.5. (c) The level of several mDA neuronal markers was examined in total RNAs from the isolated ventral midbrain tissue of wt and ak embryos at E12.5.
As both DAT and VMAT genes are known to play critical roles in DA neuron physiology, function and related behavioral effects (Fon et al. 1997; Rocha et al. 1998; Jones et al. 1999), we decided to further investigate the regulation of DAT and VMAT2 gene expression in the absence of Pitx3. First, we performed additional RT-PCR analysis for several mDA neuronal genes using total RNAs independently isolated from captured mDA neurons of E12.5 embryos. This analysis confirmed that the levels of VMAT2 and DAT mRNAs were dramatically reduced in mDA neurons of ak embryos compared with those of wt embryos (Fig. 1b). On the contrary, there was no significant difference in the level of TH, GTPCH and Engrailed 1 (En1) mRNAs between wt and ak embryos at E12.5 (Fig. 1b). Thus, the down-regulation of VMAT2 and DAT gene expressions in the absence of Pitx3 is a specific phenomenon. To address the possibility that these gene expression changes are biased during the LCM procedure, we next dissected the whole ventral midbrain area from wt and ak E12.5 embryos and prepared total RNAs from these tissues. Quantitative RT-PCR experiments on these RNAs resulted in a very similar result to that of laser captured tissues, confirming that VMAT2 and DAT genes are down-regulated in the absence of Pitx3 (Fig. 1c).
VMAT2 and DAT gene expression, but not TH gene expression, is greatly diminished in mDA neurons of ak mice during early embryonic development
To address whether VMAT2 and DAT gene expression is affected in vivo at the cellular level, in situ hybridization experiment was performed. The level of VMAT2 mRNA was mostly diminished in the ventral mDA neurons of ak mice at E12.5, compared with that of the age-matched wt mice (Fig. 2a). Interestingly, absence of Pitx3 in ak mice did not affect VMAT2 expression in serotonergic neurons of the dorsal raphe nuclei (Fig. 2a). This observation is congruent with the fact that Pitx3 is not expressed in the raphe nuclei. Thus, our results strongly suggest that the mechanism(s) responsible for VMAT2 gene expression is different between mDA and serotonergic neurons and that its expression in mDA neurons is specifically affected by Pitx3. In contrast, there was no noticeable difference in TH gene expression in the ventral midbrain area between wt and ak E12.5 embryos (Fig. 2a, panels iii and vi; also see Fig. S1). We found the same pattern of differential VMAT2 gene expression in E14.5 embryos of ak mice; its expression was largely reduced in ventral mDA neurons, but not in the serotonergic neurons (Fig. 2b).
Fig. 2.
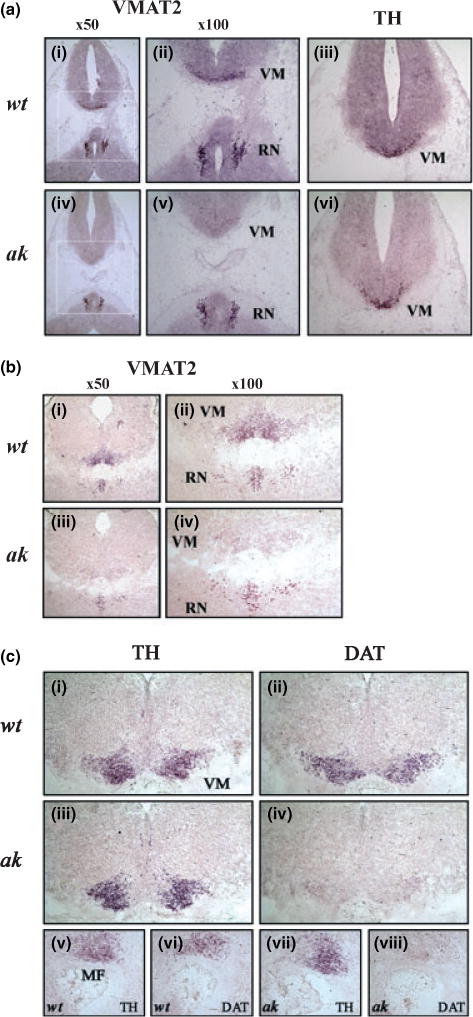
Expression of vesicular monoamine transporter 2 (VMAT2) (a, b) and dopamine transporter (DAT) (c) mRNAs in E12.5 (a) and E14.5 (b, c) embryos of wild-type (wt) and aphakia (ak) mice was examined by in situ hybridization. (a) Coronal sections of wt E12.5 (i–iii) and ak E12.5 (iv–vi) embryos were hybridized with antisense riboprobes for VMAT2 (i, ii, iv, v) and tyrosine hydroxylase (TH) (iii, vi). Panels ii and v are higher magnification of boxed areas in panels i and iv, respectively. (b) Coronal sections of wt E14.5 (i, ii) and ak E14.5 (iii, iv) embryos were hybridized with antisense riboprobes for VMAT2. Panels ii and v are higher magnification of panels i and iii, respectively. (c) Expressions of TH (i, iii, v, vii) and DAT (ii, iv, vi, viii) genes in the ventral midbrain of wt (i, ii, v, vi) and ak (iii, iv, vii, viii) mice at E14.5 were examined by in situ hybridization. Panels i–iv are coronal sections and panels v–viii are sagital sections. VM, ventral midbrain; MF, mesencephalic flexure; RN, raphe nuclei.
Dopamine transporter is considered to be a late marker of developing mDA neurons (Perrone-Capano et al. 1994). Although DAT mRNA was detected at E12.5 by RT-PCR (Fig. 1), it was not readily detected by the conventional in situ hybridization method (data not shown). As such, we used E14.5 embryos to examine the expression level of DAT gene by in situ hybridization. The mRNA levels of DAT as examined by both coronal and sagittal sections were largely diminished in mDA neurons (Fig. 2c). However, it is noteworthy that some noticeable level of DAT mRNA was still remaining in the ventral midbrain of ak mice (Fig. 2c, d and h), suggesting that other factor(s) might be also involved in the basal expression of DAT gene. In summary, our in vivo gene expression analysis demonstrates that VMAT2 and DAT gene expression is impaired during early development of mDA neurons of ak embryos.
Down-regulation of VMAT2 and DAT gene expression is sustained in mDA neurons of adult ak mice
We next sought to determine whether expression of VMAT2 and DAT genes is later restored or continues to be down-regulated in mDA neurons of adult ak mice. Toward this end, we performed in situ hybridization of brain sections of 9-week-old mice. At this stage, we could observe comparable signals for TH mRNA in both wt and ak mice (Fig. 3a). Given that DA neurons in the SNpc (A9 neurons) are largely diminished in ak mice, these TH positive neurons are most likely VTA DA neurons (A10 neurons). Interestingly, similar to embryonic stages, VMAT2 and DAT gene expressions were greatly reduced in ak mice as compared with age-matched wt mice (Fig. 3a). On the other hand, the level of VMAT2 mRNA was not affected by Pitx3 ablation in noradrenergic neurons of the locus coeruleus (Fig. 3b-ii and iv). This in situ hybridization result was further corroborated by RT-PCR experiments using total RNAs isolated from laser captured adult (9-week-old) VTA neurons. As shown in Fig. 3c, VMAT2 and DAT mRNA levels were greatly diminished in the adult ak VTA neurons, compared with their wt counterpart. In contrast, the expression levels of TH, GTPCH, En1 and c-ret genes were shown to be comparable in the ak VTA neurons to those of wt mice, demonstrating that VMAT2 and DAT gene expression is specifically reduced in the adult VTA neurons of ak mouse. Taken together, our results suggest that the effects of Pitx3 on the expression of VMAT2 and DAT genes are not transient, but are still evident in 9-week-old adult mice.
Fig. 3.
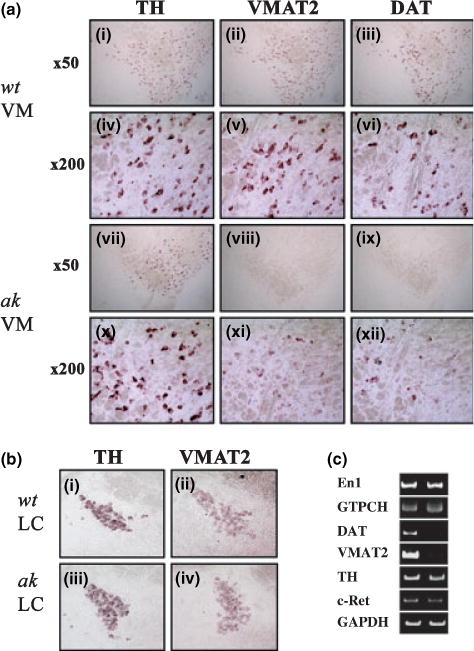
Expression of vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) genes is diminished in midbrain dopamine neurons of adult aphakia (ak) mice. (a) Expression of TH (i, iv, vii, x), VMAT2 (ii, v, viii, ix) and DAT (iii, vi, ix, xii) genes was analyzed in the ventral midbrain of adult wild-type (wt) (i–vi) and ak (vii–xii) mice. Panels iv, v, vi, x, xi and xii are higher magnifications of panels i, ii, iii, vii, viii and ix, respectively. (b) The level of TH (i, iii) and AT2 (ii, iv) mRNAs in the locus coeruleus of adult mice were examined by in situ hybridization. (c) RT-PCR analyses of several midbrain dopamine neuronal markers in the total RNAs isolated from laser captured adult VTA neurons. VM, ventral midbrain; LC, locus coeruleus.
Forced expression of Pitx3 up-regulates VMAT2 and DAT gene transcription in mouse embryonic stem cells
To further substantiate the idea that Pitx3 regulates VMAT2 and DAT gene expression, we next attempted a gain-of-function approach. Toward this goal, we speculated that mESCs are a useful tool because of their pluripotent differentiation capability. DA neuronal derivation in vitro from the mESCs were performed using the well-established 5-stage differentiation procedure (Lee et al. 2000; Chung et al. 2005) (Fig. 4a and b). To deliver an exogenous gene into mESC-derived neural precursors, we used retroviral vectors which normally transduce the majority of the cells (> 90%; Fig. 4c). Using the retroviral system, we delivered Pitx3 gene into cells of neural precursor stage which corresponds to the 16th day of the whole differentiation procedure (Fig. 4a). Forty-eight hours post-transduction, the cells continued to be differentiated for seven more days and were examined for the change of VMAT2 and DAT mRNA levels by both quantitative (Fig. 4d) and real time PCR analyses (Fig. 4e and f). This experiment showed that Pitx3 overexpression robustly increased mRNA levels of both VMAT2 and DAT genes, compared with GFP-retroviral transduction.
Fig. 4.
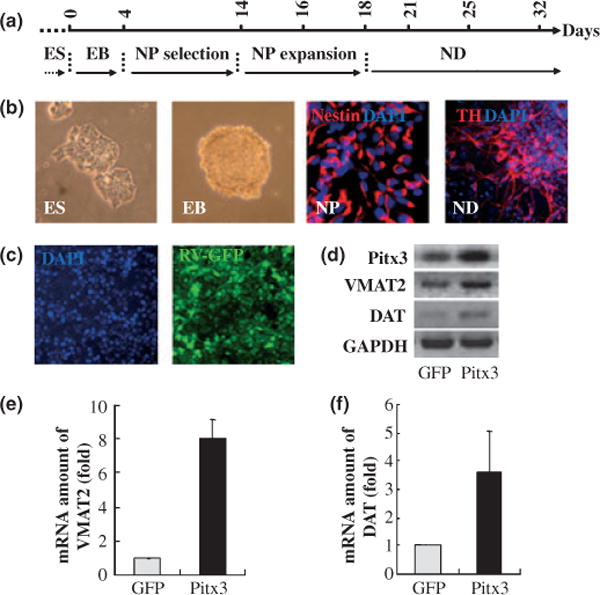
Over-expression of Pitx3 induces transcription of endogenous vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) genes during in vitro differentiation of mouse embryonic stem cells (mESCs). Total RNAs were isolated from neuronal cells after in vitro differentiation of the Pitx3-over-expressing mES cells and then RT-PCR was performed. (a) Diagram of in vitro differentiation of mESCs using the 5-stage method. ES, embryonic stem cells; EB, embryoid bodies; NP, neural precursors; ND, neuronal differentiation. (b) Representative images of each stage during mESC differentiation. (c) mESC-derived NPs are efficiently transduced with GFP-expressing retroviruses (about 90%). (e, f) VMAT2 and DAT mRNA levels are analyzed by semi- and real-time PCR analyses. Pitx3 over-expression leads to a robust increase of VMAT2 and DAT transcript levels, while the level of GAPDH transcript was not affected. Quantification based on real-time RT-PCR shows the fold change of endogenous transcripts between cells transfected with GFP control (white bars, control) and Pitx3 transfected cells (black bars, Pitx3). Relative expression of each mRNA was normalized according to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA.
Pitx3 directly binds to putative binding motifs in the 5′ upstream regions of the VMAT2 and DAT gene promoters
Our gain-of-function results prompted us to hypothesize that Pitx3 directly activates transcription of VMAT2 and DAT genes. To determine if Pitx3 directly binds to the promoter region(s) of these target genes and activate their transcription, we first determined the consensus Pitx3 binding motif by the random oligonucleotide selection procedure in vitro (Blackwell et al. 1990). Although the Pitx3-related Drosophila bicoid protein is known to bind to the ‘5-TAATCC-3’ motif (Hanes et al. 1994), the consensus binding site for Pitx3 has yet to be determined. For the random oligonucleotide selection procedure, we performed gel mobility shift experiments using a mixture of in vitro translated Pitx3 protein and the 63bp-long oligonucleotides with 14 degenerative nucleotides inside (5′-GACTACGTCGACAGCGAATTCAGA(N)14 CTCGGGATCCATGCTCAGTAGACAG-3′). The Pitx3-oligonucleotide complexes on the gel were isolated and oligonucleotides within the complexes were amplified by PCR with primers flanking the internal degenerative nucleotide sequences. The PCR-amplified oligonucleotides obtained were used for another round of the selection procedure and this procedure was repeated four times. The sequencing analyses of the oligonucleotides obtained after five rounds of the selection procedure showed that Pitx3 recognizes 5′-(C/G)NTAATCC(A/C)-3′ as the consensus binding motif (Fig. S3). Using this sequence motif, we next performed in silico analyses to identify potential Pitx3 binding motifs in the VMAT2 gene regulatory regions using the available genomic data base ECR Browser and rVista (http://www.dcode.org). Analysis of the 26 kb upstream promoter region as well as the first exon and intron areas of the VMAT2 gene, revealed seven potential binding sites with less than one nucleotide deviation from the consensus motif (Fig. 5a and b). Among these sites, we analyzed three potential sites (sites 2, 3 and 5) by ChIP assay. We designed oligonucleotide primers to amplify each putative Pitx3 binding site. mESCs were transduced with retroviral vectors expressing HA-tagged Pitx3, subsequently differentiated for 1 week, and Chip assay was carried out with anti-HA antibody or rabbit IgG (control). This ChIP analysis revealed that site 2 is bound in vivo by Pitx3 (Fig. 5c). Notably, this site 2 is highly conserved among different species, in particular between mouse and rat (Fig. 5d). Similarly, our in silico analysis identified five potential Pitx3-binding sites in the 5′ upstream sequence of DAT gene (Fig. 5e and f), although these sequences did not showed significant sequence homologies among species. When we tested the two most proximal sequences (sites 1 and 2) using the ChIP analysis, site 2 was found to interact with Pitx3 in vivo (Fig. 5g).
Fig. 5.
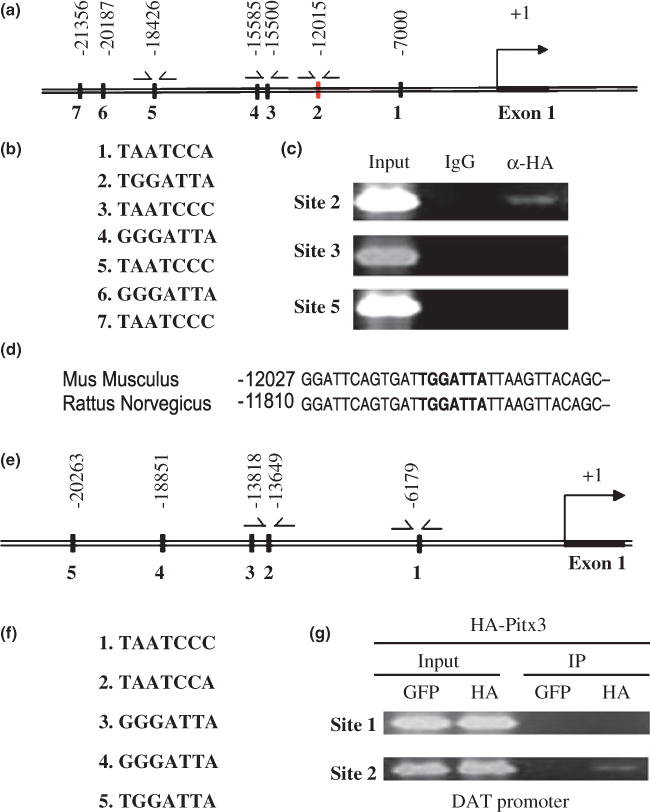
Pitx3 specifically binds to putative binding motifs in the 5′ upstream promoter of the vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) gene. (a, e) Seven and five putative Pitx3-binding sites are identified in the 5′ promoter region of the VMAT2 and DAT genes, respectively. (b, f) Their nucleotide sequences are shown in 5′ to 3′ directions. (c, g) Chromatin immunoprecipitation analysis indicates that Pitx3 interacts with both VMAT2 and DAT gene promoters in vivo. The protein–DNA complexes were immunoprecipitated using antibodies against anti-HA. As a negative control, rabbit IgG was used. (d) Comparison of nucleotide sequences flanking the site 2 of the VMAT2 gene shows that they are highly conserved between mouse and rat.
Discussion
Since its discovery in 1997 (Semina et al. 1997; Smidt et al. 1997), Pitx3 has been suspected as a transcription factor implicated in the development and/or survival of mDA neurons. It was not until 2003 that the clear involvement of Pitx3 in mDA neuronal development has been demonstrated in vivo by several groups including ours (Hwang et al. 2003; van den Munckhof et al. 2003; Nunes et al. 2003; Smidt et al. 2004). Our previous report has also shown that heterologous expresion of Pitx3 in mESCs increases A9-like DA neuronal population (Chung et al. 2005), further supporting its role in early development of mDA neurons. In addition to its role during early development, Pitx3 may also control important characteristics of mature mDA neurons as is supported by its continued expression throughout adulthood. However, at this moment, we do not know much about the functional role of Pitx3 and its underlying molecular mechanisms. This poor understanding is, at least in part, because of the limited number of known downstream target genes of Pitx3.
In this study, we sought to identify genes whose expression is down-regulated by Pitx3 ablation by comparing gene expression profile of mDA neurons from ak mice to that from wt mice. We report the identification of more than 240 genes that are down-regulated at least twofold in the mDA neurons of ak mice, many of which are implicated in diverse functions such as neurogenesis, transcriptional regulation and neuronal migration. In particular, we found that the VMAT2 and DAT genes, which critically regulate DA neurotransmission, are in the list of highly regulated genes. Furthermore, we present in vivo evidence that the expression levels of both VMAT2 and DAT genes are also diminished not only in developing but also in mature (adult) mDA neurons when Pitx3 is deficient. This result clearly showed that VMAT2 and DAT genes are the downstream genes of Pitx3 in vivo.
The reduction of VMAT2 and DAT gene expression in Pitx3-deficient ak mice is evident from E12.5 and lasts at least 9 weeks after birth. This reduction is a specific and regulated process, but not because of non-specific down-regulation of genes occurring in degenerating cells. Our conclusion is supported by the following observations: (i) several mDA neuronal markers tested are not affected by Pitx3 ablation (Figs 1 and 2); (ii) although only a portion of mDA neurons are degenerating during the early development of ak mice, the reduction of VMAT2 and DAT gene expression occurs all the mDA neurons in the ventral midbrain area; (iii) this reduction is also detected in the surviving mDA neurons in adult ak mice (Fig. 3).
In line with the involvement of Pitx3 in the regulation of VMAT2 gene expression, VMAT2 gene expression is detectable from E12 (Schutz et al. 1998) in the rat brain, the time point when Pitx3 is also expressed. At this time point, Nurr1, another factor implicated in the regulation of VMAT2 gene expression, is also present in the mDA neurons. Nurr1 was shown to be absolutely required for the induction of both VMAT2 and DAT genes, because Nurr1−/−) mice show no expression of the two genes (Hermanson et al. 2003; Smits et al. 2003). Our data suggest that Nurr1 by itself is not sufficient for the full expression of both VMAT2 and DAT genes in the ventral midbrain, because ak mice display significant reduction of VMAT2 and DAT gene expression even in the presence of normal levels of Nurr1. This result indicates that robust expression of VMAT2 and DAT genes can be detected when both Nurr1 and Pitx3 coexist and lack of either component diminishes the induction of VMAT2 and DAT gene expression. This observation can be explained by the synergistic interaction between Nurr1 and Pitx3, as suggested in a recent study of over-expression of Pitx3 and Nurr1 in embryonic stem cells (Martinat et al. 2006). In contrast to Nurr1, a recognizable minimal expression can be detected in the absence of Pitx3 (Figs 1–3), suggesting that, Nurr1 and/or other factor(s) could be responsible for this residual expression.
It is noted that DAT and VMAT2 genes are expressed in other brain regions beyond mDA neurons where Pitx3 is not normally expressed. For example, DA neurons in hypothalamus and frontal cortex that normally do not express Pitx3 exhibit prominent DAT gene expression (Shimada et al. 1992; Roth and Elsworth 1995). This suggests that different activating mechanisms of the DAT gene exist in these neurons. We also detected robust expression of VMAT2 gene in both serotonergic (Fig. 2) and noradrenergic (Fig. 3) neurons and, as expected, the expression levels in these neuronal types were not affected by Pitx3 ablation, further suggesting that independent mechanisms of gene activation exist for the VMAT2 and DAT genes in different subtypes of neurons.
In addition to the critical role in DA neurotransmission, VMAT2 and DAT also play an important role in the cell survival and protection of DA neurons against neurotoxins. In line with this notion, it has been suggested that the expression level of DAT and VMAT2 genes may determine the susceptibility of individual DA neurons to a variety of toxic substances (Miller et al. 1999). Furthermore, expression levels of both VMAT2 and DAT genes are significantly reduced in post-mortem SNpc of PD patients compared with controls (Uhl et al. 1994; Harrington et al. 1996). These observations also corroborate well with recent findings that mutations or altered expression of Nurr1 or Pitx3 are associated with PD (Le et al. 2003, 2008; Grimes et al. 2006; Fuchs et al. 2009).
It is highly probable that both Nurr1 and Pitx3 are critically involved in the development, physiological function and/or survival of mDA neurons (Fig. 6). Recently, many downstream target genes of Nurr1 have been identified and shown to be involved in many important aspects of DA neuronal development and function, such as determination of neurotransmitter identity, DA neurotransmission, cell survival and maintenance, as well as DA neuronal differentiation (Saucedo-Cardenas et al. 1998; Sakurada et al. 1999; Iwawaki et al. 2000; Wallen et al. 2001; Hermanson et al. 2003, 2006; Joseph et al. 2003; Kim et al. 2003; Smits et al. 2003; Luo et al. 2007; Volpicelli et al. 2007). In contrast, only a few downstream targets of Pitx3 have been reported yet. TH has been suggested as a gene regulated by Pitx3, but further clarification is needed because of inconsistent reports among various research groups (Cazorla et al. 2000; Smidt et al. 2000; Maxwell et al. 2005; Martinat et al. 2006; Messmer et al. 2007). Recently, Smidt and his colleagues elegantly showed that aldehyde dehydrogenase 2 (Ahd2 or Aldh1a1) involved in the synthesis of retinoic acid is directly regulated by Pitx3 (Jacobs et al. 2007). The present study will further contribute to our understanding about the potential role of Pitx3 in the regulatory cascade of mDA neuronal function, physiology and perhaps survival/maintenance. Notably, our results indicate that Pitx3 and Nurr1 may affect DA neuronal function and survival through the common downstream pathway by merging with the same target genes, including VMAT2 and DAT (Fig. 6). On the other hand, we could not detect any significant difference in the expression of TH and c-Ret genes, the genes known to be regulated by Nurr1, by the ablation of Pitx3 (Figs 1 and 3). Taken together, it appears that Pitx3 and Nurr1 play their own roles in the normal function, physiology and, maintenance of mDA neurons through common as well as distinct target genes. In support of our conclusions, a recent report demonstrated that both Pitx3 and Nurr1 recognize the same areas of many promoters in the genome (Jacobs et al. 2009). Further unveiling of the downstream target genes of Pitx3 and Nurr1 will provide us with a more comprehensive and detailed mechanistic understanding about the regulatory cascade of these critical transcription factors and their cross-talks for their control of the development and physiological function of mDA neurons and may lead to novel therapeutic approaches of associated brain disorders.
Fig. 6.
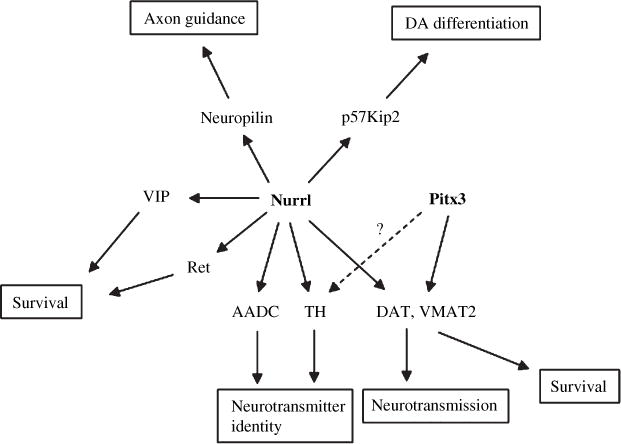
Pitx3, in addition to Nurr1, is regulating the expression of vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) genes critically involved in DA neurotransmission of midbrain dopamine neurons. DAT and VMAT2 are also implicated in the survival/vulnerability of midbrain dopamine neurons against neurotoxins.
Supplementary Material
Figure S1. Distribution of TH-positive neurons in E12.5 (a–c: rostral to caudal parts) embryos of wt and ak mice.
Figure S2. A series of coronal sections of both wt E14.5 and ak E14.5 embryos was compared for the distribution of TH-positive neurons.
Figure S3. The consensus motif for Pitx3 binding was determined by oligonucleotide selection method.
Table S1. The list of genes that are expressed more two-fold in wt mice than in Pitx3-deficient ak mice at E12.5 stage.
Acknowledgments
This work was supported by NIH grants MH48866, DC006501, Michael J Fox Foundation for Parkinson’s Research, and International Grants from Brain and Stem Cell Research Centers (SC5130 & SC5170) funded by the Ministry of Education, Science and Technology, the Republic of Korea.
Abbreviations used
- ak
aphakia
- ChIP
chromatin immunoprecipitation
- DA
dopamine
- DAT
dopamine transporter
- GTPCH
GTP cyclohydrolase I
- LCM
laser capture microdissection
- mDA
midbrain DA
- mESCs
mouse embryonic stem cells
- PBS
phosphate-buffered saline
- PD
Parkinson’s disease
- SNpc
substantia nigra pars compacta
- TH
tyrosine hydroxylase
- VMAT2
vesicular monoamine transporter 2
- VTA
ventral tegmental area
- wt
wild-type
Footnotes
The authors declare no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article:
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Castillo SO, Baffi JS, Palkovits M, Goldstein DS, Kopin IJ, Witta J, Magnuson MA, Nikodem VM. Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol Cell Neurosci. 1998;11:36–46. doi: 10.1006/mcne.1998.0673. [DOI] [PubMed] [Google Scholar]
- Cazorla P, Smidt MP, O’Malley KL, Burbach JP. A response element for the homeodomain transcription factor Ptx3 in the tyrosine hydroxylase gene promoter. J Neurochem. 2000;74:1829–1837. doi: 10.1046/j.1471-4159.2000.0741829.x. [DOI] [PubMed] [Google Scholar]
- Chung S, Hedlund E, Hwang M, Kim DW, Shin BS, Hwang DY, Jung Kang U, Isacson O, Kim KS. The homeodomain transcription factor Pitx3 facilitates differentiation of mouse embryonic stem cells into AHD2-expressing dopaminergic neurons. Mol Cell Neurosci. 2005;28:241–252. doi: 10.1016/j.mcn.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Fon EA, Pothos EN, Sun BC, Killeen N, Sulzer D, Edwards RH. Vesicular transport regulates monoamine storage and release but is not essential for amphetamine action. Neuron. 1997;19:1271–1283. doi: 10.1016/s0896-6273(00)80418-3. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Mueller JC, Lichtner P, et al. The transcription factor PITX3 is associated with sporadic Parkinson’s disease. Neurobiol Aging. 2009;30:731–738. doi: 10.1016/j.neurobiolaging.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Grimes DA, Han F, Panisset M, Racacho L, Xiao F, Zou R, Westaff K, Bulman DE. Translated mutation in the Nurr1 gene as a cause for Parkinson’s disease. Mov Disord. 2006;21:906–909. doi: 10.1002/mds.20820. [DOI] [PubMed] [Google Scholar]
- Hanes SD, Riddihough G, Ish-Horowicz D, Brent R. Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol. 1994;14:3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington KA, Augood SJ, Kingsbury AE, Foster OJ, Emson PC. Dopamine transporter (Dat) and synaptic vesicle amine transporter (VMAT2) gene expression in the substantia nigra of control and Parkinson’s disease. Brain Res Mol Brain Res. 1996;36:157–162. doi: 10.1016/0169-328x(95)00278-z. [DOI] [PubMed] [Google Scholar]
- Hermanson E, Joseph B, Castro D, et al. Nurr1 regulates dopamine synthesis and storage in MN9D dopamine cells. Exp Cell Res. 2003;288:324–334. doi: 10.1016/s0014-4827(03)00216-7. [DOI] [PubMed] [Google Scholar]
- Hermanson E, Borgius L, Bergsland M, Joodmardi E, Perlmann T. Neuropilin1 is a direct downstream target of Nurr1 in the developing brain stem. J Neurochem. 2006;97:1403–1411. doi: 10.1111/j.1471-4159.2006.03829.x. [DOI] [PubMed] [Google Scholar]
- Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain Res Mol Brain Res. 2003;114:123–131. doi: 10.1016/s0169-328x(03)00162-1. [DOI] [PubMed] [Google Scholar]
- Hwang DY, Fleming SM, Ardayfio P, Moran-Gates T, Kim H, Tarazi FI, Chesselet MF, Kim KS. 3,4-dihydroxyphenylalanine reverses the motor deficits in Pitx3-deficient aphakia mice: behavioral characterization of a novel genetic model of Parkinson’s disease. J Neurosci. 2005;25:2132–2137. doi: 10.1523/JNEUROSCI.3718-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Kohno K, Kobayashi K. Identification of a potential nurr1 response element that activates the tyrosine hydroxylase gene promoter in cultured cells. Biochem Biophys Res Commun. 2000;274:590–595. doi: 10.1006/bbrc.2000.3204. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Smits SM, Noorlander CW, von Oerthel L, van der Linden AJ, Burbach JP, Smidt MP. Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development. 2007;134:2673–2684. doi: 10.1242/dev.02865. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136:531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- Jones SR, Gainetdinov RR, Hu XT, Cooper DC, Wightman RM, White FJ, Caron MG. Loss of autoreceptor functions in mice lacking the dopamine transporter. Nat Neurosci. 1999;2:649–655. doi: 10.1038/10204. [DOI] [PubMed] [Google Scholar]
- Joseph B, Wallen-Mackenzie A, Benoit G, Murata T, Joodmardi E, Okret S, Perlmann T. p57(Kip2) cooperates with Nurr1 in developing dopamine cells. Proc Natl Acad Sci USA. 2003;100:15619–15624. doi: 10.1073/pnas.2635658100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Kim CH, Hwang DY, Seo H, Chung S, Hong SJ, Lim JK, Anderson T, Isacson O. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem. 2003;85:622–634. doi: 10.1046/j.1471-4159.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Le WD, Xu P, Jankovic J, Jiang H, Appel SH, Smith RG, Vassilatis DK. Mutations in NR4A2 associated with familial Parkinson disease. Nat Genet. 2003;33:85–89. doi: 10.1038/ng1066. [DOI] [PubMed] [Google Scholar]
- Le W, Pan T, Huang M, Xu P, Xie W, Zhu W, Zhang X, Deng H, Jankovic J. Decreased NURR1 gene expression in patients with Parkinson’s disease. J Neurol Sci. 2008;273:29–33. doi: 10.1016/j.jns.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Luo Y, Henricksen LA, Giuliano RE, Prifti L, Callahan LM, Federoff HJ. VIP is a transcriptional target of Nurr1 in dopaminergic cells. Exp Neurol. 2007;203:221–232. doi: 10.1016/j.expneurol.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Martinat C, Bacci JJ, Leete T, et al. Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proc Natl Acad Sci USA. 2006;103:2874–2879. doi: 10.1073/pnas.0511153103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell SL, Ho HY, Kuehner E, Zhao S, Li M. Pitx3 regulates tyrosine hydroxylase expression in the substantia nigra and identifies a subgroup of mesencephalic dopaminergic progenitor neurons during mouse development. Dev Biol. 2005;282:467–479. doi: 10.1016/j.ydbio.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Messmer K, Remington MP, Skidmore F, Fishman PS. Induction of tyrosine hydroxylase expression by the transcription factor Pitx3. Int J Dev Neurosci. 2007;25:29–37. doi: 10.1016/j.ijdevneu.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Miller GW, Gainetdinov RR, Levey AI, Caron MG. Dopamine transporters and neuronal injury. Trends Pharmacol Sci. 1999;20:424–429. doi: 10.1016/s0165-6147(99)01379-6. [DOI] [PubMed] [Google Scholar]
- van den Munckhof P, Luk KC, Ste-Marie L, Montgomery J, Blanchet PJ, Sadikot AF, Drouin J. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130:2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci USA. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Fan S, Li X, Fan X, Ming M, Sun Z, Le W. Overexpression of pitx3 upregulates expression of BDNF and GDNF in SH-SY5Y cells and primary ventral mesencephalic cultures. FEBS Lett. 2007;581:1357–1361. doi: 10.1016/j.febslet.2007.02.054. [DOI] [PubMed] [Google Scholar]
- Perrone-Capano C, Tino A, di Porzio U. Target cells modulate dopamine transporter gene expression during brain development. Neuroreport. 1994;5:1145–1148. doi: 10.1097/00001756-199405000-00031. [DOI] [PubMed] [Google Scholar]
- Riddle R, Pollock JD. Making connections: the development of mesencephalic dopaminergic neurons. Brain Res Dev Brain Res. 2003;147:3–21. doi: 10.1016/j.devbrainres.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Rocha BA, Fumagalli F, Gainetdinov RR, Jones SR, Ator R, Giros B, Miller GW, Caron MG. Cocaine self-administration in dopamine-transporter knockout mice. Nat Neurosci. 1998;1:132–137. doi: 10.1038/381. [DOI] [PubMed] [Google Scholar]
- Roth RH, Elsworth JD. Biochemical pharmacology of midbrain dopamine neurons. In: Bloom FE, Kupfer DJ, editors. Psychopharmacology: The Fourth Generation of Progress. Raven; New York: 1995. pp. 227–243. [Google Scholar]
- Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development. 1999;126:4017–4026. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz B, Schafer MK, Eiden LE, Weihe E. Vesicular amine transporter expression and isoform selection in developing brain, peripheral nervous system and gut. Brain Res Dev Brain Res. 1998;106:181–204. doi: 10.1016/s0165-3806(97)00196-x. [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6:2109–2116. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- Shimada S, Kitayama S, Walther D, Uhl G. Dopamine transporter mRNA: dense expression in ventral midbrain neurons. Brain Res Mol Brain Res. 1992;13:359–362. doi: 10.1016/0169-328x(92)90220-6. [DOI] [PubMed] [Google Scholar]
- Smidt MP, van Schaick HS, Lanctot C, Tremblay JJ, Cox JJ, van der Kleij AA, Wolterink G, Drouin J, Burbach JP. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci USA. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Smits SM, Bouwmeester H, Hamers FP, van der Linden AJ, Hellemons AJ, Graw J, Burbach JP. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- Smits SM, Smidt MP. The role of Pitx3 in survival of midbrain dopaminergic neurons. J Neural Transm. 2006;70:57–60. doi: 10.1007/978-3-211-45295-0_10. [DOI] [PubMed] [Google Scholar]
- Smits SM, Ponnio T, Conneely OM, Burbach JP, Smidt MP. Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur J Neurosci. 2003;18:1731–1738. doi: 10.1046/j.1460-9568.2003.02885.x. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Walther D, Mash D, Faucheux B, Javoy-Agid F. Dopamine transporter messenger RNA in Parkinson’s disease and control substantia nigra neurons. Ann Neurol. 1994;35:494–498. doi: 10.1002/ana.410350421. [DOI] [PubMed] [Google Scholar]
- Volpicelli F, Caiazzo M, Greco D, Consales C, Leone L, Perrone-Capano C, D’Amato LC, Porzio U. Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J Neurochem. 2007;102:441–453. doi: 10.1111/j.1471-4159.2007.04494.x. [DOI] [PubMed] [Google Scholar]
- Wallen AA, Castro DS, Zetterstrom RH, Karlen M, Olson L, Ericson J, Perlmann T. Orphan nuclear receptor Nurr1 is essential for Ret expression in midbrain dopamine neurons and in the brain stem. Mol Cell Neurosci. 2001;18:649–663. doi: 10.1006/mcne.2001.1057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Distribution of TH-positive neurons in E12.5 (a–c: rostral to caudal parts) embryos of wt and ak mice.
Figure S2. A series of coronal sections of both wt E14.5 and ak E14.5 embryos was compared for the distribution of TH-positive neurons.
Figure S3. The consensus motif for Pitx3 binding was determined by oligonucleotide selection method.
Table S1. The list of genes that are expressed more two-fold in wt mice than in Pitx3-deficient ak mice at E12.5 stage.


