Abstract
The live-attenuated yellow fever 17D virus is one of the most outstanding human vaccines ever developed. It induces efficacious immune responses at a low production cost with a well-established manufacture process. These advantages make the YF17D virus attractive as a vector for the development of new vaccines. At the beginning of vector development studies, YF17D was genetically manipulated to express other flavivirus prM and E proteins, components of the viral envelope. While these 17D recombinants are based on the substitution of equivalent YF17D genes, other antigens from unrelated pathogens have also been successfully expressed and delivered by recombinant YF17D viruses employing alternative strategies for genetic manipulation of the YF17D genome. Herein, we discuss these strategies in terms of possibilities of single epitope or larger sequence expression and the main properties of these replication-competent viral platforms.
Keywords: Yellow Fever 17D virus-vaccines-immune responses-expression vector-insertion sites-live recombinant
The Virus and the Vaccine
The yellow fever virus is the prototype of the Flavivirus genus within the family Flaviviridae, which includes about 70 viruses. The flaviviruses are small (40–60 nm), spherical, enveloped, with a single-stranded RNA genome. The majority of these viruses is transmitted by arthropodes, as mosquitoes or ticks, and is therefore called arboviruses (“arthropod-borne viruses”) causing important diseases in man and animals.
In 1927, the yellow fever virus was isolated in the Rhesus monkey (Macaca mulatta), through the inoculation of blood from an African patient named Asibi.1 When subcutaneously inoculated in monkeys, it results in death in 95% of the animals after 4 to 7 d with high viremia rates in the blood of these animals. The serial passage of the Asibi YF virus strain in different types of cultivated tissues led to the identification of the 17D strain at passage 176, to 17DD at passage 195 and to 17D-204 at passage 204. The 17DD strain was cultivated afterwards until passage 243 and underwent 43 extra passages in chicken embryos (vaccine at passage 287). The 17D-204 strain, in turn, gave rise to the different seed viruses used in several countries. The 17D-204 and 17DD viruses are the 2 substrains of the 17D virus nowadays adopted to produce the world’s YF vaccine. These 2 strains have accumulated genotypic and phenotypic differences due to the independent serial passages.2,3 However, both are equally immunogenic and safe for man.4
Immunological Aspects of Vaccination with the Yellow Fever 17D Virus
The attenuated Yellow Fever 17D (YF17D) vaccine has been widely adopted for the immunization of man for over 70 y.5 A single primary dose of YF17D not only can promote effective and safe seroconversion in more than 90% of the vaccinees but can also provide immunity for over 40 y.6,7 Although YF17D represents an enormous success in terms of human protection against yellow fever, the cellular and molecular mechanisms by which it can activate a polyvalent and long-lived immune response still remains to be fully elucidated.
General immune response to YF17D is mainly characterized by the stimulation of a broad range of innate immune responses, which will ultimately result in the production of high titers of neutralizing antibodies and robust YF-specific human effector T cells and memory CD8+ T cell responses.8 In addition, YF17D vaccination induces lymphocytes that produce a balanced mix of both TH1 and TH2 cytokines.9,10
Dendritic cells (DCs) have been shown to play a key role in the initiation of an innate immune response to YF17D.10,11 The YF17D virus infects and activates multiple subsets of DCs, via Toll-like receptors (TLRs), including TLRs 7, 8, and 9, to elicit proinflammatory cytokines interleukin (IL)-12p40, IL-6, and interferon (IFN)-α.10 Additionally, myeloid DCs also express the cytosolic nucleic acid-sensing receptors retinoic acid-inducible gene (RIG)-I-like (RIG-I) and melanoma differentiation associated gene-5 (MDA5) which will activate signal transduction pathways that trigger the production of type I IFNs.12 Moreover, the mammalian target of the rapamycin (mTOR) signaling pathway was also implicated in regulating TLR7/9-dependent IFN-α/β production by plasmacytoid DCs.13
The role of other cell types, like Natural Killer (NK) cells and monocytes, in the YF17D innate immune response has been suggested by systems biology studies.14 It was demonstrated that a subset of genes involved in NK cells and monocytes are upregulated in the peripheral blood of vaccinated individuals. These cells might play a role in virus clearance. The activation of innate immunity cell types, such as NK and gamma-delta T cells, results in the early production of gamma interferon in man, monkeys, and mice which may be of importance to the overall adaptive immune response.15-17
The activation and maturation of different subsets of DCs is likely to contribute to the marked adaptive immune response after vaccination with the YF17D virus.18 Analyzing microarrays on PBMCs of vaccinated humans, the most frequently activated gene was EIF2AK4 (GCN2) which is known to be related to the formation of stress granules.19,20 Besides, upregulation of TNFRSF17, a key gene whose activation is predictive for the magnitude of neutralizing antibody response, has also been identified.
T CD8+ lymphocytes are important components for the control of viral infections. The YF17D-specific effector CD8+ T cell response is broad and complex. In 1998, Reinhardt and colleagues21 first demonstrated that YF17D-vaccinated patients presented high levels of T CD8+ lymphocytes in the peripheral blood. In the following years, CD8+ T cell-specific epitopes were identified in mice, rhesus monkeys, and man, implicating the E and NS3 proteins as the most important targets for cytotoxic response to the YF17D virus.22-24
After human immunization with YF17D, it was also possible to detect effector CD8+ T cells in peripheral blood, as demonstrated by the upregulation of CD38, HLA-DR, and Ki67, as well as by the downregulation of Bcl-2 in these cells.8 The presence of effector CD8+ cells dramatically increases, reaching a peak at day 15-post vaccination. After this period of time, the gradual differentiation of these effector cells into memory cells starts. These observations were further confirmed in a complete study of activation and differentiation kinetics of various cell types of both the innate and adaptive immune systems implicated in the immune response to YF17D25. A more detailed phenotypic analysis provided further evidence for YF17D-driven activation and differentiation of CD8+ T cells, demonstrating the functional profile alteration of these lymphocytes in composition as these cell types mature from effector to memory. During the course of this transition, this cell type becomes less polyfunctional, correlating with the profile of a memory cell type.26 The same work showed that CD4+ and FoxP3+ regulatory T cell (Treg) compartments all respond to the YF17D virus, displaying a kinetic profile of activation which precedes the CD8+ T cell response. This observation suggests that the fast CD4+ T cell response might be related to the pronounced ability of DCs to present the YF17D virus antigens. The presence of CD4+ T cells secreting Th1 type cytokines has been related to higher titers of neutralizing antibodies.18 It is possible that the efficient CD4+ T cell response is crucial for the development of a high-quality humoral response to the vaccine. Concordantly, the generation of neutralizing antibodies is a hallmark of protective antiviral immunity against the YF17D virus.27,28 Neutralizing antibodies directed against structural protein E is believed to be the most critical in terms of protection.29,30 These neutralizing antibodies may persist for 40 y post vaccination, which exhibits the ability of the virus to elicit an efficient immune response that will result in a long-lived humoral response.
Development of the Live-Attenuated YF17D Virus as an Expression Vector
Live-attenuated viral vaccines, like yellow fever, polio, measles, mumps, and rubella are very efficient immunogens as they promote durable immunity in man. In fact, they are very appropriate for a large-scale production having a well-established production methodology, quality control procedures, and low cost. Thus, adoption as a replication competent vector to express heterologous antigens has been attempted with the purpose of developing new recombinant human vaccines. Since live-attenuated vaccine viruses are proliferation-competent vectors, it is expected that immunization results in viral replication in the host, spreading the viral antigen mass throughout many cells and tissues usually leading to a long-lasting and efficient protection based on a polyvalent immunity.8,31
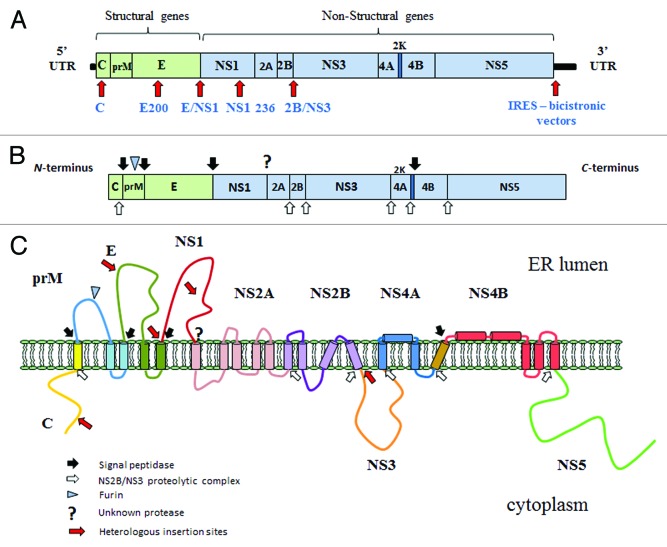
The efficacious immune responses spurred the interest in the manipulation of the YF17D virus as a vector for the development of new human vaccines. Translation of the positive-stranded viral genomic RNA in infected cells results in the synthesis of a polyprotein precursor of 3411 amino acids, which when proteolytically processed, generates 10 viral proteins (Fig. 1) from the amino- to carboxi-terminus of the precursosr polyprotein: C; prM/M; E; NS1; NS2A; NS2B; NS3; NS4A; NS4B and NS5. The first 3 are viral structural proteins that form the virus particle together with the RNA molecule, denominated capsid (C, 12–14kDa), membrane (M of 8 kDa, its precursor prM of 18–22 kDa), and envelope (E, 52–54 kDa). These 3 genes are encoded in the first quarter of the genome. The remainder of the genome encodes the non-structural proteins (NS), numbered from 1 to 5 (NS1 to NS5) in accordance with the order of synthesis. This knowledge formed the basis for the development of an infectious cDNA clone from which the YF17D virus is regenerated through in vitro transcription and RNA transfection technologies.32
Figure 1. Insertion of heterologous sequences into the YF 17D genome. (A) YF genome is composed of a single positive single-stranded RNA containing 10 862 nucleotides. The major portion of the genome consists of an open reading frame flanked by a 5` and 3` untranslated region. The heterologous sequence insertion sites that have been described so far for recovering recombinant YF virus are indicated by a red arrow; (B) The translation of this large ORF produces a precursor polyprotein of 3411 amino acid residues, which are mainly co- and post-translationally processed at specific sites by the viral NS2B-NS3 proteolytic complex (white arrows) and signal peptidase (black arrows) producing the virion (structural proteins) and replicase elements (non-structural proteins). (C) Topological arrangement of YF proteins and the regions in which were possible to express heterologous sequences, indicated by red arrows. On the cytoplasmatic side, the viral proteins are processed by the viral NS2B/NS3 proteolytic complex (white arrows) whereas on the ER side by signal peptidases, furin, and an unknown protease that cleaves the carboxi-terminus of NS1.
Since then, various recombinant live attenuated YF viruses have been developed by different approaches. Indeed, there are various possibilities to express heterologous antigens by the YF17D vaccine virus. First, the use of YF as an expression platform consists of the creation of chimeric flaviviruses in which the genes coding for prM/M and E proteins of the attenuated Yellow Fever 17D virus are substituted by the corresponding genes of the Japanese Encephalitis (JE), West Nile (WN), and dengue (DEN) viruses. This approach was feasible, in part, due to the similarity of the flavivirus genes and also because these proteins are functionally essential for virus proliferation. The second approach is the insertion of short sequences coding for antigenic epitopes into the YF17D genome having been employed into different intragenic as well as intergenic sites (Fig. 1). The YF17D vector system was also developed for the expression of larger gene fragments (Fig. 1). In this regard, 3 main alternatives of expression of larger polypeptides were tentatively established: (1) the insertion of heterologous genes into the E/NS1 intergenic region, (2) insertion into the C gene, and (3) the creation of bicistronic viral RNA molecules.
Replacement of YF17D Structural Genes with those of other Flaviviruses
Based on the original description of chimeric dengue viruses by Bray and Lai,33 new YF17D recombinant viruses have been constructed in which the prM-E genes of YF were substituted for the equivalent genes of other flaviviruses including Japanese encephalitis,34 West Nile,35 and the 4 dengue serotypes.36 Although the prM-E genes may vary among diverse flaviviruses by over 40% of the encoded amino acids, chimeric 17D viruses are still viable. The gene fusion is made at the signal peptidase cleavage site between C and prM and the signal peptidase site at the E and NS1 junction.37 Despite the high-fidelity of YF17D virus RNA polymerase during viral RNA replication, the introduction of mutations can be detected.38 Mutations noted at the structural protein level would be related to adaptation of the flavivirus envelope and prM-M protein to the YF17D capsid protein toward virus assembly. In addition, mutations observed elsewhere in the genome could be due to adaptation to growth in cell culture. This underlying genetic variation may be the basis for the observed variability of these chimeric YF17D viruses with regard to phenotypic properties such as growth rates in animals and cultured cells, attenuation to experimental animals (mice and monkeys), and immunogenicity.39
For more than 10 years, YF17D has been developed as a viral vector to express other flavivirus proteins.39 These recombinant live attenuated chimeric viruses have been tested in man to different extents, and due to the inexistence of any alternative, particular attention has been devoted to the immunogenicity and safety of the YF17D-DV tetravalent vaccine.40-46 The live attenuated chimeric 17D-DEN tetravalent vaccine candidate from Sanofi-Pasteur constitutes the most tested dengue vaccine possibility. The chimeric viruses proved to be genetically and phenotypically stable,47 less virulent when compared with the yellow fever vaccine 17D and unable to infect mosquitoes orally.39 It has displayed adequate safety and immunogenicity in clinical studies in flavivirus naive and immune subjects. Early in 2011, the vaccine was administered to over 6000 children and adults from endemic and non-endemic areas with no questions regarding its safety.41,42,46
A phase IIb clinical trial in 2669 Thai schoolchildren with the tetravalent vaccine showed poor efficacy (30.3%) of protection against febrile DEN infections.45 While vaccination reduced incidence of DEN1, DEN3, and DEN4 by 90%, the protection against DEN2 was low (9.2%), downgrading overall protection. There is a need for monitoring longer-term duration of immunity and protection. Data from an efficacy (phase III) trial of the YF17D-DEN-based tetravalent candidate vaccine should serve for the utility of such technological platform.
In this regard, it is noteworthy that YF17D-JE chimeric virus containing the prM-E genes of the SA14-14-2 virus strain has been developed. This strain of JE virus is used in some Asian countries as live attenuated vaccine and the use of its structural genes in the construction of the YF17D recombinant virus was expected to yield a bona-fide vaccine candidate. Indeed, clinical trials have shown that YF17D-JE is safe and immunogenic with a single dose providing long-lasting immunity to wild-type strains of JE virus.48-51 This recombinant technology-based vaccine was licensed for human use in Australia (Imojev—Sanofi-Pasteur). Accordingly, YF17D-WN chimeric viruses have also undergone clinical tests. Studies on human subjects based on age and dose demonstrated that the vaccine was well tolerated and immunogenic in younger adults (18–40 y) as well as patients 50 y of age or older.52 This YF17D-WN chimeric vaccine denominated ChimeriVax-WN02 displayed a highly attenuated phenotype associated with the presence of 3 mutations into the E protein of WN strain NY99 causing a reduction of neurovirulence. However, the equine WNV vaccine based on the ChimeriVax technology, expressing a wild type WN, was licensed by the USDA in 2006 and recalled in 2010 due to acute side effects including fatality among vaccinated horses.53
Insertion of Foreign Epitope Coding Sequences into the YF Genome
The first approach describing the expression of short epitopes by a replication-competent YF17D virus was conceived by McAllister et al. (2000)54 (Table 1). In this study, a short ovalbumin immunodominant cytotoxic T cell epitope (SIINFEKL) was expressed at the junction of the NS2B and NS3 genes.54 The rational of this methodology was based on endowing both sides of the heterologous sequence with the viral protease recognition motif. Consequently, the insertion was cleaved off the polyprotein due to the duplication of the viral protease NS2B/3 cleavage site. The expression of small epitopes by this methodology seems to not considerably interfere with the maturation of viral proteins, since the viral proliferation properties of this kind of recombinant viruses are similar to the vaccine counterpart.54-57 However, the foreign sequence cloning into the NS2A-NS2B, NS3-NS4A, and NS4A-NS4B junctions did not allow the recovery of a viable virus. Interestingly, the same YF cloning strategy was employed in a subsequent work to express a Plasmodium yoelli CD8+ T cell epitope.57 Both of these studies demonstrated that YF17D recombinant viruses do protect vaccinated mice against lethal challenge with malignant melanoma cells or the malaria parasite. However, in recent studies for evaluating the capacity of distinct recombinant YF17D viral vectors to induce a protective immune response against the Trypanosoma cruzi protozoan parasite, the immunodominant CD8+ T cell epitope TEWETGQI of the ASP-2 (Amastigote Surface Protein -2) cloned and expressed between NS2B and NS3 led only to marginal protection after the T.cruzi challenge.56,58
Table 1. Expression of small heterologous sequences by the YF viral platform.
| Insertion: viral genomic region | Heterologous sequence (origin) | Regimen of immunization, if any | References |
|---|---|---|---|
| NS2B/NS3 | SIINFEKL (chicken ovalbumin) | 2 doses (mice) | MacAllister et al., 2000.54 |
| SYVPSAEQIL (CSP of Plasmodium yoelii) | 1 dose or with boost with irradiaded sporozoite (mice) | Tao et al., 2005.57 | |
| TEWETGQI (ASP-2 of Trypanosoma cruzi) | 2 doses (mice) | Nogueira et al., 2011.58 | |
| GILGFVFTL (influenza matrix protein) | Human Dendritic cells | Barba-Spaeth et al., 2005.55 | |
| E protein at fg loop | TEWETGQI(ASP-2 of Trypanosoma cruzi) | 2 doses (mice) | Nogueira et al., 2011.58 |
| DYENDIEKKI, (NANP)3, and SYVPSAEQI (CS proteins from P. falciparum and P.yoelii) | 2 doses (mouse and rhesus monkeys) | Bonaldo et al., 200562 and 2006.61 | |
| NS1 at 236 residue | modified influenza A M2e epitope (SLLTEVETPIRNEWGSRSNDSSD) | 2 doses (mice) | Rumyantsev et al., 2010.63 |
Another approach for the expression of defined epitopes by the YF17D recombinant virus was based on the use of the major envelope protein of flaviviruses, the E protein. Since this protein is the major target of the humoral immune response presenting 180 monomers at the surface of mature virions, the expression of heterologous humoral epitopes in this context would be appropriate to elicit antibodies specific to the foreign epitope. Additionally, several epitopes related to T-cell response have also been mapped onto the E protein.24,59 Thus, based on three-dimensional structure modeling of the YF E protein it was possible to identify 2 expression sites permissive to heterologous insertions localized at the viral envelope surface at amino acid positions 154 and 200.60,61 A number of recombinant YF17D viruses that express T cell and humoral epitopes of the Plasmodium spp CS protein inserted at the fg loop (site at amino acid 200 of the E protein) have been constructed. These viruses were stable through serial passages in cultured vertebrate cells and also retained attenuation for mice and monkeys.58,60,62 In addition, the recombinant virus expressing a B-cell epitope from the malarial parasite Plasmodium falciparum induced antibodies against the CS protein after mouse and monkey vaccination.60,62 The epitope expression in the E protein is an interesting approach, since it was recently reported that this protein elicits dominant immune responses corresponding either to MHC class I or class II antigens.8,22,59
Another possibility for epitope expression was developed by means of a transposon-mediated technique to randomly insert and express influenza A virus epitopes into the yellow fever virus NS1 protein.63 Intriguingly, by selecting recombinant viruses with a methodology combining plaque purification with immunostaining, only one single insertion site was obtained at NS1–236 in which the influenza A epitope was expressed, strongly suggesting that NS1 is not tolerant in terms of alternative insertion points. Immunization of mice with this recombinant virus was not sufficient to afford protection against challenge with the mouse- adapted influenza A/H1N1 virus.
Expression of Larger Fragments by the YF17D Virus
Despite the importance of epitope insertion in establishing a number of methodologies to manipulate the YF genome and accessing the immunological potential of these recombinant viruses, it was of interest to improve the vectorial capacity of the YF17D virus, since the expression of larger gene fragments could allow the expression of a more complex antigenic repertoire and, consequently, increase the chances to induce a more appropriate and polyvalent immune response. Basically, 3 different methodologies approached the expression of proteins or fragments thereof.
One of the possibilities to express heterologous sequences, in general up to 2.0 kb in length, into the YF17D genome consists of the insertion between the genes that code for the E and NS1 proteins. This is mainly based on the fact that in the flavivirus genomic organization, the intergenic E/NS1 region represents a functional shift from the structural to non-structural genes. So, larger heterologous insertions might be better accommodated in this region inducing fewer disturbances in the virus cycle as compared with other sites. This method is similar, theoretically, to the insertion between genes that encode proteins cleaved by viral protease. However, the cleavage between E and NS1 is accomplished by a cellular enzyme (signal peptidase, ref. 64) present in the ER membrane. Consequently, the cleavage sites and other structural elements needed for viral viability are different, and their presence should be taken into account when designing the expression cassette at the E and NS1 intergenic region. In most of the recombinant YF17D viruses bearing insertions at the E-NS1 intergenic region (Table 2), the recombinant proteins were fused at their amino-termini with a duplication of the codons for the first 9 amino acids of YF NS1 and at their carboxi-termini with whole or part of the YF E protein stem-anchor domain, or an equivalent sequence from another flavivirus.56,65,66 A similar approach was employed to improve the genetic stability of Lassa virus glycoproteins expressed by recombinant YF17D viruses in this genomic site.67,68 Interestingly, a variant of this general methodology was described in the expression of the HIV clade B consensus Gag p24 sequence by the YF17D virus. In this case, the heterologous protein was directly expressed in frame with the YF E protein and fused to the single transmembrane anchoring segment of the Sindbis virus E2 protein. Hence, this motif is expected to display a role as a signal peptide to the NS1 translocation to the ER lumen. Even though significant humoral and cellular immune responses against p24 were elicited by the 17D recombinant virus, this construct was genetically unstable, resulting in the specific deletion of the p24 gene.9 The viral instability might be related to a decrease of viral fitness imposed by some constraints on either the viral genome or the precursor polyprotein that could have been provoked by the construct design. However, it is reasonable to suppose that in some instances the origin of the viral instability could be related to the primary function of the heterologous sequences cloned into the viral genome, such as the presence of secondary structural RNA elements that might interact with viral RNA or the expression of heterologous proteins that might disturb processes involved in viral infection. Actually, we attributed the genetic instability of a recombinant YF virus expressing a fragment of the SIV Gag precursor protein to the presence of an IRES element localized at the 3′end of gene encoding the SIV Matrix protein.69 We managed to substantially improve the genetic stability of this recombinant YF17D virus by introducing mutations in the IRES element.70
Table 2. Expression of heterologous proteins at the E-NS1 expression site of YF17D virus.
| Foreign gene | Immunization | Reference | |
|---|---|---|---|
| reporter protein | EGFP | 2 doses (mice) | Bonaldo and Galler, 200585; Bonaldo et al., 2007.65 |
| SIV | Gag 45–269 | boost in rhesus monkeys primed with rBCG | Bonaldo et al., 201071 |
| Gag 44–84 | prime with rYF and boost with rAdenovirus 5 (rhesus monkey) | Martins et al., 2013.72 | |
| Gag 76–123 | |||
| Gag 142–186 | |||
| Gag 250–415 | |||
| Vif 1–110 | |||
| Vif 102–214 | |||
| Nef 45–210 | |||
| T.cruzi | ASP-2 261–380 | 2 doses (mice) | Nogueira et al., 2013.56 |
| HIV | p24 | 2 doses (mice) | Franco et al., 2010.9 |
| Lassa Virus | GPC | 1 dose (guinea pigs) | Bredenbeek et al., 2006.67 |
| GP1 | 2 doses (mice and guinea pigs) | Jiang et al., 2011.68 | |
| GP2 | 2 doses (mice and guinea pigs) | Jiang et al., 2011.68 | |
The foreign antigens expressed by the recombinant YF17D virus in the E/NS1 region appear to remain cell-associated as described elsewhere.56,65,66 The heterologous proteins are indeed retained in the ER compartment due to the presence of anchor transmembrane motifs at their carboxi-terminus. Although these recombinant viruses elicited humoral immune responses against the foreign antigens, hypothetically, the accumulation of antigens inside the infected cells could drive the immune response to be mainly cellular.
Recently, studies performed in Indian rhesus macaques, in which recombinant YF17D viruses expressing fragments of SIV proteins, demonstrated the ability of YF17D vector to elicit cytotoxic T lymphocyte responses specific to recombinant antigens. .71-73 In one of these experiments, animals expressing the Mamu-B*08 allele, a rhesus model for an HLA-B*27 mediated elite controller, controlled the replication of SIV after vaccination with a heterologous immunization regimen with recombinant YF17D viruses as a prime dose and recombinant Adenoviruses 5 as a booster dose, both expressing fragments of Vif and Nef. These animals exhibited a high frequency of CD8+ T cells detected in blood, lymph nodes, and colon that were specific to 3 different Mamu-B*08 restricted epitopes. In order to better evaluate the efficacy of replicative-competent recombinant YF vectors, 7 YF17D viruses expressing fragments of the SIVmac239 Gag, Nef, and Vif proteins were utilized to vaccinate rhesus monkeys with different MHC haplotypes.72 Despite the detection of replication of YF viruses in rhesus monkeys after vaccination, in general, they only elicited low levels of SIV- specific cellular immune responses, which considerably improved only after a recombinant Adenovirus type-5 vector booster. However, an increase of CD4+ T cellular responses was only observed after heterologous YF17D/rAd5 immunization, but not after Ad5 vaccination only, advocating a role of recombinant YF17D viruses to prime virus-specific T-cell responses in heterologous prime-boost immunizations. Interestingly, the comparison of this YF17D vector to the recombinant Adenovirus 5 and Modified Vaccinia Ankara viruses, all of them expressing the HIV p24 protein, indicated that recombinant YF17D viruses induced better balanced CD8+ and CD4+ T cell immune responses in the mouse model.9 Furthermore, in terms of cytokine release from splenocytes in these animals, a more polyfunctional pattern was detected. Other studies confirmed the capacity of this YF E/NS1 viral platform to elicit a T cell-specific response against other antigens.31,56,67,68,74,75 These results point to a need of optimization of vaccination regimens as well as an improvement of the YF E/NS1 insertion platform immunogenicity, which should be considered since the yellow fever vaccine displays outstanding immunological properties as a human immunogen. Another possibility to exploit the expression of foreign proteins by the live-attenuated YF17D virus consists of the insertion of sequences into the C gene. This method takes into account the presence of a conserved hairpin RNA structure (CS) in the gene coding for the C protein that mediates the genome cyclization required for RNA replication.76 Thus, in this model, the heterologous gene is flanked at its 5′ end by a truncated capsid sequence of 75 nucleotides containing the CS sequence and at its 3′ end with sequences coding for the foot-mouth disease virus (FMDV) 2A peptide and the mammalian ubiquitin to ensure efficient cleavage from the downstream YF. Initially, this platform was employed to express a fragment of the circumsporozoite protein (CS) from the murine malarial parasite Plasmodium yoelii.77 A single immunization of naïve mice with recombinant YF17D resulted in robust production of IFN-γ by CD8+ T-cells and IFN-γ and IL-2 by CD4+ T-cells. When administered in a prime-boost regimen in conjunction with a boost dose of irradiated sporozoites, the vaccination protected mice from a challenge with sporozoites of P.yoelii. This YF vector was also utilized to promote the expression of the HIV p24 sequence.9 Immunogenicity studies in mice indicated that this recombinant virus induced antibodies against p24 and p24-specific CD8+ IFN-T-cell responses. In both constructs the major concern was related to the viral genetic stability, since the heterologous genes coding for the CS protein and p24 were completely deleted in the fourth and sixth serial passages suggesting that this strategy deserves some improvement to stabilize the recombinant viruses.
The last method to express larger heterologous sequences by the YF17D virus was based on the natural length variation of the 3′ untranslated region of flaviviruses78 that could accommodate the insertion of genetic cassettes containing internal ribosomal entry sites (IRES).79 These bicistronic viruses were developed for other flaviviruses to express reporter proteins.74,80-82 Nevertheless, these bicistronic flaviviruses were considered to be very unstable, since soon after few rounds of viral replication, the heterologous IRES-bearing cassettes suffered extensive deletions.74,75
Concluding Remarks
The insertion of heterologous sequences into the YF17D genome is not an obvious task, since heterologous inserts do not provide any selective advantage to the virus and in general, they indeed cause a reduction of viral fitness. This is due in part not only to the rather short YF genome size but also to interference in interactions among viral and cellular proteins, such as those that take place during the translation of the RNA, its replication and virus particle formation. Such interferences certainly contribute to the genetic instability for several foreign genes and poor immunogenicity of some recombinant YF17D viruses.
A very important aspect in establishing a strategy for heterologous expression specifically at intergenic sites is related to the topology of viral proteins in the endoplasmic reticulum membrane. It is well known that the polyprotein precursor traverses the ER membrane at various points being proteolytically processed in the ER lumen by cellular signal peptidases and in the cytoplasmic side by the viral NS2B/NS3 protease. Protein secretion and processing require the presence of functional motifs that allow the sorting to the appropriate compartment, the proteolytic processing to generate the mature viral proteins and their involvement in the replication complex. Therefore, the strategy to express a given antigen will depend upon its physical-chemical properties, cell trafficking, and the type of immune response expected against the antigen. It is conceivable that improving antigen processing will depend at least in part upon the appropriate cellular location.
One of the major concerns about this first generation of recombinant YF17D viruses is the low immunogenicity. Perhaps the increment in the ability to properly induce the immune system could be afforded by means of the introduction of mutations into the viral genome that promotes higher viral replication capacity. In this regard the NS4B (Leu52Phe) mutation introduced in a dengue virus 2 vector expressing different reporter proteins conferred a dramatic enhancement in virus infectivity and production in cell cultures and in mouse infectiousness that was consistent with the properties of the parental virus.83,84
Additionally, it was recently demonstrated that it is possible to increase the immunogenicity of recombinant YF vaccines through the use of formulations containing distinct platforms expressing heterologous antigens. Hence, a simultaneous administration of different recombinant YF viruses expressing a repertoire of antigens from the ASP-2 of Trypanosoma cruzi was considerably more immunogenic than the single administration of each virus promoting protection against a lethal challenge of the protozoan parasite in one of the mouse models.56
All in all, despite the initial results pointing to a need for improved vectorial properties, especially the genetic stability of the insertion, one must be aware that the 17D recombinant viruses generated so far are only the first generation. Considering that this vaccine virus has exceptional characteristics as a human immunogen, the improvement of such a platform is certainly warranted.
Glossary
- List of abreviations
YF, yellow fever
Acknowledgments
The authors declare that they did not receive any funding for the preparation of this review. The authors would like to thank to Mitchell Raymond Lishon, native of Chicago, Illinois, USA—U.C.L.A. 1969, for English revision of this review.
Disclosure of Potential Conflicts of Interest
The authors have declared that the insertion methodology into the YF E/NS1 region is the subject of a patent application having as authors M.C.B. and R.G. and the Fundação Oswaldo Cruz.
References
- 1.Strode GK. Yellow fever. New York: McGraw-Hill Book Company, 1951. [Google Scholar]
- 2.Galler R, Post PR, Santos CN, Ferreira II. . Genetic variability among yellow fever virus 17D substrains. Vaccine 1998; 16:1024 - 8; http://dx.doi.org/ 10.1016/S0264-410X(97)00278-8; PMID: 9682354 [DOI] [PubMed] [Google Scholar]
- 3.Marchevsky RS, Freire MS, Coutinho ES, Galler R. . Neurovirulence of yellow fever 17DD vaccine virus to rhesus monkeys. Virology 2003; 316:55 - 63; http://dx.doi.org/ 10.1016/S0042-6822(03)00583-X; PMID: 14599790 [DOI] [PubMed] [Google Scholar]
- 4.Camacho LA, de Aguiar SG, Freire MdaS, Leal MdaL, do Nascimento JP, Iguchi T, Lozana JA, Farias RH, Collaborative Group for the Study of Yellow Fever Vaccines. . Reactogenicity of yellow fever vaccines in a randomized, placebo-controlled trial. Rev Saude Publica 2005; 39:413 - 20; http://dx.doi.org/ 10.1590/S0034-89102005000300012; PMID: 15997317 [DOI] [PubMed] [Google Scholar]
- 5.Monath TP, Barrett AD. . Pathogenesis and pathophysiology of yellow fever. Adv Virus Res 2003; 60:343 - 95; http://dx.doi.org/ 10.1016/S0065-3527(03)60009-6; PMID: 14689698 [DOI] [PubMed] [Google Scholar]
- 6.Lefeuvre A, Marianneau P, Deubel V. . Current Assessment of Yellow Fever and Yellow Fever Vaccine. Curr Infect Dis Rep 2004; 6:96 - 104; http://dx.doi.org/ 10.1007/s11908-996-0005-9; PMID: 15023271 [DOI] [PubMed] [Google Scholar]
- 7.Poland JD, Calisher CH, Monath TP, Downs WG, Murphy K. . Persistence of neutralizing antibody 30-35 years after immunization with 17D yellow fever vaccine. Bull World Health Organ 1981; 59:895 - 900; PMID: 6978196 [PMC free article] [PubMed] [Google Scholar]
- 8.Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, Murali-Krishna K, Mahar PL, Edupuganti S, Lalor S, et al. . Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 2008; 28:710 - 22; http://dx.doi.org/ 10.1016/j.immuni.2008.02.020; PMID: 18468462 [DOI] [PubMed] [Google Scholar]
- 9.Franco D, Li W, Qing F, Stoyanov CT, Moran T, Rice CM, Ho DD. . Evaluation of yellow fever virus 17D strain as a new vector for HIV-1 vaccine development. Vaccine 2010; 28:5676 - 85; http://dx.doi.org/ 10.1016/j.vaccine.2010.06.052; PMID: 20600494 [DOI] [PubMed] [Google Scholar]
- 10.Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. . Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med 2006; 203:413 - 24; http://dx.doi.org/ 10.1084/jem.20051720; PMID: 16461338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulendran B, Oh JZ, Nakaya HI, Ravindran R, Kazmin DA. . Immunity to viruses: learning from successful human vaccines. Immunol Rev 2013; 255:243 - 55; http://dx.doi.org/ 10.1111/imr.12099; PMID: 23947360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childs KS, Randall RE, Goodbourn S. . LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PLoS One 2013; 8:e64202; http://dx.doi.org/ 10.1371/journal.pone.0064202; PMID: 23671710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B. . Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol 2008; 9:1157 - 64; http://dx.doi.org/ 10.1038/ni.1645; PMID: 18758466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR 3rd, Castro E, et al. . Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 2008; 205:3119 - 31; http://dx.doi.org/ 10.1084/jem.20082292; PMID: 19047440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neves PC, Rudersdorf RA, Galler R, Bonaldo MC, de Santana MG, Mudd PA, Martins MA, Rakasz EG, Wilson NA, Watkins DI. . CD8+ gamma-delta TCR+ and CD4+ T cells produce IFN-γ at 5-7 days after yellow fever vaccination in Indian rhesus macaques, before the induction of classical antigen-specific T cell responses. Vaccine 2010; 28:8183 - 8; http://dx.doi.org/ 10.1016/j.vaccine.2010.09.090; PMID: 20939995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva ML, Martins MA, Espírito-Santo LR, Campi-Azevedo AC, Silveira-Lemos D, Ribeiro JG, Homma A, Kroon EG, Teixeira-Carvalho A, Elói-Santos SM, et al. . Characterization of main cytokine sources from the innate and adaptive immune responses following primary 17DD yellow fever vaccination in adults. Vaccine 2011; 29:583 - 92; http://dx.doi.org/ 10.1016/j.vaccine.2010.08.046; PMID: 20732465 [DOI] [PubMed] [Google Scholar]
- 17.Neves PCC, Santos JR, Tubarão LN, Bonaldo MC, Galler R. . Early IFN-gamma production after YF 17D vaccine virus immunization in mice and its association with adaptive immune responses. PLoS One 2013; 8:e81953; http://dx.doi.org/ 10.1371/journal.pone.0081953; PMID: 24324734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulendran B. . Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol 2009; 9:741 - 7; PMID: 19763148 [DOI] [PubMed] [Google Scholar]
- 19.Richter JD, Sonenberg N. . Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005; 433:477 - 80; http://dx.doi.org/ 10.1038/nature03205; PMID: 15690031 [DOI] [PubMed] [Google Scholar]
- 20.Ron D, Walter P. . Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8:519 - 29; http://dx.doi.org/ 10.1038/nrm2199; PMID: 17565364 [DOI] [PubMed] [Google Scholar]
- 21.Reinhardt B, Jaspert R, Niedrig M, Kostner C, L’age-Stehr J. . Development of viremia and humoral and cellular parameters of immune activation after vaccination with yellow fever virus strain 17D: a model of human flavivirus infection. J Med Virol 1998; 56:159 - 67; http://dx.doi.org/; PMID: 9746073 [DOI] [PubMed] [Google Scholar]
- 22.Mudd PA, Piaskowski SM, Neves PC, Rudersdorf R, Kolar HL, Eernisse CM, Weisgrau KL, de Santana MG, Wilson NA, Bonaldo MC, et al. . The live-attenuated yellow fever vaccine 17D induces broad and potent T cell responses against several viral proteins in Indian rhesus macaques--implications for recombinant vaccine design. Immunogenetics 2010; 62:593 - 600; http://dx.doi.org/ 10.1007/s00251-010-0461-0; PMID: 20607226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Co MD, Kilpatrick ED, Rothman AL. . Dynamics of the CD8 T-cell response following yellow fever virus 17D immunization. Immunology 2009; 128:Suppl e718 - 27; http://dx.doi.org/ 10.1111/j.1365-2567.2009.03070.x; PMID: 19740333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Most RG, Harrington LE, Giuggio V, Mahar PL, Ahmed R. . Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology 2002; 296:117 - 24; http://dx.doi.org/ 10.1006/viro.2002.1432; PMID: 12036323 [DOI] [PubMed] [Google Scholar]
- 25.Kohler S, Bethke N, Böthe M, Sommerick S, Frentsch M, Romagnani C, Niedrig M, Thiel A. . The early cellular signatures of protective immunity induced by live viral vaccination. Eur J Immunol 2012; 42:2363 - 73; http://dx.doi.org/ 10.1002/eji.201142306; PMID: 22733156 [DOI] [PubMed] [Google Scholar]
- 26.Blom K, Braun M, Ivarsson MA, Gonzalez VD, Falconer K, Moll M, Ljunggren HG, Michaëlsson J, Sandberg JK. . Temporal dynamics of the primary human T cell response to yellow fever virus 17D as it matures from an effector- to a memory-type response. J Immunol 2013; 190:2150 - 8; http://dx.doi.org/ 10.4049/jimmunol.1202234; PMID: 23338234 [DOI] [PubMed] [Google Scholar]
- 27.Camacho LA, Freire MdaS, Leal MdaL, Aguiar SG, Nascimento JP, Iguchi T, Lozana JdeA, Farias RH, Collaborative Group for the Study of Yellow Fever Vaccines. . Immunogenicity of WHO-17D and Brazilian 17DD yellow fever vaccines: a randomized trial. Rev Saude Publica 2004; 38:671 - 8; http://dx.doi.org/ 10.1590/S0034-89102004000500009; PMID: 15499438 [DOI] [PubMed] [Google Scholar]
- 28.Julander JG, Trent DW, Monath TP. . Immune correlates of protection against yellow fever determined by passive immunization and challenge in the hamster model. Vaccine 2011; 29:6008 - 16; http://dx.doi.org/ 10.1016/j.vaccine.2011.06.034; PMID: 21718741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandriss MW, Schlesinger JJ, Walsh EE. . Immunogenicity of a purified fragment of 17D yellow fever envelope protein. J Infect Dis 1990; 161:1134 - 9; http://dx.doi.org/ 10.1093/infdis/161.6.1134; PMID: 1693159 [DOI] [PubMed] [Google Scholar]
- 30.Lobigs M, Dalgarno L, Schlesinger JJ, Weir RC. . Location of a neutralization determinant in the E protein of yellow fever virus (17D vaccine strain). Virology 1987; 161:474 - 8; http://dx.doi.org/ 10.1016/0042-6822(87)90141-3; PMID: 2446422 [DOI] [PubMed] [Google Scholar]
- 31.Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, et al. . The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol 2009; 183:7919 - 30; http://dx.doi.org/ 10.4049/jimmunol.0803903; PMID: 19933869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice CM, Grakoui A, Galler R, Chambers TJ. . Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biol 1989; 1:285 - 96; PMID: 2487295 [PubMed] [Google Scholar]
- 33.Bray M, Lai CJ. . Construction of intertypic chimeric dengue viruses by substitution of structural protein genes. Proc Natl Acad Sci U S A 1991; 88:10342 - 6; http://dx.doi.org/ 10.1073/pnas.88.22.10342; PMID: 1682924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers TJ, Nestorowicz A, Mason PW, Rice CM. . Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J Virol 1999; 73:3095 - 101; PMID: 10074160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arroyo J, Miller C, Catalan J, Myers GA, Ratterree MS, Trent DW, Monath TP. . ChimeriVax-West Nile virus live-attenuated vaccine: preclinical evaluation of safety, immunogenicity, and efficacy. J Virol 2004; 78:12497 - 507; http://dx.doi.org/ 10.1128/JVI.78.22.12497-12507.2004; PMID: 15507637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guirakhoo F, Arroyo J, Pugachev KV, Miller C, Zhang ZX, Weltzin R, Georgakopoulos K, Catalan J, Ocran S, Soike K, et al. . Construction, safety, and immunogenicity in nonhuman primates of a chimeric yellow fever-dengue virus tetravalent vaccine. J Virol 2001; 75:7290 - 304; http://dx.doi.org/ 10.1128/JVI.75.16.7290-7304.2001; PMID: 11462001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caufour PS, Motta MC, Yamamura AM, Vazquez S, Ferreira II, Jabor AV, Bonaldo MC, Freire MS, Galler R. . Construction, characterization and immunogenicity of recombinant yellow fever 17D-dengue type 2 viruses. Virus Res 2001; 79:1 - 14; http://dx.doi.org/ 10.1016/S0168-1702(01)00273-8; PMID: 11551641 [DOI] [PubMed] [Google Scholar]
- 38.Pugachev KV, Guirakhoo F, Ocran SW, Mitchell F, Parsons M, Penal C, Girakhoo S, Pougatcheva SO, Arroyo J, Trent DW, et al. . High fidelity of yellow fever virus RNA polymerase. J Virol 2004; 78:1032 - 8; http://dx.doi.org/ 10.1128/JVI.78.2.1032-1038.2004; PMID: 14694136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy B, Guirakhoo F, Barban V, Higgs S, Monath TP, Lang J. . Preclinical and clinical development of YFV 17D-based chimeric vaccines against dengue, West Nile and Japanese encephalitis viruses. Vaccine 2010; 28:632 - 49; http://dx.doi.org/ 10.1016/j.vaccine.2009.09.098; PMID: 19808029 [DOI] [PubMed] [Google Scholar]
- 40.Capeding RZ, Luna IA, Bomasang E, Lupisan S, Lang J, Forrat R, Wartel A, Crevat D. . Live-attenuated, tetravalent dengue vaccine in children, adolescents and adults in a dengue endemic country: randomized controlled phase I trial in the Philippines. Vaccine 2011; 29:3863 - 72; http://dx.doi.org/ 10.1016/j.vaccine.2011.03.057; PMID: 21477675 [DOI] [PubMed] [Google Scholar]
- 41.Dayan GH, Garbes P, Noriega F, Izoton de Sadovsky AD, Rodrigues PM, Giuberti C, Dietze R. . Immunogenicity and safety of a recombinant tetravalent dengue vaccine in children and adolescents ages 9-16 years in Brazil. Am J Trop Med Hyg 2013; 89:1058 - 65; http://dx.doi.org/ 10.4269/ajtmh.13-0304; PMID: 24189367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dayan GH, Thakur M, Boaz M, Johnson C. . Safety and immunogenicity of three tetravalent dengue vaccine formulations in healthy adults in the USA. Vaccine 2013; 31:5047 - 54; http://dx.doi.org/ 10.1016/j.vaccine.2013.08.088; PMID: 24021313 [DOI] [PubMed] [Google Scholar]
- 43.Guirakhoo F, Kitchener S, Morrison D, Forrat R, McCarthy K, Nichols R, Yoksan S, Duan X, Ermak TH, Kanesa-Thasan N, et al. . Live attenuated chimeric yellow fever dengue type 2 (ChimeriVax-DEN2) vaccine: Phase I clinical trial for safety and immunogenicity: effect of yellow fever pre-immunity in induction of cross neutralizing antibody responses to all 4 dengue serotypes. Hum Vaccin 2006; 2:60 - 7; http://dx.doi.org/ 10.4161/hv.2.2.2555; PMID: 17012873 [DOI] [PubMed] [Google Scholar]
- 44.Qiao M, Shaw D, Forrat R, Wartel-Tram A, Lang J. . Priming effect of dengue and yellow fever vaccination on the immunogenicity, infectivity, and safety of a tetravalent dengue vaccine in humans. Am J Trop Med Hyg 2011; 85:724 - 31; http://dx.doi.org/ 10.4269/ajtmh.2011.10-0436; PMID: 21976579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, et al. . Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 2012; 380:1559 - 67; http://dx.doi.org/ 10.1016/S0140-6736(12)61428-7; PMID: 22975340 [DOI] [PubMed] [Google Scholar]
- 46.Villar LA, Rivera-Medina DM, Arredondo-García JL, Boaz M, Starr-Spires L, Thakur M, Zambrano B, Miranda MC, Rivas E, Dayan GH. . Safety and immunogenicity of a recombinant tetravalent dengue vaccine in 9-16 year olds: a randomized, controlled, phase II trial in Latin America. Pediatr Infect Dis J 2013; 32:1102 - 9; http://dx.doi.org/ 10.1097/INF.0b013e31829b8022; PMID: 24067553 [DOI] [PubMed] [Google Scholar]
- 47.Mantel N, Girerd Y, Geny C, Bernard I, Pontvianne J, Lang J, Barban V. . Genetic stability of a dengue vaccine based on chimeric yellow fever/dengue viruses. Vaccine 2011; 29:6629 - 35; http://dx.doi.org/ 10.1016/j.vaccine.2011.06.101; PMID: 21745519 [DOI] [PubMed] [Google Scholar]
- 48.Monath TP, McCarthy K, Bedford P, Johnson CT, Nichols R, Yoksan S, Marchesani R, Knauber M, Wells KH, Arroyo J, et al. . Clinical proof of principle for ChimeriVax: recombinant live, attenuated vaccines against flavivirus infections. Vaccine 2002; 20:1004 - 18; http://dx.doi.org/ 10.1016/S0264-410X(01)00457-1; PMID: 11803060 [DOI] [PubMed] [Google Scholar]
- 49.Chokephaibulkit K, Sirivichayakul C, Thisyakorn U, Sabchareon A, Pancharoen C, Bouckenooghe A, Gailhardou S, Boaz M, Feroldi E. . Safety and immunogenicity of a single administration of live-attenuated Japanese encephalitis vaccine in previously primed 2- to 5-year-olds and naive 12- to 24-month-olds: multicenter randomized controlled trial. Pediatr Infect Dis J 2010; 29:1111 - 7; http://dx.doi.org/ 10.1097/INF.0b013e3181f68e9c; PMID: 20856164 [DOI] [PubMed] [Google Scholar]
- 50.Monath TP, Guirakhoo F, Nichols R, Yoksan S, Schrader R, Murphy C, Blum P, Woodward S, McCarthy K, Mathis D, et al. . Chimeric live, attenuated vaccine against Japanese encephalitis (ChimeriVax-JE): phase 2 clinical trials for safety and immunogenicity, effect of vaccine dose and schedule, and memory response to challenge with inactivated Japanese encephalitis antigen. J Infect Dis 2003; 188:1213 - 30; http://dx.doi.org/ 10.1086/378356; PMID: 14551893 [DOI] [PubMed] [Google Scholar]
- 51.Nasveld PE, Ebringer A, Elmes N, Bennett S, Yoksan S, Aaskov J, McCarthy K, Kanesa-thasan N, Meric C, Reid M. . Long term immunity to live attenuated Japanese encephalitis chimeric virus vaccine: randomized, double-blind, 5-year phase II study in healthy adults. Hum Vaccin 2010; 6:1038 - 46; http://dx.doi.org/ 10.4161/hv.6.12.13057; PMID: 21150279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biedenbender R, Bevilacqua J, Gregg AM, Watson M, Dayan G. . Phase II, randomized, double-blind, placebo-controlled, multicenter study to investigate the immunogenicity and safety of a West Nile virus vaccine in healthy adults. J Infect Dis 2011; 203:75 - 84; http://dx.doi.org/ 10.1093/infdis/jiq003; PMID: 21148499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Filette M, Ulbert S, Diamond M, Sanders NN. . Recent progress in West Nile virus diagnosis and vaccination. Vet Res 2012; 43:16; http://dx.doi.org/ 10.1186/1297-9716-43-16; PMID: 22380523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McAllister A, Arbetman AE, Mandl S, Peña-Rossi C, Andino R. . Recombinant yellow fever viruses are effective therapeutic vaccines for treatment of murine experimental solid tumors and pulmonary metastases. J Virol 2000; 74:9197 - 205; http://dx.doi.org/ 10.1128/JVI.74.19.9197-9205.2000; PMID: 10982366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barba-Spaeth G, Longman RS, Albert ML, Rice CM. . Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J Exp Med 2005; 202:1179 - 84; http://dx.doi.org/ 10.1084/jem.20051352; PMID: 16260489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nogueira RT, Nogueira AR, Pereira MC, Rodrigues MM, Neves PC, Galler R, Bonaldo MC. . Recombinant yellow fever viruses elicit CD8+ T cell responses and protective immunity against Trypanosoma cruzi. PLoS One 2013; 8:e59347; http://dx.doi.org/ 10.1371/journal.pone.0059347; PMID: 23527169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao D, Barba-Spaeth G, Rai U, Nussenzweig V, Rice CM, Nussenzweig RS. . Yellow fever 17D as a vaccine vector for microbial CTL epitopes: protection in a rodent malaria model. J Exp Med 2005; 201:201 - 9; http://dx.doi.org/ 10.1084/jem.20041526; PMID: 15657290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nogueira RT, Nogueira AR, Pereira MC, Rodrigues MM, Galler R, Bonaldo MC. . Biological and immunological characterization of recombinant Yellow Fever 17D viruses expressing a Trypanosoma cruzi Amastigote Surface Protein-2 CD8+ T cell epitope at two distinct regions of the genome. Virol J 2011; 8:127; http://dx.doi.org/ 10.1186/1743-422X-8-127; PMID: 21418577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Melo AB, Nascimento EJ, Braga-Neto U, Dhalia R, Silva AM, Oelke M, Schneck JP, Sidney J, Sette A, Montenegro SM, et al. . T-cell memory responses elicited by yellow fever vaccine are targeted to overlapping epitopes containing multiple HLA-I and -II binding motifs. PLoS Negl Trop Dis 2013; 7:e1938; http://dx.doi.org/ 10.1371/journal.pntd.0001938; PMID: 23383350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonaldo MC, Garratt RC, Caufour PS, Freire MS, Rodrigues MM, Nussenzweig RS, Galler R. . Surface expression of an immunodominant malaria protein B cell epitope by yellow fever virus. J Mol Biol 2002; 315:873 - 85; http://dx.doi.org/ 10.1006/jmbi.2001.5258; PMID: 11812154 [DOI] [PubMed] [Google Scholar]
- 61.Bonaldo MC, Garratt RC, Freire MS, Galler R. . Expression of foreign protein epitopes at the surface of recombinant yellow fever 17D viruses based on three-dimensional modeling of its envelope protein. Cell Biochem Biophys 2006; 44:313 - 24; http://dx.doi.org/ 10.1385/CBB:44:3:313; PMID: 16679518 [DOI] [PubMed] [Google Scholar]
- 62.Bonaldo MC, Garratt RC, Marchevsky RS, Coutinho ES, Jabor AV, Almeida LF, Yamamura AM, Duarte AS, Oliveira PJ, Lizeu JO, et al. . Attenuation of recombinant yellow fever 17D viruses expressing foreign protein epitopes at the surface. J Virol 2005; 79:8602 - 13; http://dx.doi.org/ 10.1128/JVI.79.13.8602-8613.2005; PMID: 15956601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rumyantsev AA, Zhang ZX, Gao QS, Moretti N, Brown N, Kleanthous H, Delagrave S, Guirakhoo F, Collett MS, Pugachev KV. . Direct random insertion of an influenza virus immunologic determinant into the NS1 glycoprotein of a vaccine flavivirus. Virology 2010; 396:329 - 38; http://dx.doi.org/ 10.1016/j.virol.2009.10.033; PMID: 19913267 [DOI] [PubMed] [Google Scholar]
- 64.Auclair SM, Bhanu MK, Kendall DA. . Signal peptidase I: cleaving the way to mature proteins. Protein Sci 2012; 21:13 - 25; http://dx.doi.org/ 10.1002/pro.757; PMID: 22031009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonaldo MC, Mello SM, Trindade GF, Rangel AA, Duarte AS, Oliveira PJ, Freire MS, Kubelka CF, Galler R. . Construction and characterization of recombinant flaviviruses bearing insertions between E and NS1 genes. Virol J 2007; 4:115; http://dx.doi.org/ 10.1186/1743-422X-4-115; PMID: 17971212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trindade GF, Santana MG, Santos JR, Galler R, Bonaldo MC. . Retention of a recombinant GFP protein expressed by the yellow fever 17D virus in the E/NS1 intergenic region in the endoplasmic reticulum. Mem Inst Oswaldo Cruz 2012; 107:262 - 72; http://dx.doi.org/ 10.1590/S0074-02762012000200017; PMID: 22415267 [DOI] [PubMed] [Google Scholar]
- 67.Bredenbeek PJ, Molenkamp R, Spaan WJ, Deubel V, Marianneau P, Salvato MS, Moshkoff D, Zapata J, Tikhonov I, Patterson J, et al. . A recombinant Yellow Fever 17D vaccine expressing Lassa virus glycoproteins. Virology 2006; 345:299 - 304; http://dx.doi.org/ 10.1016/j.virol.2005.12.001; PMID: 16412488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang X, Dalebout TJ, Bredenbeek PJ, Carrion R Jr., Brasky K, Patterson J, Goicochea M, Bryant J, Salvato MS, Lukashevich IS. . Yellow fever 17D-vectored vaccines expressing Lassa virus GP1 and GP2 glycoproteins provide protection against fatal disease in guinea pigs. Vaccine 2011; 29:1248 - 57; http://dx.doi.org/ 10.1016/j.vaccine.2010.11.079; PMID: 21145373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B. . A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res 2010; 38:1367 - 81; http://dx.doi.org/ 10.1093/nar/gkp1109; PMID: 19969542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santana MGV, Neves PCC, Santos JR, Lima NS. . SANTOS AAC, SANTOS JR, Watkins DI, Galler R, Bonaldo MC. Improved genetic stability of recombinant yellow fever 17D virus expressing a lentiviral Gag gene fragment. Virology In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonaldo MC, Martins MA, Rudersdorf R, Mudd PA, Sacha JB, Piaskowski SM, Costa Neves PC, Veloso de Santana MG, Vojnov L, Capuano S 3rd, et al. . Recombinant yellow fever vaccine virus 17D expressing simian immunodeficiency virus SIVmac239 gag induces SIV-specific CD8+ T-cell responses in rhesus macaques. J Virol 2010; 84:3699 - 706; http://dx.doi.org/ 10.1128/JVI.02255-09; PMID: 20089645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martins MA, Bonaldo MC, Rudersdorf RA, Piaskowski SM, Rakasz EG, Weisgrau KL, Furlott JR, Eernisse CM, Veloso de Santana MG, Hidalgo B, et al. . Immunogenicity of seven new recombinant yellow fever viruses 17D expressing fragments of SIVmac239 Gag, Nef, and Vif in Indian rhesus macaques. PLoS One 2013; 8:e54434; http://dx.doi.org/ 10.1371/journal.pone.0054434; PMID: 23336000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, et al. . Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 2012; 491:129 - 33; http://dx.doi.org/ 10.1038/nature11443; PMID: 23023123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deas TS, Binduga-Gajewska I, Tilgner M, Ren P, Stein DA, Moulton HM, Iversen PL, Kauffman EB, Kramer LD, Shi PY. . Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J Virol 2005; 79:4599 - 609; http://dx.doi.org/ 10.1128/JVI.79.8.4599-4609.2005; PMID: 15795246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou G, Xu HY, Qing M, Wang QY, Shi PY. . Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res 2011; 91:11 - 9; http://dx.doi.org/ 10.1016/j.antiviral.2011.05.001; PMID: 21575658 [DOI] [PubMed] [Google Scholar]
- 76.Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH. . Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol 2003; 77:2265 - 70; http://dx.doi.org/ 10.1128/JVI.77.3.2265-2270.2003; PMID: 12525663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stoyanov CT, Boscardin SB, Deroubaix S, Barba-Spaeth G, Franco D, Nussenzweig RS, Nussenzweig M, Rice CM. . Immunogenicity and protective efficacy of a recombinant yellow fever vaccine against the murine malarial parasite Plasmodium yoelii. Vaccine 2010; 28:4644 - 52; http://dx.doi.org/ 10.1016/j.vaccine.2010.04.071; PMID: 20451637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mutebi JP, Rijnbrand RC, Wang H, Ryman KD, Wang E, Fulop LD, Titball R, Barrett AD. . Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3′ noncoding region. J Virol 2004; 78:9652 - 65; http://dx.doi.org/ 10.1128/JVI.78.18.9652-9665.2004; PMID: 15331698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andino R, McAllister A. Recombinant Bicistronic Flaviviruses and Methods of Use Thereof USA: University of California, 2001:28. [Google Scholar]
- 80.Kaptein SJ, De Burghgraeve T, Froeyen M, Pastorino B, Alen MM, Mondotte JA, Herdewijn P, Jacobs M, de Lamballerie X, Schols D, et al. . A derivate of the antibiotic doxorubicin is a selective inhibitor of dengue and yellow fever virus replication in vitro. Antimicrob Agents Chemother 2010; 54:5269 - 80; http://dx.doi.org/ 10.1128/AAC.00686-10; PMID: 20837762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pierson TC, Diamond MS, Ahmed AA, Valentine LE, Davis CW, Samuel MA, Hanna SL, Puffer BA, Doms RW. . An infectious West Nile virus that expresses a GFP reporter gene. Virology 2005; 334:28 - 40; http://dx.doi.org/ 10.1016/j.virol.2005.01.021; PMID: 15749120 [DOI] [PubMed] [Google Scholar]
- 82.Yun SI, Kim SY, Rice CM, Lee YM. . Development and application of a reverse genetics system for Japanese encephalitis virus. J Virol 2003; 77:6450 - 65; http://dx.doi.org/ 10.1128/JVI.77.11.6450-6465.2003; PMID: 12743302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grant D, Tan GK, Qing M, Ng JK, Yip A, Zou G, Xie X, Yuan Z, Schreiber MJ, Schul W, et al. . A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J Virol 2011; 85:7775 - 87; http://dx.doi.org/ 10.1128/JVI.00665-11; PMID: 21632767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A, Rice CM. . Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc Natl Acad Sci U S A 2012; 109:14610 - 5; http://dx.doi.org/ 10.1073/pnas.1212379109; PMID: 22908290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonaldo MC, Galler R. Method for the Production of Recombinant Virus, DNA Constructs, Recombinant Virus and Vaccine Compositions BRAZIL: Fundação Oswaldo Cruz, 2005:93. [Google Scholar]