Abstract
OBJECTIVE
To determine the role of unit-based transmission that accounts for cases of early Clostridium difficile infection (CDI) during hospitalization for allogeneic stem cell transplant.
SETTING
Stem cell transplant unit at a tertiary care cancer center.
METHODS
Serially collected stool from patients admitted for transplant was screened for toxigenic C. difficile through the hospital stay and genotyping was performed by multilocus sequence typing. In addition, isolates retrieved from cases of CDI that occurred in other patients hospitalized on the same unit were similarly characterized. Transmission links were established by time-space clustering of cases and carriers of shared toxigenic C. difficile strains.
RESULTS
During the 27-month period, 1,099 samples from 264 patients were screened, 69 of which had evidence of toxigenic C. difficile; 52 patients developed CDI and 17 were nonsymptomatic carriers. For the 52 cases, 41 had evidence of toxigenic C. difficile on the first study sample obtained within a week of admission, among which 22 were positive within the first 48 hours. A total of 24 sequence types were isolated from this group; 1 patient had infection with the NAP1 strain. A total of 11 patients had microbiologic evidence of acquisition; donor source could be established in half of these cases.
CONCLUSIONS
Most cases of CDI after stem cell transplant represent delayed onset disease in nonsymptomatic carriers. Transmission on stem cell transplant unit was confirmed in 19% of early CDI cases in our cohort with a probable donor source established in half of the cases.
Clostridium difficile is the most common healthcare-associated infection in the United States.1 In hospitalized patients, the risk of C. difficile infection (CDI) in persons with hematologic malignancy and in hematopoietic stem cell transplant (SCT) recipients is among the highest. Cumulative incidence of CDI in the first year after allogeneic SCT has been reported to be 12.5% to 21% across various studies, a risk that is 15 to 25 times greater than for all hospitalized patients (0.85%).2–10 In these reports, the timing of CDI after SCT is strikingly similar; more than half of all cases occur early, around the time of conditioning and within the first month after transplant.5–8,11
Many transplant units struggle to control the high healthcare-associated rates of CDI, and clusters and outbreaks are frequently encountered.12,13 Transmission patterns of C. difficile in acute care settings from geographically defined populations have been elucidated by the application of various fingerprinting techniques. By characterizing CDI cases detected by a low-sensitivity enzyme immunoassay test, Walker et al14 showed that 37% of all C. difficile cases on a renal transplant unit could be attributed to ward-based transmission, much higher than the overall 23% suspected transmission among all hospitalized patients in this study. Their findings suggest that transmission of C. difficile in complex high-risk populations may be more frequent; however, a similar approach has not yet been applied to study this risk in SCT units.
The frequent occurrence of early CDI among SCT recipients offers a unique opportunity to examine the role of unit-based transmission, an extremely important aspect to recognize before effectual interventions for CDI prevention can be established.
Our study was designed to determine the frequency of unit-based transmission that led to cases of early CDI (day −10 to day +40) in adult patients hospitalized for allogeneic SCT, a short and well-defined period during which most cases of CDI occur. Serial stool samples from patients admitted for transplant were screened for C. difficile and genotyping was performed by multilocus sequence typing (MLST). In addition, toxigenic C. difficile isolates retrieved from other hospitalized patients with shared contact on the transplant unit were similarly characterized. Links between cases were established by integrating relevant surveillance, clinical, and molecular epidemiologic information.
METHODS
Study Design and Patient Population
We performed analysis of patients admitted for allogeneic transplant to the SCT unit (transplant unit) at Memorial Sloan-Kettering Cancer Center (MSKCC) from October 1, 2010, through December 31, 2012, and sought to determine transmission among incident cases of C. difficile infection or colonization among this cohort.
MSKCC is a 471-bed tertiary cancer care facility in New York City. The adult SCT unit at MSKCC is composed of a 29-bed unit, performing approximately 150 allogeneic SCTs per year during the study period and with approximately 8,000 patient-days annually. Only autologous and allogenic transplant recipients are admitted to this unit. All patients are admitted to a private single room and routinely placed under protective isolation (mask and gloves). All cases diagnosed with C. difficile are placed under contact precautions (gown, gloves, soap and water handwashing) until diarrhea resolves and the patient has completed at least 7 days of therapy.
Detection of Cases of CDI and Asymptomatic Colonization With Toxigenic C. difficile on Transplant Unit
A case of CDI was defined by clinical history compatible with CDI (more than 3 diarrheal stools over a 24-hour period) and a positive laboratory test. A case of colonization was defined by isolation of toxigenic C. difficile from a nonsymptomatic patient. Cases of C. difficile colonization and infection were identified from the following sources:
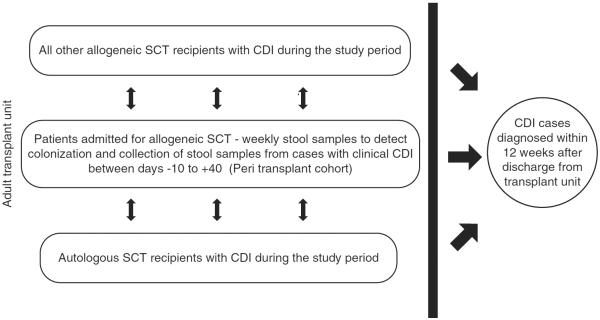
Screening of consecutive stool samples collected from 264 adult patients who enrolled in an institutional fecal bio specimen protocol (described in previous studies15) and who underwent allogeneic SCT in the specified study period. Briefly, the fecal bio specimen protocol includes stool samples obtained at weekly intervals starting from day 10 before allogeneic SCT and continuing throughout the initial transplant hospitalization. For the purposes of this study, this group of patients will be referred to as the peri transplant cohort.
- All other adult transplant recipients hospitalized on the SCT unit at MSKCC within the same period who were diagnosed with CDI during hospitalization or within 3 months after discharge. These cases were identified by querying the infection control database (AICE). Cases were categorized as follows:
-
–Community acquired; CDI diagnosed within 48 hours of admission.
-
–Hospital onset (HO); CDI diagnosed more than 48 hours after admission
-
–Community onset but healthcare facility–associated (CO-HCFA); CDI that occurred within 3 months after discharge from transplant unit
-
–
Recurrence was defined as a CDI episode that occurred 2 weeks after the index event. Banked stool samples or isolates from identified cases (source 2) were retrieved for further analysis from the repository of all C. difficile–positive stool samples that is maintained at MSKCC. For the purposes of this study, this group of patients will be referred to as other cohort.
Laboratory Methods
Diagnostic testing for CDI was performed with Gen Xpert C. difficile polymerase chain reaction assay (Cepheid).16 Stool consistency is recorded at the time of testing and reporting but there is no formal rejection policy. For the peri transplant cohort, screening for toxigenic C. difficile was performed by culture as previously described.5 Colonies morphologically consistent with C. difficile were confirmed using ProDisc (Remel). Toxigenic strains were genotyped by MLST, as previously described.17
Clinical Data
Unit-based transmission was determined by creating a time-space cluster map of cases with the same genotype (sequence type [ST]) on the SCT unit. Directional and nondirectional transmission was determined among shared ST cases with any hospitalizations that overlapped, within 8 weeks of each other, or in the 3 months after discharge. For cases where colonization was detected before infection, the first date of C. difficile isolation was considered to infer potential transmission. If toxigenic C. difficile was detected on a sample within 48 hours of admission, the case was not considered nosocomial, regardless of the timing of CDI. For multiple potential transmissions, the case that was closest in time was considered the donor. These estimates of infectious and incubation periods were derived from previous studies and current Centers for Disease Control and Prevention surveillance definitions.14,18,19
The MSKCC Institutional Review Board granted a waiver of authorization for study of existing data.
Statistical Analysis
Descriptive statistics were used to describe the characteristics and outcomes of the study population. Frequencies and proportions were reported for categorical variables and categorical proportions were compared using the χ2 or Fisher exact tests. All analyses were performed using SAS, version 9.2 (SAS Institute).
RESULTS
Colonization and Infection With C. difficile in the Peri Transplant Cohort
Over the 27-month study period (October 1, 2010, through December 31, 2012), a total of 1,099 stool samples from 264 patients admitted for allogeneic SCT were screened. For 175 of the 264 patients, the first stool sample was obtained within 3 calendar days after admission. Among the 264 patients, 69 (26.1%) had evidence of toxigenic C. difficile on at least 1 stool sample. Fifty-two patients (19.7%) developed CDI in the peri transplant period (day −10 to day +40). An additional 17 patients (6.4%) who were colonized with toxigenic C. difficile did not develop CDI within this timeframe. Two patients with colonization and 4 with CDI had a previous history of CDI.
In 61 (88%) of 69 patients, toxigenic C. difficile was isolated from a stool sample obtained before stem cell infusion. There were no significant differences in clinical characteristics of patients with C. difficile colonization or infection and those without any evidence of C. difficile in the peri transplant period except for myeloablative conditioning (Table 1). Three (5.8%) of the 52 patients diagnosed with CDI relapsed within the peri transplant period and 8 (15.4%) had a recurrent episode within 6 months. Five (29.4%) of the 17 patients with C. difficile colonization developed infection within 6 months of transplant. In 3 colonized patients, the colonization was transient, with bacteria isolated on a single specimen only (all had multiple samples tested).
TABLE 1.
Clinical and Demographic Characteristics of Patients in the Peri Transplant Cohort (Clostriaium difficile Colonization; C. difficile Infection; No Evidence of Toxigenic C. difficile)
| Variable | Nonsymptomatic colonization with C. difficile in peri transplant period, no. (%)a (N= 17) | C. difficile infection in peri transplant period, no. (%)b (N= 52) | Patients with no evidence of colonization or clinical infection, no. (%) (N=195) | P valuec (C. difficile colonization or infection vs none) |
|---|---|---|---|---|
| Male sex | 5 (29) | 32 (62) | 117 (60) | .36 |
| Transplant indication | >.99 | |||
| First transplant | 17 (100) | 50 (96) | 188 (96) | |
| Second transplant | 0 (0) | 2 (4) | 7 (4) | |
| Disease | ||||
| Leukemia (acute) | 11 (65) | 32 (62) | 82 (42) | |
| Leukemia (chronic) | 1 (6) | 5 (10) | 12 (6) | |
| Lymphoma | 4 (24) | 7 (13) | 44 (23) | |
| MDS | 1 (6) | 5 (10) | 37 (19) | |
| Multiple Myeloma | 0 (0) | 3 (6) | 17 (9) | |
| Other | 0 (0) | 0 (0) | 3 (2) | |
| Transplant Type | ||||
| Cord | 3 (18) | 8 (15) | 30 (15) | .52 |
| T-cell depleted | 7 (41) | 31 (60) | 94 (48) | |
| Unmodified | 7 (41) | 13 (25) | 71 (36) | |
| TBI | 9 (53) | 28 (54) | 85 (44) | .15 |
| Ablative | <.01 | |||
| Myeloablative | 15 (88) | 43 (83) | 129 (66) | |
| Non-myeloablative | 2 (12) | 9 (17) | 66 (34) | |
| HLA Match | ||||
| Matched, related | 7 (41) | 22 (42) | 56 (29) | .08 |
| Matched, unrelated | 5 (29) | 14 (27) | 78 (40) | |
| Mismatched, unrelated | 5 (29) | 16 (31) | 61 (31) |
NOTE. HLA, human leukocyte antigen; MDS, myelodysplastic syndrome; TBI, total body irradiation.
Patients had ≥1 study sample that was toxin-B positive during the peri transplant (−10 to +40) period but did not have any clinical episodes of C. difficile during that period.
Patients had ≥1 clinical episode of C. difficile during the peri transplant period (−10 to +40) irrespective of positivity (or toxin B positivity) of study samples.
P values based on χ2 and Fisher exact test.
Relatedness by MLST in the Peri Transplant Cohort
Overall, among the 69 patients (26.1%) who had detectable C. difficile in a stool sample in the peri transplant cohort, samples from 66 patients were successfully genotyped by MLST, including those with C. difficile colonization (n =17) and infection (n= 49). A total of 24 MLST types (STs) were isolated from this cohort. The most frequent types were ST-2 (n= 13) followed by ST-42 (n =7). ST-1, which correlates with the NAP1 strain, was isolated from a single patient. Multiple samples positive for toxigenic C. difficile from the same patient yielded identical MLST types. One person with C. difficile colonization detected on day −1 subsequently developed clinical CDI on day +31 with a discordant strain. For the remaining patients in whom colonization was detected before infection, strains remained identical.
Cases of CDI in The Peri Transplant Period Attributed to Unit-Based Transmission
The overall rate of HO-CDI on the SCT unit during the study period was 3.6 per 1,000 patient-days (Figure 1). To study the contribution of unit-based transmission to incident C. difficile in the peri transplant cohort, we identified all cases of C. difficile on the SCT unit or in persons who had recently been hospitalized on this unit. The schematic for sampling is shown in Figure 2. Overall, 116 patients were identified; 84 (72.4%) were HO-CDI, including 52 cases from the peri transplant cohort described above and an additional 32 patients outside the cohort (other) who were admitted to the unit during the same period. Fifteen (12.9%) of the 116 cases were patients with CO-HCFA C. difficile. These included patients hospitalized on the transplant unit during the study period who were subsequently diagnosed with CDI within 12 weeks after discharge. The remaining 17 cases (14.7%) were the colonized patients from the peri transplant cohort who did not develop active disease.
FIGURE 1.
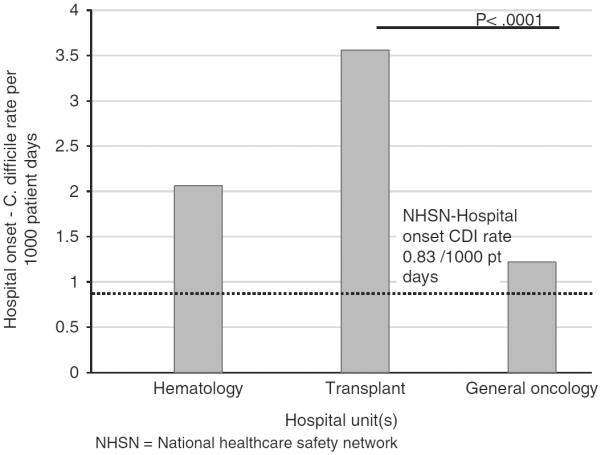
Hospital onset rates of Clostridium difficile infection (CDI) among patients admitted to hematology (leukemia and lymphoma) unit, transplant unit (adult stem cell transplant), and general oncology units (solid tumor). NHSN, National Healthcare Safety Network.
FIGURE 2.
Schematic of sampling to screen for Clostridium difficile (peri transplant) and recognize cases (other) among patients hospitalized on the transplant unit. CDI, C. difficile infection; SCT, stem cell transplant.
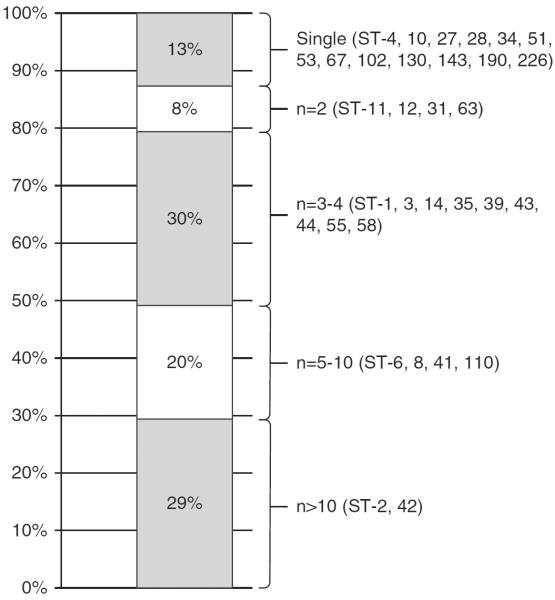
Stool samples from 102 of the 116 patients with CDI or colonization could be retrieved and were successfully genotyped, yielding 32 MLST types. The frequency of ST types is shown in Figure 3. From these 102 isolates, 13 strains (12.7%) were isolated from 1 patient each (Figure 3). For the remaining 89 cases, the temporospatial relationship for shared strains was mapped over the study period as shown in Figure 4.
FIGURE 3.
Frequency of multilocus sequence types isolated from 102 patients admitted to the transplant unit during the study period (October 1, 2010– December 31, 2012). ST, sequence type.
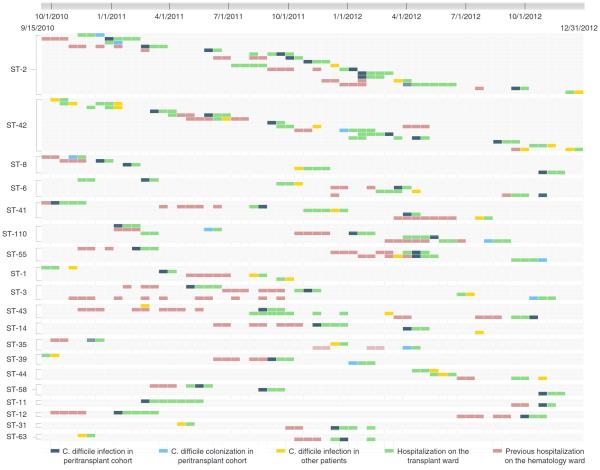
FIGURE 4.
Time-space cluster map of shared sequence types from patients (represented on each row) with Clostridium difficile infection and colonization on the transplant unit (y-axis depicts unique multilocus sequence types). The x-axis (top) shows the duration of the study period; each small box approximates a 2-week interval. Color codes for hospitalization on transplant and leukemia units and timing of C. difficile isolation are shown at the bottom of the figure.
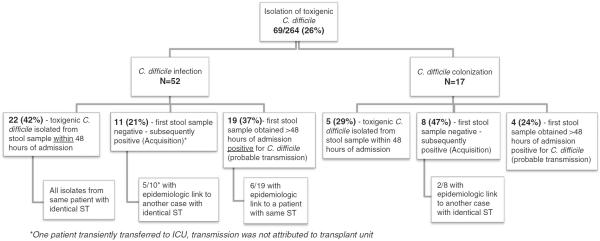
Next, genotyping data was integrated with clinical surveillance for the 69 patients in the peri transplant cohort to determine whether transmission had occurred and to identify possible links between cases (Figures 4 and 5). Twenty-seven (39%) of the 69 patients in the peri transplant cohort had toxigenic C. difficile isolated on the first study sample obtained within 48 hours of admission; these cases were not considered to be acquired on the SCT unit regardless of timing of CDI. Nineteen patients (28%) with toxigenic C. difficile had initially tested negative, strongly suggestive of transmission. In the remaining 23 cases (33%), C. difficile was isolated on the first sample obtained more than 3 calendar days but within 7 days of admission to the unit, with no preceding negative sample. Epidemiologic links could be established in only 6 (9%) of these cases, which were considered as probable transmission.
FIGURE 5.
Flowchart to depict integration of screening for Clostridium difficile, clinical surveillance, and genotyping to characterize preexisting C. difficile colonization, new acquisition, and cases of probable transmission. ICU, intensive care unit; ST, sequence type.
Transmission from cases of asymptomatic C. difficile colonization was suspected in 2 cases (Figure 4: ST-2; row 1 and ST-42; row 9). Eight cases of HO-CDI and 2 cases of CO-HCFA outside of the peri transplant study population occurred in persons epidemiologically linked with peri transplant cases of CDI (Figure 4: CO-HCFA cases are ST-6; row 4 and ST-42; row 8).
DISCUSSION
Our study elucidates the risk, timing, and dynamics of C. difficile transmission among persons admitted to undergo allogeneic SCT. Overall, by Centers for Disease Control and Prevention/ National Healthcare Safety Network criteria 94% of all cases of CDI among the peri transplant cohort were detected more than 48 hours after hospitalization and considered nosocomial acquisitions on the transplant unit. Yet our data that integrates clinical surveillance and spatial links with genotype from colonized and infected persons with C. difficile found little evidence to attribute cases to transmission on the transplant unit. Only 33% (17/52; 11 definite and 6 probable) of cases with the comprehensive sampling and inclusion of CO-HCFA cases could be explained by hospital-based transmission (Figure 5). Our sampling did not include preadmission screening for C. difficile; however, most persons (88%) were found to be colonized or with CDI before stem cell infusion, suggesting a preexisting nonsymptomatic carrier state.
This study highlights the importance of sampling methods to accurately interpret C. difficile transmission; screening for C. difficile at admission and frequently through the hospital stay is crucial to precisely ascertain connection between cases. Our data indicate that results derived only from genotyping of clinical isolates will erroneously omit, as well as overcall, transmission links.20,21 Among the 52 patients in the peri transplant cohort with clinical CDI, we were able to conclusively establish acquisition in only 11 (21%) [Figure 5]. If stool samples had not been obtained on admission, an additional 38 cases of CDI in this cohort would be considered nosocomial on the basis of clinical surveillance only (total, 49 [94%] of 52 cases). Inferring transmission from genotyping of clinical isolates, as has been done in previous studies, would have suggested transmission in 18 (35%) of the 52 cases.
The high incidence of early CDI, long length of stay with an overall high expected healthcare-related exposure, and frequent use of antibiotics are familiar characteristics among transplant patients. If the findings of our study suggesting lack of C. difficile transmission are replicated in other similar settings, it would justify consideration of screening for C. difficile in this complex and unique population. It is likely that most CDI among transplant candidates is due to healthcare-related exposure; however, a delay in recognition can misdirect surveillance and control efforts. Active screening strategies would allow accurate surveillance of HO-CDI and effective allocation of resources with a system-based approach.
Connectivity between facilities plays an important role in the epidemiology and spread of C. difficile, a phenomenon most prominently recognized as late-onset CDI in patients who transition care between long-term care facilities and acute care hospitals.22–24 Our data support that the epidemiology of CDI in the transplant setting demonstrates a similar pattern—as referrals to a higher and more specialized level of care are made, asymptomatic colonization with C. difficile may already have occurred during the pretransplant phase of illness. These findings have a substantial impact on healthcare-associated infection reporting and prevention as well as interhospital comparisons because more than half of all new admissions for SCT at specialized oncology centers can be external transfers from diverse geographic and healthcare areas.
Our findings also suggest that control strategies on SCT units should have a comprehensive approach to prevent occurrence of active disease in those already colonized and should institute measures to prevent on-unit transmission due to the high colonization pressure. Multifaceted strategies with careful attention to contact precautions (gowns, gloves, and hand hygiene), environmental disinfection, antimicrobial stewardship, and safely administered microbial therapeutics are likely to be more effective than any single intervention alone.
There are several other notable findings, of which the most striking is the lack of occurrence of NAP-1 strain (ST-1), which remains the most prevalent C. difficile strain in the United States. Previous studies at our institution among the same population had demonstrated the dominant presence of NAP-1 among transplant recipients during the years 2006–2008.5 This finding may indicate that new acquisition due to NAP-1 strain may be on a decrease or that transplant recipients do not constitute a substantial reservoir for this strain.25 Overall, the dominant strains in our study population are very similar to what has recently been reported by the Centers for Disease Control and Prevention's EIP.1 We also found transient colonization with toxigenic strains among our patients: 11 of 17 colonized patients did not have a history of CDI and did not develop it during follow-up.
Our study has several important limitations. First, serial sampling was limited to patients admitted for allogeneic transplants only. Autologous transplant recipients and patients later in the transplant course to the same unit were not systematically sampled and we may have missed transmission events originating from colonized persons among these. It is important to note that the incidence of C. difficile in these groups is lower and distributed more evenly over several months. All CO-HCFA and HO-CDI events among these persons were included. We used a standardized fingerprinting approach, albeit with limited discriminatory power, and have potentially overestimated transmission links. Higher resolution fingerprinting tools, such as next-generation sequencing, will eventually be applied to conclusively establish transmission links between cases; however, the technology is not fully standardized yet. We detected significant diversity among C. difficile isolates in our study population using MLST, and we think application of a more discriminatory genotyping method would not change the overall relevance of our findings but would be valuable to confirm or debunk suspected instances of transmission. Environmental surveillance cultures were not performed during this study; however, spatial links were closely examined. Finally, we used culture to screen for C. difficile colonization, not higher-sensitivity methods such as polymerase chain reaction assay; screening of multiple sequential samples improved the overall sensitivity of C. difficile detection. However, owing to this factor and sampling of only 66% of patients within 48 hours of admission, we may have underestimated the occurrence of colonization.
In summary, C. difficile epidemiology is highly complex and population specific. Despite the high nosocomial rates of CDI on the transplant unit during the study period, preexisting C. difficile colonization was found in most cases and only modest evidence of transmission among truly incident cases was detected. Although current surveillance methods are undoubtedly beneficial on a regional and national level, characterizing nosocomial transmission requires a deeper epidemiologic analysis, ideally facilitated by interfacility collaboration, robust sampling, and routine integration of fingerprinting techniques. This information will eventually guide effective practices to categorize cases, control infection, and curtail spread of C. difficile in high-risk settings.
ACKNOWLEDGMENTS
We thank Jennifer Brite, PhD, and Lauren Richardson, BS, for assistance with epidemiologic analysis.
Footnotes
Financial support. None reported.
Potential conflicts of interest. All authors report no conflicts of interest relevant to this article.
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo R, Greenberg A, Stone CD. Outcomes of Clostridium difficile infection in hospitalized leukemia patients: a nationwide analysis. Infect Control Hosp Epidemiol. 2015:1–8. doi: 10.1017/ice.2015.54. [DOI] [PubMed] [Google Scholar]
- 3.Alonso CD, Kamboj M. Clostridium difficile infection (CDI) in solid organ and hematopoietic stem cell transplant recipients. Curr Infect Dis Rep. 2014;16:414. doi: 10.1007/s11908-014-0414-0. [DOI] [PubMed] [Google Scholar]
- 4.Kamboj M, Son C, Cantu S, et al. Hospital-onset Clostridium difficile infection rates in persons with cancer or hematopoietic stem cell transplant: a C3IC network report. Infect Control Hosp Epidemiol. 2012;33:1162–1165. doi: 10.1086/668023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamboj M, Xiao K, Kaltsas A, et al. Clostridium difficile infection after allogeneic hematopoietic stem cell transplant: strain diversity and outcomes associated with NAP1/027. Biol Blood Marrow Transplant. 2014;20:1626–1633. doi: 10.1016/j.bbmt.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinnebrew MA, Lee YJ, Jenq RR, et al. Early Clostridium difficile infection during allogeneic hematopoietic stem cell transplantation. PLOS ONE. 2014;9:e90158. doi: 10.1371/journal.pone.0090158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso CD, Treadway SB, Hanna DB, et al. Epidemiology and outcomes of Clostridium difficile infections in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2012;54:1053–1063. doi: 10.1093/cid/cir1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willems L, Porcher R, Lafaurie M, et al. Clostridium difficile infection after allogeneic hematopoietic stem cell transplantation: incidence, risk factors, and outcome. Biol Blood Marrow Transplant. 2012;18:1295–1301. doi: 10.1016/j.bbmt.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Trifilio SM, Pi J, Mehta J. Changing epidemiology of Clostridium difficile-associated disease during stem cell transplantation. Biol Blood Marrow Transplant. 2013;19:405–409. doi: 10.1016/j.bbmt.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Lessa FC, Gould CV, McDonald LC. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis. 2012;55:65–70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang AM, Marini BL, Frame D, Aronoff DM, Nagel JL. Risk factors for recurrent Clostridium difficile infection in hematopoietic stem cell transplant recipients. Transpl Infect Dis. 2014;16:744–750. doi: 10.1111/tid.12267. [DOI] [PubMed] [Google Scholar]
- 12.Blot E, Escande MC, Besson D, et al. Outbreak of Clostridium difficile-related diarrhoea in an adult oncology unit: risk factors and microbiological characteristics. J Hosp Infect. 2003;53:187–192. doi: 10.1053/jhin.2002.1356. [DOI] [PubMed] [Google Scholar]
- 13.Hu C, Sunday R, Bruminhent J, et al. Investigation of a Clostridium difficile cluster by multilocus sequence typing in a bone marrow transplant unit. Am J Infect Control. 2014;42:691–693. doi: 10.1016/j.ajic.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Walker AS, Eyre DW, Wyllie DH, et al. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLOS Med. 2012;9:e1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babady NE, Stiles J, Ruggiero P, et al. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol. 2010;48:4519–4524. doi: 10.1128/JCM.01648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald LC, Coignard B, Dubberke E, et al. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. 2007;28:140–145. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 19.Sethi AK, Al-Nassir WN, Nerandzic MM, Bobulsky GS, Donskey CJ. Persistence of skin contamination and environmental shedding of Clostridium difficile during and after treatment of C. difficile infection. Infect Control Hosp Epidemiol. 2010;31:21–27. doi: 10.1086/649016. [DOI] [PubMed] [Google Scholar]
- 20.Curry SR, Muto CA, Schlackman JL, et al. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis. 2013;57:1094–1102. doi: 10.1093/cid/cit475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre DW, Cule ML, Wilson DJ, et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med. 2013;369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerrero DM, Nerandzic MM, Jury LA, Chang S, Jump RL, Donskey CJ. Clostridium difficile infection in a Department of Veterans Affairs long-term care facility. Infect Control Hosp Epidemiol. 2011;32:513–515. doi: 10.1086/659765. [DOI] [PubMed] [Google Scholar]
- 23.Han A, Jump RL. Discrepancies between surveillance definition and the clinical incidence of Clostridium difficile infection in a Veterans Affairs long-term care facility. Infect Control Hosp Epidemiol. 2014;35:1435–1436. doi: 10.1086/678434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45:992–998. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 25.Black SR, Weaver KN, Jones RC, et al. Clostridium difficile outbreak strain BI is highly endemic in Chicago area hospitals. Infect Control Hosp Epidemiol. 2011;32:897–902. doi: 10.1086/661283. [DOI] [PubMed] [Google Scholar]