Significance
Pectin and alginate are polysaccharides found in the cell walls of plants and brown algae, respectively. These polysaccharides largely consist of chains of uronates, which can be metabolized by bacteria through a pathway of enzymatic steps to the key metabolite 2-keto-3-deoxygluconate (KDG). Understanding the metabolism of these sugars is important because pectin degradation is used by many plant-pathogenic bacteria during infection, and both pectin and alginate represent abundant sources of carbohydrate for the production of biofuels. Here we demonstrate that KdgF, a protein of previously unknown function, catalyzes the linearization of unsaturated uronates from both pectin and alginate. Furthermore, we show that KdgF contributes to efficient production of KDG and a bacterium’s ability to grow on uronates.
Keywords: uronate, ring opening, tautomerization, pectin, alginate
Abstract
Uronates are charged sugars that form the basis of two abundant sources of biomass—pectin and alginate—found in the cell walls of terrestrial plants and marine algae, respectively. These polysaccharides represent an important source of carbon to those organisms with the machinery to degrade them. The microbial pathways of pectin and alginate metabolism are well studied and essentially parallel; in both cases, unsaturated monouronates are produced and processed into the key metabolite 2-keto-3-deoxygluconate (KDG). The enzymes required to catalyze each step have been identified within pectinolytic and alginolytic microbes; yet the function of a small ORF, kdgF, which cooccurs with the genes for these enzymes, is unknown. Here we show that KdgF catalyzes the conversion of pectin- and alginate-derived 4,5-unsaturated monouronates to linear ketonized forms, a step in uronate metabolism that was previously thought to occur spontaneously. Using enzyme assays, NMR, mutagenesis, and deletion of kdgF, we show that KdgF proteins from both pectinolytic and alginolytic bacteria catalyze the ketonization of unsaturated monouronates and contribute to efficient production of KDG. We also report the X-ray crystal structures of two KdgF proteins and propose a mechanism for catalysis. The discovery of the function of KdgF fills a 50-y-old gap in the knowledge of uronate metabolism. Our findings have implications not only for the understanding of an important metabolic pathway, but also the role of pectinolysis in plant-pathogen virulence and the growing interest in the use of pectin and alginate as feedstocks for biofuel production.
Polysaccharides, such as those found as biomass in marine algae and terrestrial plants, comprise a vast sink of photosynthetically fixed carbon and a potentially enormous source of energy to organisms with the appropriate metabolic systems to unlock it. Pectin, a complex polyuronate largely made up of the uronate d-galacturonate connected by α-(1,4) linkages, is one such polysaccharide. The principal reservoir of pectin is the primary cell wall of terrestrial plants and, as such, its degradation and metabolism by microbes is an important component of the natural turnover of biomass, infection by plant pathogens, digestion of dietary fiber in the mammalian gut, and the utilization of biomass as feedstocks for biofuels or other valuable products (1). Similarly, alginate, another polyuronate that comprises β-linked d-mannuronate and l-guluronate (in varying lengths and arrangements), is a major cell wall component of brown macroalgae (seaweed). Alginate can comprise up to 40% of the dry weight of the algae, making the recycling of this photosynthetically fixed carbon a significant aspect of the ocean carbon cycle. The prevalence of both polysaccharides in biomass has made them important considerations in the production of ethanol from marine algae and terrestrial plant feedstocks (e.g., refs. 2–4).
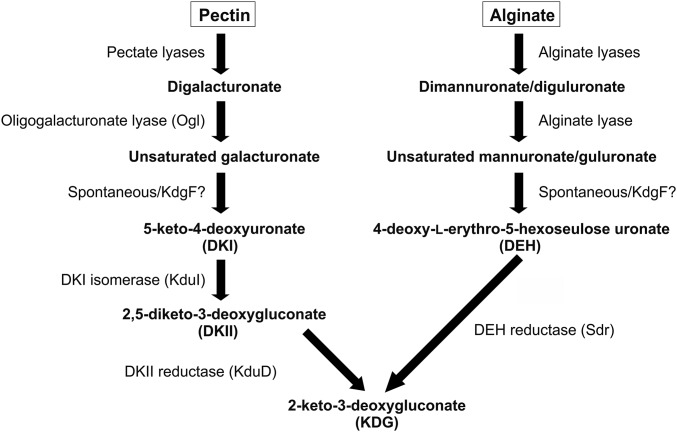
The microbial pathways responsible for the metabolism of pectin and alginate are considered well understood and essentially parallel, with both resulting in the production of the key metabolite 2-keto-3-deoxygluconate (KDG) (Fig. 1). For both polysaccharides, the typical process is initiated by endoacting polysaccharide lyases (PLs), which cleave the polysaccharide backbone using a β-elimination mechanism, thus producing oligouronates with 4,5-unsaturated nonreducing ends. After being imported into the microbe, these products are completely degraded to 4,5-unsaturated monouronates by oligouronate-specific PLs (1). In pectinolytic organisms, the conversion of 4,5-unsaturated galacturonate (ΔGalUA) to KDG occurs via three steps and the two intermediates 5-keto-4-deoxyuronate (DKI) and 2,5-diketo-3-deoxygluconate (DKII). The final two steps are catalyzed by an isomerase (KduI) and a reductase (KduD) (5). In alginolytic organisms, 4,5-unsaturated mannuronate (ΔManUA) and guluronate (ΔGulUA) are converted to KDG via two steps and the intermediate 4-deoxy-l-erythro-5-hexoseulose uronate (DEH), with the second step catalyzed by DEH reductase (4) (Fig. 1). The two pathways converge at KDG, which in turn is processed into pyruvate and 3-phosphoglyceraldehyde via the Entner–Doudoroff pathway, ultimately providing energy in a form an organism can readily use.
Fig. 1.
Schematic depiction of the major steps in the degradation of unsaturated uronates from pectin and alginate.
On the basis of these well-defined metabolic pathways, genes encoding the hallmark enzymes of pectinolytic and alginolytic pathways can be easily identified in a large number of microbial genomes. A general comparison of these predicted enzymes, which are often encoded at a single distinct genomic locus, reveals the consistent cooccurrence and colocalization of a small ORF called kdgF in both pectinolytic and alginolytic systems (SI Appendix, Fig. S1) (6). kdgF was initially identified within the pectin degradation locus of Dickeya dadantii (formerly Erwinia chrysanthemi) where it was observed to be regulated by the pectin degradation repressor protein KdgR (5). Although disruption of kdgF in D. dadantii did not prevent growth on polygalacturonate (PGA), it did result in lower induction of pectate lyases by PGA and reduced maceration of potato tubers. Thus, KdgF is now annotated as a cupin-like protein involved in pectin degradation (InterPro IPR025499). The specific role of KdgF in pectin metabolism has not been uncovered, nor has the reason for the occurrence of KdgF homologs in alginate-processing loci been addressed.
Examination of the pectin and alginate metabolism pathways reveals that a known enzyme catalyzes each step, with the exception of the conversion of ΔGalUA and ΔManUA/ΔGulUA into DKI and DEH, respectively (Fig. 1). The initial conversion of the 4,5-unsaturated monouronates is assumed to occur spontaneously and proceed by opening of the pyranose ring followed by an enol–keto tautomerization (4, 7–9). The timescales of monosaccharide mutarotation (which include a ring opening) and known enol–keto tautomerizations are on the order of minutes to hours (10–12), making the likely timescale of the proposed spontaneous conversion of 4,5-unsaturated monouronates too large to be feasibly incorporated into an efficient metabolic pathway. Given that KdgF is the only conserved component of pectinolytic and alginolytic pathways without a known function, we hypothesized that this protein in fact catalyzes the generation of DKI or DEH. In this study, we use a combination of enzyme activity assays, X-ray crystallography, mutagenesis, and NMR to demonstrate that KdgF from both pectinolytic and alginolytic loci catalyzes the initial conversion step of 4,5-unsaturated monouronates. Furthermore, we show that KdgF from a pectinolytic system contributes to efficient production of KDG in a reconstituted in vitro pathway and in the context of microbial growth on PGA in vivo.
Results
KdgF Catalyzes Double Bond Depletion in Unsaturated Monouronates.
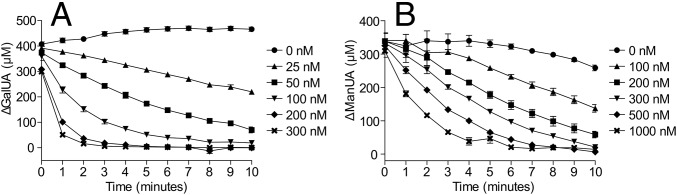
For this study, we initially focused on the pectinolytic locus from Yersinia enterocolitica (SI Appendix, Fig. S1), which is one of the most thoroughly structurally and functionally characterized pectinolytic systems, with only the function of the KdgF component (YeKdgF) being unknown (e.g., refs. 1, 13, and 14). As the predicted substrate of KdgF spontaneously converts at a significant rate, we could not assay the activity of recombinant YeKdgF on ΔGalUA directly. Instead, a linked assay was used where the oligogalacturonate lyase from Y. enterocolitica (YeOgl) was incubated with digalacturonate [α-d-GalUA-(1–4)-d-GalUA] to produce 1 mole of ΔGalUA per mole of digalacturonate (SI Appendix, Fig. S2). The formation of ΔGalUA by YeOgl can be monitored by absorbance of the newly formed 4,5-double bond of the enol product at 230 nm (13). Thus, we reasoned that the predicted production of the linearized ketone tautomer of the monosaccharide by YeKdgF could be monitored by a decrease in the absorbance after addition of the protein. Indeed, the addition of YeKdgF to the samples after the YeOgl reactions had reached equilibrium (judged by a stable absorbance reading) resulted in a rapid decrease in the A230nm in a manner that was dependent upon the concentration of YeKdgF and, therefore, consistent with the catalyzed depletion of the double bond in ΔGalUA (Fig. 2A). In the presence of 200 nM YeKdgF, the catalyzed rate of double bond depletion was ∼30-fold greater than the fastest spontaneous rate observed.
Fig. 2.
Activity of YeKdgF and HaKdgF on unsaturated uronates. Double bond depletion in unsaturated monouronates in the presence of YeKdgF or HaKdgF was followed at 230 nm using a lyase-coupled assay (SI Appendix, Fig. S2). (A) Concentration-dependent activity of YeKdgF on ΔGalUA. (B) Concentration-dependent activity of HaKdgF on ΔManUA. Error bars, where visible, represent the SEM (n = 3).
To assess the activity of KdgF on ΔManUA, we performed a parallel experiment using dimannuronate [β-d-ManUA-(1–4)-d-ManUA], the oligoalginate lyase Alg17c from Saccharophagus degradans (15) to produce ΔManUA and HaKdgF from the alginate processing locus of a Halomonas sp. isolated from brown algae (SI Appendix, SI Methods). HaKdgF also catalyzed double bond depletion in a concentration-dependent manner; however, slightly higher concentrations of enzyme were required (Fig. 2B).
Thus, YeKdgF and HaKdgF are both able to convert the products of their respective upstream enzymes. These enzymes share >50% amino acid sequence identity, whereas ΔGalUA and ΔManUA differ only by being C2 epimers, suggesting that the enzymes may be cross-specific. Therefore, we tested the activity of YeKdgF and HaKdgF on the products of Alg17c and YeOgl, respectively, which represent their nonnatural substrates. Both enzymes were able to catalyze double bond depletion in their alternative substrate and have roughly equivalent abilities to do so (SI Appendix, Fig. S3). Due to the nature of the lyase-coupled assay, we were unable to determine Michaelis–Menten parameters for the different enzyme–substrate combinations; however, comparison of initial velocities estimated from curves generated under the same experimental conditions suggest that the enzymes have similar activities on each of the substrates and both have a more than threefold preference for ΔGalUA over ΔManUA (SI Appendix, Table S1).
YeKdgF Catalyzes the Conversion of ΔGalUA to DKI.
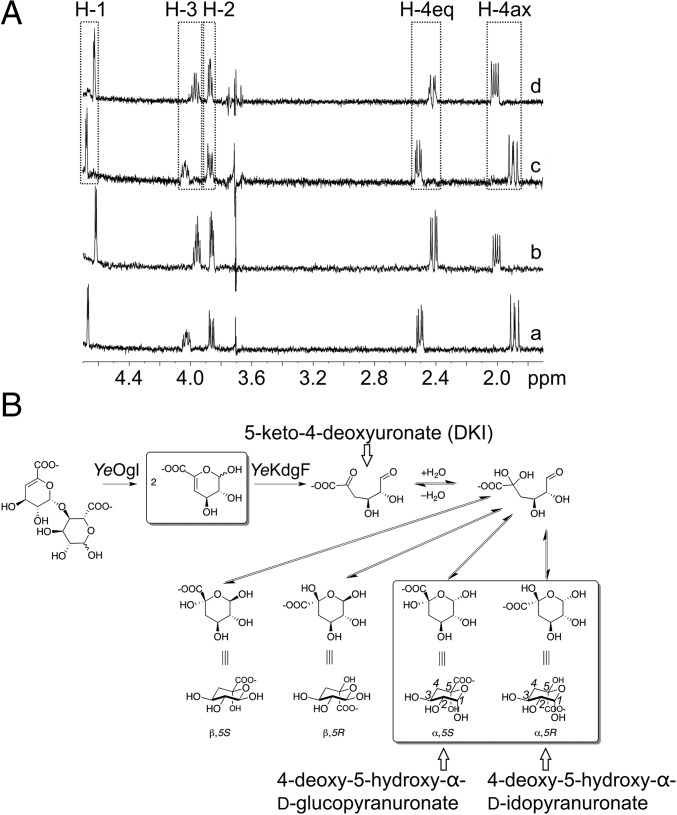
The results of our enzyme assay are consistent with YeKdgF and HaKdgF catalyzing double bond depletion within unsaturated monouronates, which we presume to be a result of the enol–ketone tautomerization that occurs after ring opening. In the case of ΔGalUA, the reported product of this conversion step, and therefore the expected product of YeKdgF, is DKI (Fig. 1). To provide support for this, we used NMR to compare the products of spontaneous conversion of ΔGalUA and the YeKdgF catalyzed reaction (Fig. 3). For this experiment, we used 4,5-unsaturated digalacturonate (ΔGalUA2), as cleavage of this disaccharide by YeOgl produces two molecules of ΔGalUA. An overlay of the 1D selective total correlation spectroscopy (TOCSY) NMR subspectra for ΔGalUA2 treated with YeOgl and YeKdgF, or YeOgl alone, reveals that both reactions produce the same two isomeric products (Fig. 3A). From their resonances, coupling patterns and chemical shifts, we identified these two final products as isomers of a pyranose, closed-ring form of DKI (Fig. 3B and SI Appendix). We propose that formation of pyranose rings occurs after spontaneous hydration of the electron-poor ketone carbonyl before cyclization of the hydrate onto the aldehyde. Two of the four possible diastereomers are formed (Fig. 3B). The small C1(H)–C2(H) coupling constant of 3.7 Hz in both products indicates that they are both α-anomers; no β-anomeric resonances were seen (16). All chemical shifts and coupling constants (SI Appendix, Tables S2 and S3) are consistent with the products being 4-deoxy-5-hydroxy-α-d-glucopyranuronate and 4-deoxy-5-hydroxy-α-d-idopyranuronate.
Fig. 3.
One-dimensional selective TOCSY NMR determination of the product of YeKdgF. (A) One-dimensional selective TOCSY NMR subspectra of the products produced from ΔGalUA2 by the action of either YeOgl and YeKdgF (a and b) or YeOgl alone (c and d). In both cases, two products were observed: 4-deoxy-5-hydroxy-α-D-idopyranuronate (a and c) and 4-deoxy-5-hydroxy-α-D-glucopyranuronate (b and d). Coupling constants for the two observed products and detailed explanation of structural assignments are given in SI Appendix. Dashed boxes indicate the peaks for H-1, H-2, H-3, H-4 axial, and H-4 equatorial, which correspond to the carbon numbering shown on the two observed products in B. (B) Proposed reaction scheme for the action of YeOgl and YeKdgF upon ΔGalUA2 under NMR observation. The spontaneous and YeKdgF-catalyzed ring opening is followed by two rapid steps: hydration of the electron-poor keto-acid carbonyl and ring closure to form the two pyranose α-anomers shown. No evidence for β-anomeric products is present. The boxed species are observed directly by NMR.
KdgF Is Important to KDG Production and Growth on Polygalacturonate.
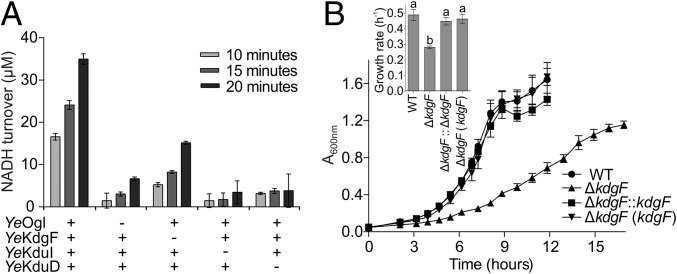
Our data are consistent with the production of DKI by YeKdgF from the product of YeOgl catalysis. In the Y. enterocolitica system, the YeKdgF-catalyzed generation of DKI would provide the substrate for downstream processing by the isomerase YeKduI and the NADH-dependent reductase YeKduD into KDG (5). To test this, we established a coupled assay in which the oxidation of NADH by YeKduD is linked to the sequential processing of digalacturonate by YeOgl, YeKdgF, and YeKduI (Fig. 4A). When all four enzymes were present, we observed time-dependent turnover of NADH. The omission of YeKdgF resulted in a significant decrease in NADH turnover at each time point (P = 0.0201, paired t test, n = 3) of 57–69%. In comparison, omission of either YeOgl, YeKduI, or YeKduD resulted in an 81–93% decrease in NADH turnover. The oxidation of NADH in the absence of KdgF is consistent with the spontaneous production of DKI, whereas the dramatic increase of NADH turnover in the presence of YeKdgF reveals the important role of this protein in generating DKI and increasing flux through the reconstituted pathway.
Fig. 4.
KdgF contributes to KDG production and growth on polygalcturonate. (A) Linked assay in which the production of KDG (and concomitant oxidation of NADH) is dependent on the sequential processing of digalacturonate by YeOgl, YeKdgF, YeKduI, and YeKduD. Data shown indicate the effect of omitting one of the four enzymes. Error bars represent the SEM (n = 3). (B) ΔkdgF mutant of E. coli ATCC 25922 displays a reduced growth rate on PGA compared with wild-type and complemented strains (either kdgF reinserted into the genome or provided on a plasmid). Error bars represent the SEM (n = 4). The Inset bar chart indicates the mean growth rate determined for each strain from the growth curves shown. Lowercase letters above bars indicate statistically significant differences between means as determined by a one-way ANOVA with Tukey’s test (P ≤ 0.01).
The positive effect of YeKdgF on the rate of KDG production suggests that KdgF would contribute toward efficient pectin catabolism in vivo. Most strains of Escherichia coli cannot grow on PGA as the sole carbon source as they contain only a subset of the uronate utilization genes found in Y. enterocolitica (6). However, some strains, including American Type Culture Collection (ATCC) 25922, contain an additional uronate utilization locus, which includes kdgF (SI Appendix, Fig. S1). We produced KdgF from ATCC 25922 (EcKdgF) recombinantly and confirmed that it catalyzes conversion of ΔGalUA in vitro (SI Appendix, Fig. S4). We then tested the ability of ATCC 25922 to grow on PGA and found that it could do so if the medium was supplemented with the endoacting polygalacturonate lyase YePL2A (14) to promote generation of oligogalacturonides (examination of the genome of ATCC 25922 reveals that it lacks its own endoacting PL). A marker-less kdgF deletion mutant of ATCC 25922 displayed a >40% reduction in growth rate on YePL2A-treated PGA compared with wild-type ATCC 25922 and complemented strains (Fig. 4B; 0.49⋅h−1 for wild type vs. 0.28⋅h−1 for ΔkdgF, P ≤ 0.001, one-way ANOVA with Tukey’s test). There was no statistically significant difference in lag time between the strains under these conditions (P > 0.05, one-way ANOVA with Tukey’s test). This decrease in growth rate is consistent with our in vitro coupled enzyme data where in the absence of KdgF, KDG production was not completely abolished but was decreased to a basal level of spontaneous ΔGalUA conversion.
X-Ray Crystallographic Analysis of KdgF.
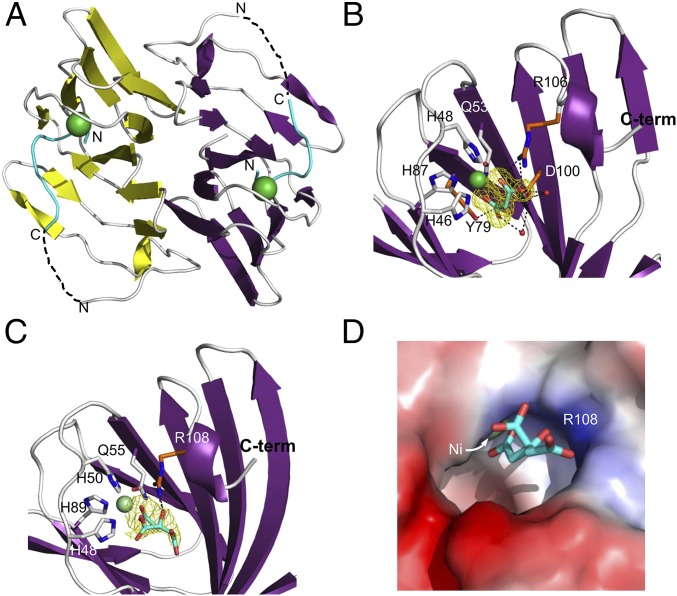
To provide insight into the molecular details behind the activity of KdgF, we determined the X-ray crystal structures of YeKdgF and HaKdgF. The initial crystal form of YeKdgF in the spacegroup P212121 yielded diffraction data to 1.5-Å resolution and the structure was solved by molecular replacement to give a model with two molecules of the protein in the asymmetric unit. Each YeKdgF monomer adopts the canonical β-barrel cupin fold (Fig. 5A). The two molecules of YeKdgF modeled in the asymmetric unit were related by C2 noncrystallographic symmetry (Fig. 5A) and, as determined by PISA (Proteins, Interfaces, Structures and Assemblies) analysis, associated via an extensive surface comprising 1689 Å2 of interface area, 14 hydrogen bonds, six salt bridges, and a predicted binding energy of −30 kcal/mole, indicating this is the biological assembly for YeKdgF.
Fig. 5.
X-ray crystal structures of YeKdgF and HaKdgF. (A) Cartoon representation of the YeKdgF dimer (PDB 5FPX). Dashed lines approximate what is a likely disordered and therefore unmodeled region. (B) Structure of YeKdgF in spacegroup P3221 showing the ordered C terminus and bound malonate molecule (PDB 5FPZ). (C) Structure of HaKdgF with a citrate molecule adjacent to the metal atom (PDB 5FQ0). In B and C the bound ligands (light blue sticks) and their Fo–Fc electron density maps (contoured at 3σ; yellow mesh) are shown. Residues coordinating the bound metal atom (gray sticks) and interacting with the bound ligands (orange sticks) are shown. (D) The solvent accessible surface of the KdgF metal binding site, as represented by HaKdgF in complex with citrate. In A–D, a bound metal that was modeled as a Ni2+ atom is shown (green sphere).
The active site of each monomer contained a metal atom coordinated by three histidines and a glutamine. Analysis of YeKdgF crystals by X-ray fluorescence excitation yielded emission peaks at 7.5 keV and 8.3 keV, which match most closely the KL and KM edges of Ni (SI Appendix, Fig. S5A). A multiwavelength anomalous dispersion (MAD) scan of the crystals at the nickel edge revealed anomalous scattering from nickel, and anomalous difference maps from data collected at the peak wavelength for anomalous scattering by this metal localized the anomalous signal to the metal atom associated with the protein (SI Appendix, Fig. S5 B and C). Furthermore, the refined coordination bond geometry and distances were consistent with Ni2+, as was the refined B factor when the atom was modeled as a Ni2+.
The N terminus of a sequence corresponding to the six-histidine tag used for purification of YeKdgF was found associated with the metal atom (SI Appendix, Fig. S6), likely resulting in the C-terminal sequence of KRDDFL being disordered. We explored an alternate crystal form obtained in the spacegroup P3221, which contained a single monomer in the asymmetric unit; a dimer generated by the crystallographic twofold axis was the same as that observed in the P212121 crystal form. This structure revealed an ordered and complete C terminus that flanks the metal binding site (Fig. 5B). Electron density for a malonate molecule derived from the tacsimate in the crystallization condition was found adjacent to the metal atom. The modeled malonate was bound by the metal atom, R106, and water mediated interactions with Y79 and D100 (Fig. 5B).
We also solved the structure of HaKdgF to 2.35-Å resolution. The protein crystallized with two dimers of HaKdgF in the asymmetric unit, with each dimer having a root mean square deviation of 0.5 Å with the YeKdgF dimer over all of the Cαs (SI Appendix, Fig. S7). Electron density consistent with a citrate molecule derived from the crystallization condition was found adjacent to the metal atom in each of the monomers (Fig. 5C). Unlike the malonate in YeKdgF, where each of the carboxylate groups interacted with the metal atom, only a single carboxylate of the citrate in HaKdgF was coordinated by the metal. R108 made a hydrogen bond with the central hydroxyl group of the citrate (Fig. 5C).
For both YeKdgF and HaKdgF, the metal binding sites sit at the bottom and to the side of a distinct pocket in the surface of the protein, with the malonate and citrate molecules accommodated in the pocket (Fig. 5D). The dimensions of the opening to the pocket are ∼10 Å × 10 Å, whereas it has a maximum depth of ∼12 Å, and thus of roughly sufficient size to accommodate binding of either a closed ring uronate (∼6-Å maximum dimension) or open ring uronate (∼8-Å maximum dimension).
Identification of the KdgF Active Site.
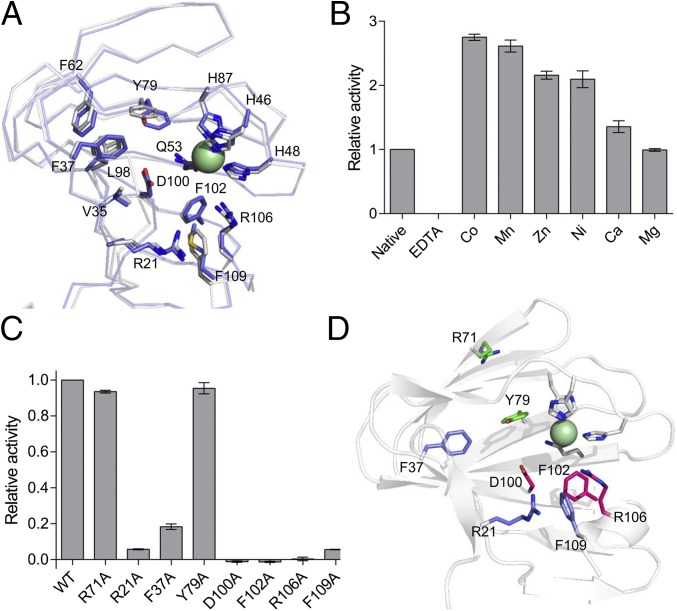
The amino acid residues lining the pocket housing the metal binding site in YeKdgF are highly conserved with HaKdgF (Fig. 6A) and indeed the KdgF family (SI Appendix, Fig. S8). Thus, this pocket of the protein is an obvious candidate for the active site. To probe the role of the metal in catalysis, YeKdgF was depleted of metal and then reconstituted by the addition of exogenous metals and assayed. The enzyme proved to be inactive when metal depleted, whereas several metals, including Ni2+, were able to restore activity to levels higher than the untreated enzyme (Fig. 6B). This points to the importance of the metal in the activity of YeKdgF, whereas the greater activity of the reconstituted enzymes suggests that the untreated enzyme may have had only partial occupation of the metal binding site. Due to the presence of Ni2+ during the immobilized metal affinity chromatography purification of YeKdgF, we are unable to say whether Ni2+ is the biological metal, although it is clear the enzyme can function when bound to several different metal ions.
Fig. 6.
Metal dependence of YeKdgF and identification of the active site. (A) Overlay of YeKdgF and HaKdgF structures showing the conserved residues that line the proposed active site pocket. Residue numbering corresponds to YeKdgF. (B) Metal dependence of YeKdgF activity. YeKdgF was treated with EDTA and then dialyzed against buffer containing different metals (or no metal). Initial rate of activity is shown relative to native YeKdgF. (C) Catalytic activity of alanine mutants of YekdgF. A number of residues that line the proposed active site pocket were mutated to alanine and the mutant proteins assayed for activity. Initial rate of activity is shown relative to wild type. In B and C, error bars represent the SEM (n = 4). (D) Positons of mutated residues shown on the YeKdgF structure and color coded according to the effect of the alanine substitution on activity: magenta, no activity; light purple, reduced activity; and green, no effect on activity.
We continued analysis of the active site by making alanine substitution mutants of seven residues in YeKdgF that are conserved with HaKdgF and project into the putative active site cleft. An R71A mutation was included as a control, as its position in the structure of YeKdgF would not be expected to affect activity. Of the seven proposed active site mutants, three of them (D100A, F102A, and R106A) displayed no catalysis of double bond depletion (Fig. 6 C and D); reactions were allowed to proceed for at least 30 min but still showed no activity above background (SI Appendix, Fig. S9). Three further mutants (R21A, F37A, and F109A) exhibited reduced activity, whereas the R71A and Y79A mutants displayed activity comparable with wild type (Fig. 6 C and D). All mutant proteins were confirmed to be folded using differential scanning fluorimetry (SI Appendix, Fig. S10). Taken in concert with the metal depletion data, this supports the identification of the cleft housing the metal binding site as the active site of YeKdgF and likely all KdgF proteins.
Discussion
The steps for the enzymatic conversion of ΔManUA acid to KDG were delineated by a series of elegant experiments over 50 y ago (8, 17). Subsequently, these steps have been found to be largely paralleled in the enzymatic conversion of ΔGalUA acid to KDG (9, 18). However, the assumption has been that bacteria rely on spontaneous conversion of the 4,5-unsaturated monouronate to DKI or DEH (4, 8, 9); yet this does not seem to be the case. In vitro, both YeKdgF and HaKdgF were able to deplete the 4,5-unsaturated monouronate products of oligouronate lyase reactions, and the reaction products generated by YeKdgF were determined to have NMR properties consistent with DKI. Including YeKdgF in a reconstituted pathway increased the activity of the final enzyme in the pathway, KduD, which generates KDG, revealing the ability of YeKdgF to produce the DKI substrate of KduI and its overall importance to the efficiency of the pathway. Finally, KdgF contributed to the in vivo capacity of an E. coli strain to grow on enzymatically fragmented polygalacturonate. Thus, the results of our experiments support the conclusion that YeKdgF and HaKdgF, components of pectinolytic and alginolytic pathways, respectively, catalyze the ketonization of 4,5-unsaturated uronates, thereby filling an over 50-y-old gap in the knowledge of these metabolic pathways.
Although the pathways of depolymerizing pectins and alginate to 4,5-unsaturated uronates are procedurally parallel, microbes must deploy multiple enzymes belonging to different carbohydrate-active enzyme families to depolymerize the two different polysaccharides (Fig. 1); interestingly, KdgF is the only component conserved between the pectin and alginate utilization pathways. Bioinformatic analysis of KdgF reveals that it is most prevalent among members of the Proteobacteria, to which Y. enterocolitica and Halomonas sp. belong. However, KdgF homologs are also widely dispersed among other, diverse phyla of bacteria (such as Bacteroidetes, Spirochaetes, and Firmicutes) and some Archaea. The composition and arrangement of pectin and alginate utilization loci can vary greatly between organisms, but kdgF is typically associated with a hallmark enzyme, such as a PL or a KDG-processing enzyme (SI Appendix, Fig. S11). The perpetual occurrence of kdgF in pectinolytic and alginolytic prokaryotes (despite the variety in genomic context), its wide distribution among different phyla of Bacteria and Archaea, and its position as the only conserved component between the pectin and alginate utilization pathways underlines its importance in efficient metabolism of uronates. Intriguingly, there are a small number of organisms, such as the cyanobacterium Chroococcidiopsis thermalis and the archaeon Pyrobaculum aerophilum, whose genomes encode for a predicted KdgF but no known PLs or other enzymes associated with uronate metabolism [according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database] (19), leaving their putative functions unclear.
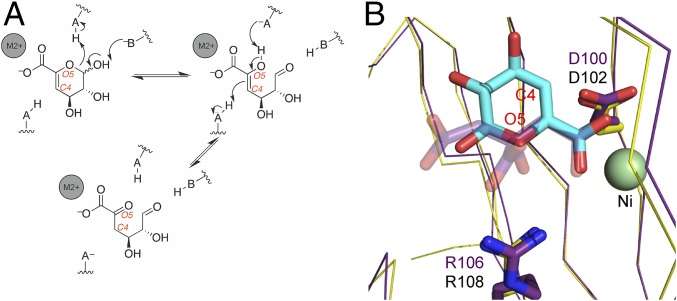
The conversion of 4,5-unsaturated uronates to the ketonized linear form requires an opening of the ring and an enol–keto tautomerization. Given that the spontaneous opening of pyranose rings occurs on the order of minutes, whereas some enol–keto tautomerizations happen on an analogous timescale, it is possible that KdgF catalyzes both the ring opening and the enol–keto tautomerization (10–12). Catalyzed pyranose ring-opening reactions, as exemplified by galactose mutarotase, occur through protonation of the endocyclic O5 and deprotonation of the C1 hydroxyl group by separate amino acid sidechains acting as acid and base catalysts, respectively (20). Thus, by analogy, we propose that KdgF catalyzes a similar ring opening to generate the linear enol form of the 4,5-unsaturated uronates (Fig. 7A). Likewise, we anticipate the potential enol–keto tautomerization catalyzed by KdgF to be similar to that performed by macrophage migration inhibitory factor on phenylpyruvate (21). In our proposed ketonization reaction, the newly formed O5 enol hydroxyl of the linearized sugar would be deprotonated followed by protonation of C4 to generate the final ketone product (Fig. 7A). In this reaction, the role of the divalent metal is most likely to coordinate the substrate carboxylate, thereby stabilizing the interaction and orienting the substrate. The ability of the metal to neutralize the charge of the substrate carboxylate may also play a role in catalysis.
Fig. 7.
Proposed catalytic mechanism of KdgF. (A) Schematic depiction of the two proposed reactions catalyzed by KdgF. The first step is a ring opening, the second an enol–keto tautomerization. Hypothetical acid/base catalysts that would be present on the enzyme are indicated. Gray spheres represent the divalent metal. (B) Overlay of YeKdgF (yellow) with the citrate-bound HaKdgF (purple). ΔGalUA (light-blue sticks) was overlapped with the citrate molecule (transparent purple sticks) via its carboxylate and the nickel-coordinated carboxylate of the citrate to approximate a possible binding mode of ΔGalUA. The proposed aspartate and arginine catalytic residues are shown as sticks.
Some support for the proposed reaction scheme is provided by our structural and mutagenesis studies. Mutagenesis of F102, D100, and R106 revealed these amino acids to be indispensable for the catalytic activity of YeKdgF, with the sidechains of the latter two residues having chemical properties consistent with the ability to act as a general acid and/or base. To approximate the possible initial binding mode of ΔGalUA, we structurally overlapped YeKdgF with HaKdgF and then overlapped the carboxylic acid of ΔGalUA with the metal-bound carboxylic acid of the HaKdgF-bound citrate molecule (Fig. 7B). From this we find that O5 and C4 of ΔGalUA come in proximity (within 3–4 Å) to R106 and D100, respectively, in YeKdgF (R108 and D102 in HaKdgF; Fig. 7B). The alternate orientation of ΔGalUA (i.e., the pyranose ring rotated 180° around the C5–C6 bond) brings O5 and C4 of ΔGalUA in proximity to D100 and R106, respectively. Thus, consistent with their general importance to the catalytic activity of KdgF, the sidechains of these residues may be appropriately positioned relative to key atoms in the substrate to perform as acid and/or base catalysts. However, further work is necessary to detail the participation of these sidechains in the molecular basis of KdgF catalysis. Notably, our proposed mode of uronate binding focuses recognition and catalytic reactions on the C4–C5–C6–O5 portion of the substrate, which is planar by virtue of the sp2-hybridized C5. This conformation would be shared by all 4,5-unsaturated monouronates, perhaps explaining the interchangeable activity of YeKdgF and HaKdgF on the C2 epimers ΔGalUA and ΔManUA and, by extrapolation, giving them postulated activity on the ΔGulUA that also results from alginate depolymerization.
Here we have shown that KdgF is a central component of uronate metabolism and that it contributes to efficient production of the key metabolite KDG from lyase-catalyzed depolymerization of pectin and alginate. Given our ever-increasing demand for energy, and the role that both pectin and alginate have as components of feedstocks for biofuel production, the discovery of the role of KdgF in efficient uronate metabolism has the potential to be quite impactful. A number of studies have focused on the engineering of microbial platforms for efficient ethanol production from alginate but have overlooked the role of KdgF. In a study by Wargacki et al. (4), an alginate utilization locus from Vibrio splendidus was integrated into E. coli and the importance of individual genes assessed by deletion and growth assays. Due to the unknown function of kdgF at that time, the importance of this gene for growth on alginate (and subsequent production of ethanol) was not evaluated. Given the growth defect on PGA that we observed following kdgF deletion, we anticipate that a similar phenotype would be observed with growth on alginate upon deletion of kdgF from the V. splendidus locus. Due to its unknown function, KdgF was also omitted from the engineering of a synthetic yeast platform for ethanol production from brown macroalgae (3); in light of our findings, we would expect its inclusion to increase the efficiency of ethanol production. Finally, our findings explain the decrease in virulence observed upon disruption of kdgF in D. dadantii more than 20 y ago (5). The purpose and environment of uronate metabolism in microbes can vary—from carbon source utilization to virulence, from terrestrial and marine environments to the mammalian gut—but KdgF appears to be a maintained and central component of this pathway.
Materials and Methods
Genes encoding for KdgFs, YeKduI and YeKduD, were amplified from genomic DNA and cloned into pET28a for expression. All proteins, with the exception of Alg17c (16), were expressed and purified as previously described (22). KdgF enzyme assays were performed with 1 mM digalacturonate or dimannuronate, 0.5 µM YeOgl or Alg17c, and varying concentrations of KdgF [an equivalent volume of 20 mM Tris⋅HCl (pH 8.0) was added to the 0-nM KdgF reactions]. Unsaturated uronates were detected at 230 nm and absorbance values converted to micromolar using an extinction coefficient of 5,200 M−1⋅cm−1 (7) following subtraction of the blank (initial absorbance of digalacturonate or dimannuronate). Metal dependence was assayed essentially as described by McLean et al. (22). The linked assay contained 1 mM digalacturonate, 200 µM NADH, and 0.5 µM each enzyme. Activity was followed at 340 nm. 1H NMR spectra were recorded on a Bruker AV-500 spectrometer at 500 MHz with a double presaturation to eliminate the water and Tris⋅HCl resonances. ΔGalUA2 (1 mM) was digested with 1 µM YeKdgF and/or 1 µM YeOgl. When the spectra stabilized (indicating that the reactions had reached completion), 1D selective TOCSY subspectra were obtained. Deletion of kdgF was performed using the plasmid pKOV-unstuff (23). Complementation was achieved either by reinsertion of kdgF at the site of deletion or by providing kdgF in trans. The contribution of kdgF to growth on PGA was assessed in minimal medium containing 0.4% (wt/vol) PGA and 0.5 µM YePL2A. YeKdgF and HaKdgF were crystallized by the hanging drop method following optimization of the crystallization condition. All structures were solved by molecular replacement. Data collection and refinement statistics can be found in SI Appendix, Table S4. (For additional details, see SI Appendix, SI Methods).
Supplementary Material
Acknowledgments
We thank Benjamin Kerr and Satish Nair for providing materials and the Genome Sciences Centre of the British Columbia Cancer Agency for performing the genome sequencing reactions and genome assembly. We thank the staff at the Canadian Light Source (CLS) where diffraction data were collected. The CLS is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. This research was supported by Natural Sciences and Engineering Research Council of Canada Discovery Grant FRN 04355. A.B.B. acknowledges the support of an E. W. R. Steacie Memorial Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.S.T. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5FPX, 5FPZ, and 5FQ0). The sequences reported in this paper have been deposited in the GenBank database (accession nos. KU156827 and KU697803–KU697810).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524214113/-/DCSupplemental.
References
- 1.Abbott DW, Boraston AB. Structural biology of pectin degradation by Enterobacteriaceae. Microbiol Mol Biol Rev. 2008;72(2):301–316. doi: 10.1128/MMBR.00038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards MC, Doran-Peterson J. Pectin-rich biomass as feedstock for fuel ethanol production. Appl Microbiol Biotechnol. 2012;95(3):565–575. doi: 10.1007/s00253-012-4173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enquist-Newman M, et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature. 2014;505(7482):239–243. doi: 10.1038/nature12771. [DOI] [PubMed] [Google Scholar]
- 4.Wargacki AJ, et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012;335(6066):308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 5.Condemine G, Robert-Baudouy J. Analysis of an Erwinia chrysanthemi gene cluster involved in pectin degradation. Mol Microbiol. 1991;5(9):2191–2202. doi: 10.1111/j.1365-2958.1991.tb02149.x. [DOI] [PubMed] [Google Scholar]
- 6.Rodionov DA, Gelfand MS, Hugouvieux-Cotte-Pattat N. Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other gamma-proteobacteria. Microbiology. 2004;150(Pt 11):3571–3590. doi: 10.1099/mic.0.27041-0. [DOI] [PubMed] [Google Scholar]
- 7.Shevchik VE, Condemine G, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of pectin catabolism in Erwinia chrysanthemi 3937. J Bacteriol. 1999;181(13):3912–3919. doi: 10.1128/jb.181.13.3912-3919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preiss J, Ashwell G. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J Biol Chem. 1962;237:309–316. [PubMed] [Google Scholar]
- 9.Preiss J, Ashwell G. Polygalacturonic acid metabolism in bacteria. I. Enzymatic formation of 4-deoxy-L-threo-5-hexoseulose uronic acid. J Biol Chem. 1963;238:1571–1583. [PubMed] [Google Scholar]
- 10.Huang L, et al. Study on the kinetics of keto-enol tautomerism of p-hydroxyphenylpyruvic acid using capillary electrophoresis. J Chromatogr A. 2007;1175(2):283–288. doi: 10.1016/j.chroma.2007.10.078. [DOI] [PubMed] [Google Scholar]
- 11.Wertz PW, Garver JC, Anderson L. Anatomy of a complex mutarotation. Kinetics of tautomerization of α-d-galactopyranose and β-d-galactopyranose in water. J Am Chem Soc. 1981;103(13):3916–3922. [Google Scholar]
- 12.Zhou CC, Hill DR. The keto-enol tautomerization of ethyl butylryl acetate studied by LC-NMR. Magn Reson Chem. 2007;45(2):128–132. doi: 10.1002/mrc.1930. [DOI] [PubMed] [Google Scholar]
- 13.Abbott DW, Gilbert HJ, Boraston AB. The active site of oligogalacturonate lyase provides unique insights into cytoplasmic oligogalacturonate β-elimination. J Biol Chem. 2010;285(50):39029–39038. doi: 10.1074/jbc.M110.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbott DW, Boraston AB. A family 2 pectate lyase displays a rare fold and transition metal-assisted β-elimination. J Biol Chem. 2007;282(48):35328–35336. doi: 10.1074/jbc.M705511200. [DOI] [PubMed] [Google Scholar]
- 15.Park D, Jagtap S, Nair SK. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J Biol Chem. 2014;289(12):8645–8655. doi: 10.1074/jbc.M113.531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jongkees SA, Withers SG. Glycoside cleavage by a new mechanism in unsaturated glucuronyl hydrolases. J Am Chem Soc. 2011;133(48):19334–19337. doi: 10.1021/ja209067v. [DOI] [PubMed] [Google Scholar]
- 17.Preiss J, Ashwell G. Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-L-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-D-gluconic acid. J Biol Chem. 1962;237:317–321. [PubMed] [Google Scholar]
- 18.Preiss J, Ashwell G. Polygalacturonic acid metabolism in bacteria. II. Formation and metabolism of 3-deoxy-D-glycero-2, 5-hexodiulosonic acid. J Biol Chem. 1963;238:1577–1583. [PubMed] [Google Scholar]
- 19.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thoden JB, Kim J, Raushel FM, Holden HM. The catalytic mechanism of galactose mutarotase. Protein Sci. 2003;12(5):1051–1059. doi: 10.1110/ps.0243203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamps SL, Taylor AB, Wang SC, Hackert ML, Whitman CP. Mechanism of the phenylpyruvate tautomerase activity of macrophage migration inhibitory factor: Properties of the P1G, P1A, Y95F, and N97A mutants. Biochemistry. 2000;39(32):9671–9678. doi: 10.1021/bi000373c. [DOI] [PubMed] [Google Scholar]
- 22.McLean R, et al. Functional analyses of resurrected and contemporary enzymes illuminate an evolutionary path for the emergence of exolysis in polysaccharide lyase family 2. J Biol Chem. 2015;290(35):21231–21243. doi: 10.1074/jbc.M115.664847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsey HA, Gallie J, Taylor S, Kerr B. Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature. 2013;494(7438):463–467. doi: 10.1038/nature11879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.