We have read the article “Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade” (1) with interest and were astonished by the high number of genes horizontally transferred into the tardigrade genome. Still, we were surprised by the reported genome size of >200 Mbp, which is in stark contrast to a previously published size of ∼78 Mbp determined by the same group (2).
To investigate this difference, we reestimated the genome size based on the Illumina read datasets k-mer spectra. Averaged, the estimated genome size was (109 18) Mbp and thereby in close agreement with the experimental estimates. Close examination of the k-mer spectra revealed a substantial number of k-mers with a coverage much lower than the true genome peak(s), pointing to a substantial amount of contamination. Because contaminations can impede the assembly process dramatically, we set out to reduce them upfront. First, we identified those k-mers that are present in all Illumina read datasets (trusted k-mers). Next, we extracted all Moleculo reads covered by at least 95% with trusted k-mers. Indeed, only 9.6% of the k-mers were supported by all read datasets. Still, these recovered 90% of the Moleculo dataset, providing an expected genome coverage of 60-fold.
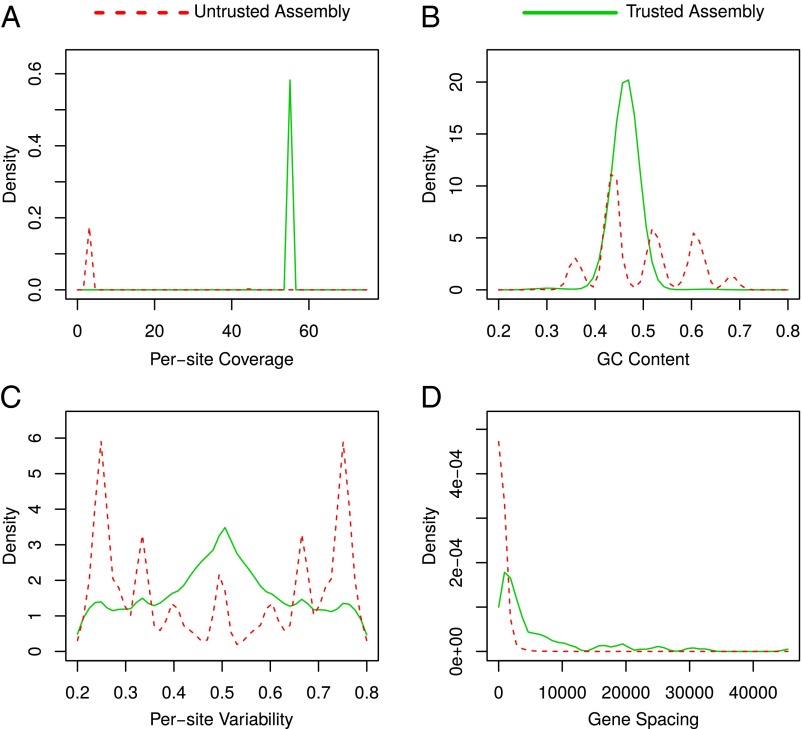
We then assembled the trusted and the untrusted Moleculo reads separately. The trusted dataset assembled into 126 Mbp (N50 17 Kbp), an assembly size that fits with our previous estimates and is in agreement with results of an independent genome project (3). The untrusted dataset resulted in an assembly of 39 Mbp (N50 110 Mbp), showing a suspiciously high number of large contigs (1.1 Mbp to 4.7 Mbp). In total, the untrusted assembly encoded 38,305 genes, of which 5,576 were almost identical (identity ≥99%, expected value ≤1 ) to 3,641 genes predicted by ref. 1. Of those, 2,200 had reciprocal best hits in 1,501 genes that were flagged horizontal gene transfer (HGT)-derived by ref. 1. Comparing structural features revealed that both assemblies are dramatically different in their GC spectra, their per-site coverage, and their per-site variability, as well as their gene spacing (Fig. 1).
Fig. 1.
(A) Per-site coverage of trusted and untrusted assembly based on mappings of Moleculo reads. Contigs from the untrusted assembly generally don’t share the coverage of the trusted, most likely nuclear, genome. (B) GC content of trusted and untrusted assembly estimated using sliding window approach. The untrusted assembly contains multiple peaks pointing toward contig subpopulations with different GC content. (C) Per-site variability of trusted and untrusted assembly, which can serve as ploidy proxy. The untrusted variability spectrum seems distorted and contains a multitude of different peaks, whereas the trusted assembly shows a typical diploid spectrum. (D) Length distribution of intergenetic regions. Intragenetic regions are significantly larger in the trusted assembly than in the untrusted.
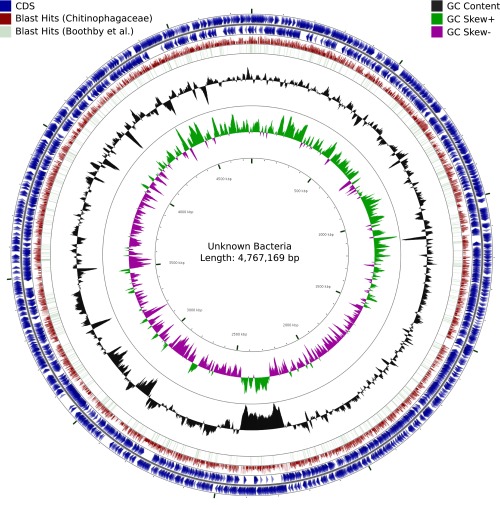
Closer inspection of the largest contigs revealed that they strongly resemble complete bacterial genomes (Fig. 2), with up to 4,783 genes on a single contig. For us, it seems highly unlikely that the genome of Hypsibius dujardini contains continuous parts of bacterial sequences in the size of up to 4.7 Mbp, coding for several thousand genes, with different structural properties than the rest of the eukaryotic genome. Rather, we see this as strong evidence for a dramatic bacterial contamination in the assembly.
Fig. 2.
Circular map of an unknown bacterial genome probably belonging to the Chitinophagaceae drawn with CGView. Tracks 1 and 2 (blue) indicate GeneMark-S annotated genes on forward and reverse strand. Track 3 (red) visualizes regions of homology to a set of 30,844 Chitinophagaceae proteins downloaded from UniProtKB. Track 4 (green) shows homology between GeneMark-S predicted proteins and the published protein set of ref. 1.
Admittedly, bacterial contamination can hardly be avoided when sequencing complete animals. Still, finding noneukaryotic genes in a eukaryotic background does not necessarily point to HGT, especially without a proper quality control of input data and further experimental evidence. We thus suggest that the published high rate of HGT in the genome of H. dujardini is an artifact of sample preparation rather than a biological signal.
Footnotes
The authors declare no conflict of interest.
Data deposition: All scripts, settings, and methodological details are available at https://github.com/greatfireball/hypsibius_genome_revised.
References
- 1.Boothby TC, et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci USA. 2015;112(52):15976–15981. doi: 10.1073/pnas.1510461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabriel WN, et al. The tardigrade Hypsibius dujardini, a new model for studying the evolution of development. Dev Biol. 2007;312(2):545–559. doi: 10.1016/j.ydbio.2007.09.055. [DOI] [PubMed] [Google Scholar]
- 3.Koutsovoulos G, et al. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc Natl Acad Sci USA. 2016;113(18):5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]