Significance
Since the discovery of Mimivirus in 2003, many new giant viruses have been isolated and characterized, now that the definition of viruses excludes their ability to pass through the finest available filters. At least one of these viruses can cause serious infections. Here we describe the structure of faustovirus, the prototypic member of a new family of large double-stranded DNA viruses that are homologs of the Asfarviridae. Contrary to other large viruses, faustovirus has a double-protein shell surrounding the genomic material, something never described before for a DNA virus. In addition, the gene that encodes the major capsid protein shows an unusual organization, stretching over 17,000 bp in the genome while encoding for only 652 amino acids.
Keywords: faustovirus, structure, double-protein shell, major capsid protein
Abstract
Many viruses protect their genome with a combination of a protein shell with or without a membrane layer. Here we describe the structure of faustovirus, the first DNA virus (to our knowledge) that has been found to use two protein shells to encapsidate and protect its genome. The crystal structure of the major capsid protein, in combination with cryo-electron microscopy structures of two different maturation stages of the virus, shows that the outer virus shell is composed of a double jelly-roll protein that can be found in many double-stranded DNA viruses. The structure of the repeating hexameric unit of the inner shell is different from all other known capsid proteins. In addition to the unique architecture, the region of the genome that encodes the major capsid protein stretches over 17,000 bp and contains a large number of introns and exons. This complexity might help the virus to rapidly adapt to new environments or hosts.
The discovery of Mimivirus in 2002 (1) broke with the long-held assumption that viruses are small particles that can be isolated from samples by filtering through small pore sizes. Although larger viruses had been described before, the combination of less stringent selection criteria with new hosts enabled the discovery of a broad range of large viruses that had previously been ignored. Amoebae have been shown to be versatile hosts for the isolation of new viruses from environmental samples. A recent study (2) described the isolation of faustovirus by using a new amoebal host, Vermamoeba verformis, which is associated with the human environment.
Faustovirus shows distant homology to African swine fever virus but probably constitutes its own family within the growing proposed order of Megavirales. The virus has a double-stranded DNA (dsDNA) genome of 466,000 base pairs that encodes 451 predicted ORFs, about two-thirds of which do not have any known homologs. Faustovirus propagates in amoebae and forms a viral factory that produces hundreds of viral particles in an infection cycle that usually lasts about 24 h and ends with the complete lysis of the host cell. The virus forms icosahedrally shaped particles with a diameter of about 2,400 Å.
Here we describe the structure of the mature faustovirus capsid as determined by cryo-electron microscopy (cryoEM). Furthermore, we identified the genetic region encoding the major capsid protein (MCP) and also solved the crystal structure of this protein, generating a pseudoatomic model of the outer shell, which shows features previously not seen in large dsDNA viruses.
Genomic Organization of the Faustovirus MCP Gene
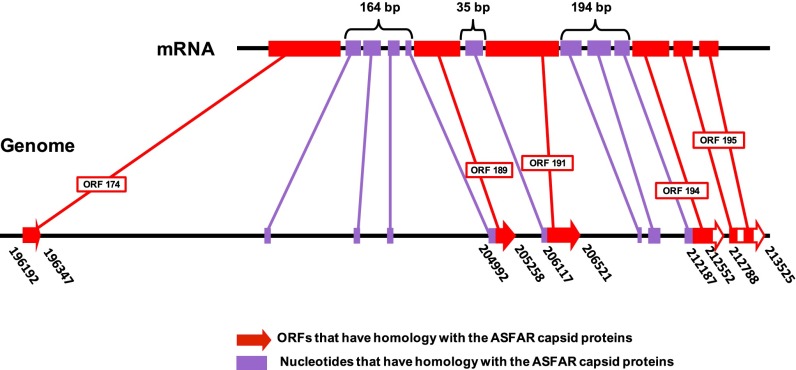
Sequencing and preliminary bioinformatic analysis of the faustovirus genome (2) showed a region of ∼17,000 bp with five putative ORFs that are homologous to parts of the MCP of African swine fever virus. To identify which regions are contributing to the capsid protein of faustovirus, 2D gel electrophoresis was used to isolate the MCP for mass spectrometric analyses. The 645-amino-acid sequence of the MCP was determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-MS), corresponding to an accumulated length of 1,935 bp. The protein sequence was aligned with the originally translated ORFs located among the 17,000 bp-long region that had initially been identified in the genome. The genomic organization of the MCP gene was thus determined. The coding sequence of the MCP has 11 exons and 10 introns (Fig. 1). Exons 1, 5, 6, 9, and 10 and 11 cover the original predicted ORFs 174, 181, 191, 194, and 195, respectively. It is worth noting that the central part of ORF 195 has introns. In addition, eight small fragments between predicted ORFs are transcribed (Fig. 1).
Fig. 1.
Overview of the genomic region in faustovirus that encodes the MCP. The top part shows the final mRNA. The bottom part shows the genomic region, with the predicted MCP ORFs shown in red and additional fragments found in final mRNA in purple. Primers used to sequence additional fragments are listed in Table S1.
Bioinformatic analysis of the genome showed that faustovirus shared a large number of orthologs and protein homologs with asfaviruses (2). However, the architecture of the MCP gene of faustovirus is clearly different from the asfaviruses and other nucleocytoplasmic large dsDNA viruses (NCLDVs).The gene that encodes the MCP in asfarvirus, B646L, is in the form of a single fragment of 1,941 bp (3, 4). In faustovirus, the genomic organization of the MCP gene is novel and, to our knowledge, unique in the virus world. The capsid protein is encoded by 11 exons with large introns (Fig. 1). The presence of introns and exons has been observed in other giant viruses but not in asfaviruses. In Acanthamoeba polyphaga Mimivirus, L425, the gene that encodes the capsid protein, is segmented into three coding sequences that are separated by two introns. These introns are excised during the transcription and/or maturation of the mRNA (5).
Structure of the Faustovirus MCP
The MCP of faustovirus was isolated from purified virions by heat extraction. The protein crystalized in two different crystal forms, both in space group P21 but with different unit cell dimensions. The Matthews coefficient indicated that the asymmetric unit contained six monomers for the smaller crystal form and 12 monomers for the larger one. A self-rotation function showed the presence of two and four threefold peaks, indicating the presence of two and four capsomers in the asymmetric unit, respectively, for the two crystal forms. The structure of the smaller crystal form was solved by molecular replacement using PHASER (6) with the structure of the Paramecium bursaria Chlorella virus 1 (PBCV-1) capsomer as a search model (PDB ID code 1M3Y). The larger crystal form was then solved by molecular replacement using the model from the smaller crystal form as a search model.
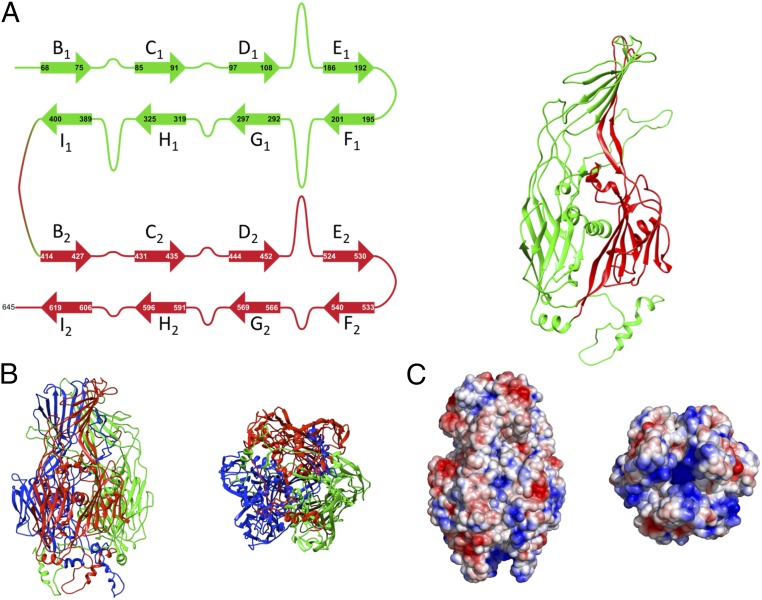
Almost all of the 645 amino acids of the faustovirus MCP were visible in the crystal structures, with the exception of a few amino acids, probably disordered, close to the C terminus. The MCP forms a homotrimer with each monomer consisting of two sequential jelly-roll folds. Similar pseudohexameric capsomer structures can be found in many other viral capsid proteins including adenovirus (7), the phage PRD1 (8), PBCV-1 (9), the virophage Sputnik (10), and the scaffolding protein of vaccinia virus (11, 12). A single jelly-roll fold consists of eight antiparallel β-strands that are named from B to I along the polypeptide sequence. The two sheets that form the opposite sides of a β-barrel consist of strands BIDG and CHEF.
As is the case for other double jelly-roll capsid proteins, the DE and FG loops in both jelly-roll fold domains have large insertions that form the crown of the capsomers and are oriented toward the outside of the virus (Fig. 2). In the faustovirus MCP, this crown is formed by about 200 residues, which form a seven-stranded antiparallel β-sheet with an open half-barrel shape. In addition, two short antiparallel β-strands, connected by a 40 amino acid-long hairpin loop, interlock the seven-stranded β-sheet structures from neighboring monomers at the bottom of the crown structure. The arrangement of the β-strands from the three different monomers gives the crown domain an overall right-handed screw-like structure and is significantly different from the decorations seen in other double jelly-roll capsomers.
Fig. 2.
The faustovirus MCP. (A) A faustovirus MCP monomer is shown with the two jelly-roll domains colored in green and red, respectively. A schematic overview shows the locations of the major insertions that form the crown of the capsid protein. Contrary to many other double jelly-roll proteins, the faustovirus MCP has a large insertion in the H1I1 loop that stabilizes the MCP trimer. (B) Side and bottom view of the faustovirus MCP. The individual monomers are colored in red, green, and blue. The crown domains as well as the helices extending from the bottom of the capsomer are visible. (C) Surface charge representation of the faustovirus MCP colored from blue (–3 kT/e) to red (3 kT/e) and shown in the same orientation as in B. The bottom part of the MCP that is facing the inside of the virus shows a predominantly positive charge, but the helices extending from the bottom carry no dominant charge.
Furthermore, the faustovirus crown is the second largest insertion domain described to date—the biggest being in adenovirus with almost 500 amino acids. A search for comparable domains using secondary structural matching (13) found no structural homologs with known function. The faustovirus MCP crown domain is yet another example of how a common structural fold can be used to accommodate a wide range of structural decorations (7–12). It is currently unclear whether the crown domain that forms the surface of the virus plays a role in host recognition or cell entry.
The N terminus of all known double jelly-roll capsid proteins, including the virus protein VP2–VP3 complex in picornaviruses, is on the inside of the capsomer. The N termini in double jelly-roll structures, including faustovirus, contributes to the stability of the homotrimer by interacting with the nearest counterclockwise neighboring subunit. In addition, the faustovirus MCP has two alpha-helices located close to the N terminus that extend by several Angstrom away from the capsid base. These helices interact with the inner shell at points located around the twofold axis of the virus (Fig. 3C). It is possible that this unique structural feature plays a role in the assembly process of the virus.
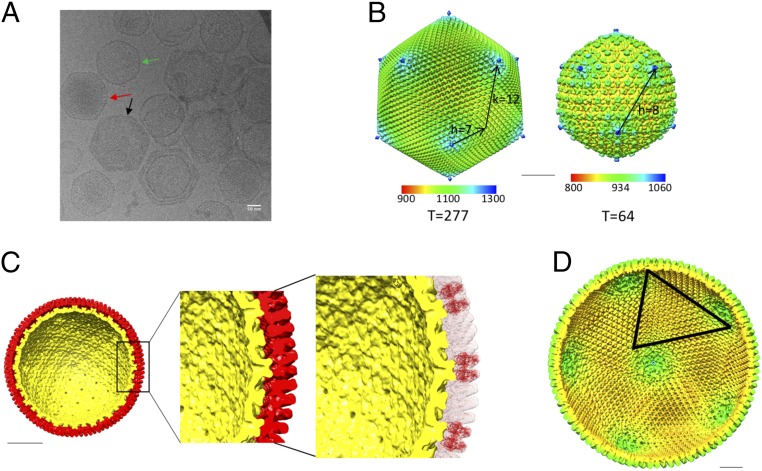
Fig. 3.
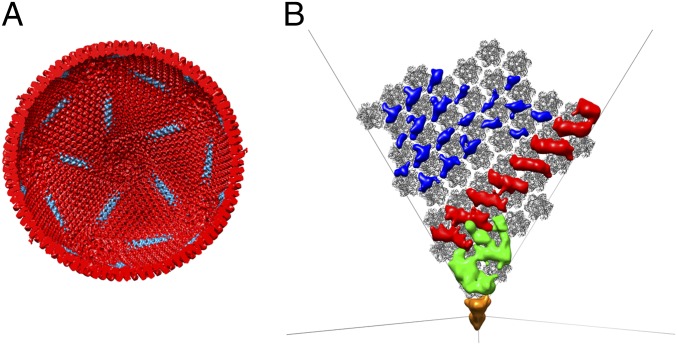
Structure of the faustovirus virion. (A) Micrograph showing the different faustovirus maturation stages. Empty inner shells (green), filled inner shells (red), and complete virions (black) are highlighted with arrows. (B) Reconstructions of the mature virion and the inner shell colored by radial distance (radial color scale bar in Å). Their T numbers including the h and k vectors are indicated. (C) Cross-section viewed down the icosahedral fivefold axis of the reconstructions of the outer shell (red) and the inner shell (yellow). The enlarged area highlights the contact between the inner and the outer shell. A fit of the MCP into the outer shell density shows that the protein interacts with the inner shell at several protrusions. (D) Cross-section viewed down the icosahedral fivefold axis of the outer shell colored by radius. The borders of one trisymmetron are highlighted and correspond to the position of the cement proteins. (Scale bars, 50 nm.)
The capsomers are about 75 Å in diameter, a size that is comparable to many other double jelly-roll capsid proteins, such as PBCV-1 or PRD1. However, the thickness of the capsomer exceeds 120 Å, making it almost twice as thick as the PBCV-1 MCP and placing it en par with the adenovirus hexon protein. This overall size distribution also contributes to the unique shape of the faustovirus virion (Fig. 3).
The surface charge at the bottom of the capsomer is overall positive and comparable to the one seen in PBCV-1, PRD1, and vaccinia virus, all of which have an internal membrane located underneath the MCP. However, the long helices extending underneath the capsid protein have an overall neutral charge, with positive patches on the inside and negative patches on the outside. The extreme tip of the helices in the crystal structure is composed of a mix of uncharged and charged polar amino acids (C33LRAKQGFS41), making it unlikely that this part of the capsid protein is embedded in a membrane.
Virus Structure
CryoEM images of the purified virus showed particles of distinct sizes and shapes (Fig. 3). The larger diameter particles have a spiky appearance with a distinct icosahedral shape, whereas the smaller particles have a rather smooth surface. The larger spiky virions measure about 2,600 Å in diameter, whereas the smaller smooth particles are between 1,600 and 1,900 Å in diameter. The 3D cryoEM reconstructions were determined for both types of particles assuming icosahedral symmetry. These reconstructions had a resolution of 15 Å and 26 Å for the larger and smaller particles, respectively, as determined by the Fourier shell correlation between two independently reconstructed particle subsets. These resolutions are comparable to other reconstructions of large dsDNA viruses, as these viruses are generally not as homogenous as smaller molecular assemblies. In addition, their large size can produce flexibility. This is especially true for the inner capsid, which varies significantly in size. The reconstruction of the larger spiky particles clearly showed additional density located beneath the outer capsid shell, resembling the smaller particles. The raw EM data showed occasionally some broken spiky particles (Fig. 3), indicating that the smaller particles are contained in the larger spiky particles. The results of the reconstruction confirmed that the structure of the small smooth particles is similar to the inner capsid structure found in the larger spiky particles. Thus, faustoviruses are composed of two concentric protein shells.
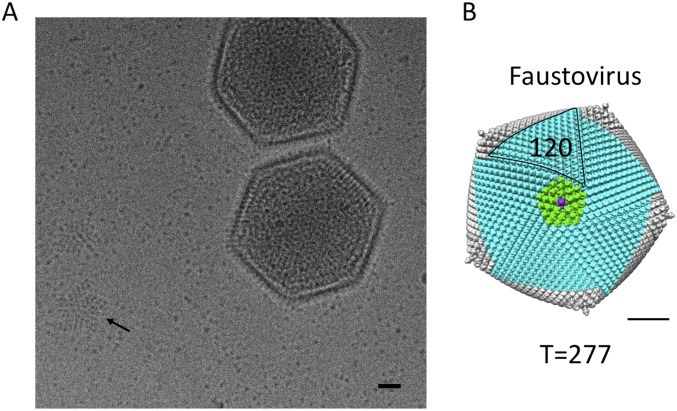
Because of the distinct features of the outer shell and the available high-resolution structure of the MCP, it is clear that the outer shell of the virus corresponds to the abundant fausto virion protein whose structure is reported here. The protein is arranged in an icosahedral fashion with h = 7 and k = 12, giving a triangulation number of 277 (14). Although the organization of the pseudohexagonal capsomeres is consistent with icosahedral symmetry as suggested by Caspar and Klug (14), the capsomer at the fivefold vertices has fivefold, not pseudo-sixfold symmetry. In faustovirus, like in all other known large dsDNA viruses [e.g., PBCV1, Sputnik, Chilo iridescent virus (CIV), PRD1, and more], the capsomere at the fivefold vertices is a different protein, homologous with the MCP but consisting of only a single jelly roll. Therefore, in faustovirus, the icosahedral asymmetric unit is composed of one-fifth of a pentameric capsomer and 46 pseudohexameric capsomers, giving 46 × 6 plus 1 (277) jelly-roll structures in the outer capsid.
As described by Wrigley in 1969 (15), most large dsDNA viruses can be decomposed into 20 trisymmetrons and 12 pentasymmetrons. The organization of the faustovirus outer capsid follows the same principles as can be seen in the 3D reconstruction and in some micrographs of broken particles (Fig. S1). The pentasymmetrons are located around the icosahedral fivefold axes and in faustovirus are composed of one pentameric capsomer and 30 pseudohexameric capsomers. The trisymmetrons are centered around the icosahedral threefold axis and in faustovirus consist of 120 trimers. The orientations of the capsomers within one trisymmetron are the same but differ by 60° between capsomers in neighboring trisymmetrons, giving rise to an easily observable cleavage line between them. The arrangement of penta- and trisymmetrons is comparable to the composition observed in CIV (16), PBCV-1 (9), and the marine algal virus PpV01 (17). All these viruses, including faustovirus, have a pentasymmetron of the same size, but the number of subunits per trisymmetron increases from 55 for CIV, 66 for PBCV-1, and 91 for PpV01 (17, 18) to 120 for faustovirus (Fig. S1). The constant size of the pentasymmetron implies that this size is ideal to relieve the strain induced around the fivefold axis for icosahedral viruses with diameters between about 1,500 Å and 2,600 Å, a hypothesis that has been described for other large viruses (17, 18). The cleavage lines between neighboring tri- and pentasymmetrons are reinforced by additional “cement” proteins (19, 20) that are located underneath the capsid protein (Figs. 3 and 4). Although it is impossible to segment individual proteins in a 15 Å resolution map, the presence of cement proteins has been observed in the higher resolution structures of adenovirus (21), PRD-1 (22), PBCV-1 (23), and CIV (16). Additional densities similar to the cement proteins observed in these viruses are present in faustovirus and presumably serve to maintain the stability of the outer shell.
Fig. S1.
Trisymmetrons and pentasymmetrons in faustovirus. (A) High defocus micrograph of mature faustovirus particles. A pentasymmetron from a destroyed virus particle that is visible in the background is marked with an arrow. (B) Schematic representation of the arrangement of trisymmetrons and pentasymmetrons in faustovirus. The pentasymmetron at one fivefold axis is shown in green, and the central pentamer is highlighted in purple. The trisymmetrons surrounding the pentasymmetron are shown in cyan, and one trisymmetron is highlighted. (Scale bars, 50 nm.)
Fig. 4.
Cement proteins in faustovirus. (A) Cross-section of the outer shell. The approximate area where the cement proteins are located is highlighted in cyan. (B) Difference map between the MCP of faustovirus in one asymmetric unit and the outer shell. The density for the proteins that are located underneath the capsid layer are highlighted in blue, red, and green. The different colors were chosen to group proteins that are likely the same based on their shape and location. Because the central pentamer was not present in the model used to generate the difference map, it is visible in the difference map (orange). The outline of the icosahedron is shown in black lines in the background.
To the best of our knowledge, this is the first time that a large dsDNA virus has been observed to have both an outer and inner protein capsid. Multiple capsid layers can be found in the much smaller dsRNA T = 13 laevo rotaviruses (24, 25) and in phi6 (26). A comparison between the spiky virus reconstruction and the reconstruction of the smooth inner capsid shows that the structure of the inner shell does not change significantly upon addition of the outer capsid layer. The inner protein shell contacts the outer shell at protrusions located on the icosahedral twofold axes of the inner particle. This interaction might be the nucleation point at which the outer shell assembles around the inner shell. Because icosahedral symmetry was applied during the reconstruction, it is not possible to discern if this interaction is unique for one of the twofold axes or if all of the twofold axes are equivalent.
The inner capsid shows a regular array of sixfold hillocks arranged in a hexagonal array within the icosahedral asymmetric unit and smaller protrusions that are located between the major spikes (Fig. 3B). The center-to-center distance between the hexameric hillocks is ∼300 Å. Assuming that the hillock represents one hexameric capsomer, the inner shell of the virus would have T = 16 quasiicosahedral symmetry, with h = 4 and k = 0.
The center-to-center distance between hexamers that form a viral capsid is exceedingly constant from one virus to another. For instance, the center-to-center distance for capsids based on the double jelly-roll structure is 75 Å, whereas the center-to-center distance for capsids assembled from HK97 folds is 140 Å. Thus, the 300 Å center-to-center distance between hexamers in the inner faustovirus capsid cannot be either of these folds and is therefore a structure not previously found in virus capsids.
The inner capsid shell seems to be very flexible. Particles that are filled with dsDNA have a larger diameter in cryoEM images (Fig. 3) in comparison with particles containing no genetic material (Fig. 3). The empty inner core has an average diameter of 1,600 Å and expands to almost 1,900 Å when filled with DNA. This resembles the expansion that can be seen during the assembly and subsequent maturation of HK97 (27), which expands by 100 Å during this process. Furthermore, the assembly process of HSV-1 also involves the structural rearrangement of the pentons and hexons in a T = 16 icosahedral capsid, although without any major expansion of the capsid shell (28).
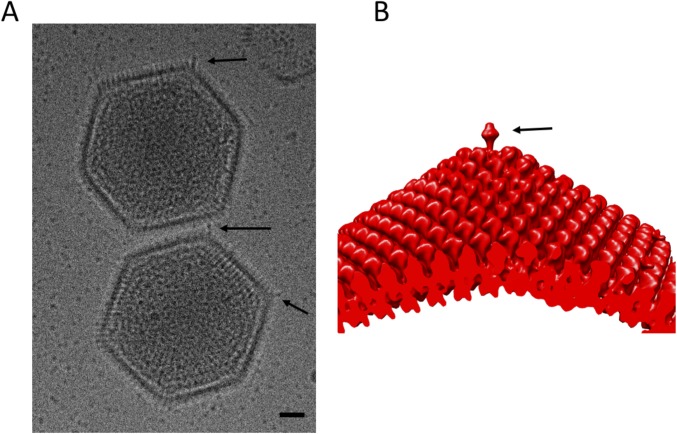
The reconstruction of the spiky particles showed the presence of long, thick fibers that are attached to each of the fivefold vertices (Fig. 3 and Fig. S2). The fibers extend at least 50 Å away from the base of the pentamer but might be significantly longer. Their inherent flexibility makes it difficult to establish the dimensions of these features in the cryoEM reconstruction of the spiky particles. Because icosahedral symmetry was applied during the reconstruction, it is possible that the fibers are not located at all fivefold vertices. However, fibers are visible on multiple vertices in high-defocus micrographs (Fig. S2), implying that the observed fibers are indeed present on most or all fivefold vertices. Apart from the fibers located at the fivefold vertices, the reconstruction also shows the presence of three fibers that are attached to capsomers close to the center of each trisymmetron.
Fig. S2.
Faustovirus fibers. (A) High defocus micrograph of mature faustovirus particles, comparable to the one shown in Fig. S1A. Fibers present on two mature faustovirus particles are marked with arrows. (B) Side view of the reconstruction of the outer capsid shell. The fiber present at the fivefold axis is marked with an arrow. (Scale bar, 50 nm.)
The presence of fibers that decorate the surface of the virus has been observed for other large dsDNA viruses. PBCV-1 has one fiber associated with one specific capsomer per trisymmetron that is probably important for the initial attachment of the virus to host cells (23, 29). CIV has one fiber associated with each capsomer, decorating the whole capsid, but the function of these fibers is currently unknown (16).
Membranes
Many large dsDNA viruses have an internal membrane that surrounds the genomic material and interacts with the capsid protein shell at certain anchor points. In contrast to this, the cryoEM structures of the mature faustovirus virion did not show any density with features that can be attributed to a double-layer membrane either between the outer and inner protein shells or between the inner shell and the genome. The inner capsid touches the outer MCP shell at several positions, leaving no space for any continuous membrane surrounding the inner capsid shell. There are also no indications in the cryoEM reconstructions of an internal membrane inside the inner capsid shell. However, it is possible that such an internal membrane would be too flexible and/or have a different shape than the surrounding material, which would render it invisible in the symmetry-averaged reconstructions. To verify these observations, the purified virus particles were analyzed by MS to detect the presence of lipids. These experiments showed the presence of small but significant amounts of lipids (Figs. S3 and S4) in the sample. However, the amount of lipids is reduced by a factor of 5 in virus preparations harvested from a gradient versus particles that were washed extensively in PBS. This indicates that either small amounts of lipid contamination were still present in the sample after purification or there is an inner membrane that is not visible in the cryoEM reconstructions.
Fig. S3.
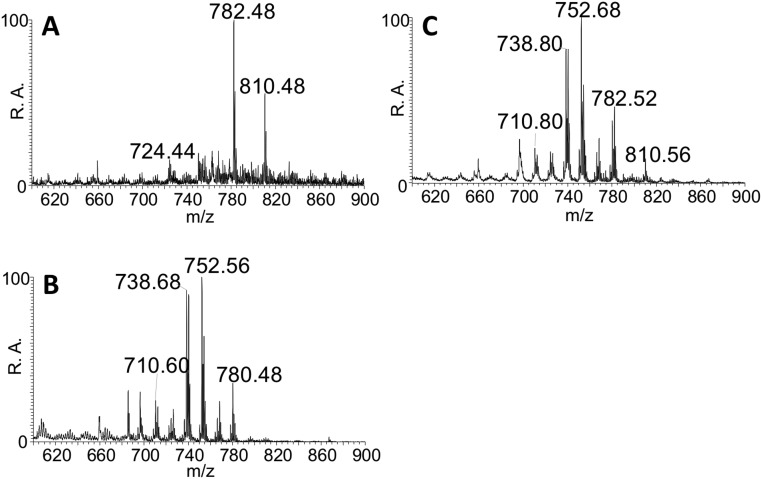
Positive ion mode mass spectra of (A) a 125 µg/mL porcine brain lipid extract, (B) a faustovirus sample, and (C) a solution of 12.5-µg/mL porcine extract and faustovirus analyzed by relay electrospray ionization. All three mass spectra showed the presence of even-numbered peaks in the m/z 700–900 range. Several of the peaks corresponded to phosphotidylcholines as determined by MS/MS.
Fig. S4.
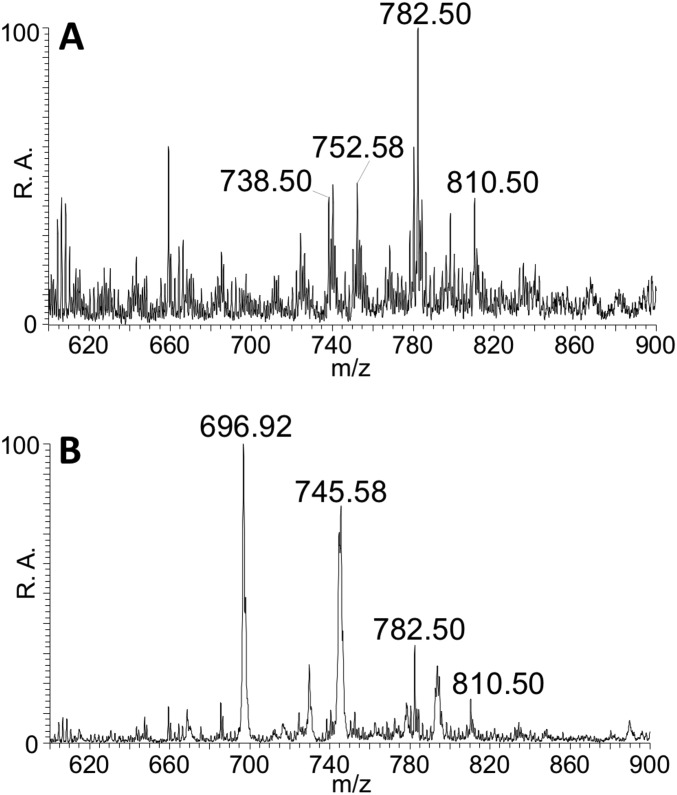
The mass spectra of (A) a filtered faustovirus sample and (B) a filtered faustovirus sample after being run through a density gradient. Both spectra were obtained by relay electrospray ionization in positive ion mode.
Conclusions
The presence of large dsDNA viruses in human environments is likely to not yet have completely recognized consequences. For example, members of the Mimiviridae have been isolated from patients (30, 31), and PBCV-1 has been associated with alterations in human cognitive functions (32). Faustovirus is, to our knowledge, the first large dsDNA virus for which a double-layered protein capsid has been observed. The outer shell is primarily made of a double jelly-roll fold capsid protein, a structural feature that can be found in many other dsDNA viruses. However, the complexity of the associated genome region is unprecedented in what are considered simple organisms like viruses. It is not clear if this organization is beneficial to the virus in exchanging parts of its MCP gene with other available genetic material to adapt itself more rapidly to different environments. The inner shell is constructed of one or more proteins that are unlike other common virus capsids and maybe defines a new structural protein fold. The observation that a major percentage of lipids can be removed after running a faustovirus sample through a density gradient suggests that the detected lipids are more likely to be contaminants in the sample than a defined membrane structure inside of the capsid shell. Instead, the internal membrane that usually encapsidates the viral genome in many other NCLDVs has been replaced by an additional protein layer. It remains to be seen if this phenomenon is a unique feature of faustovirus or a case of divergent evolution, where some members of the NCLDVs acquired a membrane to protect their genetic material and others developed a protein-based secondary protection layer.
Faustovirus probably assembles by filling the internal capsid shell with DNA, which then expands. Once all of the genetic material has been loaded into this first compartment, the second layer is acquired, presumably in the form of the preassembled trisymmetrons and pentasymmetrons that are subsequently stabilized together by the cement proteins. The latter interact with the inner shell at defined points and may act as nucleation centers for the outer shell assembly.
Materials and Methods
Cell Culture for Capsid Sequencing.
Vermamoeba vermiformis cells (strain CDC 19) were grown at 28 °C with peptone–yeast–glucose extract. After 24 h, cells were infected with faustovirus E12 and then incubated at 32 °C. At 6 h after infection, infected cells were harvested and centrifuged at 2,087 × g for 30 min. The pellets were resuspended in 1 mL of PBS and stored at –20 °C.
RNA and DNA Extraction.
Viral DNA was extracted by using the automated EZ1 Virus Mini-Kit v.2 (Qiagen GmbH) according to the manufacturer’s instructions. Extractions were performed using 200 μL of faustovirus culture. Viral DNA extracts were eluted in 100 µL.
Viral RNA was extracted using the RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions. Briefly, 600 μL of buffer RLT was added to 100 µL of the sample. The mixture was homogenized and transferred into a QIAshredder spin column. The column was centrifuged at 16,000 × g for 2 min. Then 600 μL of ethanol [70% (vol/vol)] was added, and the mixture was transferred into an RNeasy column and centrifuged for 15 s at 8,000 × g. Subsequently 700 µL of buffer RW1 to the column and centrifuged it at 8,000 × g for 15 s. The eluate was removed, and 500 µL of buffer RPE was added to the column and centrifuged at 8,000 × g for 15 s.
After the eluate was removed, 500 µL of RPE buffer was again added to the column and centrifuged twice: a first centrifugation at 8,000 × g for 2 min to remove the eluate and a second at 16,000 × g for 1 min to dry the column. At the last stage of the extraction, 30 µL of RNase-free water buffer was added to the column, followed by a centrifugation at 8,000 × g for 1 min to elute the RNA. TurboDNase enzyme was used to digest free DNA present in the extract. This step was carried out for 4 h at 37 °C. RNA extract was purified using the RNeasy miniElute Cleanup kit (Qiagen) according to manufacturer’s method. Briefly, 3.5 RLT buffer volumes were added to the extracted RNA. Then 2.5 volumes of ethanol [100% (vol/vol)] were added into the mixture. The entire solution was transferred to an RNeasy MinElute spin column and spun at 9,400 × g for 15 s. After removing the eluate, 500 μL of RPE buffer was added into the column and centrifuged for 15 s at 9,400 × g. We added 500 µL of ethanol [80% (vol/vol)] to the column twice and centrifuged it for 2 min at 9,400 × g and for 5 min at maximum speed, respectively. Finally, the elution was performed in 30 µL of RNA-free water by centrifuging at maximum speed for 1 min. RNA was quantified by spectrophotometry using RiboGreen.
Primer Design, Reverse Transcription, and PCR.
Primers for PCR amplification and sequencing (Table S1) were designed using PrimerQuest in the area containing the capsid sequences predicted by the preliminary bioinformatic study (2).
FV12_174_Int_fwd and FV12_195_fin_rev primers were used to amplify the complete cDNA (Table S1). Total RNA of faustovirus E12 was reverse-transcribed to cDNA with the SuperScript vilo cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions in a reaction volume of 20 µL [4 µL of 5× buffer, 2 µL of 10× enzyme mixture, 9 µL of DNase/RNase-free water (Gibco), and 5 µL of RNA extract]. The RT reaction was performed in the automated thermal cycler PTC-200 (MJ Research) as follows: 25 °C for 10 min, 42 °C for 60 min, and 85 °C for 5 min.
PCRs were performed with a PTC-200 automated thermal cycler (MJ Research) in a final volume of 25 µL of PCR mixture containing 2.5 µL of 10× buffer (Qiagen), 0.5 µL of each primer (10 pM; Eurogentec), 2.5 µL of each dNTP at 2 mM (euromedex), 1 µL of MgCl2 at 1.5 mM (Qiagen), 0.25 µL of HotStar Taq polymerase (Qiagen), 13 µL of Dnase/Rnase-free distilled water (Gibco), and 5 µL of DNA template or cDNA template. An initial 15-min denaturation at 95 °C was followed by 40 cycles of 1-min denaturation at 95 °C, 30-s annealing at 62 °C, and 2-min extension at 72 °C. Amplification was completed by a 5-min holding at 72 °C. Negative controls consisting of the PCR mixture without DNA template were added in each PCR run. PCR products were purified with the PCR filter plate Millipore NucleoFast 96 PCR kit as recommended by the manufacturer (Macherey–Nagel). PCR controls were performed directly on RNA extract using the primers FSV174fwd and FSV195frev to control DNA contamination.
Sequencing and Sequence Analysis.
Sequencing reactions were performed using the Big-Dye Terminator, version 1.1, Cycle Sequencing Kit DNA in accordance with the manufacturer’s instructions (Applied Biosystems). All PCR products were sequenced in both directions with the same primers as those used for PCRs in a 2720 Thermal Cycler (Applied Biosystems), with an initial 1-min denaturation step at 96 °C, followed by 25 cycles of 10-s denaturation at 96 °C, 20-s annealing at 55 °C, and 4-min extension at 60 °C. Sequencing products were purified using MultiScreen 96-well plates (Merck Millipore), containing 5% (vol/vol) of Sephadex G-50 (Sigma-Aldrich), and sequences were analyzed on an ABI PRISM 31309 Genetic Analyzer (Applied Biosystems). The sequences were edited using the ChromasPro software (version 1.42; Technelysium Pty Ltd.). Multiple alignments of nucleic acid sequences were performed with the muscle program (Mega5.1. software). The nucleic acid sequences were translated into protein sequences using web.expasy.org/cgi-bin/translate/dna_aa. The protein sequences were aligned by Clustalw2.1 (Mega5.1. software). All BLAST searches were carried out on blast.ncbi.nlm.nih.gov/BlastAlign.cgi.
Proteomic Analyses.
The capsid protein of faustovirus E12 was extracted and analyzed by SDS/PAGE, followed by Western blotting according to the protocol described by Azza et al. (5). Identification of the peptide sequence was performed by matrix-assisted laser desorption ionization time-of-flight MS and nano–LC-MS/MS as described by Reteno et al. (2).
Virus Propagation and Purification.
Faustovirus was propagated in V. vermiformis cells (strain CDC 19) according to established protocols (2). In summary, amoebae were grown to confluence in peptone–yeast–glucose extract and infected with an inoculum of faustovirus at an MOI of 100. After complete cell lysis, the supernatant was filtered through a 0.45-µm filter (Millipore). The filtrate was subsequently spun at 10,000 × g for 30 min, washed twice with PBS, and the final pellet resuspended in 1 mL PBS. To separate complete virions from empty particles, 500 µL of virus suspension was loaded on top of a 12-mL discontinuous gradient of 40–60% (wt/vol) iodixanol in PBS and spun at 100,000 × g for 24 h. The band containing the mature virus particles was harvested from the gradient and buffer exchanged into PBS. Samples were stored at 4 °C.
Isolation and Purification of the MCP.
The MCP of faustovirus was isolated by heating the virus to 70 °C for 20 min. Insoluble proteins were separated from soluble capsid proteins by centrifugation at 16,000 × g for 20 min. The soluble supernatant was diluted in buffer A (50 mM Tris, pH 9.2) and loaded onto a 5 mL HiTrap Q HP ion exchange column (GE Healthcare). The capsid protein was subsequently eluted by applying a gradient from 0% to 100% buffer B (50 mM Tris, pH 9.2, 2 M NaCl) over 15 CV. The protein was over 95% pure after this purification step, as estimated from an SDS/PAGE gel. The fractions containing the MCP were concentrated to 10 mg/mL and subsequently used for crystallization.
Crystallization and Data Collection.
Crystallization conditions were screened in a 96-well format using a variety of commercial screens from Hampton Research and Emerald BioSystems. Initial hits were obtained in conditions that contained ammonium sulfate in the precipitant solution. After optimization, about 100 µm-size crystals were grown by vapor diffusion in a hanging drop setup at 20 °C. An equal volume of 10 mg/mL protein solution was mixed with the well solution containing 1.6 M ammonium sulfate and 10% (wt/vol) dioxane. Crystals grew within 10–14 d.
Diffraction data were collected at beamline 23ID-D at the Advanced Photon Source. The datasets (Table S2) were integrated and scaled using XDS (33).
Structure Determination.
Two different crystal forms were observed, both with space group P21 but with different unit cell dimensions. The smaller crystal had a unit cell of a = 77.3 Å, b = 390.7 Å, c = 79.5 Å, and α = 90.0, β = 112.9, and γ = 90.0, whereas the larger crystal form had unit cell dimensions of a = 86.38 Å, b = 390.8 Å, c = 130.1 Å, and α = 90.0, β = 91.7, and γ = 90.0. The structure of the smaller crystal form was solved by molecular replacement using PHASER (6) with the structure of the PBCV-1 capsomer as a search model (PDB ID code 1M3Y). The initial map allowed model building using a combination of automated methods and manual model building in COOT in combination with refinement using phenix.refine (34) from the PHENIX software suite (35).
The larger crystal form was subsequently solved by molecular replacement using the model from the smaller crystal form as a search model.
The final values for Rwork/Rfree were 0.203/0.248 and 0.205/0.252 for the small and large crystal forms, respectively (Table S2).
Fig. 2 and the surface charge representation in this figure were prepared using Pymol (36) and the APBS plugin (37).
CryoEM Data Collection and Processing.
A volume of 2 µL purified faustovirus particles were applied to C-flat holey carbon grids (Ted Pella) and plunge-frozen into liquid ethane using a CryoPlunge 3 (Gatan) with a blot time of 5 s. Micrographs were collected in a semiautomated fashion on a FEI Titan Krios electron microscope at a nominal magnification of 47,000× on a Gatan Ultrascan camera using Leginon (38). The electron dose was adjusted to ∼20 e−/Å2, and the defocus range was set between 1.5 and 3.5 µm. Particles were boxed from micrographs using e2boxer.py from the EMAN2 software package (39). Reconstruction was performed using the jspr software suite (40). The final reconstruction used 1,188 and 10,915 particles for the reconstruction of the small and large virus capsids, respectively. The resolution of the reconstructions was 26 Å and 15 Å, respectively, for both particle sets, as judged by a Fourier shell correlation of 0.143 between two independently reconstructed subsets, which were kept from the beginning of the reconstruction procedure. Figs 3 and 4 and Figs. S1 and S2 were prepared using CHIMERA (41).
Sample Preparation for Lipid Analysis.
A 25-mg/mL porcine brain lipid extract in chloroform (Avanti Polar Lipids) was obtained for use as an internal standard for the analysis of the faustovirus sample. Standard solutions of 125 µg/mL and 12.5 µg/mL were made by diluting the extract into 2:1 methanol:chloroform. Methanol was obtained from Sigma and chloroform from Macron.
To extract the lipids from the PBS, the faustovirus sample was diluted 9× into a solution of 2:1 methanol:chloroform. Both the extracted virus and standard solutions were vortexed for 3 min before analysis. The supernatant of each solution was collected and analyzed to avoid any precipitate. A mixture of the two solutions was made by mixing 10 µL of the original virus sample, 10 µL of 125 µg/mL porcine standard, and 80 µL of 2:1 methanol:chloroform, yielding a total concentration of 12.5 µg/mL standard and a 10× dilution of the virus.
To determine if the lipids resulted from cellular membrane fragments of the host, two additional faustovirus samples were analyzed. The first sample was similar to the sample analyzed previously. The second sample, in addition to a filtration step, was run through a density gradient and charge-exchanged to further remove lipid contaminants. A 10× dilution using 2:1 methanol:chloroform was made for each sample. A 12.5 µg/mL porcine lipid brain extract was added to each sample as an internal standard. All virus samples analyzed were diluted 10× from the originally purified sample (see above).
MS for Lipid Analysis.
The faustovirus sample and the porcine standard were analyzed using relay electrospray ionization in the positive ion mode (42). Briefly, 10 µL of solution was loaded into nanospray emitters (Sutter Instruments) with 5-µm tip diameters. The emitters were pulled using a P-97 micropipette tip puller (Sutter Instruments). The emitters were placed about 5 mm away from the inlet of a Thermo Fisher LTQ mass spectrometer (Thermo Scientific). A piezoelectric plasma discharge gun (Milty) was used as an ion source. Positive ions were generated by compressing the trigger of the gun. The gun was placed at a distance of about 5 mm away from the nanospray emitters. As the ions from the gun landed into or onto the emitters, electrospray plumes formed for as long as the trigger was compressed (∼2 s). The mass spectrometer parameters used are listed in Table S3.
Phospholipid Characterization.
It is known that phosphotidylcholines ionize readily in positive ion mode. Phosphotidylcholines also have readily identifiable mass-to-charge ratios, most of which are even numbered. The structures of several phosphotidylcholines were determined by using MS/MS to obtain fragmentation profiles and comparing the results to an online database (43). Typical fragmentation patterns of phosphotidylcholines were obtained by consulting the scientific literature (44).
Supplementary Material
Acknowledgments
We thank Vidya Mangala Prasad, Moh Lan Yap, and Erica Zbornik for helpful discussions; Sheryl Kelly for help in submission of the manuscript; the staff at the Purdue CryoEM facility for their help with data collection; and the staff at the beamline 23 GM/CA-CAT of the Advanced Photon Source, Argonne National Laboratory, for help with the data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. This work was supported by National Institutes of Health Grant AI011219 (to M.G.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron density maps have been deposited in the EM Data Bank (accession nos. EMD-8144 and EMD-8145). The X-ray structures and the fitted models have been deposited in the Protein Data Bank (PDB ID codes 5J7O, 5J7U, and 5J7V).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523999113/-/DCSupplemental.
References
- 1.La Scola B, et al. A giant virus in amoebae. Science. 2003;299(5615):2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Reteno DG, et al. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol. 2015;89(13):6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salas ML, Andrés G. African swine fever virus morphogenesis. Virus Res. 2013;173(1):29–41. doi: 10.1016/j.virusres.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Yáñez RJ, et al. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208(1):249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 5.Azza S, Cambillau C, Raoult D, Suzan-Monti M. Revised Mimivirus major capsid protein sequence reveals intron-containing gene structure and extra domain. BMC Mol Biol. 2009;10:39. doi: 10.1186/1471-2199-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rux JJ, Kuser PR, Burnett RM. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77(17):9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín CS, et al. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9(10):917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 9.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99(23):14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, et al. Structure of Sputnik, a virophage, at 3.5-Å resolution. Proc Natl Acad Sci USA. 2012;109(45):18431–18436. doi: 10.1073/pnas.1211702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyun J-K, et al. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog. 2011;7(9):e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19(7):1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 15.Wrigley NG. An electron microscope study of the structure of Sericesthis iridescent virus. J Gen Virol. 1969;5(1):123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- 16.Yan X, et al. The capsid proteins of a large, icosahedral dsDNA virus. J Mol Biol. 2009;385(4):1287–1299. doi: 10.1016/j.jmb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X, Chipman PR, Castberg T, Bratbak G, Baker TS. The marine algal virus PpV01 has an icosahedral capsid with T=219 quasisymmetry. J Virol. 2005;79(14):9236–9243. doi: 10.1128/JVI.79.14.9236-9243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinkovits RS, Baker TS. A tale of two symmetrons: Rules for construction of icosahedral capsids from trisymmetrons and pentasymmetrons. J Struct Biol. 2010;170(1):109–116. doi: 10.1016/j.jsb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Martín C, et al. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat Struct Biol. 2002;9(10):756–763. doi: 10.1038/nsb837. [DOI] [PubMed] [Google Scholar]
- 20.Fuller S. A PRD1 by another name? Structure. 2005;13(12):1738–1740. doi: 10.1016/j.str.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Reddy VS, Nemerow GR. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci USA. 2014;111(32):11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrescia NGA, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432(7013):68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc Natl Acad Sci USA. 2011;108(36):14837–14842. doi: 10.1073/pnas.1107847108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105(6):1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan J, et al. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc Natl Acad Sci USA. 2009;106(11):4225–4230. doi: 10.1073/pnas.0812071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huiskonen JT, et al. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14(6):1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Veesler D, et al. Maturation in action: CryoEM study of a viral capsid caught during expansion. Structure. 2012;20(8):1384–1390. doi: 10.1016/j.str.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143(2):222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Van Etten JL, Lane LC, Meints RH. Viruses and viruslike particles of eukaryotic algae. Microbiol Rev. 1991;55(4):586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanspauwen MJ, et al. Infections with mimivirus in patients with chronic obstructive pulmonary disease. Respir Med. 2012;106(12):1690–1694. doi: 10.1016/j.rmed.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 31.La Scola B, Marrie TJ, Auffray J-P, Raoult D. Mimivirus in pneumonia patients. Emerg Infect Dis. 2005;11(3):449–452. doi: 10.3201/eid1103.040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yolken RH, et al. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc Natl Acad Sci USA. 2014;111(45):16106–16111. doi: 10.1073/pnas.1418895111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrödinger, LLC 2010. The PyMOL Molecular Graphics System, Version 1.7.6 (Schrödinger, LLC, New York)
- 37.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol. 2014;1117(Chapter 19):401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Li A, Hollerbach A, Luo Q, Cooks RG. On-demand ambient ionization of picoliter samples using charge pulses. Angew Chem Int Ed Engl. 2015;54(23):6893–6895. doi: 10.1002/anie.201501895. [DOI] [PubMed] [Google Scholar]
- 43.Sud M, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35(Database issue):D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu F-F, Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: The fragmentation processes. J Am Soc Mass Spectrom. 2003;14(4):352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.