Significance
Methane emission from the ruminant livestock sector—a by-product from enteric fermentation of plant biomass in the ruminant digestive system—is produced by methanogenic archaea and represents not only a significant amount of anthropogenic greenhouse gases contributing to climate change but also an energy loss and a reduction in feed efficacy. The present study elucidates the development and the unique mode of action of the highly specific inhibitor 3-nitrooxypropanol (3-NOP), which is targeting the nickel enzyme methyl-coenzyme M reductase in rumen archaea that catalyzes the methane-forming reaction. At the very low effective concentrations recently applied in vivo (dairy and beef cattle), 3-NOP appears to inhibit only methanogens and thus to be attractive for development as a feed supplement.
Keywords: methanogenesis, methyl-coenzyme M reductase, enteric methane, greenhouse gas, climate change
Abstract
Ruminants, such as cows, sheep, and goats, predominantly ferment in their rumen plant material to acetate, propionate, butyrate, CO2, and methane. Whereas the short fatty acids are absorbed and metabolized by the animals, the greenhouse gas methane escapes via eructation and breathing of the animals into the atmosphere. Along with the methane, up to 12% of the gross energy content of the feedstock is lost. Therefore, our recent report has raised interest in 3-nitrooxypropanol (3-NOP), which when added to the feed of ruminants in milligram amounts persistently reduces enteric methane emissions from livestock without apparent negative side effects [Hristov AN, et al. (2015) Proc Natl Acad Sci USA 112(34):10663–10668]. We now show with the aid of in silico, in vitro, and in vivo experiments that 3-NOP specifically targets methyl-coenzyme M reductase (MCR). The nickel enzyme, which is only active when its Ni ion is in the +1 oxidation state, catalyzes the methane-forming step in the rumen fermentation. Molecular docking suggested that 3-NOP preferably binds into the active site of MCR in a pose that places its reducible nitrate group in electron transfer distance to Ni(I). With purified MCR, we found that 3-NOP indeed inactivates MCR at micromolar concentrations by oxidation of its active site Ni(I). Concomitantly, the nitrate ester is reduced to nitrite, which also inactivates MCR at micromolar concentrations by oxidation of Ni(I). Using pure cultures, 3-NOP is demonstrated to inhibit growth of methanogenic archaea at concentrations that do not affect the growth of nonmethanogenic bacteria in the rumen.
Since the agricultural and industrial revolution 200 y ago, the methane concentration in the atmosphere has increased from less than 0.6 to 1.8 ppm. The present concentration is only 0.45% of that of CO2, but because methane has a 28- to 34-fold higher global warming potential than CO2 on a 100-y horizon, it contributes significantly to global warming (1). On the other hand, the lifetime of atmospheric methane is relatively short relative to CO2. Accordingly, the climate response to reductions of methane emissions will be relatively rapid. Thus, measures targeting methane emissions are considered paramount to mitigate climate change (2).
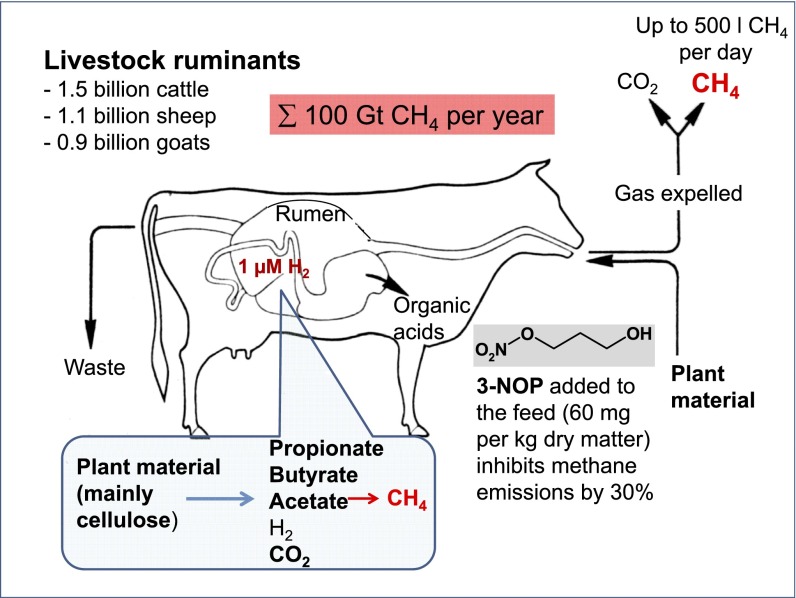
One of the main anthropogenic sources of atmospheric methane are ruminants (cattle, sheep, goats), the number of which has grown in parallel with the world population. Presently, there are about 1.5 billion cattle, 1.1 billion sheep, and 0.9 billion goats raised by humans (3). Ruminants emit about 100 million tons of methane per year, which corresponds to ∼20% of global methane emissions (4).
In the rumen (Fig. 1), plant material is fermented by anaerobic bacteria, protozoa, fungi, and methanogenic archaea in a trophic chain, predominantly yielding acetate, propionate, butyrate, CO2, and methane with H2 as intermediate (5, 6). Whereas organic acids are absorbed and metabolized by the animals, methane escapes the rumen into the atmosphere via eructation and breathing of the animals. The generation of methane by methanogenic archaea in the intestine of domestic ruminants lessens feed efficacy, as up to 12% of the gross energy ingested by the animal is lost this way (7).
Fig. 1.
Methane formation in the rumen of a dairy cow and its inhibition by 3-nitrooxypropanol (3-NOP). The H2 concentration in the rumen fluid is near 1 µM (≙140 Pa = 0.14% in the gas phase).
Methane (CH4) formation is the main H2 sink in the rumen. It is formed by methanogenic archaea at the bottom of the trophic chain mainly from carbon dioxide (CO2) and hydrogen (H2) (Fig. 1). However, the methane eructated by ruminants contains only minute amounts of H2; the concentration of dissolved H2 in the rumen is near 1 µM (8), equivalent to a H2 partial pressure of near 140 Pa. Because at 1 µM, H2 formation from most substrates in the rumen is exergonic (9), the low H2 concentration indicates that H2 is consumed in the rumen by the methanogens more rapidly than it is formed by other microorganisms (10). The H2 concentration increases substantially only when methane formation from H2 and CO2 is specifically inhibited by more than 50% (10, 11). Already a small increase in the H2 concentration (8) leads to both down-regulation of H2-generating pathways (12) and up-regulation of H2-neutral and H2-consuming pathways such as propionate formation, resulting in additional energy supply to the host animal (13–15). Thus, the H2 concentration stays constant, although its consumption by methanogens is partially inhibited in the rumen.
The amount of methane formation per unit of ingested feedstuff can differ significantly between individual animals as it is a heritable trait (16). Understanding these differences has been the scientific motivation to pursue the development of selective inhibitors of methanogenesis that are nontoxic to animals (17, 18). Only recently, a compound has been described that apparently can both substantially decrease CH4 and increase propionate productions in the rumen without compromising animal performance and health (19). It is the small molecule 3-nitrooxypropanol (3-NOP) (chemical structure shown in Fig. 1) that has been found to persistently decrease enteric methane emissions from sheep (20), dairy cows (21), and beef cattle (22) without apparent negative side effects (19). 3-NOP, given to high-producing dairy cows at 60 mg/kg feed dry matter (Fig. 1), not only decreased methane emissions by 30% but also increased body weight gain significantly without negatively affecting feed intake nor milk production and composition (19).
Methane formation in methanogenic archaea is catalyzed by methyl-coenzyme M reductase (MCR), involving methyl-coenzyme M and coenzyme B as substrates (Fig. 2A). MCR is a nickel enzyme in which the nickel is bound in a tetrapyrrole derivative named cofactor F430 (23, 24). This nickel-containing cofactor has to be in the Ni(I) oxidation state for the enzyme to be active. Because the redox potential Eo′ of the F430(Ni2+)/ F430(Ni1+) couple is −600 mV, the enzyme is very susceptible to inactivation by oxidants (23, 24). MCR has been well characterized by high-resolution X-ray structures (25–27) and EPR spectroscopy (28) with either substrates or products bound.
Fig. 2.
Binding of 3-NOP to methyl-coenzyme M reductase (MCR) as suggested by molecular docking. The crystal structure of inactive isoenzyme I from M. marburgensis was used in the docking experiments (25). (A) MCR-catalyzed reaction. CH3-S-CoM, methyl-coenzyme M; CoM-S-S-CoB, heterodisulfide of coenzyme M and coenzyme B; HS-CoB, coenzyme B. (B) 3-NOP in the active site with its nitrate group in electron transfer distance to Ni(I) of F430 and its hydroxyl group interacting via a water molecule with Arg120. (C) 3-NOP in the active site with its hydroxyl group in coordination distance to Ni(I) of F430 and its nitrate group interacting with Arg120. (D) Methyl-coenzyme M (CH3-S-CoM) in the active site with its thioether sulfur in electron transfer distance to Ni(I) and its sulfonate group interacting with both a water molecule and Arg120. The molecules 3-NOP, CH3-S-CoM, and coenzyme B (HS-CoB) are drawn as ball-and-stick models in orange and F430 in light gray highlighting nitrogen in blue, oxygen in red, sulfur in yellow, and nickel(I) as a green sphere. The position of methyl-coenzyme M obtained via docking is almost identical to that found via EPR measurements of active MCR (28).
The molecular shape of 3-NOP (Fig. 1) is similar to that of methyl-coenzyme M (Fig. 2A). This fact and the moderate oxidation potential of 3-NOP suggested that inhibition of methanogenesis in ruminants is achieved by targeting the active site of MCR, for which we now provide experimental evidence. We start by describing how the development of 3-NOP was facilitated by molecular modeling.
Results
In silico, in vitro, and in vivo studies were performed to show that 3-NOP specifically targets MCR.
In Silico Studies.
Based on the structure and properties of MCR, inhibitors were developed with the aid of 3D pharmacophore-based virtual screening and molecular docking studies focusing on analogs of methyl-coenzyme M as lead structure. The inhibitor was designed to be both uncharged, allowing cell penetration by diffusion, and a moderate oxidant facilitating the oxidation of Ni(I) in the active site of MCR. These considerations led to a series of potential candidates that best fit into the active site of MCR. Among these, 3-NOP turned out to be the most promising. Based on the docking scores (Methods), two preferred sets of binding poses for the nitrate ester were identified, both occurring with similar probability, whereas other potential binding modes scored significantly lower. Strikingly, one of the binding poses of 3-NOP (Fig. 2B) was found to be very similar to that of the natural ligand methyl-coenzyme M (Fig. 2D). The nitrate-ester group of 3-NOP, which can be reduced easily (29), is positioned in electron transfer distance to Ni(I) of F430 and the hydroxyl group of 3-NOP interacts via a water molecule with Arg120 that is responsible for coordinating the sulfonate group of methyl-coenzyme M. In the second binding pose, the nitrate group of 3-NOP interacts with Arg120, whereas the hydroxyl group of 3-NOP is located close to the Ni(I) (Fig. 2C).
In Vitro Studies.
Purified MCR was found to be inactivated by 3-NOP as indicated by a rapid decrease in the rate of methane formation in the presence of the nitrate ester, confirming our in silico studies. Only 0.1 µM 3-NOP was required to completely inactivate MCR within several minutes of exposure (Fig. 3A).
Fig. 3.
Inactivation of MCR by 3-NOP and nitrite. The experiments were performed with purified isoenzyme I from Methanothermobacter marburgensis. (A) Effect of 3-NOP on the MCR activity; (B) Effect of 3-NOP on the EPR signals MCRred1 (78 µM) and MCRox1 (4 µM) (30). The spectrum (Inset) remaining after complete MCR inactivation with 3-NOP (100 µM) is that of MCRox1 (see In Vitro Studies). (C) Formation of nitrite and nitrate upon inactivation of MCR with 3-NOP (50 mM). (D) Effect of nitrite and nitrate on the EPR signal MCRred1. The 0.3-mL assays contained about 1 mM coenzyme M that was added to MCR during purification and storage to stabilize its activity (Methods).
MCR inactivation was also monitored by EPR spectroscopy. Untreated enzyme exhibits a characteristic EPR spectrum as shown in Fig. 3B (black trace). The observed spectrum is the result of a superposition of two MCR states: 95% MCRred1 [the active Ni(I) form of the enzyme], and 5% MCRox1 [an inactive Ni(III) form] (30). After complete inactivation by 3-NOP (5 mM), only the signal corresponding to the 5% MCRox1 was detected (red trace). Apparently, the MCRred1 signal was completely quenched, implying that Ni(I) in MCRred1 was oxidized by 3-NOP to an EPR silent Ni(II) oxidation state. Because quenching of the MCRred1 signal [oxidation of Ni(I) to Ni(II)] is a one-electron process, one would expect that somewhere in the sample a free electron is formed, which should be detectable in EPR. Indeed, radical species could be detected but their signal intensity was very low. When there are multiple possible reactions and/or the electron is hydrated by water, such a free electron is very difficult to detect.
Fig. 3B also displays the change of the EPR signal intensity over the course of titrating active MCR with 3-NOP. The EPR signal decreased with increasing concentrations of 3-NOP. Less than a 20 µM concentration of 3-NOP was required for a 50% quench of the 78 µM MCRred1 signal. After complete inactivation, about 0.2 mol of nitrite and 0.7 mol of nitrate per mol of quenched MCRred1 were found in the samples (Fig. 3C), indicating that 3-NOP was at least partly reduced to nitrite. Interestingly, nitrite was also found to inactivate purified MCR via Ni(I) oxidation at very low concentrations (Fig. 3D). However, the rate of inactivation in the presence of nitrite was slower by several orders of magnitude (minutes vs. milliseconds) compared with 3-NOP. The ability of both 3-NOP and its reduction product nitrite to inactivate MCR makes 3-NOP a unique tandem-charge warhead inhibitor (31) and explains why substoichiometric amounts of 3-NOP were required to oxidize Ni(I) in the active site of MCR to Ni(II) (Fig. 3B). [A tandem-charge weapon is a projectile that has two stages of detonation (31).] On the other hand, both sodium nitrate (Fig. 3D) and 1,3-propanediol (see below) had no effect on the EPR spectra of MCR at concentrations up to 10 mM.
It is reasonable to assume that 1,3-propanediol is another main reaction product. We have proven the formation of 1,3-propanediol in vivo by radiolabeling experiments (see In Vivo Inhibition of Methanogenesis), but in vitro quantification by NMR or GC-MS turned out to be difficult due to insufficient sensitivity: based on our experiments, less than 1 mol of 1,3-propanediol was formed per mole of inactivated MCR.
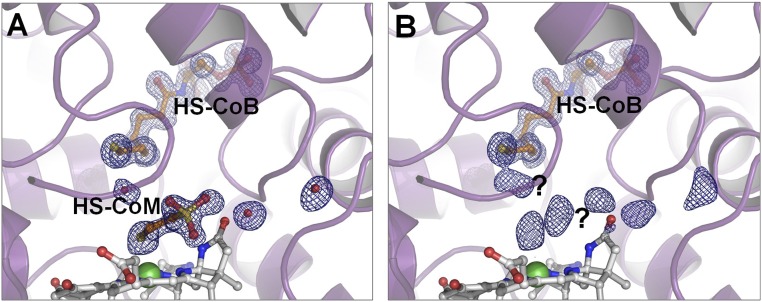
We were curious to see whether the products of 3-NOP reduction could be identified in the crystal structure of MCR, which was inactivated by 3-NOP in vivo. Indeed, structural comparison of the untreated, active MCR (Fig. 4A) with the enzyme inactivated in vivo (Fig. 4B) revealed differences that suggest entrapment of reduction products of 3-NOP in the active site. Although in Fig. 4A the continuous electron density can be assigned to HS-CoM, in Fig. 4B there are only electron density blobs that might be due to nitrite and 1,3-propanediol (see In Vitro Studies) bound nonspecifically in the active site, which makes their accurate modeling impossible.
Fig. 4.
View into the active site of MCR crystallized before (A) and after (B) in vivo inactivation by 3-NOP. The crystal structures were resolved to 1.25-Å resolution. Coenzyme B (HS-CoB) (at an occupancy near 100%) and coenzyme M (HS-CoM) (at an occupancy near 80%) are drawn as ball-and-stick models in orange and F430 in light gray with nickel highlighted as a green sphere. The secondary structure of the protein is cartooned in light purple. Three water molecules are shown as red spheres. The 2Fo-mFc map is contoured at 1σ in dark-blue mesh. For an interpretation, see In Vitro Studies.
After purification, the nickel in MCR (Fig. 4 A and B) was in the Ni(II) oxidation state and contained coenzyme M (Fig. 4A) and coenzyme B tightly bound (Fig. 4 A and B). When MCR, as prepared for Fig. 4A, was dialyzed against 5 mM 3-NOP for 24 h at 60 °C to try to remove the bound coenzyme M and coenzyme B and was then crystallized, the crystal structure was found to be almost identical to that shown in Fig. 4A and this was confirmed by data collection at the sulfur K-edge. This is consistent with previous findings that the ligand bound to Ni(II) does not exchange with substrate analogs outside of inactivated MCR, probably because the bound coenzyme B blocks the substrate entrance to the active site (26).
In Vivo Inhibition of Methanogenesis.
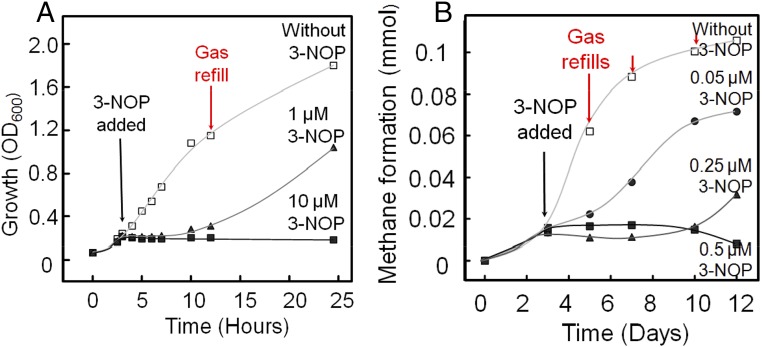
In a subsequent series of experiments, the specificity of this inhibitor toward selected representatives of different methanogenic and nonmethanogenic cultures was evaluated. We first investigated its effect on growth with the model organism Methanothermobacter marburgensis. Upon addition of 3-NOP (final concentration of 10 µM) to cultures of M. marburgensis, both growth and methanogenesis almost immediately stopped (Fig. 5A). At a 10-fold lower concentration of 3-NOP (1 µM), complete inhibition was also observed, but after 5 h, growth and methane formation resumed. It is known that methanogens contain a repair system that can reactivate MCR in a H2-, ATP-, and chaperone-dependent reduction process (32–34). Thus, inhibition of methanogenesis is reversible in vivo.
Fig. 5.
Inhibition of growth of two methanogenic archaea on H2 (80%) and CO2 (20%) in the presence of 3-NOP. (A) Methanothermobacter marburgensis grown at 65 °C in a 25-mL batch culture with a 100-mL gas phase; (B) Methanobrevibacter ruminantium grown at 37 °C in a 5-mL batch culture with a 10-mL gas phase.
Inhibition of both growth and methane production by 3-NOP was also observed with methanogens from the rumen (Fig. 5B) and from other environments (Table S1): Methanobrevibacter ruminantium (<0.25 µM; Fig. 5B), Methanobacterium smithii (1 µM), Methanobrevibacter millerae (1 µM), Methanobacterium bryantii (1 µM), Methanothermobacter wolfeii (<1 µM), Methanosphaera stadtmanae (>1 µM), Methanomicrobium mobile (>10 µM), and Methanosarcina barkeri (>10 µM). At the 3-NOP concentrations indicated, inhibition was only transient (as shown in Fig. 5A for 1 µM 3-NOP and in Fig. 5B for 0.25 µM). As a control, the effect of 3-NOP (100 µM) on the growth of nonmethanogenic rumen bacteria was assessed: Ruminococcus albus, Ruminococcus flavefaciens, Selenomonas ruminantium, Streptococcus bovis, Fibrobacter succinogenes, Anaerovibrio lipolytica, Prevotella bryantii, Prevotella ruminicola, Megasphaera elsdenii, Butyvibrio fibriosolvens, Clostridium aminophilum, and Escherichia coli (Table S1). Growth of none of these cultures was negatively affected by the nitrate ester (100 µM). The effect of 3-NOP is thus highly specific toward methanogenic archaea.
Table S1.
List of methanogenic archaea and rumen bacteria, culture media, and culture sources
| Strain | Species | Culture medium | Source |
| Archaea | |||
| 16643 | Methanobrevibacter millerae | 119 | DSMZ |
| 861 | Methanobacterium smithii | 119 | DSMZ |
| 863 | Methanobacterium bryantii | 119 | DSMZ |
| 1093 | Methanobrevibacter ruminantium | 119 | DSMZ |
| 800 | Methanosarcina barkeri | 120 | DSMZ |
| 1539 | Methanomicrobium mobile | 161 | DSMZ |
| 3091 | Methanosphaera stadtmanae | 322 | DSMZ |
| 2133 | Methanothermobacter marburgensis | Schönheit et al. (66) | DSMZ |
| 2970 | Methanothermobacter wolfeii | Schönheit et al. (66) | DSMZ |
| Bacteria | |||
| S85 | Fibrobacter succinogenes | M8 | UA-RINH |
| SY3 | Ruminococcus albus | M8 | UA-RINH |
| OO7 | Ruminococcus flavefaciens | M8 | UA-RINH |
| JW11 | Butyrivibrio fibrisolvens | M8 | UA-RINH |
| Z108 | Selenomonas ruminantium | M8 | UA-RINH |
| 23 | Prevotella ruminicola | M8 | AU-IBERS |
| B14 | Prevotella bryantii | M8 | UA-RINH |
| ES1 | Streptococcus bovis | M8 | UA-RINH |
| J1 | Megasphaera elsdenii | M8 | UA-RINH |
| 5S | Anaerovibrio lipolytica | M8 | UA-RINH |
| ATCC 49906 | Clostridium aminophilum | M8 | UA-RINH |
| 35 | Escherichia coli | LB | UV-CECT |
| 45 | Bacillus subtilis | LB | UV-CECT |
AU-IBERS, Aberystwyth University, Institute of Biological, Environmental, and Rural Science; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany); UA-RINH, University of Aberdeen–Rowett Institute of Nutrition and Health; UV-CECT, University of Valencia, Colección Española de Cultivos Tipo.
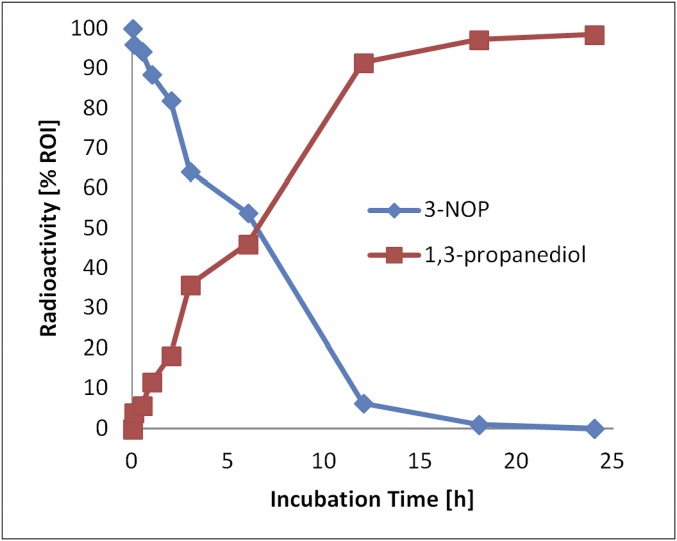
We also tested whether 3-NOP is metabolized by the rumen microorganisms and, if yes, to what product(s). Samples of actively fermenting cow rumen fluid were anaerobically incubated with 14C-labeled 3-NOP at 38 °C and analyzed in several time intervals for 14C-labeled product formation. Under the experimental conditions chosen, almost all of the added 3-NOP (0.036 µmol/2 mL = 18 µM) was converted to 1,3-propanediol after 24 h as revealed by radio-HPLC (Fig. S1). 14C-labeled products other than 1,3-propanediol were not found in significant amounts. No conversion of 3-NOP was observed under aerobic conditions.
Fig. S1.
Conversion of 3-NOP to 1,3-propanediol in ex situ rumen fluid under strictly anoxic conditions. The gas phase was 100% CO2, and the temperature was 38 °C. The rumen fluid (0.4 mL) was diluted with 1.6 mL of buffer A (pH 6.5) and supplemented with 0.05 g of feed dry matter (50% grass silage and 50% compound feed). At time 0, 3-nitrooxy[3-14C]propane-1-ol was added (0.036 µmol/10 mL, 28 µCi/µmol), and after the time intervals indicated, samples were analyzed for 14C radioactivity via radio-HPLC. The radioactivity cochromatographed with 3-NOP (21-min retention time) and with 1,3-propanediol (5-min retention time). No conversion was observed under aerobic conditions. For quantitative analysis, peak integration was performed with the Berthold Radiostar 4.6.0.0 software. Peaks in the radioactivity chromatogram were integrated after background subtraction. The sum of integrated peaks were set to 100%. ROI, region of interest.
Discussion
3-NOP, whose development was guided by in silico methods, was shown to inhibit methanogenesis, in vitro and in vivo, by targeting MCR and oxidizing its active site Ni(I). Nitrate, nitrite, and most probably 1,3-propanediol were formed alongside MCR inactivation. Nitrite, in turn, was also found to effectively inactivate MCR. At the low concentrations of 3-NOP and of nitrite required to completely inhibit methanogenesis, these compounds are not toxic to animals (19, 35, 36). 1,3-Propanediol (37) and nitrite (38) are commonly occurring intermediates in the rumen.
The amount of 3-NOP, which has to be added to the feed to obtain 30% inhibition of methanogenesis (60 mg per kg dry matter ≙ 25 µM, assuming the dry matter content of the rumen to be 5%), is considerably higher than predicted from our pure culture studies. There are two likely explanations why the steady-state concentration of 3-NOP in the rumen is lower than calculated. It is known that 3-NOP is reduced to nitrite and 1,3-propanediol by rumen bacteria as shown for cow rumen fluid in Fig. S1. For example, Enterobacter species contain an aliphatic nitroester reductase, closely related to the old yellow enzyme, that catalyzes this reaction using NADPH as electron donor (29). Furthermore, 3-NOP can easily diffuse through membranes. As a result, the compound is quickly distributed throughout the animal where it is denitrated, primarily in the liver (39).
In the past, two other specific inhibitors of MCR have been described, namely bromoethane sulfonate (BES) and bromopropane sulfonate (BPS). Both compounds exert their inhibitory effect in vitro at low concentrations by inactivation of MCR, BES at an IC50 of 4 µM (concentration required for 50% inhibition) and BPS at an IC50 of 0.05 µM (40–42). The mechanism of inactivation has been shown to be an electrophilic attack of the bromo compounds on the Ni(I) resulting in its alkylation and oxidation (43). Because of the negatively charged sulfonate group of BES and BPS, both inhibitors cannot freely diffuse through the cytoplasmic membrane of methanogens and are therefore generally poor inhibitors of methanogenesis in vivo (44). Generally, more than 1 mM BES or BPS are required to inhibit growth and methanogenesis. An exception appears to be M. ruminantium with an in vivo IC50 of 1 µM for BES (42, 44). This rumen methanogen requires coenzyme M as vitamin (45, 46) and contains a coenzyme M transporter (47), by which most probably also BES (48) but not BPS (44) is actively carried across the cell membrane. However, the unfavorable toxicological profile of BES, a result of its alkylating potential, prevents it from being authorized as a feed additive for ruminants (42).
In conclusion, 3-NOP inhibits enteric methane emissions from ruminants by specific inactivation of the enzyme MCR via a tandem-charge warhead inhibition mechanism. Both the detailed understanding of this mechanism and 3-NOP’s high specificity are likely to promote the nitrate ester as a ruminant feed supplement. More importantly, it promises to be a valuable research tool to study the role of methanogenesis in rumen fermentation and in other methanogenic biotopes such as the intestinal tract of termites and paddy field soils (4). There is a caveat, however: The 3-NOP concentration required for inhibiting growth of Methanosarcina barkeri on methanol and H2 and of Methanomicrobium mobile on H2 and CO2 was almost 100 times higher than required to inhibit growth of M. ruminantium on H2 and CO2. The reason for this difference in activity is presently unclear.
Methods
Sources of chemicals, culture sources, used growth media, growth assays, and the methods for the determination of 1,3-propanediol, nitrate, and nitrite are described in SI Methods (49–51).
Molecular Modeling.
The pharmacophore generation protocol in LigandScout 3.1 (Inte:Ligand GmbH) was used to generate the pharmacophore model directly from the receptor–ligand interactions as revealed in the 1HBN PDB structure (52).
Molecular modeling was performed with Schrödinger Software Suites v2010-2015 (Schrödinger release 2010; Maestro 9.1; Schrödinger, LLC) and Glide (version 5.6; Schrödinger, LLC) was used for virtual screening and flexible ligand–receptor docking (53). Docking scores and rankings of binding poses were obtained using complex scoring algorithms as implemented in Glide SP and Glide XP as part of the Schrodinger software suite (54–56). Fig. 2 B–D was generated and rendered with PyMOL (The PyMOL Molecular Graphic System, version 1.8.0; Schrödinger, LLC).
MCR Purification.
The enzyme was purified from Methanothermobacter marburgensis. The methanogen contains two MCR isoenzymes designated MCR I and MCR II that have similar properties (57). Both nickel enzymes have an apparent molecular mass of near 280 kDa, are composed of three different subunits in an α2, β2, γ2 configuration, and harbor two interlinked active sites with each a F430 as prosthetic group (25). For the studies here, only MCR I was used.
For activity and EPR measurements, MCR was purified from M. marburgensis grown exponentially in a 12-L fermenter containing 10 L of culture gassed with 80% H2/20% CO2/0.01% H2S at a rate of 10 L per min and stirred at a rate of 1,000 rpm. When the culture had reached an OD of ∼4, gassing was switched to 100% H2 and the culture was cooled to 4 °C and harvested, yielding about 80 g of cells (wet mass). From the cells, MCR I was anaerobically purified in the presence of 10 mM coenzyme M as described previously (58). The protein concentration was determined by measuring the absorbance of oxidized enzyme (MCRsilent) at 420 nm using ε = 44,000 M−1⋅cm−1 for a molecular mass of 280,000 Da.
For crystal structure determination, MCR was purified from M. marburgensis grown exponentially in two 2-L fermenters containing 1.5 L of culture gassed with 80 H2/20% CO2/0.01% H2S at a rate of 0.75 L per min and stirred at a rate of 1,000 rpm. When the culture had reached an OD of ∼4, 3-NOP (0.5 mM) was added to one of the cultures and after another 30 min of continuous gassing and stirring both cultures were cooled down to 4 °C and harvested separately by centrifugation, yielding 16 g of cells (wet mass) in the case of the untreated culture and 13 g in the case of the 3-NOP treated culture. The cell pellets thus obtained were directly used for anaerobic purification of MCR I in the absence of coenzyme M essentially as described previously (58), with the exception that, in the case of the cells treated with 3-NOP, the lysis buffer contained 0.5 mM 3-NOP before harvest. About 20 mg of native MCR I and about 16 mg of the 3-NOP inactivated MCR I were thus obtained.
MCR Activity Assay (Fig. 3B).
The assay of methane formation from methyl-coenzyme M and coenzyme B was performed in a closed 7.5-mL bottle containing 400 µL of assay solution and 100% N2 as gas phase. The solution contained 50 mM Tris⋅HCl, pH 7.6, 10 mM methyl-coenzyme M, 1 mM coenzyme B, 10 mM DTT (to regenerate coenzyme B from CoM-S-S-CoB; Fig. 2A), and purified MCR. DTT was used as an electron source instead of titanium(III) citrate plus cobalamin to regenerate coenzyme B, because titanium(III) citrate spontaneously reduces 3-NOP. The reaction was started by the addition of enzyme. At the times indicated in Fig. 3A, 100-µL gas samples were withdrawn from the head space and analyzed for methane gas chromatographically with flame ionization detection (Buck Scientific; model 910).
MCR EPR Signal Determination (Fig. 3).
MCR, purified as described above, was supplemented with 3-NOP, sodium nitrate, or sodium nitrite (final volume, 0.3 mL) and incubated for 15 min at room temperature in an EPR tube that was subsequently frozen in liquid nitrogen. Continuous-wave (CW) EPR spectra were measured at X-band (9-GHz) frequency on a Bruker EMX spectrometer fitted with an ER-4119-HS (high-sensitivity) perpendicular-mode cavity. Measurements at 77 K were performed by fitting the cavity with a liquid nitrogen finger Dewar. All spectra were recorded with a field modulation frequency of 100 kHz, modulation amplitude of 0.6 mT, and frequency of 9.386 GHz. The EPR signal intensities were determined by measuring the respective EPR-active species under nonsaturating conditions. Because the signals represent the first derivative of the absorption-type signal, the spectra were double integrated, and the surface area of each signal, corrected for the presence of two nickel sites per MCR, was compared with that of a 10 mM copper perchlorate standard (10 mM CuSO4; 2 M NaClO4; 10 mM HCl). The values obtained this way were compared with the known enzyme concentration, which was set to 100%. When more than one signal is present, each signal is simulated, the compound spectrum is reproduced, and the double-integration value of each individual component is obtained. The BioEPR package was used for spectral simulation and double integration of the signals (59). Only the intensities of the paramagnetic species (red1 and ox1) could be determined as described. The amount of the EPR-inactive form, MCRsilent, was assigned as the difference between the concentration of MCR present and the concentration of the paramagnetic species.
MCR Crystal Structure Determination (Fig. 4).
Crystallization was performed at 8 °C under aerobic conditions using the sitting-drop method in 24-well plates (CombiClover Junior Plate; Jena Bioscience). To 1 µL of crystallization solution [25% (vol/vol) PEG 400, 300 mM MgCl2, 200 mM NaCl, and 100 mM Hepes, pH 7.5], 2 µL of MCR I solution (30 mg/mL) was added and mixed. Brick-shaped yellow crystals appeared after 20 h. The crystals were flash-frozen directly in liquid nitrogen. The frozen crystals were analyzed by X-ray diffraction (Table S2). The data were processed with iMosflm and scaled with SCALA in the CCP4 suite (60). The crystal structures were solved by molecular replacement using the MCR I structure from M. marburgensis as a template (PDB ID code 3POT) with Molrep from CCP4 (61). The rigid-body refinement was first executed with REFMAC5 (62), and then the model was manually rebuilt with COOT (63) and further refined with PHENIX (64); noncrystallographic symmetry and refinement with hydrogen in the riding position were applied. The final model was validated through the MolProbity server (molprobity.biochem.duke.edu). Fig. 4 was generated and rendered with PyMOL (Schrödinger, LLC).
Table S2.
Data collection and refinement statistics for the structure of MCR I isolated from M. marburgensis treated with 3-NOP
| Data collection and refinement | Data |
| Synchroton | SLSX10SA |
| Wavelength, Å | 0.97128 |
| Space group | P21 |
| Resolution, Å | 38.79–1.25 (1.32–1.25) |
| Cell dimension | |
| a, b, c, Å | 82.2, 118.1, 122.6 |
| α, β, γ, ° | 90.00, 92.03, 90.00 |
| Rmerge, %* | 5.0 (26.6) |
| I/σΙ * | 14.4 (4.4) |
| Completeness, %* | 98.5 (98.1) |
| Redundancy* | 4.6 (4.4) |
| Refinement | |
| Resolution, Å | 38.79–1.25 |
| No. of reflections | 633,010 |
| Rwork/Rfree, %† | 10.52/12.37 |
| No. of atoms | |
| Protein | 19,740 |
| Ligands/ions | 191 |
| Solvent | 2,609 |
| Average B factors, Å2 | 15.54 |
| Molprobity clash score, all atoms | 2.05 (98th percentile) |
| Ramachandran plot | |
| Favored regions, % | 97.5 |
| Outlier regions, % | 0.08 |
| rmsd bond length, Ň | 0.012 |
| rmsd bond angle, °‡ | 1.522 |
| PDB ID code | 5G0R |
SLS, Swiss Light Source.
Values relative to the highest resolution shell are within parentheses.
Rfree was calculated as the Rwork for 5% of the reflections that were not included in the refinement.
rmsd, root-mean-square deviation.
SI Methods
3-NOP and MCR Substrates.
3-NOP was synthesized from 1,3-propanediol and nitric acid (49). Coenzyme M, methyl-coenzyme M, and coenzyme B were synthesized as described (50, 51).
3-Nitrooxy[3-14C]propane-1-ol (1.021 MBq/µmol = 27.58 µCi/µmol) was obtained from Selcia Limited, and 3-nitrooxy[1,2,3-13C]propane-1-ol from DSM Nutritional Products.
Culture Sources.
The methanogenic archaea and bacteria investigated and the media used for their growth are listed in Table S1. Media for methanogenic archaea were prepared anaerobically under a gas atmosphere of 80% H2 and 20% CO2 and adjusted to pH 6.7 (https://www.dsmz.de/). M8 medium for bacteria was prepared under a gas atmosphere of 100% CO2 (65).
Testing the Effect of 3-NOP on the Growth of Methanothermobacter marburgensis (Fig. 5A).
The archaeon was grown in 125-mL glass bottles containing 25 mL of mineral salts medium (66) and closed with a rubber stopper. The 100-mL gas phase was 80% H2/20% CO2 at 1.6 × 105 Pa pressure. At time 0 (Fig. 5), 1.5 mL of an actively growing culture was used as inoculum. Incubation was at 65 °C under continuous rapid shaking. Growth was followed by measuring the increase in optical density (OD) at 600 nm in 1-mL samples. After about 2.5 h, when an OD of 0.2 was reached, 3-NOP was added at the concentrations indicated.
Testing the Effect of 3-NOP on the Growth of Methanobrevibacter ruminantium (Fig. 5B) and of Other Microorganisms.
Archaea.
Methanogenic archaea other than M. marburgensis were grown in 5 mL of medium at 37 °C in 15-mL Hungate tubes sealed with butyl rubber septum and aluminum seals. The 10-mL gas phase was 80% H2/20% CO2 at 2 × 105 Pa pressure. In the case of M. stadtmanae and M. barkeri, H2 and methanol (100 mM) was the energy source. After inoculation with 0.3 mL of an actively growing preculture, the tubes were horizontally placed in a shaking incubator at 37 °C at 120 rpm. Growth was followed by CH4 production using a flame ionization-detector GC. In brief, a subsample of 0.5 mL of gas from head space in each tube was taken and then injected manually in the GC (HP Hewlett 5890, Packard Series II) using a 1-mL Sample-Lock syringe (Hamilton). The concentration of CH4 was determined using a standard curve generated by injecting different volumes of pure CH4 before and after the injection of samples. Methane was measured over 12 d every 2 d coinciding with new pressurization of head space to 2 bar with 80:20 H2:CO2 gas mixture. To test the effect of 3-NOP on methanogenesis, three consecutive 5-mL batches of incubations were conducted. In each batch, the following treatments were used in triplicate: (i) blanks, only media; (ii) controls, cultures in the absence of inhibitor; (iii) 3-NOP added to a final concentration of 0.025, 0.25, 0.5, 5, 50, 100, 250, or 500 µM. The 3-NOP was added on day 2 after culture inoculation.
Bacteria.
Bacteria were grown in 7 mL of medium at 37 °C in 15-mL sealed Hungate tubes. The 8-mL gas phase was 100% CO2 at 2 × 105 Pa pressure. After inoculation with 0.5 mM of an actively growing preculture, the tubes were horizontally placed in a shaking incubator at 37 °C at 120 rpm. Bacterial growth was monitored over 32 h by measuring OD at 600 nm. 3-NOP (100 µM) was delivered in the culture straight after inoculation.
Testing the Metabolism of 3-NOP in Rumen Fluid.
The assays were performed at 38 °C under strictly anoxic conditions in 10-mL bottles closed with a rubber stopper, filled with 100% CO2 and 1.6 mL of buffer A at pH 6.5 composed per liter of 3.5 g of K2HPO4, 2.6 g of NH4H2PO4, 0.143 g of MgSO4×7H2O, 9.8 g of NaHCO3, and 0.125 mL of trace element solution. After equilibration of buffer A with the gas phase, first 0.05 g of feed dry matter (50% grass silage and 50% compound feed) and then 0.4 mL of strained fresh cow rumen fluid were added. The reaction was started by the addition of 14C-labeled 3-NOP (0.036 µmol, 28 µCi/µmol). During incubation for 24 h, NaOH was stepwise added to keep the pH at about 6.2. At the times indicated in Fig. S1, microbial activity was stopped by addition of 0.3 mL of 1 M HCl, and samples were analyzed for 14C-3-NOP and 14C-1,3-propanediol via radio-HPLC [see Determination of 1,3-Propanediol (Fig. S1)].
Determination of 1,3-Propanediol (Fig. S1).
Samples from the ex situ rumen fluid experiment (see Testing the Metabolism of 3-NOP in Rumen Fluid) were centrifuged for 10 min at 14,100 × g, and 50 µL of the supernatant (corresponding to about 44,000 dpm) were analyzed at room temperature by radio-HPLC: Column, Phenomenex Aqua C18, 5µ, 125 Å, 250 × 4.6 mm (Phenomenex); guard column, LiChrospher 100 RP-18e, 5µ, (Merck); flow, 1 mL/min; gradient, solvent A, 10 mM ammonium formate in H2O, pH 3.5; solvent B, CH3CN: 0 min 0% B, 5 min 0% B, 5.3 min 5% B, 20 min 5% B, 30 min 100% B, and 50 min 100% B; detectors: Jasco MD-1510 DAD, at 227 nm, and Berthold Radioflow Detector LB 509 B with YG 150 solid scintillation cell. Typical retention times for 3-NOP and 1,3-propanediol were 21 and 5 min, respectively. The compound eluting with a retention time of 5 min was identified by cochromatography and by 13C-NMR/1H-NMR spectroscopy to be 1,3-propanediol. To increase the spectroscopic sensitivity, 13C-3-NOP was added to the 14C-3-NOP at time 0 in the rumen fluid experiment (see Testing the Metabolism of 3-NOP in Rumen Fluid).
Determination of Nitrite and Nitrate (Fig. 3).
MCR (0–80 µM) and 3-NOP (50 mM) were anaerobically incubated for 15 min at room temperature and then assayed for nitrite and nitrate. The two anions were quantitated with the Nitrate/Nitrite Colorimetric Assay Kit (no. 780001) from Cayman Chemical Company. The kit allows for the direct detection of nitrite using the Griess reagent or the total amount of nitrate and nitrite after conversion of nitrate to nitrite by nitrate reductase and subsequent detection by the Griess reagent. For the detection of nitrite, 100 µL of each sample was taken and mixed with 33 µL of Griess reagent [mixture of sulfanilamide (reagent 1) and N-(1-naphthyl) ethylenediamine (reagent 2)] and 867 µL of water. The formation of the azo product of reagent 1 and 2 was measured at a wavelength of 540 nm and compared with a standard curve to calculate the amount of nitrite. For the detection of both compounds, 120 µL of the 3-NOP/MCR mixtures was taken and mixed with 15 µL of cofactor mix (including NADPH as reductant) and 15 µL of nitrate reductase mix. The sample was incubated for 3 h at room temperature. At this point, all nitrate was converted into nitrite. A volume of 100 µL of these samples were taken and mixed with 33 µL of Griess reagent and 867 µL of water. The amount of nitrite detected was corrected for the amount present without the enzymatic conversion to calculate the amount of nitrate formed. The 0 µM MCR samples showed that 3-NOP by itself does not give a detectable amount of nitrate or nitrite.
Acknowledgments
We thank Peter Livant (Auburn University), Elisabeth Jimenez (Consejo Superior de Investigaciones Cientificas), John Wallace (University of Aberdeen), and Jamie Newbold (Aberystwyth University) for the use of the gas chromatograph, for the in vitro culture work, and for providing stock pure cultures, respectively; Ulrich Ermler and the staff of the PXII beam line at Swiss Light Source (Villigen, Switzerland) for helping with the X-ray data collection; and David Rinaldo (Schrödinger, LLC) for molecular modeling support.
Footnotes
Conflict of interest statement: The authors working at Auburn University and at the Max Planck Institute in Marburg (E.C.D., T.W., S.S., D.P., B.C., and R.K.T.) declare no competing financial interests. The authors affiliated with Estation Experimental del Zaidin (D.R.Y.-R.) or DSM Nutritional Products (S.D., R.R., R.T.S., and M.K.) have filed patent applications for nitrate esters as inhibitors of rumen methanogenesis.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5G0R).
See Commentary on page 6100.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600298113/-/DCSupplemental.
References
- 1.Myhre G, et al. Anthropogenic and natural radiative forcing. In: Stocker TF, et al., editors. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge Univ Press; Cambridge, UK: 2013. pp. 659–740. [Google Scholar]
- 2.Shindell D, et al. Simultaneously mitigating near-term climate change and improving human health and food security. Science. 2012;335(6065):183–189. doi: 10.1126/science.1210026. [DOI] [PubMed] [Google Scholar]
- 3.Robinson TP, et al. Mapping the global distribution of livestock. PLoS One. 2014;9(5):e96084. doi: 10.1371/journal.pone.0096084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ Microbiol Rep. 2009;1(5):285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- 5.Wolin MJ. Fermentation in the rumen and human large intestine. Science. 1981;213(4515):1463–1468. doi: 10.1126/science.7280665. [DOI] [PubMed] [Google Scholar]
- 6.Zinder SH. Physiological ecology of methanogens. In: Ferry JG, editor. Methanogenesis: Ecology, Physiology, Biochemistry and Genetics. Chapman and Hall; New York: 1993. pp. 128–206. [Google Scholar]
- 7.Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;73(8):2483–2492. doi: 10.2527/1995.7382483x. [DOI] [PubMed] [Google Scholar]
- 8.Hungate RE. Hydrogen as an intermediate in the rumen fermentation. Arch Mikrobiol. 1967;59(1):158–164. doi: 10.1007/BF00406327. [DOI] [PubMed] [Google Scholar]
- 9.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ungerfeld EM. Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: A meta-analysis. Front Microbiol. 2015;6:37. doi: 10.3389/fmicb.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, et al. A mathematical model to describe in vitro kinetics of H2 gas accumulation. Anim Feed Sci Technol. 2013;184(1-4):1–16. [Google Scholar]
- 12.Zheng Y, Kahnt J, Kwon IH, Mackie RI, Thauer RK. Hydrogen formation and its regulation in Ruminococcus albus: Involvement of an electron-bifurcating [FeFe]-hydrogenase, of a non-electron-bifurcating [FeFe]-hydrogenase, and of a putative hydrogen-sensing [FeFe]-hydrogenase. J Bacteriol. 2014;196(22):3840–3852. doi: 10.1128/JB.02070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen PH. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim Feed Sci Technol. 2010;160(1-2):1–22. [Google Scholar]
- 14.Ungerfeld EM, Kohn RA. The role of thermodynamics in the control of ruminal fermentation. In: Sejrsen K, Hvelplund TMO, Nielsen MO, editors. Ruminant Physiology: Digestion, Metabolism and Impact of Nutrition on Gene Expression, Immunology and Stress. Academic; Wageningen, The Netherlands: 2008. pp. 55–86. [Google Scholar]
- 15.Denman SE, Martinez Fernandez G, Shinkai T, Mitsumori M, McSweeney CS. Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front Microbiol. 2015;6:1087. doi: 10.3389/fmicb.2015.01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodds KG, Auvray B, Newman SA, McEwan JC. Genomic breed prediction in New Zealand sheep. BMC Genet. 2014;15(92):92. doi: 10.1186/s12863-014-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wedlock DN, Janssen PH, Leahy SC, Shu D, Buddle BM. Progress in the development of vaccines against rumen methanogens. Animal. 2013;7(Suppl 2):244–252. doi: 10.1017/S1751731113000682. [DOI] [PubMed] [Google Scholar]
- 18.Hristov AN, et al. Special topics—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J Anim Sci. 2013;91(11):5045–5069. doi: 10.2527/jas.2013-6583. [DOI] [PubMed] [Google Scholar]
- 19.Hristov AN, et al. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc Natl Acad Sci USA. 2015;112(34):10663–10668. doi: 10.1073/pnas.1504124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Fernández G, et al. Effects of ethyl-3-nitrooxy propionate and 3-nitrooxypropanol on ruminal fermentation, microbial abundance, and methane emissions in sheep. J Dairy Sci. 2014;97(6):3790–3799. doi: 10.3168/jds.2013-7398. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds CK, et al. Effects of 3-nitrooxypropanol on methane emission, digestion, and energy and nitrogen balance of lactating dairy cows. J Dairy Sci. 2014;97(6):3777–3789. doi: 10.3168/jds.2013-7397. [DOI] [PubMed] [Google Scholar]
- 22.Romero-Perez A, et al. Sustained reduction in methane production from long-term addition of 3-nitrooxypropanol to a beef cattle diet. J Anim Sci. 2015;93(4):1780–1791. doi: 10.2527/jas.2014-8726. [DOI] [PubMed] [Google Scholar]
- 23.Thauer RK. Biochemistry of methanogenesis: A tribute to Marjory Stephenson. Microbiology. 1998;144(Pt 9):2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 24.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6(8):579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 25.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Crystal structure of methyl-coenzyme M reductase: The key enzyme of biological methane formation. Science. 1997;278(5342):1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 26.Ermler U. On the mechanism of methyl-coenzyme M reductase. Dalton T. 2005;2005(21):3451–3458. doi: 10.1039/b506697b. [DOI] [PubMed] [Google Scholar]
- 27.Cedervall PE, et al. Structural analysis of a Ni-methyl species in methyl-coenzyme M reductase from Methanothermobacter marburgensis. J Am Chem Soc. 2011;133(15):5626–5628. doi: 10.1021/ja110492p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinderberger D, et al. Coordination and binding geometry of methyl-coenzyme M in the red1m state of methyl-coenzyme M reductase. J Biol Inorg Chem. 2008;13(8):1275–1289. doi: 10.1007/s00775-008-0417-0. [DOI] [PubMed] [Google Scholar]
- 29.Nivinskas H, et al. Reduction of aliphatic nitroesters and N-nitramines by Enterobacter cloacae PB2 pentaerythritol tetranitrate reductase: Quantitative structure-activity relationships. FEBS J. 2008;275(24):6192–6203. doi: 10.1111/j.1742-4658.2008.06744.x. [DOI] [PubMed] [Google Scholar]
- 30.Duin EC, et al. Spectroscopic investigation of the nickel-containing porphinoid cofactor F(430). Comparison of the free cofactor in the (+)1, (+)2 and (+)3 oxidation states with the cofactor bound to methyl-coenzyme M reductase in the silent, red and ox forms. J Biol Inorg Chem. 2004;9(5):563–576. doi: 10.1007/s00775-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 31. Wikipedia Contributors (2016) Tandem-charge. Wikipedia, The Free Encyclopedia. Available at https://en.wikipedia.org/wiki/Tandem-charge. Accessed April 15, 2016.
- 32.Prakash D, Wu Y, Suh SJ, Duin EC. Elucidating the process of activation of methyl-coenzyme M reductase. J Bacteriol. 2014;196(13):2491–2498. doi: 10.1128/JB.01658-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhner CH, Lindenbach BD, Wolfe RS. Component A2 of methylcoenzyme M reductase system from Methanobacterium thermoautotrophicum delta H: Nucleotide sequence and functional expression by Escherichia coli. J Bacteriol. 1993;175(10):3195–3203. doi: 10.1128/jb.175.10.3195-3203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Dorchak AE, Ragsdale SW. In vivo activation of methyl-coenzyme M reductase by carbon monoxide. Front Microbiol. 2013;4:69. doi: 10.3389/fmicb.2013.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgavi DP, Forano E, Martin C, Newbold CJ. Microbial ecosystem and methanogenesis in ruminants. Animal. 2010;4(7):1024–1036. doi: 10.1017/S1751731110000546. [DOI] [PubMed] [Google Scholar]
- 36.Scott RS, Frame SR, Ross PE, Loveless SE, Kennedy GL. Inhalation toxicity of 1,3-propanediol in the rat. Inhal Toxicol. 2005;17(9):487–493. doi: 10.1080/08958370590964485. [DOI] [PubMed] [Google Scholar]
- 37.Kubiak P, et al. Physiological predisposition of various Clostridium species to synthetize 1,3-propanediol from glycerol. Process Biochem. 2012;47(9):1308–1319. [Google Scholar]
- 38.Asanuma N, Yokoyama S, Hino T. Effects of nitrate addition to a diet on fermentation and microbial populations in the rumen of goats, with special reference to Selenomonas ruminantium having the ability to reduce nitrate and nitrite. Anim Sci J. 2015;86(4):378–384. doi: 10.1111/asj.12307. [DOI] [PubMed] [Google Scholar]
- 39.Govoni M, Tocchetti P, Lundberg JO. Metabolism and pathways for denitration of organic nitrates in the human liver. J Pharmacol Exp Ther. 2013;346(1):96–104. doi: 10.1124/jpet.113.203356. [DOI] [PubMed] [Google Scholar]
- 40.Ellermann J, et al. Methyl-coenzyme-M reductase from Methanobacterium thermoautotrophicum (strain Marburg). Purity, activity and novel inhibitors. Eur J Biochem. 1989;184(1):63–68. doi: 10.1111/j.1432-1033.1989.tb14990.x. [DOI] [PubMed] [Google Scholar]
- 41.Goenrich M, et al. Probing the reactivity of Ni in the active site of methyl-coenzyme M reductase with substrate analogues. J Biol Inorg Chem. 2004;9(6):691–705. doi: 10.1007/s00775-004-0552-1. [DOI] [PubMed] [Google Scholar]
- 42.Gräwert T, et al. Inhibition of methyl-CoM reductase from Methanobrevibacter ruminantium by 2-bromoethanesulfonate. J Agric Food Chem. 2014;62(52):12487–12490. doi: 10.1021/jf505056g. [DOI] [PubMed] [Google Scholar]
- 43.Hinderberger D, et al. A nickel-alkyl bond in an inactivated state of the enzyme catalyzing methane formation. Angew Chem Int Ed Engl. 2006;45(22):3602–3607. doi: 10.1002/anie.200600366. [DOI] [PubMed] [Google Scholar]
- 44.Ungerfeld EM, Rust SR, Boone DR, Liu Y. Effects of several inhibitors on pure cultures of ruminal methanogens. J Appl Microbiol. 2004;97(3):520–526. doi: 10.1111/j.1365-2672.2004.02330.x. [DOI] [PubMed] [Google Scholar]
- 45.Taylor CD, McBride BC, Wolfe RS, Bryant MP. Coenzyme M, essential for growth of a rumen strain of Methanobacterium ruminantium. J Bacteriol. 1974;120(2):974–975. doi: 10.1128/jb.120.2.974-975.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovley DR, Greening RC, Ferry JG. Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl Environ Microbiol. 1984;48(1):81–87. doi: 10.1128/aem.48.1.81-87.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balch WE, Wolfe RS. Transport of coenzyme M (2-mercaptoethanesulfonic acid) in Methanobacterium ruminantium. J Bacteriol. 1979;137(1):264–273. doi: 10.1128/jb.137.1.264-273.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santoro N, Konisky J. Characterization of bromoethanesulfonate-resistant mutants of Methanococcus voltae: Evidence of a coenzyme M transport system. J Bacteriol. 1987;169(2):660–665. doi: 10.1128/jb.169.2.660-665.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biava M, et al. Novel analgesic/anti-inflammatory agents: Diarylpyrrole acetic esters endowed with nitric oxide releasing properties. J Med Chem. 2011;54(22):7759–7771. doi: 10.1021/jm200715n. [DOI] [PubMed] [Google Scholar]
- 50.Scheller S, Goenrich M, Thauer RK, Jaun B. Methyl-coenzyme M reductase from methanogenic archaea: Isotope effects on the formation and anaerobic oxidation of methane. J Am Chem Soc. 2013;135(40):14975–14984. doi: 10.1021/ja406485z. [DOI] [PubMed] [Google Scholar]
- 51.Kaster AK, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci USA. 2011;108(7):2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45(1):160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 53.Friesner RA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 54.Repasky MP, Shelley M, Friesner RA. 2007. Flexible ligand docking with glide. Curr Protoc Bioinformatics Chapter 8:Unit 8.12. [DOI] [PubMed]
- 55.Friesner RA, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 56.Halgren TA, et al. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 57.Bonacker LG, Baudner S, Mörschel E, Böcher R, Thauer RK. Properties of the two isoenzymes of methyl-coenzyme M reductase in Methanobacterium thermoautotrophicum. Eur J Biochem. 1993;217(2):587–595. doi: 10.1111/j.1432-1033.1993.tb18281.x. [DOI] [PubMed] [Google Scholar]
- 58.Duin EC, Prakash D, Brungess C. Methyl-coenzyme M reductase from Methanothermobacter marburgensis. Methods Enzymol. 2011;494:159–187. doi: 10.1016/B978-0-12-385112-3.00009-3. [DOI] [PubMed] [Google Scholar]
- 59.Hagen W. Biomolecular EPR Spectroscopy. CRC; Boca Raton, FL: 2009. [Google Scholar]
- 60.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30(Pt 6):1022–1025. [Google Scholar]
- 62.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Afonine PV, et al. phenix.model_vs_data: A high-level tool for the calculation of crystallographic model and data statistics. J Appl Cryst. 2010;43(Pt 4):669–676. doi: 10.1107/S0021889810015608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McSweeney C, Denman S, Roderick M. Rumen bacteria. In: Makkar HPS, McSweeney CS, editors. Methods in Gut Microbial Ecology for Ruminants. Springer; Dordrecht, The Netherlands: 2005. pp. 23–37. [Google Scholar]
- 66.Schönheit P, Moll J, Thauer RK. Growth parameters (Ks, Mu-Max, Ys) of Methanobacterium hermoautotrophicum. Arch Microbiol. 1980;127(1):59–65. doi: 10.1007/BF00403508. [DOI] [PubMed] [Google Scholar]