Significance
Strigolactone hormones regulate many plant growth and developmental processes and are particularly important in regulating growth in response to nonoptimal conditions. Plants produce a range of bioactive strigolactone-like compounds, suggesting that the biosynthesis pathway is complex. Despite this complexity, only one type of enzyme, the MORE AXILLARY GROWTH1 (MAX1) cytochrome P450, has been attributed to the diversity of strigolactones. Using transcriptomics and reverse genetics, we discovered a previously uncharacterized gene that encodes a 2-oxoglutarate and Fe(II)-dependent dioxygenase involved in strigolactone production downstream of MAX1. Studies with the corresponding mutant have shown that previously identified strigolactone-type compounds in Arabidopsis are not the major strigolactone-type shoot branching hormone in this model species.
Keywords: plant, branching, strigolactone, biosynthesis, Arabidopsis
Abstract
Strigolactones are a group of plant compounds of diverse but related chemical structures. They have similar bioactivity across a broad range of plant species, act to optimize plant growth and development, and promote soil microbe interactions. Carlactone, a common precursor to strigolactones, is produced by conserved enzymes found in a number of diverse species. Versions of the MORE AXILLARY GROWTH1 (MAX1) cytochrome P450 from rice and Arabidopsis thaliana make specific subsets of strigolactones from carlactone. However, the diversity of natural strigolactones suggests that additional enzymes are involved and remain to be discovered. Here, we use an innovative method that has revealed a missing enzyme involved in strigolactone metabolism. By using a transcriptomics approach involving a range of treatments that modify strigolactone biosynthesis gene expression coupled with reverse genetics, we identified LATERAL BRANCHING OXIDOREDUCTASE (LBO), a gene encoding an oxidoreductase-like enzyme of the 2-oxoglutarate and Fe(II)-dependent dioxygenase superfamily. Arabidopsis lbo mutants exhibited increased shoot branching, but the lbo mutation did not enhance the max mutant phenotype. Grafting indicated that LBO is required for a graft-transmissible signal that, in turn, requires a product of MAX1. Mutant lbo backgrounds showed reduced responses to carlactone, the substrate of MAX1, and methyl carlactonoate (MeCLA), a product downstream of MAX1. Furthermore, lbo mutants contained increased amounts of these compounds, and the LBO protein specifically converts MeCLA to an unidentified strigolactone-like compound. Thus, LBO function may be important in the later steps of strigolactone biosynthesis to inhibit shoot branching in Arabidopsis and other seed plants.
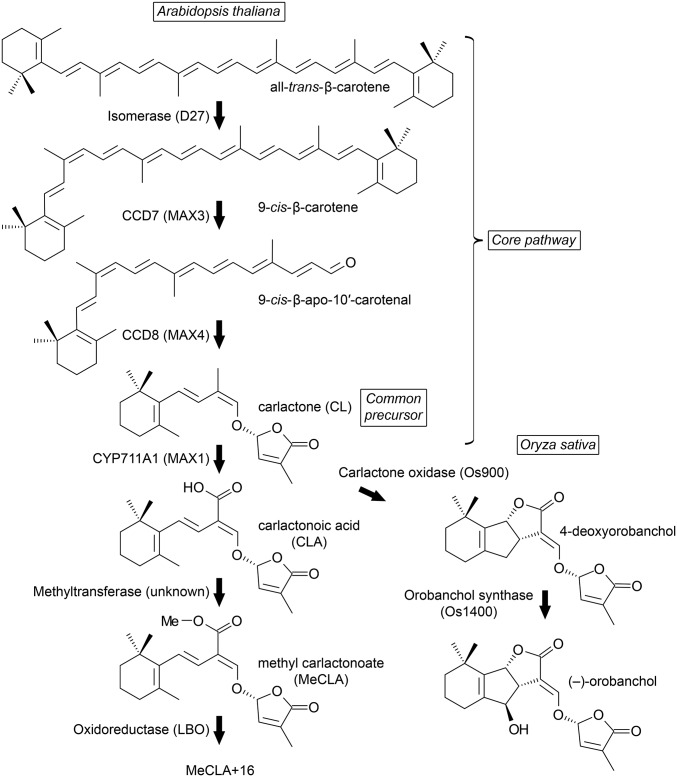
Strigolactone synthesis from β-carotene involves 9-cis/all-trans-β-carotene isomerase, carotenoid cleavage dioxygenase 7 (CCD7), and CCD8 (reviewed in ref. 1). These three proteins can sequentially convert β-carotene into carlactone (CL), an intermediate molecule so far detected in root extracts from both rice and Arabidopsis (2, 3). In Arabidopsis, the isomerase is encoded by DWARF27 (D27), whereas CCD7 and CCD8 are encoded by MORE AXILLARY GROWTH 3 (MAX3) and MAX4, respectively (Fig. 1).
Fig. 1.
The strigolactone biosynthesis pathway in Arabidopsis thaliana. An isomerase and two CCD enzymes convert β-carotene into CL, the common precursor of diverse strigolactones. In rice, CL is oxidized by two cytochrome P450 enzymes to orobanchol, which functions in the rhizosphere as a signal for mycorrhizal fungi. In Arabidopsis, CL is oxidized by the MAX1 cytochrome P450 to CLA, which is converted to MeCLA. Evidence presented in this study suggests that LBO facilitates additional processing to an unknown strigolactone-like product (MeCLA + 16 Da), which is required for the control of shoot branching.
In rice, conversion of CL to the four-ringed canonical strigolactone structure involves the addition of two oxygen atoms to C19 and closure of the B and C rings to produce 4-deoxyorobanchol (4DO; previously known as ent-2′-epi-5-deoxystrigol) (Fig. 1). This reaction is catalyzed by a member of the MAX1 family of cytochrome P450 proteins (4, 5). A second MAX1 enzyme oxidizes 4DO to make orobanchol (4). In Arabidopsis, the situation is less clear, because it has only a single MAX1 gene, and the encoded protein oxidizes C19 of CL to produce carlactonoic acid (CLA), but it does not apparently catalyze formation of B and C rings (3, 4, 6) (Fig. 1). These studies reported endogenous CLA and methyl carlactonoate (MeCLA), the production of the latter requiring an unknown methyl transferase (6) (Fig. 1). Based on interaction with receptor D14, it was proposed that MeCLA is bioactive, whereas CL and CLA are not bioactive (6). Grafting experiments in Arabidopsis suggest that both the upstream and downstream products of MAX1 are graft-transmissible: that is, the MAX1 substrate and products can move from the rootstock to the shoot (5). This finding implies that the graft-transmissible mobile products in Arabidopsis could be CL- and CLA-related products. Previous reports of other strigolactones in Arabidopsis (7, 8) have not been repeated (3, 6).
Strigolactones are thought to be perceived and cleaved by an α/β-fold hydrolase (AtD14 in Arabidopsis), possibly within a receptor complex that may include an F-box protein (MAX2 in Arabidopsis) (reviewed in ref. 1). The active complex targets proteins for degradation, such as SMAX1-like proteins (SMXL6–SMXL8), and promotes expression of transcription factor genes, such as BRANCHED1, directly in buds (9–13). The active complex is unusual in that binding with strigolactone triggers destabilization and degradation of the D14 receptor and cleavage of the strigolactone compound (14–16). Negative feedback on some strigolactone biosynthesis genes has been observed in species, including pea and Arabidopsis (reviewed in ref. 17).
Forward genetic screens identifying increased branching mutants in several species have led to identification of all of the strigolactone biosynthesis and signaling pathway components to date (reviewed in ref. 18). It is likely that additional enzymes are yet to be discovered because of the diversity of strigolactones observed in plants. Strigolactones of variable structure have recently been isolated from rice (orobanchyl acetate, 7-oxoorobanchyl acetate, and three methoxy-5-deoxystrigol isomers), oats (avenaol), and sunflower (heliolactone) (19–21). If additional enzymes are required for bioactive strigolactones, the genes involved would prove difficult to discover using forward genetics if the corresponding mutants had weak phenotypes and/or enzyme redundancy.
A common feature of MAX3 and MAX4 genes in Arabidopsis is that their expression is increased in strigolactone biosynthesis and perception mutants, and their transcript levels alter with changes in auxin levels and/or auxin treatments (17). Therefore, we postulated that additional novel strigolactone biosynthesis genes, if present, may be coexpressed with these strigolactone biosynthesis genes under specific conditions. Thus, we used an Arabidopsis microarray to identify candidate genes followed by reverse genetics to discover new branching mutants. Here, we describe a previously uncharacterized branching gene in Arabidopsis, LATERAL BRANCHING OXIDOREDUCTASE (LBO), which encodes an oxidoreductase-like enzyme of the 2-oxoglutarate and Fe(II)-dependent dioxygenase (2OGD) superfamily and is represented widely across seed plant species. Our analysis suggests that LBO plays an essential role in the production of strigolactone-like compounds downstream of MAX genes and CL. This discovery will be vital for identifying additional bioactive strigolactones and increasing our understanding of the components and regulation of the strigolactone biosynthesis pathway and hence, the levels and functions of strigolactones in plants.
Results
Transcriptomics and Reverse Genetics Reveal a Novel Branching Gene.
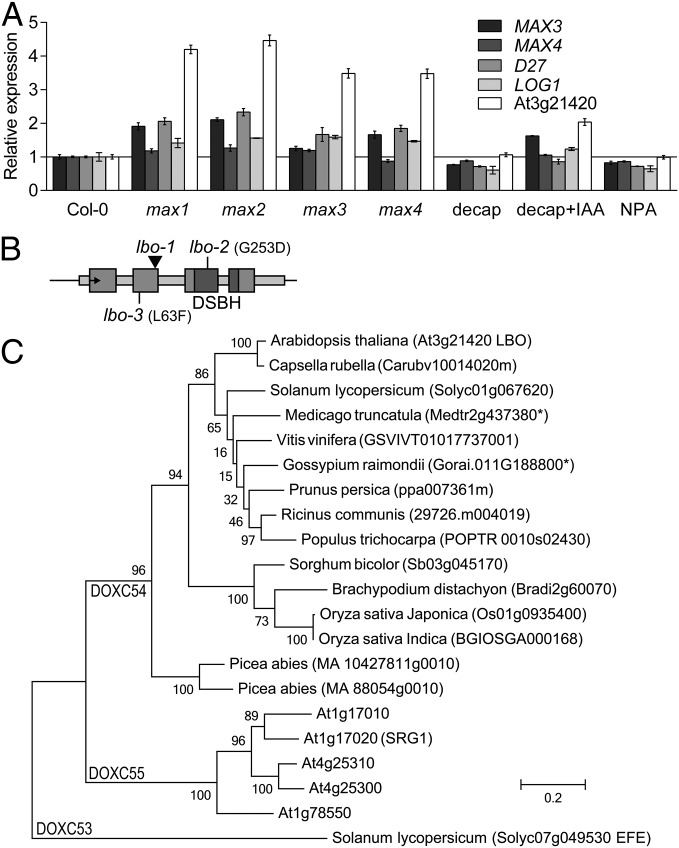
To identify gene(s) involved in strigolactone biosynthesis in Arabidopsis, we analyzed changes in gene expression in eight conditions using an Arabidopsis microarray. Compared with untreated WT plants, two conditions decreased the expression of MAX3: removal of the shoot tip (decapitation) and treatment with an auxin transport inhibitor (N-1-naphthylphthalamic acid). Five additional conditions enhanced MAX3 gene expression: mutants of max1, max2, max3, and max4 as well as the WT decapitated and treated with auxin (indole-3-acetic acid) (Fig. 2A).
Fig. 2.
Discovery of the LBO gene by reverse genetics. (A) Expression from microarray data revealed genes that followed the distinctive pattern (41) of the MAX3 gene in hypocotyls dissected from soil-grown WT, max mutants, decapitated WT (decap) with or without apical auxin [indole-3-acetic acid (IAA)] added, or WT with N-1-naphthylphthalamic acid (NPA) applied locally beneath the rosette. This group included D27, LONELY GUY1 (LOG1), and an uncharacterized gene At3g21420 (LBO). Microarray expression values are relative to untreated Col-0. Mean ± SEM (n = 2–3; 120–150 hypocotyls per replicate). (B) Diagram representing the DNA sequence of the LBO locus. The arrow points to the start codon, exons are large boxes, and introns and untranslated regions are narrow boxes. The promoter region spans 3,627 bp to the next gene, and 1,711 bp were used for GUS expression. The positions of the FLAG_119G09 line T-DNA insert (lbo-1; arrowhead), other point mutation alleles (lbo-2 and lbo-3), and the double-stranded β-helix conserved catalytic domain (DSBH) are shown. (C) A phylogenetic tree of 2OGD amino acid sequences from 2OGD clade DOXC54 (25) from prominent species reveals the similarity of At3g21420 (LBO). For reference, the Arabidopsis sequences from the next nearest clade (DOXC55) and the nearest similar sequence with reported functional characterization, ETHYLENE FORMING ENZYME (EFE) from tomato (Solanum lycopersicum) in clade DOXC53, are included. Bootstrap values are shown for each branch. Units are amino acid substitutions per site. *Existence of a splice variant.
Hypocotyl tissue just before bolting was selected for expression analysis, because cell expansion and growth have been completed in this tissue. Furthermore, the hypocotyl is physiologically relevant: it has been shown as a source of the branching inhibition signal, and changes in expression of strigolactone genes have been observed in this tissue (5, 17). Using the Cluster Analysis of Sequences bioinformatics program (22), we were able to identify genes that showed appreciable coexpression with MAX3. This analysis led to the identification of the D27 gene, which we subsequently reported (23), and LONELY GUY1 (At2g28305), which is involved in the control of shoot branching through a role in cytokinin metabolism (Fig. 2A) (24). D27 had not been previously identified from forward genetic screens in Arabidopsis. The analysis also identified an uncharacterized gene At3g21420, which we named LBO based on gene phylogeny (Fig. 2) and mutant phenotype (Fig. 3).
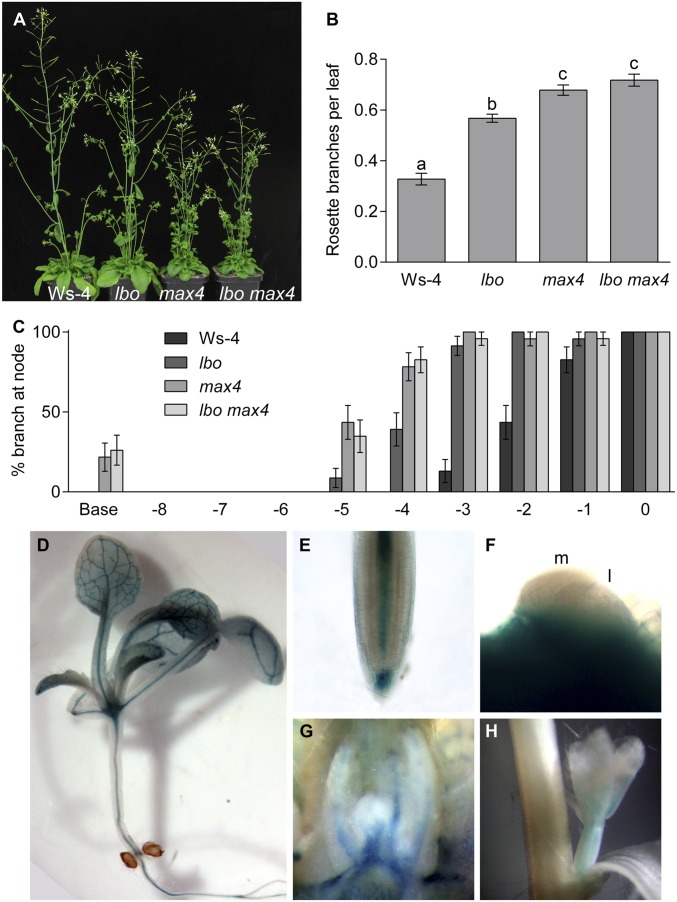
Fig. 3.
Mutant phenotype and reporter gene expression of LBO. (A) The lbo-1 mutant shoot branching phenotype was intermediate between those of max4-9 and the Ws-4 WT. The lbo-1 max4-9 double mutant appeared most similar to max4-9. (B) These relationships were reflected in the average number of branches (>5 mm) per rosette leaf. Values with different letters are significantly different from each other (P < 0.05; t test). Mean ± SEM (n = 23–24). (C) Careful examination of the occurrence of a branch in each leaf axil (node) was carried out, with the highest rosette node denoted as zero, counting backward down the rosette. This analysis revealed extra branching in the middle rosette nodes in max4-9 and lbo-1 max4-9 plants and mainly the upper middle nodes in lbo-1 compared with in the WT (Ws-4). Occasionally, branches were seen at the extreme base of the rosette, but it was not possible to determine the precise location. (D) To further characterize LBO, we generated a pLBO::GUS construct using 1,711 bp of the promoter and examined expression in whole-mount samples (Col-0). Representative lines showed GUS expression in the vasculature throughout the plant. (E) Expression extended into the stele of the root tip and was also seen in the root cap and weakly in the epidermis. (F) The meristem (m) and tiny leaves (l) of repressed buds from lower nodes did not show expression (although high expression occurred in leaf vasculature just below these tissues). (G) Larger repressed buds of higher nodes showed expression in the vasculature that reaches up into the four or five immature leaves/flowers. (H) In the inflorescence bolt, most GUS expression was observed specifically in the vasculature of cauline branches and not in the main stem.
The predicted LBO amino acid sequence is similar to 2OGDs, which comprise a family of around 130 members in Arabidopsis (25). Related 2OGDs are involved in the biosynthesis of a range of plant hormones and secondary metabolites (26). Like other 2OGDs, LBO has a predicted oxoglutarate/iron-dependent dioxygenase catalytic domain near the C terminus that contains a double-stranded β-helix core fold (Fig. 2B). Based on the crystal structure of similar proteins, the LBO catalytic domain contains hallmark amino acid residues His-235, His-293, and Asp-237 for iron binding and Arg-303 for substrate interaction (27–29).
Phylogenetic analyses suggest that LBO exists as a highly conserved and mostly single-copy gene in seed plant species (25) (Fig. 2C). LBO falls within the DOXC54 clade of 2OGDs, and sequences from the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii do not group in this clade (25), suggesting that the DOXC54 clade evolved after the divergence of lycophyte and seed plant lineages. Clades that cluster near DOXC54 display more diversity, and their functions are generally unknown (25) (Fig. 2C). The closest sequence with reported enzymatic function is ETHYLENE-FORMING ENZYME from tomato, which converts 1-aminocyclopropane-1-carboxylic acid to ethylene (Fig. 2C) (30).
Analysis of lbo Mutants.
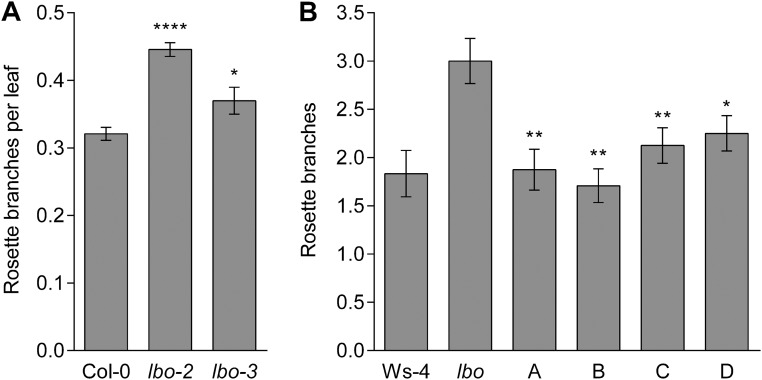
We identified one transfer-DNA (T-DNA) insertion mutant in At3g21420; FLAG_119G09 from the Versailles Arabidopsis Stock Center (31). This line is in the Wassilewskija ecotype (Ws-4) and carries a T-DNA in the second exon of At3g21420 (lbo-1) (Fig. 2B). Two additional mutant alleles [lbo-2 and lbo-3; Columbia-0 (Col-0) erecta background] were isolated from the University of California, Davis TILLING Core (32). These alleles contain a nonconservative change in the protein sequence; in lbo-2, amino acid G253 was changed to D (nucleotide G1200 to A), and in lbo-3, amino acid L63 was changed to F (nucleotide C187 to T) (Fig. 2B). Homozygous mutant plants exhibited increased rosette branches compared with their respective WTs (Fig. 3 A and B and Fig. S1A). The T-DNA in lbo-1 would be expected to severely shorten the predicted protein sequence of LBO because of a truncation before the putative double-stranded β-helix catalytic oxygenase domain. The lbo-3 mutant displays a weaker branching phenotype compared with lbo-2 (Fig. S1A). This finding indicates that the lbo-3 allele, despite altering a highly conserved amino acid, may not fully disrupt LBO function (Fig. 2B).
Fig. S1.
(A) Increased branching of lbo-2 (PB7) and lbo-3 (PB15) alleles compared with Col-0 erecta-109. Mean ± SEM (n = 15). (B) Complementation of branching by LBO. Branching in four complemented p35S::LBO lbo-1 lines showed significantly reduced branching compared with lbo-1 mutants. Mean ± SEM (n = 11–24). *P < 0.05 (t test); **P < 0.01 (t test); ****P < 0.0001 (t test).
The increased branching phenotype of lbo-1 and max4-9 was mainly observed in the middle nodes of the rosette (Fig. 3C). These branches occurred less frequently in lbo compared with max4 (Fig. 3C). This subtle but significant phenotype of lbo mutants would probably not have been observed in a forward genetics screen. Independent lbo-1 lines constitutively expressing LBO using the cauliflower mosaic virus 35S promoter showed WT levels of branching (Fig. S1B). These findings show that LBO is required for branching inhibition. Similar to strigolactone biosynthesis genes (5, 23, 33, 34), overexpression of LBO was not able to decrease branching below WT levels (Fig. S1B).
To determine the localization of LBO within the plant, LBO reporter gene lines expressing β-glucuronidase (GUS) were generated. Lines expressing pLBO::GUS suggest that LBO is expressed in the vasculature throughout the plant and in the buds and root tips (Fig. 3 D–H). Other genes involved in strigolactone production and response are also expressed in vascular tissue (5, 34, 35).
LBO May Act as a Strigolactone Biosynthesis Enzyme Downstream of MAX1.
To elucidate the role of LBO in the strigolactone pathway, we made lbo max4 double mutants. The phenotype of lbo max4 double-mutant plants seemed similar to that of max4 single mutants (Fig. 3 A–C), implying that LBO may act in the same pathway as MAX4.
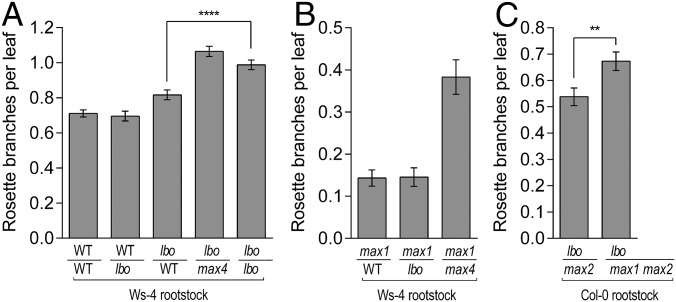
Previously, grafting has shown that strigolactone biosynthesis occurs in both the roots and shoots and that the strigolactone signal moves upward in plants (36). Therefore, to further understand the role of LBO, grafting experiments with WT and other strigolactone mutants were carried out. Grafting lbo onto the WT (Ws-4) rootstocks significantly reduced branching in lbo mutant shoots (Fig. 4A). Branching of WT (Ws-4) shoots was not affected by grafting onto lbo rootstocks, which is similar to grafting results found with other strigolactone mutants (36–38). Thus, grafting results together with LBO expression patterns suggest that LBO acts in the root and shoot to control a long-distance signal that inhibits branching.
Fig. 4.
Grafting with lbo-1. (A) lbo rootstock did not promote branching in Ws-4 shoots. Ws-4 rootstock repressed lbo shoot branching nearly to Ws-4 self-graft levels, whereas max4-9 rootstock did not repress lbo shoots. These grafting results mean that the normal enzymatic action of LBO in Ws-4 roots can produce an upwardly mobile product that can repress branching (although not completely) and that this product requires MAX4 enzyme action (i.e., MAX4 is upstream of LBO). Mean ± SEM (n = 19–36). ****P < 0.0001 (t test). (B) lbo rootstock represses branching in max1-1 (background Col-0) mutant shoots to the same extent as Ws-4 rootstock compared with max4-9. This inhibition by lbo rootstocks means that the enzyme action of MAX1 in lbo mutant rootstock produces a mobile product that can fully restore max1 branching and that this product requires MAX4 action. Mean ± SEM (n = 15–20). (C) The max2-1 rootstock reduced branching in the lbo-1 mutant shoot compared with max1-1 max2-1 double-mutant rootstock. This finding suggests that LBO (in max2) can produce a mobile branching inhibitor and that this product requires MAX1 action upstream of LBO. The rationale for using max2-1 (in Col-0) rootstock here was that impaired signaling is expected to increase production of strigolactones and mobile intermediates (17) and that max1 and max2 were not available in the Ws-4 ecotype. Mean ± SEM (n = 20–23). **P < 0.01 (t test).
Previous studies showed that mutant max1 rootstocks can fully restore branching to max3 or max4 mutant shoots but that max3 and max4 rootstocks cannot inhibit branching in max1 shoots (5). This grafting shows that the MAX1 enzyme acts downstream of MAX3 and MAX4. In addition, it confirms that MAX1 acts on a mobile substrate produced by MAX3 and MAX4. Using the same logic, our reciprocal grafting suggests that LBO acts downstream of MAX4 and MAX1. Mutant lbo rootstocks fully rescued branching in max1 shoots (Fig. 4B). However, neither max1 nor max4 rootstocks could reduce branching in lbo shoots, despite these rootstocks having the LBO enzyme (Fig. 4C). The LBO enzyme may, therefore, be required to convert a mobile product of MAX1 to a product that is either a branching inhibitor or further converted into one.
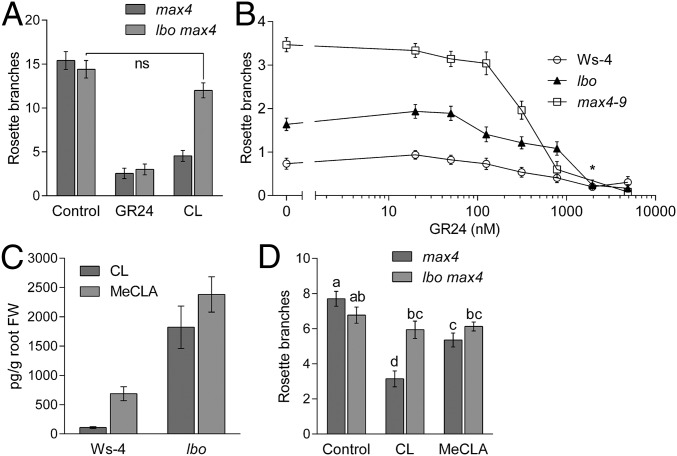
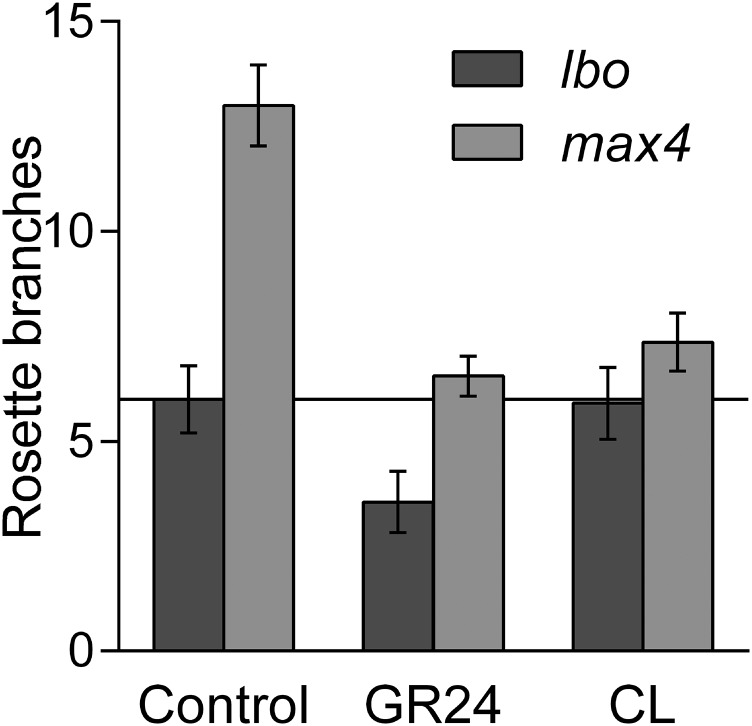
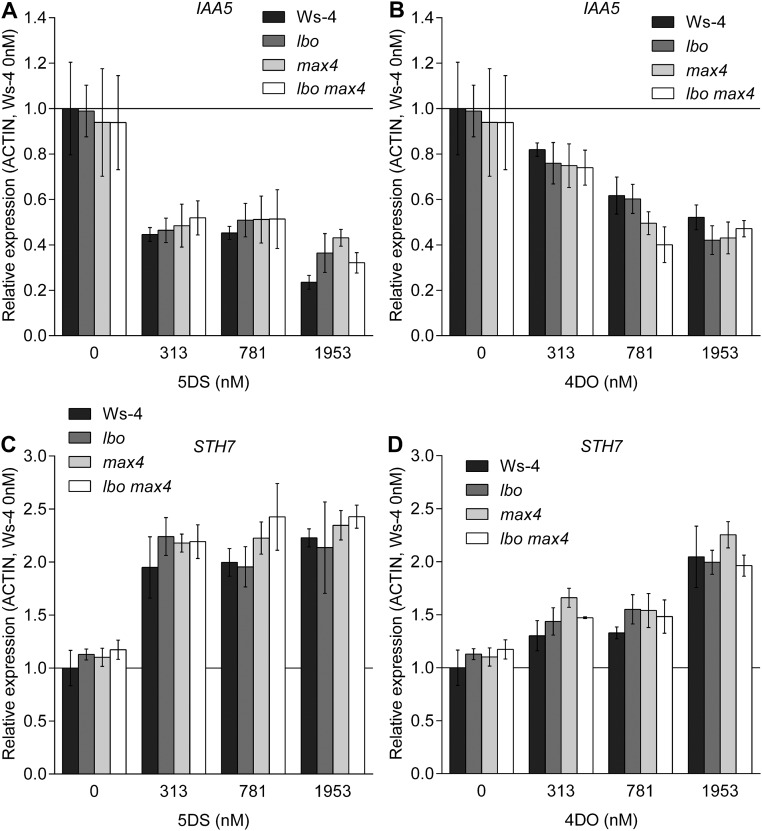
If LBO acts downstream of MAX1 in strigolactone biosynthesis, then branching in lbo mutants should be inhibited by canonical strigolactones but possibly not by CL, which is produced after the action of D27, MAX3, and MAX4 but before MAX1 activity (Fig. 1); max1 mutants respond to synthetic strigolactone (rac-GR24) and not CL (6, 39). As expected, when rac-GR24 or CL was supplied hydroponically, branching in max4 shoots was reduced (Fig. 5A and Fig. S2). The lbo mutants also responded to rac-GR24 in a concentration-dependent manner (Fig. 5B). As predicted, similarly to max1, there was no significant response to CL in lbo single mutants (Fig. S2) or lbo max4 double-mutant plants (Fig. 5A). Gene expression experiments also indicate that max4 and lbo do not differ in strigolactone sensitivity. Gene expression responses to 5-deoxystrigol or 4DO were not different among WT, lbo, and max4 lines (Fig. S3).
Fig. 5.
Strigolactone responses and quantification in lbo-1. (A) Branching in max4-9 was reduced with 1 μM rac-GR24 or 1 μM CL supplied hydroponically. In contrast, combining the lbo-1 mutation with max4-9 as a double mutant specifically inhibits just the response to CL (P > 0.05; t test). Mean ± SEM (n = 10–12). Ns, Not significant. (B) lbo-1 branching responded similarly to Ws-4 across a dose range of rac-GR24 added to agar growth medium, suggesting that lbo is not defective in strigolactone response. *No max4-9 data at this concentration. Mean ± SEM (n = 10–34). (C) CL and MeCLA were increased in lbo-1 mutant root tissue extracts compared with the WT (Ws-4). No canonical strigolactones were detected in any Arabidopsis sample. Mean ± SEM (n = 20). FW, fresh weight. (D) Ten micromolar CL or MeCLA reduced branching in max4-9 but not in the lbo-1 max4-9 double mutant. Values with different letters are significantly different from each other (P < 0.05; t test). Mean ± SEM (n = 10–16).
Fig. S2.
CL response of lbo-1. The max4-9 mutant branching phenotype was reduced in response to 1 μM rac-GR24 or 1 μM CL supplied hydroponically. Although lbo plants also responded to rac-GR24 (P < 0.05; t test), they did not respond to CL. Mean ± SEM (n = 9–12).
Fig. S3.
Gene expression response in lbo-1. Whole seedlings of lbo mutant and lbo max4 double mutant showed equal sensitivity in (A and B) the decrease of IAA5 and (C and D) the decrease of STH7 gene expression after 4 h treatment with different doses of 5-deoxystrigol (5DS) or 4DO. This result strongly suggests that lbo is not deficient in response to strigolactones, inferring that lbo may instead be defective in producing strigolactones. (A and B) IAA5 can be promoted by auxin (53) or repressed by strigolactones (54), and (C and D) SALT TOLERANCE HOMOLOG7 (STH7) can be promoted by karrikins or strigolactones (55). Mean ± SEM (n = 3; 10–15 seedlings each).
Arabidopsis MAX1 has been shown to convert CL into CLA, and then, CLA is thought to be converted by another enzyme into MeCLA (6) (Fig. 1). To clarify further that LBO acts downstream of MAX1, we examined the level of CL and MeCLA in the lbo mutant. If LBO acts downstream of MAX1, we might expect one or both of these compounds to be increased in lbo mutant plants. Indeed, lbo mutant backgrounds had increased levels of CL and MeCLA (Fig. 5C and Figs. S4 and S5).
Fig. S4.
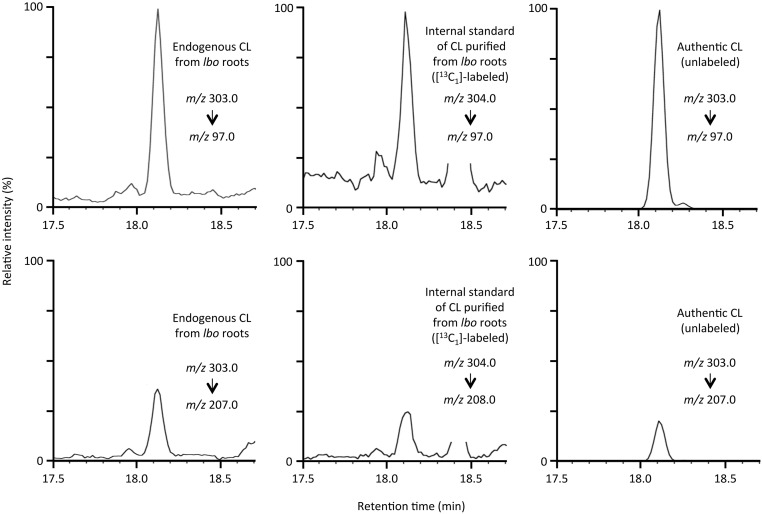
Liquid chromatography–MS/MS analysis of endogenous CL from root extracts. Selected reaction monitoring using major fragment ions of endogenous CL extracted from lbo-1 roots, [1-13CH3]–labeled (internal standard) purified from lbo-1 roots, and unlabeled authentic standard are shown.
Fig. S5.
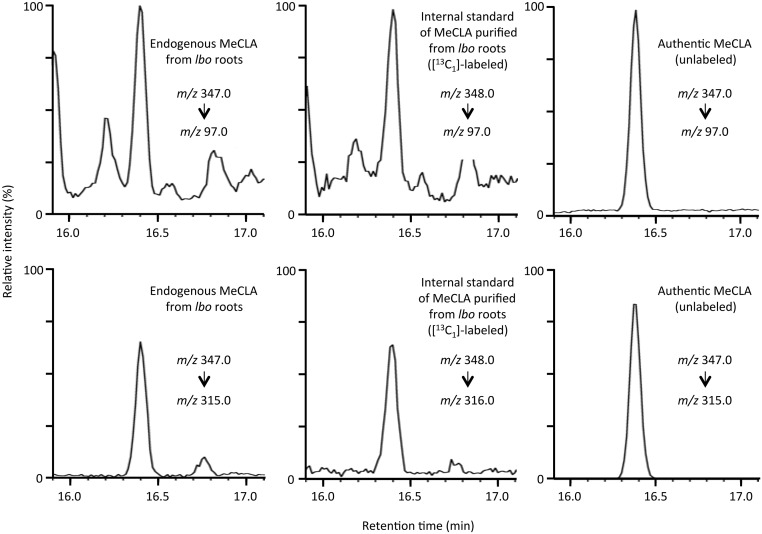
Liquid chromatography–MS/MS analysis of endogenous MeCLA from root extracts. Selected reaction monitoring using major fragment ions of endogenous MeCLA extracted from lbo-1 roots, [1-13CH3] –labeled (internal standard) purified from lbo-1 roots, and unlabeled authentic standard are shown.
Because lbo mutants have increased levels of MeCLA, we predict that the LBO enzyme acts downstream of MeCLA and that it should, therefore, have a reduced response to this compound. We treated max4 and lbo max4 double mutants with MeCLA to determine if MeCLA could reduce branching. The results show that MeCLA slightly but significantly reduced branching in max4 mutants but that it had no effect on the lbo max4 double mutant (Fig. 5D). The differences in the response of max4 to CL and MeCLA are most likely caused by CL being more stable than MeCLA, thus contributing to the greater inhibition observed with CL than with MeCLA. Nevertheless, these results coupled with the increased endogenous levels of MeCLA in the lbo mutants suggest that MeCLA is not very effective at inhibiting branching in plants. This finding is in contrast with a previous report suggesting that MeCLA is the bioactive strigolactone for shoot branching in Arabidopsis (6). Although MeCLA interacts with D14, indicating potential importance in strigolactone signaling, this study reveals that MeCLA requires the presence of LBO to be an effective branching inhibitor.
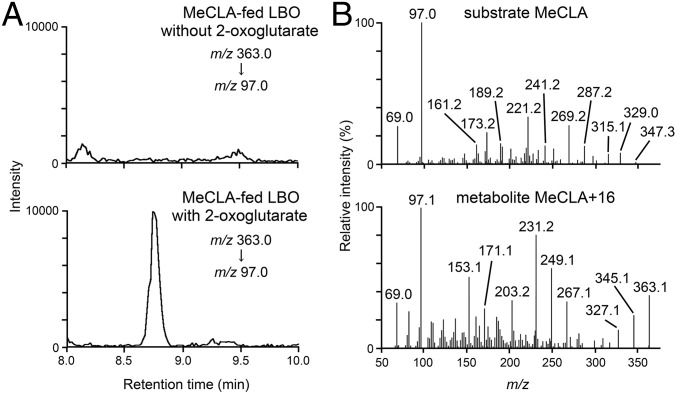
The enzymatic action of LBO was investigated by incubating the soluble protein of Escherichia coli cells expressing the LBO recombinant protein (Fig. S6). Protein extracts were incubated with CL, CLA, or MeCLA, and their levels were monitored by liquid chromatography–MS/MS; comparison was made with protein extracts from cells containing the empty vector (Fig. S7) and with or without 2-oxoglutarate (Fig. 6A). Levels of MeCLA were reduced within 15 min in the presence of LBO (Fig. 6A), whereas levels of CL or CLA were unaffected by LBO (Fig. S7). An ion [M+H]+ of m/z 363 was consistently observed from reactions with MeCLA, which correlates to MeCLA + 16 Da (Fig. 6B), and as expected for a 2OGD enzyme, the production of MeCLA + 16 Da required 2-oxoglutarate (Fig. 6A). The observed mass of MeCLA + 16 Da suggests that LBO has added an oxygen to MeCLA. However, identification of this compound has not yet been possible because of its high instability.
Fig. S6.
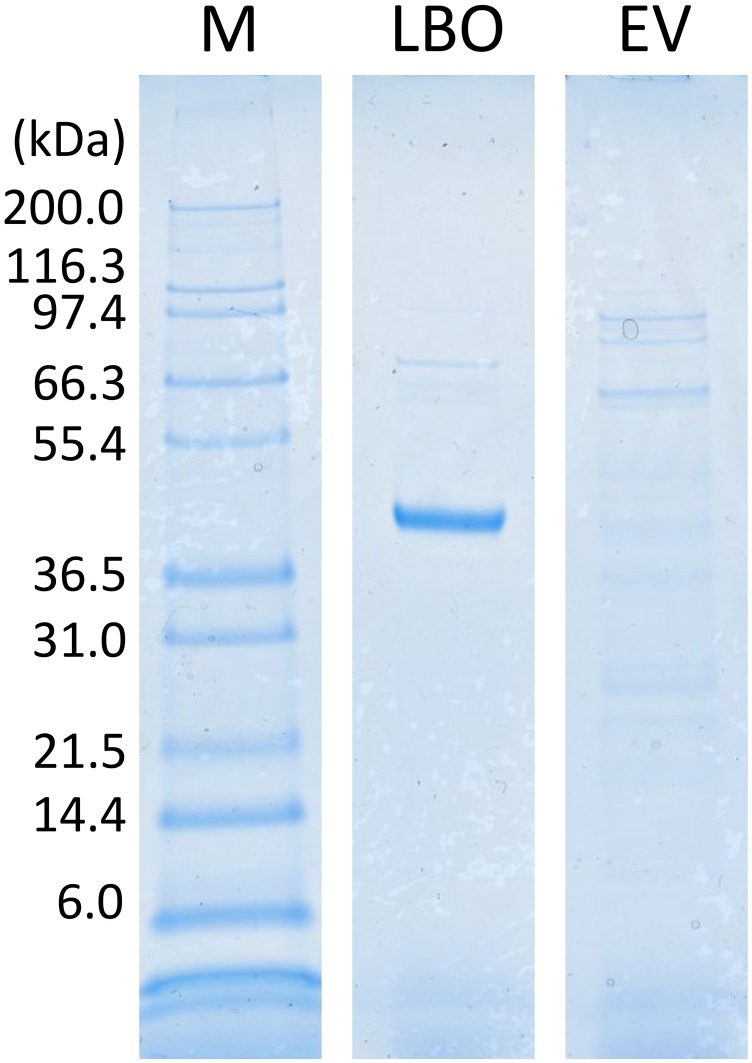
SDS/PAGE of recombinant LBO protein. Soluble protein fractions in E. coli having an LBO expression vector (LBO) and empty vector (EV) were fractionated using an affinity column for histidine-tagged proteins and then, loaded in a 4–12% (wt/vol) polyacrylamide gradient gel. The gel was stained using Coomassie Brilliant Blue. M, protein marker.
Fig. S7.
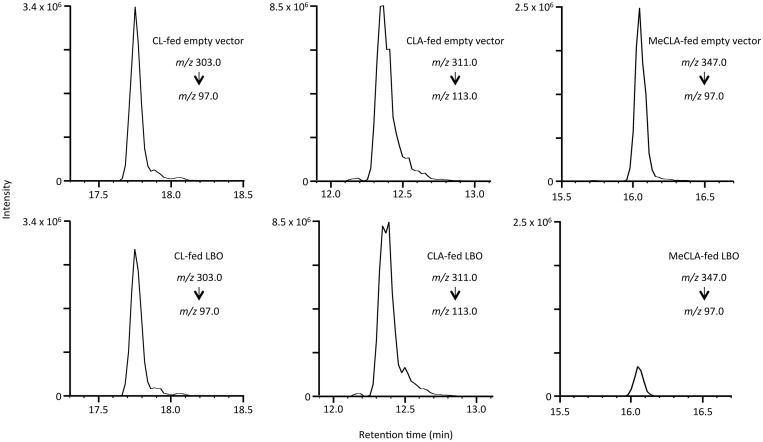
Consumption of CL, CLA, and MeCLA by recombinant LBO protein. Empty vector-transformed cells were used as a control. Substrates were incubated with soluble protein fractions prepared from E. coli having either (Upper) empty vector or (Lower) LBO, and the extracts were analyzed using liquid chromatography–MS/MS. Selected Reaction Monitoring chromatograms of (Left) CL, (Center) CLA, and (Right) MeCLA are shown.
Fig. 6.
Conversions of MeCLA to MeCLA + 16 Da (MeCLA+16) by recombinant LBO protein synthesized in E. coli. MeCLA was incubated with LBO protein for 15 min, and the extracts were analyzed using electrospray ionization (ESI)-positive mode liquid chromatography–MS/MS. (A) Selected reaction monitoring chromatograms for MeCLA-fed LBO (Upper) without or (Lower) with 2-oxoglutarate. (B) Full-scan spectra of product ions from (Upper) substrate MeCLA and (Lower) metabolite MeCLA+16. The retention times of MeCLA and MeCLA+16 are 15.9 and 8.7 min, respectively.
Discussion
Our microarray study was designed to discover potential strigolactone-related genes by coexpression with MAX3. We identified LBO as a gene with an expression pattern similar to MAX3 (Fig. 2A). The sequence identity of LBO, as an oxidoreductase-like enzyme, made it a good candidate for involvement in hormone biosynthesis. Mutational analysis and reciprocal grafting with strigolactone mutants suggest that LBO likely acts toward the end of the strigolactone biosynthesis pathway, after MAX1 which is required for CLA production. This suggestion is supported by observation of elevated levels of CLA and MeCLA in the lbo mutant (Fig. 5C) but not in max1 (6). Moreover, the lbo mutation causes a reduced response to CL and MeCLA but not to canonical strigolactones (Fig. 5D). The LBO protein consumes MeCLA but not CL or CLA and converts MeCLA to an unidentified strigolactone-like compound (Fig. 6). Together, these findings suggest a function of LBO in the strigolactone pathway downstream of MAX1 and MeCLA to produce a strigolactone-type signal(s) involved in shoot branching.
The lbo mutant may lack production of only a subset of strigolactones, because it can still produce MeCLA and at increased levels (Fig. 5C). This hypothesis would explain why the lbo mutant shows a relatively weak branching phenotype compared with max mutants and validates our approach for finding the additional strigolactone biosynthesis genes by screening gene expression. The weaker phenotype of lbo, despite increased MeCLA, implies that endogenous MeCLA is not very effective at repressing branching. In addition, the weak response of lbo mutant shoots to WT rootstocks (Fig. 4) suggests that the product(s) of LBO enzyme action may be unstable and/or poorly transported. Thus, the endogenous, LBO-dependent signals that inhibit branching may alter or enhance signaling specificity and localize activity, making it more effective at inhibiting branching compared with MeCLA. Moreover, the existence of CLA and LBO in rice (6, 25) implies that LBO-dependent signal(s) might be important for branching in a broad variety of plant species.
The 2OGD enzymes are extremely versatile and catalyze a wide range of reactions, including oxidations and hydroxylations (26). The products of LBO are not yet known, but detection of a MeCLA + 16 Da compound (Fig. 6) may mean that LBO oxidizes MeCLA to produce an active but unstable MeCLA-related compound and/or that the MeCLA + 16 Da product is further converted to a strigolactone-like compound by LBO and/or other enzymes. Recently, there have been several strigolactones with noncanonical structure found, such as avenaol (20) and heliolactone (21), and the LBO product might be related to those compounds. Moreover, CLA was detected in rice roots (6), and therefore, an MAX1 homolog(s) and LBO may also act to produce such strigolactone-like compounds in rice. Thus, the LBO-dependent signal may represent one of the strigolactones so far identified in rice (19), or perhaps, a completely novel compound. Our studies have not yet revealed compounds from plant tissue that are consistent with the MeCLA + 16 Da LBO product. However, strigolactones are difficult to detect, because they exist in very low concentrations. The effects of high instability and localized strigolactone production combined with strong feedback might render the product of LBO extremely difficult to detect in plants. The most important next step will be to try to prepare enough of this compound in vitro to identify it and then, chemically synthesize it.
The discovery of LBO as a 2OGD most likely involved in strigolactone production is an important step forward for fully elucidating the strigolactone biosynthesis pathway. Additional studies into the precise function of LBO are expected to resolve outstanding questions and provide knowledge and insight into how strigolactones function to regulate a wide range of traits in plants. This study highlights the possibility that multiple endogenous strigolactones within a single species may induce strigolactone signal transduction and response. Future studies should test whether different strigolactones regulate different processes within a single species.
Experimental Procedures
Plants were grown in standard conditions as described (40). ATH1 gene expression chip (Affymetrix) was used for transcriptomics, and after normalizing and filtering, Cluster Analysis of Sequences (41) was used to detect genes with similar expression patterns. Phylogenetic analysis was made using MEGA (42). Arabidopsis lines were from laboratory stocks, except lbo mutants, which were identified using SIGnAL (43) and provided by the French National Institute for Agricultural Research (INRA) in the Ws-4 ecotype (31) or identified and provided by Davis TILLING in the Col-0 erecta background (32). The max4-9 allele is from the Ws-4 ecotype (44). Branching was defined as total rosette branches >5 mm. Constitutive and reporter transgenes were made using the pMDC162 and pMDC32 binary vectors (45). Arabidopsis was transformed by floral dip method (46) and whole-mount GUS staining was as described (47). The abutting method was used for Arabidopsis grafting (48). Hormones were applied by direct treatment (Fig. 5D) (49), through hydroponics (Fig. 5A and Fig. S2) (39), in Phytatrays (Sigma-Aldrich) (Fig. 5B) (40), or by pipetting solutions directly onto seedlings lying flat on agar (Fig. S3). Strigolactones were produced with the following methods: rac-GR24 (50), 5-deoxystrigol (51), 4DO (51), CL (39), and MeCLA (6). Decapitation, auxin, and N-1-naphthylphthalamic acid treatments were as described (17). Differences in the level of branching between experiments can be caused by the ecotype that was used or the growth medium and conditions, which are described in the figures and SI Experimental Procedures. Quantitative RT-PCR was essentially performed as described (23) using primers listed in Table S1. Analysis of CL and MeCLA from Arabidopsis root tissues was performed as previously reported (6). LBO protein was expressed in E. coli using the pOPIN-E vector (Oxford Protein Production Facility U.K.) with a polyhistidine tag at the N terminus, and protein was purified by affinity column chromatography for histidine-tagged proteins (His GraviTrap; GE Healthcare). LBO protein reactions were based on the reported method (52) and analyzed by liquid chromatography–MS/MS as described (6). Additional details are provided in SI Experimental Procedures.
Table S1.
Primer list
| Gene | Arabidopsis Gene Identifier number | Forward | Reverse | Spans intron? |
| IAA5 | At1g15580 | GCTCTGCAAATTCTGTTCGGA | ATTCACTTTCCTTCAACGTATCATCAA | Yes |
| STH7 | At4g39070 | CATCTCCGGTTCTCTCTCACTTCT | CATTCTCTGCATAGTATTGCTCTGTC | Yes |
| ACTIN2 | At3g18780 | AGTGGTCGTACAACCGGTATTGT | GATGGCATGAGGAAGAGAGAAAC | Yes |
| ACTIN7 | At5g09810 | AGTGGTCGTACAACCGGTATTGT | GAGGAAGAGCATACCCCTCGTA | Yes |
| ACTIN8 | At1g49240 | AGTGGTCGTACAACCGGTATTGT | GAGGATAGCATGTGGAAGTGAGAA | Yes |
| LBO promoter clone | CACTACCCTAAGACCGTTGAAAG | ATTGAAAGCAGACCAAGAAAGAA | n/a | |
| LBO cDNA clone | ATGGCTCCTCTACCTATTTCTTC | TTAATTGAGAATCTTGGCAAAATC | n/a | |
| LBO WT genotype | TTGGTCTGCTTTCAATATGGC | AGCACTTCCATCAGAATGTGG | n/a | |
| lbo-1 T-DNA genotype | CAGGTGCCCACGGAATAGT | AGCACTTCCATCAGAATGTGG | n/a | |
| LBO TILLING screen | GCCTGTCATTGATTTCTCTAAGC | AAGTCCAAAGTCCTCTTTCCTTG | n/a | |
| lbo-2 PCR sequencing | GTTTGCCCTTGGTGTTCATCC | CCTCTCCTTTTCCCTGTTTGTTAC | n/a | |
| lbo-3 PCR sequencing | TTGGTCTGCTTTCAATATGGC | AGCACTTCCATCAGAATGTGG | n/a |
STH7, SALT TOLERANCE HOMOLOG7. n/a, not applicable.
SI Experimental Procedures
Plant Materials and Assays.
Arabidopsis plants were grown in standard conditions on soil as described (40), except that Ws-4 backgrounds were grown initially for 2 wk at 18 °C/16 °C day/night for 14-h-day length to delay flowering and increase rosette leaf production and branching. Strigolactone treatment is as described for direct method (Fig. 5D) (49), hydroponics with CL treatment (Fig. 5A and Fig. S2) (39), or Phytatrays (Sigma-Aldrich) with strigolactone treatments added to media (Fig. 5B) (40). Strigolactones were produced with the following methods: rac-GR24 (50), 5-deoxystrigol (51), 4DO (51), CL (39), and MeCLA (6). For short-term strigolactone treatments (Fig. S3), seedlings were grown upright on 0.5× Murashige and Skoog (MS) agar plates for 12 d. Then, 0.5× MS liquid with the specified concentration of rac-GR24, 5-deoxystrigol, 4DO, or control solution was pipetted over the seedlings lying flat for 4 h. The seedling tissues were frozen in liquid nitrogen for RNA extraction and real-time quantitative RT-PCR essentially as previously described (23) using primers listed in Table S1. Decapitation, indole-3-acetic acid (IAA; auxin; Sigma-Aldrich), and N-1-naphthylphthalamic acid (Sigma-Aldrich) treatments were as described (17). Arabidopsis lines were from laboratory stocks, except that the lbo T-DNA mutant line in the Ws-4 ecotype was identified using the online SIGnAL database (signal.salk.edu/cgi-bin/tdnaexpress) (43) and purchased from the French National Institute for Agricultural Research (INRA) (publiclines.versailles.inra.fr) (31). The lbo point mutation alleles in the Col-0 erecta background were identified using the primers listed in Table S1 and purchased from the diploid population of the University of California, Davis TILLING (tilling.ucdavis.edu/index.php/Main_Page) (32). Genotyping with gene- and T-DNA–specific primers (Table S1) combined with sequencing of PCR products confirmed the presence and homozygosity of the mutations in the lbo alleles. Branching was defined as total rosette branches >5 mm (or per rosette leaf) as near as possible to first silique senescence (about 1 wk earlier for plants in Phytatrays or hydroponics). Arabidopsis grafting was achieved by abutting young seedlings together near the top of the hypocotyl (48). Analysis of CL and MeCLA from Arabidopsis root tissues was performed as previously reported (6).
Transcriptomic, Gene Expression, and Sequence Analyses.
Microarray analysis was carried out using RNA extracted from 120 to 150 hypocotyls of 14-d-old Arabidopsis seedlings. For decapitation, the shoot meristem and young leaves were removed. For decapitation and IAA, IAA (3 mg mL−1) was supplied to the decapitated stump, and for N-1-naphthylphthalamic acid, N-1-naphthylphthalamic acid (3 mg mL−1) was applied in lanolin around the top of the hypocotyl (with no decapitation). After 24 h, hypocotyls were excised for RNA extraction, and Columbia-0 and max mutant hypocotyls were collected. Data from the ATH1 gene expression chip (Affymetrix) were normalized using the GCRMA package (gcrma: Background Adjustment Using Sequence Information; R package, version 2.32.0) and then, filtered by fitting an exponential decay curve to exclude probe set responses within background noise levels. Probe sets of genes that did not show any statistically significant difference in expression (P ≤ 0.05; ANOVA) across any of the experimental conditions were then excluded. Next, in Cluster Analysis of Sequences (41), clusters of genes with similar expression were identified. The Pearson’s correlation coefficient of probe set responses under different experimental conditions was used as the similarity metric for all pairwise comparisons. Quantitative RT-PCR analysis was carried out essentially as previously described (23) using primers listed in Table S1. LBO-related sequences were found using BLASTP online at PlantGDB (www.plantgdb.org/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). Protein structure was predicted online using Prosite (prosite.expasy.org/). Phylogenetic analyses were created using the MEGA program (www.megasoftware.net/) (42) with ClustalW alignment and Maximum Likelihood method using default settings.
Transformation of Arabidopsis.
Constitutive expression of LBO was constructed using PCR of LBO cDNA, pCR8/GW/TOPO (Invitrogen) shuttle vector, and pMDC32 binary vector (45). GUS reporter expression was constructed using genomic DNA for the LBO promoter 1,711 bp upstream of LBO, pCR8/GW/TOPO, and pMDC162 binary vector (45). Primers are listed in Table S1. Clones were confirmed error-free by Sanger sequencing at the Australian Genome Resource Facility. Arabidopsis was transformed by the floral dip method (46) and whole-mount GUS staining as described (47).
Enzyme Preparation from Escherichia coli.
For heterologous expression, LBO was cloned into the pOPIN-E vector (Oxford Protein Production Facility U.K.) with N-terminal polyhistidine tag by the UQ Protein Expression Facility (www.uq.edu.au/pef/) and transformed into Escherichia coli strain Rosetta 2(DE3)pLysS (Novagen) along with the empty vector as a control. The transformed colonies were inoculated into 200 mL LB (5 g/L yeast extract, 10 g/L Bacto Tryptone, 10 g/L sodium chloride) with carbenicilin (100 μg/mL) and grown at 37 °C in a shaking incubator (180 rpm) until the cell density reached an OD600 of 0.5–0.8. After isopropyl β-d-1-thiogalactopyranoside (1 mM) was added, the transformed E. coli were incubated at 20 °C for 14–16 h. To prepare enzyme fractions, E. coli cells were collected by centrifugation of 10,000 × g for 1 min and suspended in 4 mL 20 mM phosphate buffer (pH 7.4) containing 500 mM NaCl and 20 mM imidazole. The suspended cells were mechanically lysed by using a high-pressure homogenizer (Emulsi Flex B15; AVESTIN) and then, centrifuged at 15,000 × g for 5 min at 4 °C. The supernatant was purified through an affinity column for histidine-tagged proteins (His GraviTrap; GE Healthcare) at 4 °C.
Liquid Chromatography–MS/MS Analysis of LBO Metabolites of MeCLA.
Histidine-tagged protein fraction (1 mL) was incubated with 4.6 mM 2-oxoglutarate, 5.8 mM ascorbic acid, 0.6 mM iron ascorbate, and 40 μM substrates at 27 °C for 15 min, similar to the reported method (52). The reaction mixture was extracted: 1 mL ethyl acetate twice. The ethyl acetate phase was dried using sodium sulfate and evaporated under nitrogen gas flow. The residual solid was dissolved immediately in ethyl acetate and subjected to liquid chromatography–MS/MS analysis. Liquid chromatography–MS/MS analysis was conducted as reported previously (6).
Acknowledgments
We thank Kerry Condon, Rosanna Powell, Sathyapriya Gunaseelan, Alice Hayward, Skye Thomas-Hall, Brett Ferguson, Kemal Kazan, Louise Shuey, Tanya Waldie, and Benedicte Wenden for technical assistance and Ottoline Leyser for the supply of max1 max2 double-mutant seeds. This study was supported by Australian Research Council Grants DP110100997 and DP130103646; the Japan Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry; a Japan Science and Technology Agency CREST grant; a Japan Society for the Promotion of Science Restart Postdoctoral Fellowship; and Japan Society for the Promotion of Science KAKENHI Grant 15K07093.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper are found in The Arabidopsis Information Resource (accession no. LBO At3g21420).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601729113/-/DCSupplemental.
References
- 1.Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- 2.Alder A, et al. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 3.Seto Y, et al. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc Natl Acad Sci USA. 2014;111(4):1640–1645. doi: 10.1073/pnas.1314805111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol. 2014;10(12):1028–1033. doi: 10.1038/nchembio.1660. [DOI] [PubMed] [Google Scholar]
- 5.Booker J, et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell. 2005;8(3):443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Abe S, et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA. 2014;111(50):18084–18089. doi: 10.1073/pnas.1410801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlen W, et al. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155(2):974–987. doi: 10.1104/pp.110.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldwasser Y, Yoneyama K, Xie XA, Yoneyama K. Production of Strigolactones by Arabidopsis thaliana responsible for Orobanche aegyptiaca seed germination. Plant Growth Regul. 2008;55(1):21–28. [Google Scholar]
- 9.Braun N, et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiol. 2012;158(1):225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504(7480):401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou F, et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504(7480):406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, et al. Strigolactone signaling in arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell. 2015;27(11):3128–3142. doi: 10.1105/tpc.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soundappan I, et al. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell. 2015;27(11):3143–3159. doi: 10.1105/tpc.15.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamiaux C, et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol. 2012;22(21):2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Chevalier F, et al. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell. 2014;26(3):1134–1150. doi: 10.1105/tpc.114.122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao LH, et al. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 2015;25(11):1219–1236. doi: 10.1038/cr.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward A, Stirnberg P, Beveridge C, Leyser O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 2009;151(1):400–412. doi: 10.1104/pp.109.137646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant. 2013;6(1):18–28. doi: 10.1093/mp/sss130. [DOI] [PubMed] [Google Scholar]
- 19.Xie X, et al. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol Plant. 2013;6(1):153–163. doi: 10.1093/mp/sss139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HI, et al. Avenaol, a germination stimulant for root parasitic plants from Avena strigosa. Phytochemistry. 2014;103:85–88. doi: 10.1016/j.phytochem.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Ueno K, et al. Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry. 2014;108:122–128. doi: 10.1016/j.phytochem.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Frickey T, Lupas A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20(18):3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 23.Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 2012;159(3):1073–1085. doi: 10.1104/pp.112.196253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroha T, et al. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21(10):3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai Y, Ono E, Mizutani M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014;78(2):328–343. doi: 10.1111/tpj.12479. [DOI] [PubMed] [Google Scholar]
- 26.Farrow SC, Facchini PJ. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front Plant Sci. 2014;5:524. doi: 10.3389/fpls.2014.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilmouth RC, et al. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure. 2002;10(1):93–103. doi: 10.1016/s0969-2126(01)00695-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Ren JS, Clifton IJ, Schofield CJ. Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase--the ethylene-forming enzyme. Chem Biol. 2004;11(10):1383–1394. doi: 10.1016/j.chembiol.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 29.McDonough MA, et al. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2) Proc Natl Acad Sci USA. 2006;103(26):9814–9819. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton AJ, Bouzayen M, Grierson D. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc Natl Acad Sci USA. 1991;88(16):7434–7437. doi: 10.1073/pnas.88.16.7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunaud V, et al. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 2002;3(12):1152–1157. doi: 10.1093/embo-reports/kvf237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Till BJ, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13(3):524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bainbridge K, Sorefan K, Ward S, Leyser O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 2005;44(4):569–580. doi: 10.1111/j.1365-313X.2005.02548.x. [DOI] [PubMed] [Google Scholar]
- 34.Stirnberg P, Furner IJ, Ottoline Leyser HM. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007;50(1):80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- 35.Shen H, Luong P, Huq E. The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 2007;145(4):1471–1483. doi: 10.1104/pp.107.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foo E, Turnbull CG, Beveridge CA. Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol. 2001;126(1):203–209. doi: 10.1104/pp.126.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turnbull CG, Booker JP, Leyser HM. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 2002;32(2):255–262. doi: 10.1046/j.1365-313x.2002.01419.x. [DOI] [PubMed] [Google Scholar]
- 38.Sorefan K, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17(12):1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaffidi A, et al. Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 2013;76(1):1–9. doi: 10.1111/tpj.12265. [DOI] [PubMed] [Google Scholar]
- 40.Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009;150(1):482–493. doi: 10.1104/pp.108.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frickey T, Weiller G. Analyzing microarray data using CLANS. Bioinformatics. 2007;23(9):1170–1171. doi: 10.1093/bioinformatics/btm079. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen A, et al. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 2012;158(4):1976–1987. doi: 10.1104/pp.111.187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Brewer PB, et al. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development. 2004;131(16):4035–4045. doi: 10.1242/dev.01279. [DOI] [PubMed] [Google Scholar]
- 48.Brosnan CA, et al. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(37):14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 50.Mangnus EM, Dommerholt FJ, Dejong RLP, Zwanenburg B. Improved synthesis of strigol analog GR24 and evaluation of the biological activity of its diastereomers. J Agric Food Chem. 1992;40(7):1230–1235. [Google Scholar]
- 51.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10(12):2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251(4):533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 54.Scaffidi A, et al. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 2014;165(3):1221–1232. doi: 10.1104/pp.114.240036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson DC, et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2011;108(21):8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]