Abstract
The C-terminal alpha-helix of gp41 membrane-proximal external region (MPER; 671NWFDITNWLWYIK683) encompassing 4E10/10E8 epitopes is an attractive target for HIV-1 vaccine development. We previously reported that gp41-HR1-54Q, a trimeric protein comprised of the MPER in the context of a stable six-helix bundle (6HB), induced strong immune responses against the helix, but antibodies were directed primarily against the non-neutralizing face of the helix. To better target 4E10/10E8 epitopes, we generated four putative fusion intermediates by introducing double point mutations or deletions in the heptad repeat region 1 (HR1) that destabilize 6HB in varying degrees. One variant, HR1-Δ10-54K, elicited antibodies in rabbits that targeted W672, I675 and L679, which are critical for 4E10/10E8 recognition. Overall, the results demonstrated that altering structural parameters of 6HB can influence immunogenic properties of the MPER and antibody targeting. Further exploration of this strategy could allow development of immunogens that could lead to induction of 4E10/10E8-like antibodies.
Keywords: HIV-1, gp41, MPER, fusion intermediate
Introduction
It is widely hypothesized that a successful AIDS vaccine should induce antibodies that can neutralize a large number of HIV-1 isolates from multiple clades. However, such broadly neutralizing antibodies (bnAbs) have been observed only in a small fraction of HIV-1 infected patients (Y. Li et al., 2007; Simek et al., 2009), suggesting that the generation of these bnAbs is a complex, difficult process. Nevertheless, efforts to develop immunogens and/or vaccine strategies that can elicit bnAbs must continue.
Recent isolation and characterization of potent bnAbs from patients has helped the field of vaccine research immensely by providing better understanding of both the epitopes targeted and the unique features of antibodies that contribute to their broad neutralizing ability. Most of the bnAbs that target gp120 recognize highly conformational, non-linear epitopes that might involve tertiary and/or quaternary structures (for a review, see (Mouquet, 2014)). These epitopes include the CD4 binding site, the glycan associated V1/V2 loops, the V3 loops, and a glycan only epitope targeted by 2G12. In addition, the glycan associated bridging region between gp120 and gp41 has also been identified as a target for multiple bnAbs recently (Blattner et al., 2014; Falkowska et al., 2014; Scharf et al., 2014). In contrast, those that target gp41 recognize linear epitopes that are structurally simpler and reside in a highly conserved, ~22 amino acid long domain called the membrane proximal external region (MPER) (for a review, see (Montero et al., 2008). These bnAbs are thought to inhibit conformational changes that are critical for fusion between viral and cellular membranes. A more recent discovery of the highly potent and broadly neutralizing 10E8 mAb (Huang et al., 2012), along with previously characterized 2F5, 4E10 and Z13e1 (Purtscher et al., 1994; Stiegler et al., 2001; Zwick et al., 2001), has renewed interests in designing MPER-based immunogens.
To date, eliciting anti-MPER bnAbs through vaccination has been elusive. Several approaches have been examined, including (1) immunization with short MPER peptides either alone or coupled to carrier proteins (Decroix et al., 2001; Joyce, 2002; Liao et al., 2000; Matoba et al., 2006; McGaughey et al., 2003; Ni et al., 2004), (2) neutralizing epitopes presented on scaffolds (Correia et al., 2010; Guenaga et al., 2011; Ofek et al., 2010), (3) MPER peptides delivered on liposomes (Dennison et al., 2011; Hanson et al., 2015; Hulsik et al., 2013; Lai et al., 2014; Matyas et al., 2009; Mohan et al., 2014; Serrano et al., 2014; Venditto et al., 2013; 2014), (4) MPER containing hybrid/fusion proteins, and (5) chimeric viruses or virus like particles (Arnold et al., 2009; Benen et al., 2014; Bomsel et al., 2011; Eckhart et al., 1996; Jain et al., 2010; Kamdem Toukam et al., 2012; E. Kim et al., 2014; Luo et al., 2006; Marusic et al., 2001; Muster et al., 1995; Ye et al., 2011; Yi et al., 2013; Zhang et al., 2004). Although a handful of studies have shown induction of modest levels of neutralizing activity with limited breadth against tier 1 HIV-1 isolates (Hulsik et al., 2013; Krebs et al., 2014; Lai et al., 2014; Ye et al., 2011; Yi et al., 2013), neither the identity of antibodies responsible for neutralization nor the mechanistic nature of inhibition have been further demonstrated.
Multiple crystal structures of short MPER peptides in complex with bnAbs have been solved (Cardoso et al., 2007; Huang et al., 2012; Julien et al., 2008; Ofek et al., 2004). Despite simpler epitope structures, the difficulty in designing immunogens that can induce similar antibodies lies partly on the fact that the MPER structure in the context of a native trimeric envelope spike on virus particles remains unknown. In this regard, it is possible that antibody-bound MPER structures do not accurately represent the MPER conformation on native trimers. Furthermore, gp41 undergoes large structural changes during the fusion process (Pancera et al., 2014), and it is likely that the MPER also assumes several different conformations. Thus, studies that characterize the structural and immunological properties of MPER in context of larger gp41-based proteins are much needed.
As an initial effort, we generated a soluble gp41 construct named gp41-HR1-54Q consisting of heptad repeat regions 1 and 2 (HR1and HR2, respectively) and the MPER (Shi et al., 2010). While the HR1 and HR2 domains formed a stable six-helix bundle (6HB), much of the MPER domain remained quite flexible and free from association with the 6HB (Shi et al., 2010). Surprisingly, this protein induced strong antibody responses against a peptide that encompass 4E10/10E8 epitopes (671NWFDITNWLW680; (Habte et al., 2015). Further analyses indicated that these antibodies targeted the non-neutralizing face of the C-terminal α-helix, but partly overlapping with 4E10/10E8 epitopes. One possible reason for the preferential targeting of the non-neutralizing face of the helix could be its orientation when the MPER is presented in the context of a stable 6HB structure, which represents a post-fusion conformation. It had been suggested that MPER might be more exposed during the fusion process as gp41 undergoes conformational changes (Chakrabarti et al., 2011; de Rosny et al., 2004; Dimitrov et al., 2007; Finnegan et al., 2002; Frey et al., 2008; M. Kim et al., 2011).
Recent crystal structures of BG505 SOSIP gp140 provided partial information on the pre-fusion state of gp41 (Julien et al., 2013; Pancera et al., 2014). However, fusion intermediate structures are completely unknown. In particular, there is no information on how the MPER is oriented relative to the rest of the protein, and when and whether it make any contact with the rest of gp41 or gp120, prior to the post-fusion state. The only certainty is that HR1 and HR2 are in the process of coming together to form 6HB. As such, we took an empirical approach of generating four variants of gp41-HR1-54Q that might represent different stages of fusion process by disrupting 6HB formation in varying degrees. Biochemical, antigenic, and immunogenic properties of these putative fusion intermediates (PFIs) were characterized. Although we did not succeed in inducing bnAbs against the MPER in rabbits, the results from the study should facilitate development of improved MPER immunogens.
Results
Designing gp41-HR1-54Q variants with destabilized 6HB
The trimeric structure of gp41-HR1-54Q is stabilized by both inter- and intramolecular interactions between HR1 and HR2 (Shi et al., 2010). The exact order of molecular interactions between HR1 and HR2 that leads to 6HB formation is not known, although a leading working model suggests a zipping process along HR1-HR2 that begins at the C- and N-termini of respective domains in an anti-parallel fashion, with a central trimeric HR1 core (Markosyan et al., 2009). As such, we hypothesized that it might be possible to generate partially opened hairpin loop structures that might simulate fusion intermediates if HR1-HR1 or HR1-HR2 interactions were destabilized.
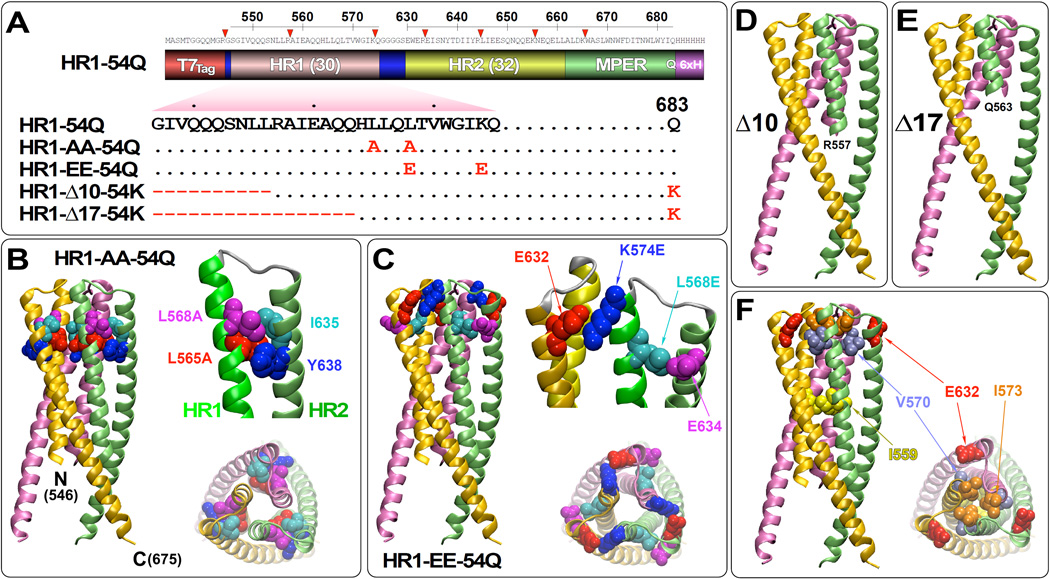
Four variants were generated by either introducing point mutations or deletions (Fig. 1A). All mutations were restricted to the HR1 only, so as to avoid altering the native conformation of the HR2 or MPER domains. The C-terminal half of HR1 was mutated by introducing two point mutations (HR1-AA-54Q and HR1-EE-54Q), whereas the N-terminal half of HR1 was mutated more drastically by deletions (HR1Δ10-54K and HR1Δ17-54K). The HR1-AA-54Q variant contained L565A and L568A mutations with an intent of weakening hydrophobic interactions with I635 and Y638 residues on HR2 (Fig. 1B). In contrast, HR1-EE-54Q contained L568E and K574E mutations designed destabilize 6HB formation by introducing intra- and inter-molecular charge-charge repulsions with E634 and E632 residues on HR2, respectively (Fig. 1C). Deleting the N-terminal 10 or 17 amino acids of HR1 is designed to allow initiation of 6HB formation, but halt the zipping process in the middle to generate structures that might resemble fusion intermediates (Figs. 1D and 1E, respectively). For these constructs with deletions, the terminal 683Q residue was reverted back to the wild type lysine as it was later reported to be critical for 10E8 binding (Huang et al., 2012). Although we do not know whether any of these constructs would mimic true fusion intermediates, they will be referred herein as putative fusion intermediates (PFIs) for simplicity.
Fig 1.
Design of putative fusion intermediates of gp41-HR1-54Q. (A) A domain structure of gp41-HR1-54Q (indicated as HR1-54Q for simplicity) consisting of the T7Tag, heptad repeat region 1 (HR1), GGGGS linker, heptad repeat region 2 (HR2), membrane-proximal external region (MPER) and the 6× His tag is shown. The HR1 domain sequence, along with the terminal 683Q residue, is indicated for gp41-HR1-54Q. Point mutations and deletions introduced into the HR1 domain to generate variants HR1-AA-54Q, HR1-EE-54Q, HR1-Δ10-54K and HR1-Δ17-54K are indicated. The terminal 683Q residue was reverted back to 683K in HR1-Δ10-54K and HR1-Δ17-54K. (B) The mutations introduced in HR1-AA-54Q (L565A and L568A) are plotted on the gp41-HR1-54Q crystal structure (pdb: 3K9A) (Shi et al., 2010) to highlight the proximity of these residues to the neighboring I635 and Y638 residues located on the HR2 domain. Structures of the unmutated amino acids are shown. (C) The mutations introduced in HR1-EE-54Q (L568E and K574E) are plotted on the gp41-HR1-54Q crystal structure to display the proximity of these residues to E632 and E634 residues. The truncations introduced at the N-terminal end of the HR1 domain are plotted onto the gp41-HR1-54Q structure simply to show the point of deletion for (D) HR1-Δ10-54K and (E) HR1-Δ17-54K. (E) As a point of reference, other residues that have been previously shown to destabilize 6HB when mutated (I559P, V570D and I573D; (Kesavardhana and Varadarajan, 2014; Sanders et al., 2002)) are indicated.
Biochemical and antigenic properties of PFIs
Similar to gp41-HR1-54Q, all of the PFIs were insoluble when expressed in E. coli. However, they could be solubilized through a urea denaturation/renaturation process as we previously described for gp41-HR1-54Q (Habte et al., 2015). Unlike gp41-HR1-54Q and PFIS containing point mutations, however, HR1Δ10-54K and HR1Δ17-54K were prone to aggregation, especially upon freeze-thawing, and appeared as polydispersed oligomers.
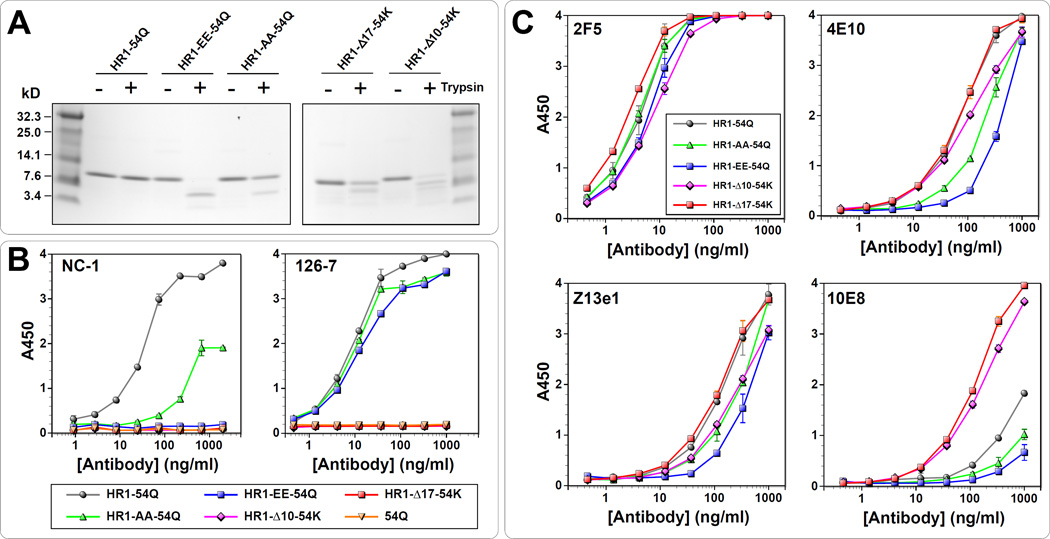
As we previously reported, gp41-HR1-54Q is highly resistant to trypsin digestion (Habte et al., 2015), presumably due to its rigid structure. To determine how mutations might affect the protein, trypsin sensitivity of the PFIs was assessed. As shown in Fig. 2A, gp41-HR1-54Q was completely resistant to trypsin digestion even after one hour. In contrast, PFIs exhibited varying degrees of trypsin sensitivity. Not surprisingly, HR1-AA-54Q was least sensitive. Unexpectedly, however, HR1-EE-54Q was most sensitive and that HR1-Δ10-54K was more sensitive than HR1-Δ17-54K. The differences in trypsin sensitivity among the PFIs suggested that they likely have folded into structures different from each other, and certainly different from gp41-HR1-54Q.
Fig 2.
Biochemical and antigenic properties of putative fusion intermediates. (A) Evaluation of trypsin sensitivity of PFIs in comparison to gp41-HR1-54Q. (B) ELISA with mAbs NC-1 and 126-7 to monitor effects of the mutation on six-helix bundle formation. gp41-HR1-54Q was used as a positive control, while another protein (gp41-54Q) that lacks the HR1 domain was used as a negative control. (C) The antigenic integrity of the variants was tested by performing ELISA with bnAbs 2F5, 4E10, Z13e1 and 10E8.
Next, the PFIs were probed with NC-1, a mouse monoclonal antibody (mAb) that recognizes post-fusion 6HB structure (Jiang et al., 1998). NC-1 recognizes amino acid residues from 643 to 655 within HR2 (Yuan et al., 2009), which is present in all four PFIs. Thus, any changes in NC-1 binding should be the result of conformational changes induced by the mutations. As shown in Fig. 2B (left panel), NC-1 binding was completely abolished for HR1-EE-54Q, HR1-Δ10-54K and HR1-Δ17-54K. Although HR1-AA-54Q could be recognized, the binding was substantially weaker than gp41-HR1-54Q. These results indicate that introduced mutations were able to disrupt formation of the post-fusion 6HB conformation.
Next, the PFIs were probed with 126-7, a human mAb (an IgG2 version of 126-6) that only recognizes a trimeric conformation of gp41 shared between both pre- and post-fusion state (Gorny et al., 1989; Robinson et al., 1991; Tyler et al., 1990; Xu et al., 1991; Yuan et al., 2009). It recognizes residues from 641 to 648 in the cluster II of gp41. As shown in Fig. 2B (right panel), gp41-HR1-54Q, HR1-AA-54Q and HR1-EE-54Q bound nearly equally to 126-7 suggesting that the trimeric conformations of these proteins were similar (at least at the 126-7 epitope). Not surprisingly, 126-7 failed to recognize both HR1-Δ10-54K and HR1-Δ17-54K. The elimination of three and five helical turns in HR1, respectively, likely prevented formation of stable trimeric HR1 core.
To examine whether epitopes recognized by MPER bnAbs remained conformationally intact and accessible on PFIs, they were probed with 2F5, Z13e1, 4E10 and 10E8 by ELISA (Fig. 2C). For 2F5, there were only minor differences between HR1-54Q and PFIs. The results were similar for Z13e1, although HR1-EE-54Q showed slightly weaker binding. The reduced binding to HR1-EE-54Q was more pronounced with 4E10 and 10E8. To a lesser extent, binding was also reduced for HR1-AA-54Q. In general, antibody binding to HR1-Δ10-54K and HR1-Δ17-54K was quite similar to gp41-HR1-54Q, except for 10E8, for which there was significantly better binding. However, this enhanced binding is most likely due to reverting back to lysine at position 683, rather than deletions themselves, since K683 is one of the critical residues that 10E8 recognizes. Taken together, these results suggest that destabilization of 6HB, depending on the approach taken, could potentially affect MPER structure, which could in turn alter conformation or accessibility of epitopes targeted by bnAbs.
Immunogenic properties of PFIs
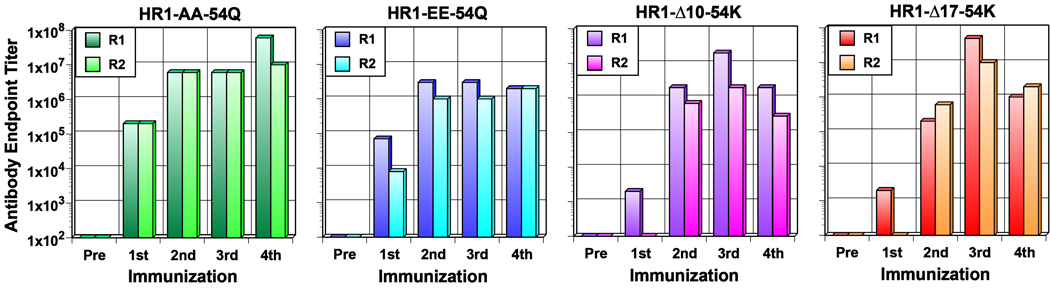
Immunogenic properties of PFIs were evaluated in rabbits as previously described for gp41-HR1-54Q (Habte et al., 2015). For this initial study, two animals were used for each of the four PFIs. Serum samples were collected two weeks after each immunization (on weeks 0, 4, 9 and 15). Antibody titers against autologous antigens were assessed by ELISA (Fig. 3). Overall, antibody responses against the PFIs were weaker than gp41-HR1-54Q, which induced end point antibody titers greater than 5×106 even after a single immunization (Habte et al., 2015). PFIs with point mutations were more immunogenic than the ones with deletions, which was more noticeable after the first immunization. The reduced antibody responses could be due in part to elimination of helper T cell epitopes, especially for the deletion mutants, in addition to altered conformations or loss of epitopes.
Fig 3.
Antibody end-point titers induced by putative fusion intermediates. Serum samples collected two weeks after each immunization were evaluated by ELISA to determine the end-point antibody titers against autologous antigens. Pre-immune serum was used as a negative control.
To better understand how mutations on PFIs altered immune responses, immunogenic linear epitopes were mapped by ELISA using overlapping biotinylated peptides as we previously described (Habte et al., 2015). Since mutations and deletions were in the HR1 domain, we focused on antibody responses directed against the HR2 and the MPER. Notwithstanding some animal-to-animal variations, the immunogenic epitope profile of HR1-AA-54Q was somewhat similar to that of gp41-HR1-54Q, with 671 peptide (671NWFDITNWLW680) being highly immunogenic in both animals. Interestingly, antibody responses against HR1-EE-54Q were directed towards N-terminus of HR2 and the C-terminus of MPER with little to no antibodies against peptides spanning the cluster II region (644RLIEESQNQQEKNEQELLAL663) that typically elicits non-neutralizing antibodies (Alam et al., 2008; Frey et al., 2010; Hioe et al., 1997). Compared to gp41-HR1-54Q, one major difference in immune responses against HR1-Δ10-54K is strong antibody responses against the N-terminal end of HR2. Although the 671 peptide remained immunogenic, peptides 629 (629MEWEREISNY638), 632 (632EREISNYTDI641) and 635 (635ISNYTDIIYR634) were clearly immunodominant. By far, the most striking change in immunogenic epitope profile was with HR1-Δ17-54K. Virtually all of the peptides, except for peptides 665, 668 and 674, were highly immunogenic in one or both of the rabbits.
Despite strong antibody responses against the 671 peptide (671NWFDITNWLW680) that contained all or most of 4E10 and 10E8 epitopes, none of the rabbit sera exhibited neutralizing activity when tested against HIV-1 pseudoviruses SF162 (tier 1A, clade B), MW965.26 (tier 1A, clade C), and MN.3 (tier 1A, clade B) in a standard TZM-bl assay.
Detailed analyses of antibodies targeting near 4E10/10E8 epitopes
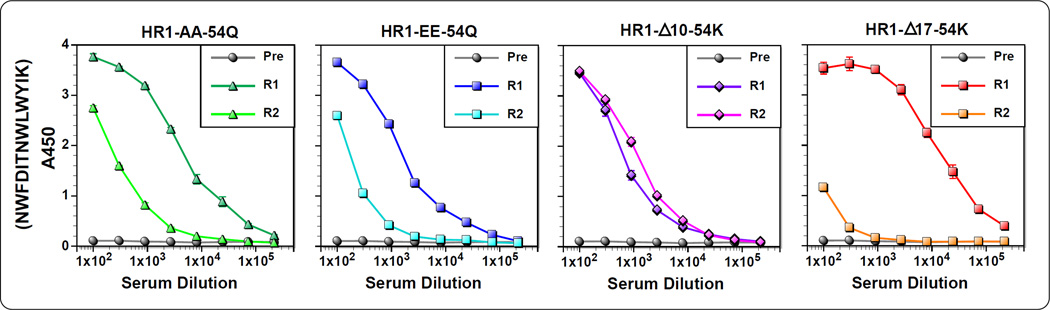
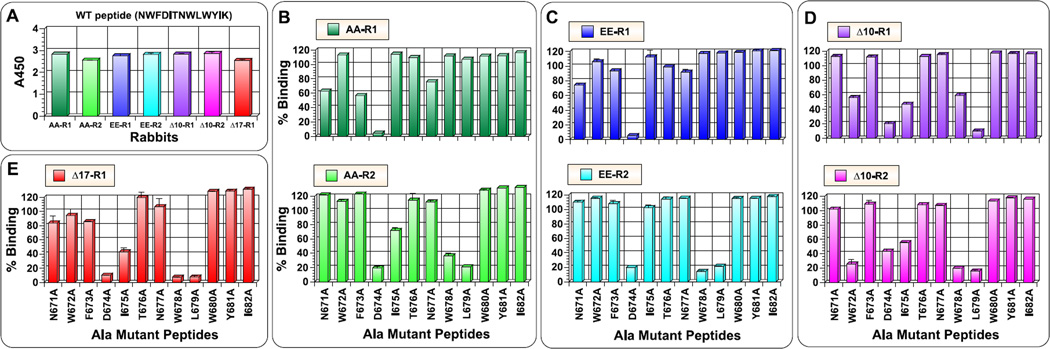
Although we did not succeed in inducing bnAbs against gp41 MPER, better characterization of antibody responses near 4E10/10E8 epitopes could facilitate designing of better immunogens. In particular, we were curious to see whether and how epitope targeting was altered for PFIs compared to gp41-HR1-54Q. Towards this goal, we conducted fine epitope mapping analyses using alanine-scanning mutants of a 13-mer peptide (671NWFDITNWLWYIK683), which we previously used to characterize antibody responses against gp41-HR1-54Q (Habte et al., 2015). Initially, antibody reactivity against the wild-type 13-mer 671 peptide was measured (Fig. 5). Interestingly, some of the antisera reacted poorly to the 13-mer peptide when compared to their reactivity against the 10-mer 671 peptide (Fig. 4). This was particularly severe with rabbit #2 immunized with HR1-Δ17-54K, and, to a lesser degree, rabbit #2 immunized with HR1-AA-54Q. Since ELISA using 10-mer peptides (Fig. 4) was done with a mixture of both N- and C-terminally biotinylated peptides, and that the 13-mer is biotinylated at the C-terminal K683 residue, it was possible that the orientation of the peptide attachment to an ELISA plate, could have affected antibody binding. However, this might not be the case since antibodies reacted strongly to the 10-mer peptide that was biotinylated at the C-terminus (data not shown). Thus, the reason for the discrepancy is unknown at the present time.
Fig 5.
Antibody titers against a wild type peptide containing C-terminal 13 amino acids. Serum samples collected after the fourth immunization was evaluated for binding biotinylated 13-mer peptide (671NWFDITNWLWYIK683) that contains 4E10/10E8 epitopes. Pre-immune serum was used as negative control.
Fig 4.
PepScan analyses using linear, overlapping peptides. Serum samples collected after the fourth immunization were evaluated for reactivity against biotinylated 10-mer peptides (a mixture of peptides biotinylated at the N-terminal and C-terminal ends) spanning both HR2 and MPER domains. The amino acid sequence of each peptide is indicated by horizontal brackets. The first and last residue in the peptide panel is indicated in red as they can differ from the immunogens. The core binding epitopes for 2F5, 4E10 and 10E8 bnAbs are also indicated. Pre-immune serum was used as a negative control.
With the caveat that we would be evaluating only a subset of antibodies targeting near 4E10/10E8 epitopes, we proceeded to characterize antibodies using the panel of 13-mer mutant peptides. All sera, except from rabbit #2 immunized with HR1-Δ17-54K, were analyzed. In doing so, serum samples were first normalized to yield comparable signals when bound to the wild-type peptide (Fig. 6A). For all rabbits tested, D674 residue was critical for antibody binding, which is likely due to a critical role it plays in maintaining a helical conformation of the peptide (Brunel et al., 2006). For HR1-AA-54Q, the two animals exhibited different antibody epitope profiles (Fig. 6B); such animal-to-animal variations have been observed with gp41-HR1-54Q also (Habte et al., 2015). Interestingly, similar patterns were also observed in animals immunized with HR1-EE-54Q (Fig. 6C). This might suggest structural similarity of the C-terminal MPER for HR1-AA-54Q and HR1-EE-54Q. For the both groups, the profile shown on the top panels resembled one of the patterns observed from animals immunized with gp41-HR1-54Q. The profile shown on the bottom panels (critical residues being D674, W678, L679, and I675 for HR1-AA-54Q) was not observed in any of the six animals immunized with gp41-HR1-54Q. Thus, the latter profile could be specific to antibody responses against PFIs. Coincidently, a similar profile was observed for a rabbit immunized with HR1-Δ17-54K (Fig. 6E). The same four residues were also critical for antibodies induced by HR1-Δ10-54K (Fig. 6D). In addition, antibodies induced by HR1-Δ10-54K also targeted W672, which may be highly significant since this residue was never targeted by antibodies induced with gp41-HR1-54Q or with any of the other PFIs. More importantly, this is one of the critical residues recognized by both 4E10 and 10E8.
Fig 6.
Detailed epitope mapping analysis of antibodies against the C-terminal 13 amino acid residues using alanine-scanning mutant peptides. (A) Serum samples after the fourth immunization were examined for binding biotinylated 13-mer peptide (671NWFDITNWLWYIK683). The analyses were done using normalized serum samples to yield comparable binding signal (AA-R1 at 1:2000 dilution; AA-R2 and EE-R2 at 1:100 dilution; EE-R1 at 1:600 dilution; Δ10-R1 at 1:300 dilution; Δ10-R2 at 1:400 dilution; and Δ17-R1 at 1:5000 dilution). (B–E) The same dilutions were tested for binding to mutant peptides. Results are shown as the percentage of binding to the wild type peptide observed in panel (A).
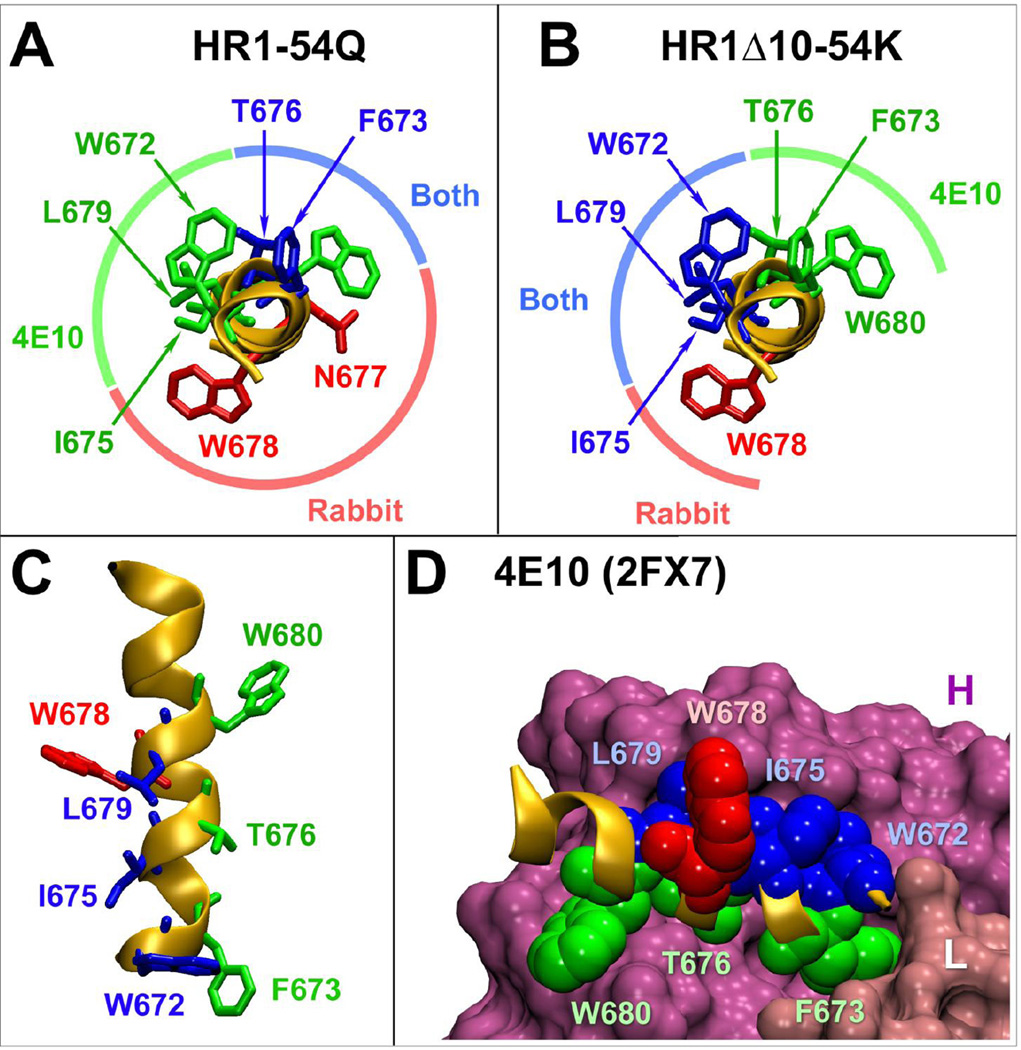
To better compare antibodies induced by gp41-HR1-54Q and HR1-Δ10-54K, amino acid residues critical for binding were visualized on a structure of a peptide co-crystallized with 4E10 (Fig. 7; Cardoso et al., 2007). This analysis revealed that antibodies induced by HR1-Δ10-54K targeted nearly the opposite face of the helix compared to those induced by HR1-54Q with W678 being targeted by both. More importantly, HR1-Δ10-54K-induced antibodies targeted three of the five most critical residues for 4E10 binding (W672, F673, I675, T676 and L679). So, although we were not able to induce nAbs, the results of our study demonstrate that it is possible to alter immunogenicity of epitopes simply by changing structural context of an immunogen.
Fig 7.
Comparison of critical binding residues for antibodies induced by HR1-Δ10-54K and gp41-HR1-54Q relative to 4E10. (A) Critical binding residues for 4E10 and antibodies induced by gp41-HR1-54Q are plotted onto the peptide co-crystalized with 4E10 (pdb: 2FX7) (Cardoso et al., 2007). Residues critical for 4E10 or rabbit antibody only are shown in green and red, respectively. Residues important for both are shown in blue. (B) Critical binding residues for antibodies induced by HR1-Δ10-54K were also plotted onto the same peptide revealing significant difference from the pattern observed with gp41-HR1-54Q. (C) A lateral view of the peptide displaying critical binding residues for 4E10 and HR1-Δ10-54K-induced antibodies. (D) Position of all the HR1-Δ10-54K critical residues in context of the 4E10 bound peptide. The heavy and light chains for the antibody are indicated as H and L.
Discussion
In previous studies, we described structural and immunological properties of gp41-HR1-54Q, which likely represents a near-post-fusion conformation of gp41 (Habte et al., 2015; Shi et al., 2010). Although antibodies elicited in rabbits by this antigen bound epitopes that partially overlap with those targeted by 4E10 and 10E8, they were largely directed against the non-neutralizing face of the helix and failed to exhibit neutralizing activity. It had been speculated that anti-MPER bnAbs primarily target fusion intermediate forms of gp41 (Chen et al., 2014; Dimitrov et al., 2007; Frey et al., 2008). However, their structures are not yet known, except for those of short peptides bound to the antibodies. As such, we decided to take an empirical approach of generating fusion intermediates with a simple assumption that they would have minimal or partial HR1-HR2 pairing and 6HB formation. We further assumed that the MPER would likely exist in a conformation that is different from the one observed on gp41-HR1-54Q. With these assumptions, four putative fusion intermediates (PFIs) were generated by introducing double point mutations or deletions into the HR1 of gp41-HR1-54Q to destabilize HR1-HR1 and HR1-HR2 interactions, and their biochemical and immunological properties were evaluated in this study.
Although we do not have structural evidence, the increased sensitivity of the PFIs to trypsin digestion and their altered reactivity to NC-1 and/or 126-7 mAbs demonstrated that the structure of 6HB on PFIs has been disrupted in varying degrees. Not surprisingly, the four PFIs revealed different immunological profiles with respect to the overall immunogenicity (i.e. total antibody titers induced; Fig. 3), immunodominance of epitopes across the HR2 and MPER (Fig. 4), and specific amino acid residues targeted by antibodies directed against the C-terminal region containing 4E10/10E8 epitopes (671NWFDITNWLWYIK683; Fig. 6). Together, these results indicate that immunogenicity of HR2 and MPER domains are highly dependent on the structural context in which they are presented.
While we did not succeed in inducing bnAbs, detailed epitope mapping analyses revealed a few important findings about antibodies induced by PFIs, HR1-Δ10-54K in particular. First, antibodies induced in both rabbits immunized with HR1-Δ10-54K targeted W672. Targeting of this residue was never observed in any of the six rabbits immunized with gp41-HR1-54Q (Habte et al., 2015; Shi et al., 2010) or in any of the animals immunized with other PFIs. Replacement of this residue with an alanine has been reported to reduce 4E10 binding by over 1000-fold, highlighting its overarching importance (Brunel et al., 2006). W672 is also critical for 10E8 binding (Huang et al., 2012). Second, animals immunized with HR1-Δ10-54K and HR1-Δ17-54K induced antibodies that bound strongly to I675 and L679, both of which line up with W672 along the same side of the helix and contribute significantly to 4E10 and 10E8 binding (Brunel et al., 2006; Cardoso et al., 2005; Huang et al., 2012). Targeting these three residues highlights a remarkable shift from the binding pattern of antibodies elicited by gp41-HR1-54Q. Third, while gaining recognition of these three residues, antibodies induced by HR1-Δ10-54K seemed to have lost recognition of F673 and T676, which were quite well recognized by antibodies induced with gp41-HR1-54Q and targeted by both 4E10 and 10E8.
One cautionary note for interpreting the results of our study is that the epitope mapping analyses shown in Fig. 6 were conducted with polyclonal antibodies. Thus, the phenotypic changes we observed are average of all antibodies that bind the peptide. Accordingly, when there are many antibodies that bind to different epitopes, we might not see significant reduction in antibody binding for mutations at any given position; this might be the case for rabbit #1 of HR1-AA-54Q and HR1-EE-54Q. Significant reduction in binding would be seen only when the antibody response is homogeneous or when all or most antibodies target same residues. In this regard, characterizing antibodies at the monoclonal level would provide a more accurate assessment.
In the absence of a crystal structure of HR1-Δ10-54K, it is hard to speculate how deleting ten residues from the N-terminal end of HR1 (with a potential contribution from K683) influenced the overall MPER conformation to promote such a major shift in antibody response. Nevertheless, our results clearly demonstrated that changes in HR1, which in turn affect stability of 6HB, significantly influence how antibodies target the MPER. Considering the difficulties in crystallizing proteins that contain the hydrophobic C-terminal ectodomain of gp41, MPER-based vaccine development may have to rely on reiterative, empirical approaches. Based on the results from this study, HR1-Δ10-54K would be an excellent starting point.
To improve the immunogen, one factor that could be adjusted is the length of HR1. In this study, we deleted 10 or 17 residues, which account for roughly 3 and 5 helical turns, respectively. As shown in Fig. 4, the antibodies induced by HR1-Δ17-54K were drastically different from all others, which we believe is due to complete disruption of 6HB, thereby rendering the protein highly flexible and inducing greater diversity of antibodies. By adding two helical turns (HR1-Δ10-54K), which likely increased the stability of 6HB, antibody responses against HR2 was largely restricted to the N-terminal end and W672 could be targeted. This raises a question as to what would happen to the antibody response if less than 10 residues were deleted. Would any of them allow targeting of F673 and T676 (as did gp41-HR1-54Q), while also targeting W672, I675 and L679?
Another factor that could be considered for improving antibody responses is to minimize immunogenicity of W678. As shown in Fig. 6, W678 was targeted on all of the PFIs as well as on gp41-HR1-54Q (Habte et al., 2015), suggesting its dominant role in determining antibody responses. Most likely, W678 is not exposed during the natural course of the fusion process, rendering these antibodies useless. Thus, preventing antibody responses against W678 could improve the chance of inducing bnAbs. Perhaps substituting W678 with a less immunogenic residue (e.g. glycine or alanine) might redirect the immune system to shift the focus away from W678 towards F673 and T676.
In all of our immunization studies, we used Zn-chitosan not only as an adjuvant, but also as an antigen delivery platform. Although all of our PFIs, as well as gp41-HR1-54Q, are soluble proteins, they all have 6×His tag at the C-terminus, which was used to affix the proteins to Zn-chitosan. Thus, the flexibility of the C-terminal end of the MPER was most likely limited. However, neither the spatial distribution nor the orientation of the MPER when it is bound to Zn-chitosan is known. More importantly, how it would affect MPER immunogenicity is completely unknown. In this regard, it would be worthwhile to evaluate immunogenicity of some of the PFIs in the context of lipid membranes (e.g. delivered on liposomes or expressed directly on the cell surface), which would better resemble the MPER structure and the microenvironment of virus particles. Although the discovery of 10E8 (Huang et al., 2012) has shown that MPER-directed antibodies do not have to interact with membranes to exhibit neutralizing activity, it is possible that anchoring the MPER onto the membrane (via a transmembrane domain with or without a cytoplasmic tail) could (1) stabilize the correct conformation of the neutralizing epitope, (2) provide rigidity and limit the mobility of the region, and/or (3) allow better orientation/exposure of the neutralizing face of the helix, thereby facilitate induction of 4E10/10E8-like antibodies.
Conclusion
Previously characterized gp41-HR1-54Q, which likely exists in a near post-fusion state, induced antibodies primarily against the non-neutralizing face of the C-terminal α-helix that contains 4E10/10E8 epitopes. To generate immunogens that might better resemble fusion intermediates, four constructs were generated by introducing double point mutations or deletions in the HR1 of gp41-HR1-54Q. Different mutations disrupted the six-helix bundle (6HB) structure in varying degrees and differentially affected immunogenicity of the MPER. Antibodies induced by one of the variants, HR1-Δ10-54K, targeted three residues critical for recognition by 4E10 and 10E8. Further exploration of this strategy could lead to development of immunogens that could elicit 4E10/10E8-like antibodies.
Materials and Methods
Cloning, Expression and Purification of PFIs
To generate PFI constructs with point mutations, the QuikChange® XL Site directed mutagenesis kit was used as per the manufacturer’s instructions using the original gp41-HR1-54Q plasmid as the template (Shi et al., 2010). For HR1-AA-54Q, the mutations L565A and L568A were introduced using the sense primer 5’-GAGGCCCAGCAGCACGCCCTGCAGGCCACCGTGTGGGGCATC-3’ and the antisense primer 5’-GATGCCCCACACGGTGGCCTGCAGGGCGTGCTGCTGGGCCTC-3’. For HR1-EE-54Q, the mutations L568E and K574E were introduced using the sense primer 5’-GCACCTGCTGCAGGAGACCGTGTGGGGCATCGAGCAGGGAGGAGG-3’ and the antisense primer 5’-CCTCCTCCCTGCTCGATGCCCCACACGGTCTCCTGCAGCAGGTGC-3’.
For the deletion variants, 10 and 17 residues were deleted from the N terminus end of the HR1 domain as shown in Fig 1A. Both constructs were synthesized from IDT (Integrated DNA Technology) in the pUC57 backbone with flanking restriction sites for BamHI and EcoRI at the 5’ and 3’ ends of the constructs, respectively. The sequence was also altered to encode terminal 683K residue instead of the 683Q as in gp41-HR1-54Q. These constructs were cloned into the pET-21a vector (Novagen; cat#69740-3) using BamHI and EcoRI. All constructs were expressed and purified similar to gp41-HR1-54Q (Shi et al., 2010). The final proteins were dialyzed into 1× PBS (pH 8.0) and stored at −80 degrees.
Trypsin sensitivity assay
All PFIs were incubated with trypsin at 1:100 (enzyme:protein) mass ratio for one hour at 37 °C. 3 µg of untreated and trypsin treated samples were then run on a Novex® 10-20% tricine gel (Thermo Fisher Scientific; cat# EC6625BOX).
Rabbit immunization
Eight New Zealand white female rabbits (2.5 to 3 kg) were purchased from Charles River (USA), housed under specific pathogen free environments and used in compliance with the animal protocol approved by IACUC of Iowa State University. Two animals were immunized with each antigen using Zn-chitosan as an adjuvant. The immunization protocol including the adjuvant preparation, antigen/adjuvant dosage, the immunization and bleeding schedule were all exactly the same as that previously described for gp41-HR1-54Q (Habte et al., 2015).
Enzyme-linked immunosorbent assay (ELISA)
All ELISAs were performed using the standard protocol described for gp41-HR1-54Q (Habte et al., 2015) except for the use of an alternate blocking buffer consisting of PBS (pH 7.5) with 2.5% skim milk and 5% calf sera. For ELISAs testing the binding of antibodies NC-1, 126-7, 2F5, 4E10, Z13e1 and 10E8, coating antigen amounts for all other antigens equimolar to 30 ng/well of gp41-HR1-54Q using the same coating conditions as described for gp41-HR1-54Q. In order to determine end point titers, all antigens were coated at 30 ng/well. The end-point ELISA titers were defined as serum dilution factor that gave readings of average + 2×SD (standard deviation) of the background as described previously (Qin et al., 2014). Coating for linear epitope mapping using 10-mer biotinylated peptides and 13-mer alanine scanning was also performed as previously described (Habte et al., 2015).
Neutralization assays
Neutralization assays were performed in TZM-bl cells as previously described (M. Li et al., 2005; Qin et al., 2014; Wei et al., 2002). Viruses tested included SF162 (tier 1A, clade B), MW965.26 (tier 1A, clade C), and MN.3 (tier 1A, clade B). Murine leukemia virus Envpseudotyped virus was used as a negative control.
Highlights.
Four gp41 MPER-based immunogens that resemble fusion intermediates were generated.
C-terminal region of MPER that contains 4E10/10E8 epitopes was highly immunogenic.
Altering 6HB structure can influence immunogenic properties of the MPER.
Induced antibodies targeted multiple residues critical for 4E10/10E8 binding.
Development of immunogens based on fusion intermediates is a promising strategy.
Acknowledgments
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 anti-gp41 mAb NC-1 from Dr. Shibo Jiang (Cat# 11482), 126-7 from Dr. Susan Zolla-Pazner (Cat# 9967), 2F5 from Dr. Hermann Katinger (Cat# 130220), 4E10 from Dr. Herman Katinger (Cat# 10091), Z13e1 from Dr. Michael Zwick (Cat# 11557) and 10E8 from Dr. Mark Connors (Cat# 12294). We would like to thank Dr. Marisa Banasik for editorial assistance. This work was supported by a grant from the NIH, NIAID (P01 AI074286) grant and funding from Iowa State University. MWC has an equity interest in NeoVaxSyn Inc., and serves as the CEO/President. NeoVaxSyn Inc. did not contribute to this work or the interpretation of the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, Xia S-M, Montefiori DC, Tomaras GD, Weinhold KJ, Karim SA, Hicks CB, Liao H-X, Robinson J, Shaw GM, Haynes BF. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol. 2008;82:115–125. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold GF, Velasco PK, Holmes AK, Wrin T, Geisler SC, Phung P, Tian Y, Resnick DA, Ma X, Mariano TM, Petropoulos CJ, Taylor JW, Katinger H, Arnold E. Broad Neutralization of Human Immunodeficiency Virus Type 1 (HIV-1) Elicited from Human Rhinoviruses That Display the HIV-1 gp41 ELDKWA Epitope. J Virol. 2009;83:5087–5100. doi: 10.1128/JVI.00184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benen TD, Tonks P, Kliche A, Kapzan R, Heeney JL, Wagner R. Development and immunological assessment of VLP-based immunogens exposing the membrane-proximal region of the HIV-1 gp41 protein. J Biomed Sci. 2014;21:79. doi: 10.1186/s12929-014-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, la Peña de AT, Cupo A, Julien J-P, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel M, Tudor D, Drillet A-S, Alfsen A, Ganor Y, Roger M-G, Mouz N, Amacker M, Chalifour A, Diomede L, Devillier G, Cong Z, Wei Q, Gao H, Qin C, Yang G-B, Zurbriggen R, Lopalco L, Fleury S. Immunization with HIV-1 gp41 Subunit Virosomes Induces Mucosal Antibodies Protecting Nonhuman Primates against Vaginal SHIV Challenges. Immunity. 2011;34:269–280. doi: 10.1016/j.immuni.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Brunel FM, Zwick MB, Cardoso RMF, Nelson JD, Wilson IA, Burton DR, Dawson PE. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J Virol. 2006;80:1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso RMF, Brunel FM, Ferguson S, Zwick M, Burton DR, Dawson PE, Wilson IA. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 2007;365:1533–1544. doi: 10.1016/j.jmb.2006.10.088. [DOI] [PubMed] [Google Scholar]
- Cardoso RMF, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Poignard P, Burton DR, Wyatt RT. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol. 2011;85:8217–8226. doi: 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Frey G, Peng H, Rits-Volloch S, Garrity J, Seaman MS, Chen B. Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. J Virol. 2014;88:1249–1258. doi: 10.1128/JVI.02664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia BE, Ban Y-EA, Holmes MA, Xu H, Ellingson K, Kraft Z, Carrico C, Boni E, Sather DN, Zenobia C, Burke KY, Bradley-Hewitt T, Bruhn-Johannsen JF, Kalyuzhniy O, Baker D, Strong RK, Stamatatos L, Schief WR. Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- de Rosny E, Vassell R, Jiang S, Kunert R, Weiss CD. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J Virol. 2004;78:2627–2631. doi: 10.1128/JVI.78.5.2627-2631.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroix N, Hocini H, Quan CP, Bellon B, Kazatchkine MD, Bouvet JP. Induction in mucosa of IgG and IgA antibodies against parenterally administered soluble immunogens. Scand J Immunol. 2001;53:401–409. doi: 10.1046/j.1365-3083.2001.00894.x. [DOI] [PubMed] [Google Scholar]
- Dennison SM, Sutherland LL, Jaeger FH, Anasti KM, Parks R, Stewart S, Bowman C, Xia S-M, Zhang R, Shen X, Scearce RM, Ofek G, Yang Y, Kwong PD, Santra S, Liao H-X, Tomaras G, Letvin NL, Chen B, Alam SM, Haynes BF. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS ONE. 2011;6:e27824. doi: 10.1371/journal.pone.0027824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry. 2007;46:1398–1401. doi: 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- Eckhart L, Raffelsberger W, Ferko B, Klima A, Purtscher M, Katinger H, Rüker F. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J Gen Virol. 1996;77(Pt 9):2001–2008. doi: 10.1099/0022-1317-77-9-2001. [DOI] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang C-H, Mcbride R, von Bredow B, Shivatare SS, Wu C-Y, Chan-Hui P-Y, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong C-H, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan CM, Berg W, Lewis GK, DeVico AL. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J Virol. 2002;76:12123–12134. doi: 10.1128/JVI.76.23.12123-12134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey G, Chen J, Rits-Volloch S, Freeman MM, Zolla-Pazner S, Chen B. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat Struct Mol Biol. 2010;17:1486–1491. doi: 10.1038/nsmb.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Dosenovic P, Ofek G, Baker D, Schief WR, Kwong PD, Karlsson Hedestam GB, Wyatt RT. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS ONE. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habte HH, Banerjee S, Shi H, Qin Y, Cho MW. Immunogenic properties of a trimeric gp41-based immunogen containing an exposed membrane-proximal external region. Virology. 2015;486:187–197. doi: 10.1016/j.virol.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MC, Abraham W, Crespo MP, Chen SH, Liu H, Szeto GL, Kim M, Reinherz EL, Irvine DJ. Liposomal vaccines incorporating molecular adjuvants and intrastructural T-cell help promote the immunogenicity of HIV membrane-proximal external region peptides. Vaccine. 2015;33:861–868. doi: 10.1016/j.vaccine.2014.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe CE, Xu S, Chigurupati P, Burda S, Williams C, Gorny MK, Zolla-Pazner S. Neutralization of HIV-1 primary isolates by polyclonal and monoclonal human antibodies. Int. Immunol. 1997;9:1281–1290. doi: 10.1093/intimm/9.9.1281. [DOI] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsik DL, Liu Y-Y, Strokappe NM, Battella S, El Khattabi M, McCoy LE, Sabin C, Hinz A, Hock M, Macheboeuf P, Bonvin AMJJ, Langedijk JPM, Davis D, Quigley AF, Aasa-Chapman MMI, Seaman MS, Ramos A, Poignard P, Favier A, Simorre J-P, Weiss RA, Verrips CT, Weissenhorn W, Rutten L. A gp41 MPER-specific Llama VHH Requires a Hydrophobic CDR3 for Neutralization but not for Antigen Recognition. PLoS Pathog. 2013;9:e1003202. doi: 10.1371/journal.ppat.1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Patrick AJ, Rosenthal KL. Multiple tandem copies of conserved gp41 epitopes incorporated in gag virus-like particles elicit systemic and mucosal antibodies in an optimized heterologous vector delivery regimen. Vaccine. 2010;28:7070–7080. doi: 10.1016/j.vaccine.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Jiang S, Lin K, Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JG. Enhancement of alpha -Helicity in the HIV-1 Inhibitory Peptide DP178 Leads to an Increased Affinity for Human Monoclonal Antibody 2F5 but Does Not Elicit Neutralizing Responses in Vitro. Implications for vaccine design. Journal of Biological Chemistry. 2002;277:45811–45820. doi: 10.1074/jbc.M205862200. [DOI] [PubMed] [Google Scholar]
- Julien J-P, Bryson S, Nieva JL, Pai EF. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J. Mol. Biol. 2008;384:377–392. doi: 10.1016/j.jmb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse P-J, Sanders RW, Moore JP, Wilson IA, Ward AB. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem Toukam D, Tenbusch M, Stang A, Temchura V, Storcksdieck Genannt Bonsmann M, Grewe B, Koch S, Meyerhans A, Nchinda G, Kaptue L, Uberla K. Targeting antibody responses to the membrane proximal external region of the envelope glycoprotein of human immunodeficiency virus. PLoS ONE. 2012;7:e38068. doi: 10.1371/journal.pone.0038068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavardhana S, Varadarajan R. Stabilizing the native trimer of HIV-1 Env by destabilizing the heterodimeric interface of the gp41 postfusion six-helix bundle. J Virol. 2014;88:9590–9604. doi: 10.1128/JVI.00494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim E, Okada K, Okada K, Kenniston T, Kenniston T, Raj VS, Raj VS, AlHajri MM, AlHajri MM, Farag EABA, Farag EABA, AlHajri F, AlHajri F, Osterhaus ADME, Osterhaus ADME, Haagmans BL, Haagmans BL, Gambotto A, Gambotto A. Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine. 2014;32:5975–5982. doi: 10.1016/j.vaccine.2014.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Sun Z-YJ, Rand KD, Shi X, Song L, Cheng Y, Fahmy AF, Majumdar S, Ofek G, Yang Y, Kwong PD, Wang J-H, Engen JR, Wagner G, Reinherz EL. Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat Struct Mol Biol. 2011;18:1235–1243. doi: 10.1038/nsmb.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs SJ, McBurney SP, Kovarik DN, Waddell CD, Jaworski JP, Sutton WF, Gomes MM, Trovato M, Waagmeester G, Barnett SJ, DeBerardinis P, Haigwood NL. Multimeric Scaffolds Displaying the HIV-1 Envelope MPER Induce MPER-Specific Antibodies and Cross-Neutralizing Antibodies when Co-Immunized with gp160 DNA. PLoS ONE. 2014;9:e113463. doi: 10.1371/journal.pone.0113463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai RPJ, Hock M, Radzimanowski J, Tonks P, Hulsik DL, Effantin G, Seilly DJ, Dreja H, Kliche A, Wagner R, Barnett SW, Tumba N, Morris L, LaBranche CC, Montefiori DC, Seaman MS, Heeney JL, Weissenhorn W. A Fusion Intermediate gp41 Immunogen Elicits Neutralizing Antibodies to HIV-1. J Biol Chem. 2014;289:29912–29926. doi: 10.1074/jbc.M114.569566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Lu Y, Xiao Y, Dierich MP, Chen Y. Induction of high level of specific antibody response to the neutralizing epitope ELDKWA on HIV-1 gp41 by peptide-vaccine. Peptides. 2000;21:463–468. doi: 10.1016/s0196-9781(00)00179-0. [DOI] [PubMed] [Google Scholar]
- Luo M, Yuan F, Liu Y, Jiang S, Song X, Jiang P, Yin X, Ding M, Deng H. Induction of neutralizing antibody against human immunodeficiency virus type 1 (HIV-1) by immunization with gp41 membrane-proximal external region (MPER) fused with porcine endogenous retrovirus (PERV) p15E fragment. Vaccine. 2006;24:435–442. doi: 10.1016/j.vaccine.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Markosyan RM, Leung MY, Cohen FS. The six-helix bundle of human immunodeficiency virus Env controls pore formation and enlargement and is initiated at residues proximal to the hairpin turn. J Virol. 2009;83:10048–10057. doi: 10.1128/JVI.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusic C, Rizza P, Lattanzi L, Mancini C, Spada M, Belardelli F, Benvenuto E, Capone I. Chimeric plant virus particles as immunogens for inducing murine and human immune responses against human immunodeficiency virus type 1. J Virol. 2001;75:8434–8439. doi: 10.1128/JVI.75.18.8434-8439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba N, Geyer BC, Kilbourne J, Alfsen A, Bomsel M, Mor TS. Humoral immune responses by prime-boost heterologous route immunizations with CTB-MPR649-684, a mucosal subunit HIV/AIDS vaccine candidate. Vaccine. 2006;24:5047–5055. doi: 10.1016/j.vaccine.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Matyas GR, Wieczorek L, Beck Z, Ochsenbauer-Jambor C, Kappes JC, Michael NL, Polonis VR, Alving CR. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS. 2009;23:2069–2077. doi: 10.1097/QAD.0b013e32832faea5. [DOI] [PubMed] [Google Scholar]
- McGaughey GB, Citron M, Danzeisen RC, Freidinger RM, Garsky VM, Hurni WM, Joyce JG, Liang X, Miller M, Shiver J, Bogusky MJ. HIV-1 Vaccine Development: Constrained Peptide Immunogens Show Improved Binding to the Anti-HIV-1 gp41 MAb. Biochemistry. 2003;42:3214–3223. doi: 10.1021/bi026952u. [DOI] [PubMed] [Google Scholar]
- Mohan T, Verma P, Rao DN. Comparative mucosal immunogenicity of HIV gp41 membrane-proximal external region (MPER) containing single and multiple repeats of ELDKWA sequence with defensin peptides. Immunobiology. 2014;219:292–301. doi: 10.1016/j.imbio.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. 2008;72:54–84. doi: 10.1128/MMBR.00020-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H. Antibody B cell responses in HIV-1 infection. Trends Immunol. 2014;35:549–561. doi: 10.1016/j.it.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Muster T, Ferko B, Klima A, Purtscher M, Trkola A, Schulz P, Grassauer A, Engelhardt OG, García-Sástre A, Palese P. Mucosal model of immunization against human immunodeficiency virus type 1 with a chimeric influenza virus. J Virol. 1995;69:6678–6686. doi: 10.1128/jvi.69.11.6678-6686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Powell R, Baskakov IV, DeVico A, Lewis GK, Wang L-X. Synthesis, conformation, and immunogenicity of monosaccharide-centered multivalent HIV-1 gp41 peptides containing the sequence of DP178. Bioorg Med Chem. 2004;12:3141–3148. doi: 10.1016/j.bmc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD. Elicitation of structure-specific antibodies by epitope scaffolds. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang G-Y, Ofek G, Stewart-Jones GBE, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, Tauer C, Berger R, Barrett N, Jungbauer A. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- Qin Y, Banasik M, Kim S, Penn-Nicholson A, Habte HH, LaBranche C, Montefiori DC, Wang C, Cho MW. Eliciting neutralizing antibodies with gp120 outer domain constructs based on M-group consensus sequence. Virology. 2014;462–463:363–376. doi: 10.1016/j.virol.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WE, Gorny MK, Xu JY, Mitchell WM, Zolla-Pazner S. Two immunodominant domains of gp41 bind antibodies which enhance human immunodeficiency virus type 1 infection in vitro. J Virol. 1991;65:4169–4176. doi: 10.1128/jvi.65.8.4169-4176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, Scheid JF, Lee JH, West AP, Chen C, Gao H, Gnanapragasam PNP, Mares R, Seaman MS, Ward AB, Nussenzweig MC, Bjorkman PJ. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano S, Araujo A, Apellániz B, Bryson S, Carravilla P, de la Arada I, Huarte N, Rujas E, Pai EF, Arrondo JLR, Domene C, Jiménez MA, Nieva JL. Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope. J Biol Chem. 2014;289:6565–6580. doi: 10.1074/jbc.M113.527747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Bohon J, Han DP, Habte H, Qin Y, Cho MW, Chance MR. Structural characterization of HIV gp41 with the membrane-proximal external region. J Biol Chem. 2010;285:24290–24298. doi: 10.1074/jbc.M110.111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker L-G, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Tyler DS, Stanley SD, Zolla-Pazner S, Gorny MK, Shadduck PP, Langlois AJ, Matthews TJ, Bolognesi DP, Palker TJ, Weinhold KJ. Identification of sites within gp41 that serve as targets for antibody-dependent cellular cytotoxicity by using human monoclonal antibodies. J Immunol. 1990;145:3276–3282. [PubMed] [Google Scholar]
- Venditto VJ, Watson DS, Motion M, Montefiori D, Szoka FC. Rational design of membrane proximal external region lipopeptides containing chemical modifications for HIV-1 vaccination. Clin. Vaccine Immunol. 2013;20:39–45. doi: 10.1128/CVI.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditto VJ, Wieczorek L, Molnar S, Teque F, Landucci G, Watson DS, Forthal D, Polonis VR, Levy JA, Szoka FC. Chemically modified peptides based on the membrane-proximal external region of the HIV-1 envelope induce high-titer, epitope-specific nonneutralizing antibodies in rabbits. 2014;21:1086–1093. doi: 10.1128/CVI.00320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JY, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Wen Z, Dong K, Wang X, Bu Z, Zhang H, Compans RW, Yang C. Induction of HIV neutralizing antibodies against the MPER of the HIV envelope protein by HA/gp41 chimeric protein-based DNA and VLP vaccines. PLoS ONE. 2011;6:e14813. doi: 10.1371/journal.pone.0014813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G, Lapelosa M, Bradley R, Mariano TM, Dietz DE, Hughes S, Wrin T, Petropoulos C, Gallicchio E, Levy RM, Arnold E, Arnold GF. Chimeric rhinoviruses displaying MPER epitopes elicit anti-HIV neutralizing responses. PLoS ONE. 2013;8:e72205. doi: 10.1371/journal.pone.0072205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Li X, Kasterka M, Gorny MK, Zolla-Pazner S, Sodroski J. Oligomer-specific conformations of the human immunodeficiency virus (HIV-1) gp41 envelope glycoprotein ectodomain recognized by human monoclonal antibodies. AIDS Res Hum Retroviruses. 2009;25:319–328. doi: 10.1089/aid.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Huang Y, Fayad R, Spear GT, Qiao L. Induction of Mucosal and Systemic Neutralizing Antibodies against Human Immunodeficiency Virus Type 1 (HIV-1) by Oral Immunization with Bovine Papillomavirus-HIV-1 gp41 Chimeric Virus-Like Particles. J Virol. 2004;78:8342–8348. doi: 10.1128/JVI.78.15.8342-8348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]