Summary
Immune responses differ between laboratory mice and humans. Chronic infection with viruses and parasites are common in humans, but are absent in laboratory mice, and thus represent potential contributors to inter-species differences in immunity. To test this, we sequentially infected laboratory mice with herpesviruses, influenza, and an intestinal helminth, and compared their blood immune signatures to mock-infected mice before and after vaccination against Yellow Fever Virus (YFV-17D). Sequential infection altered pre- and post-vaccination gene expression, cytokines, and antibodies in blood. Sequential pathogen exposure induced gene signatures that recapitulated those seen in blood from pet store-raised versus laboratory mice, and adult versus cord blood in humans. Therefore basal and vaccine-induced murine immune responses are altered by infection with agents common outside of barrier facilities. This raises the possibility that we can improve mouse models of vaccination and immunity by selective microbial exposure of laboratory animals to mimic that of humans.
Introduction
Substantial variation in human immune responses is due to environmental influences (Brodin et al., 2015; Roederer et al., 2015). Potential variables include nutritional status, different health practices, age, socioeconomic status and geographic location. In addition, the bacterial microbiome influences immune and inflammatory responses (Honda and Littman, 2012; Hooper et al., 2012). An added, but less well understood, environmental contributor to variation is the history of infection with acute and chronic pathogens, including herpesviruses and intestinal parasites (Foxman and Iwasaki, 2011; Furman et al., 2015; Salgame et al., 2013; Virgin, 2014; Virgin et al., 2009). Persistent infections change the immune response to unrelated pathogens and vaccines (Furman et al., 2015; Oldstone, 2005; Osborne et al., 2014; Reese et al., 2014; Salgame et al., 2013; Selin et al., 2006; Slifka et al., 2003; Virgin, 2014). Some chronic co-infections enhance, while others inhibit immunity to secondary challenge (Barton et al., 2007; MacDuff et al., 2015; Stelekati et al., 2014; Stelekati and Wherry, 2012). Moreover, humans are frequently infected with acute viral pathogens which may change the immune system (Foxman and Iwasaki, 2011).
There is concern that rodent models do not faithfully predict human immune responses (Mestas and Hughes, 2004; Seok et al., 2013; Takao and Miyakawa, 2015), limiting the value of this powerful model system. However, mouse models are indispensable for biomedical studies and play a significant role in the development of vaccines and therapeutics. This highlights the need for studies that identify environmental variables that might, in addition to chromosomal genetic variation, contribute to species-specific immune response differences between mice and humans. Notably, barrier-raised mice are free of many acute and chronic infections that are recognized to contribute to human immune variation (Salgame et al., 2013; Virgin, 2014). For example, people chronically infected with intestinal helminths have lower responses to vaccination with Bacillus Calmette-Guerin (BCG)(Elias et al., 2001), cholera (Cooper et al., 2001) and tetanus toxoid (Nookala et al., 2004; Sabin et al., 1996). Furthermore, chronic infection with the herpesvirus cytomegalovirus (HCMV) alters responses to human influenza vaccination (Furman et al., 2015), and infection of mice with murine CMV (MCMV) and/or a murine γ-herpesvirus (MHV68) alters bacterial immunity and reverses inherited immunodeficiency (Barton et al., 2007; MacDuff et al., 2015). We therefore sought to test the hypothesis that infection history, and in particular the presence of chronic co-infections in mice with agents similar to those commonly acquired by human children as they develop, alters basal and vaccine-induced immunity. We went on to assess the relationship of the changes we observed to gene expression differences between cord blood and adult blood in humans.
Results
Reduced antibody response in mice co-infected with multiple viruses and a helminth after YFV-17D vaccination
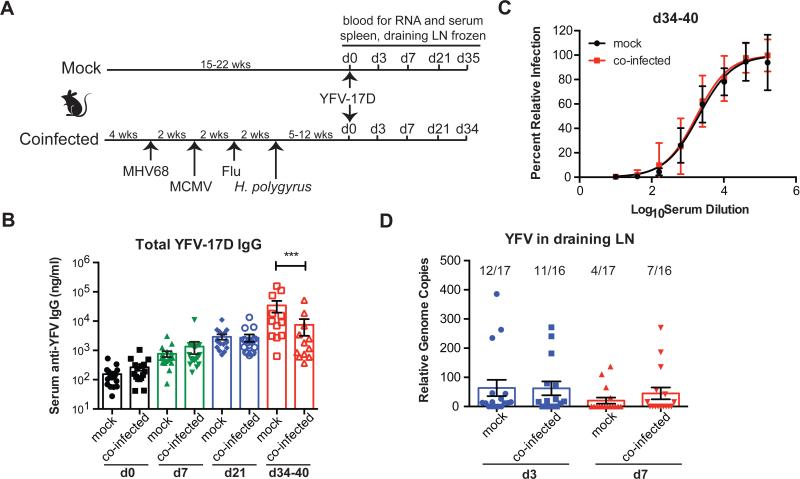
We separated 159 C57Bl/6 barrier-raised mice into four separate experiments (Figure S1B). Within each experiment half of the mice were sequentially inoculated with PBS (‘mock-infected’) and half were infected with a series of viruses and a helminth parasite starting at weaning to mimic a diverse infection history (‘co-infected’, Figure 1A, Figure S1). For co-infections we chose MHV68 (related to human Epstein-Barr virus and Kaposi's sarcoma herpesvirus) and MCMV (related to human CMV), both of which establish persistent and latent infections in mice and alter immune responses and/or gene expression in multiple organs during chronic infection (Barton et al., 2007; Canny et al., 2014; White et al., 2010). Co-infected mice were also challenged with influenza strain WSN as a representative acute respiratory viral challenge, and Heligmosomoides polygyrus, an intestinal helminth that establishes chronic infection in the small intestines of mice (Osborne et al., 2014; Reese et al., 2014). All of these pathogens are models for pathogens that commonly infect humans starting early in childhood. Five to 16 weeks after the last infection, co-infected and mock-infected mice were vaccinated with the live attenuated yellow fever vaccine YFV-17D. Total anti-YFV IgG was measured in serum following vaccination (Figure 1B). Co-infected and control mice exhibited equivalent antibody responses early after vaccination, but by day 34 total anti-YFV IgG was lower in co-infected than mock mice (Figure 1B). Total neutralizing antibody titers were similar at this time point, suggesting that co-infection changed the amount of antibody rather than the quality of the neutralizing response (Figure 1C). Viral replication of YFV-17D is very low in mice and is often undetectable in vaccinated mice (Meier et al., 2009). Using a sensitive RT-PCR assay (Mantel et al., 2008) we detected no difference in the number of mice with detectable YFV in the draining lymph node either 3 or 7 days after vaccination (Figure 1D), suggesting that antibody differences were not due to difference in virus replication. Whether the statistically significant difference we observe in total antibody to YFV-17D is biologically significant remains to be determined. Neutralizing antibody is used as a correlate of protection for YFV (Mason et al., 1973), however antibodies have other functions in controlling viruses and there is evidence these Fc-mediated functions not measured by neutralization may have important roles in virus control (Chung et al., 2015; Pulendran and Ahmed, 2011).
Figure 1. Co-infection of mice with chronic viruses and a helminth leads to reduced antibody production after vaccination with YFV-17D.
(A) Experimental outline. See experimental procedures and Figure S1 for more detail on number of mice and groups.
(B) Measurement of total anti-YFV-17D IgG from serum.
(C) Total serum neutralizing anti-YFV-17D antibody was quantified 34-40 days after vaccination.
(D) Genome copies (normalized to actin) of YFV-17D were measured in draining lymph node (LN) of mice on days 3 and 7 following vaccination. Numbers above bar are the number of mice positive for YFV genome over the number of mice analyzed. One-way ANOVA was used to calculate statistical differences *** p< 0.01. Dots represent individual animals, and bars represent averages +/− SEM.
Defining transcriptional effects of co-infections before and after vaccination
To define genome-wide transcriptional effects of co-infection on basal and vaccine-induced immunity, we harvested peripheral blood from mice prior to (day 0) and 3, 7, or 21 days after vaccination, and examined gene expression by microarray. Unsupervised analysis revealed significant differences between mock and co-infected samples, with mock samples clustering together and co-infected samples clustering separately (Figure S2). These data indicate that co-infection altered the basal state of the immune system prior to vaccination as well as gene expression patterns in blood cells following vaccination. Consistent with this, fold change scatter plots indicated minimal overlap in gene expression profiles between co-infected mice and mock-infected mice after vaccination at days 3 (rho =0.029), 7 (rho =0.11), and 21 (rho=0.06) (Figure S3). We identified genes differentially expressed between co-infected and mock mice, and performed K-means clustering to associate groups of genes that changed significantly over time during YFV infection between mock and co-infected mice. We identified five temporally distinct groups of genes differentially expressed between mock and co-infected mice. These genes contained a total of 701 genes that fall into a broad range of biological functions related to immunity, organelle function, protein catabolism, cell death, metabolism, hemostasis and other processes (Figure 2A, B).
Figure 2. Microarray analysis of blood samples of vaccinated animals reveals kinetically distinct gene expression in co-infected mice relative to mock.
(A) Heatmap of significant kinetically different genes between mock and co-infected. See Table S1.
(B) Kinetics of each cluster, represented in individual points for each of three experiments as the median expression value of all genes in a given cluster across a sample. Line represents the mean of three replicates median expression values.
(C) Heatmap of top significantly enriched GO terms in each cluster. Lack of ontological agreement between mock and co-infected high clusters suggests highly differential response to vaccine.
(D) Analysis of cytokines in serum of vaccinated animals over time in pictograms (pg)/ml. One-way ANOVA was used to calculate statistical differences * p< 0.5, ** p<0.1, *** p<0.01. Symbols are individual animals, and bars are averages of all animals +/− SEM.
Pathway analysis of five gene clusters in mock and co-infected mice
Cluster 1 and 2 contain genes that were relatively similar in expression in co-infected and mock mice prior to vaccination and were more increased in response to vaccination in mock than in co-infected mice, especially 7 and 21 days after vaccination (Figure 2A, B). Pathway analysis revealed that cluster 1 genes are involved in ubiquitination, cell cycle, NFκB signaling and innate immune response signaling (Figure 2C, Table S1). Cluster 2 includes a smaller set of genes with a similar kinetic pattern that are related to Golgi vesicle transport. Thus co-infection can substantially blunt transcriptional responses corresponding to these processes in barrier-raised mice.
Cluster 3 and 4 genes were elevated in co-infected mice prior to vaccination and were further induced by vaccination of co-infected mice at day 3 and then diminished with varying kinetics by days 7 and 21 (Figure 2A, B). These genes represent a set of processes that are induced by co-infection, but less substantially up-regulated by vaccination of mock mice. This suggests that co-infection markedly changed the basal state of the immune system and enhanced certain vaccine-induced transcriptional changes. Cluster 3 genes are involved in the response to interferon-γ (IFNγ) and IFNαβ, regulation of interleukin-1β (IL-1β) production, T cell activation, and cell death (Figure 2C, Table S1). These pathways are consistent with analysis of serum cytokine data, where elevated IFNγ and to a lesser extent IL-12p70 and IL-1β were detected in co-infected mice at day 0 prior to vaccination (Figure 2D). Cluster 4 genes are involved in amine metabolism, symbiotic interactions, and negative regulation of protein modifications (Figure 2C, Table S1).
Cluster 5 genes were lower prior to vaccination in co-infected mice and followed a kinetic pattern most similar to cluster 1 genes with the exception that these genes were induced by vaccination in mock mice earlier than observed for cluster 1 genes, peaking at day 3. These genes are involved in IL-12 production and the function of nonlymphoid blood cells such as basophils and phagocytes as well as platelets (Figure 2C, Table S1).
Leading Edge Metagene analysis and comparison of metagenes with pet store versus laboratory mice
Together these data indicate that a history of infection in barrier-raised mice with agents similar to those that commonly infect humans changes both the basal state of the immune system and the nature and the kinetics of vaccine responses. We next sought to determine how the sets of genes regulated by co-infection related to gene expression patterns observed in other biologically relevant settings. We first considered that co-infection of barrier-raised laboratory mice might induce gene expression patterns similar to those observed in mice raised in non-barrier conditions. To this end, we performed Leading Edge Metagene (LEM) analysis (Godec et al., 2016), comparing co-infected and mock at baseline, to identify sets of genes with correlated expression (metagenes) representing specific biological processes within the co-infection gene expression profile in our data sets. This analysis identified a type I interferon response-enriched metagene in co-infected mice and a naïve lymphocyte-enriched metagene in mock infected mice (Figure S4, Table S2). The significant enrichment of genes in co-infected mice in multiple pathways related to interferon signaling and innate immune responses is consistent with prior observations of the effects of latent and persistent herpesvirus infection on IFNγ expression (Barton et al., 2007; MacDuff et al., 2015).
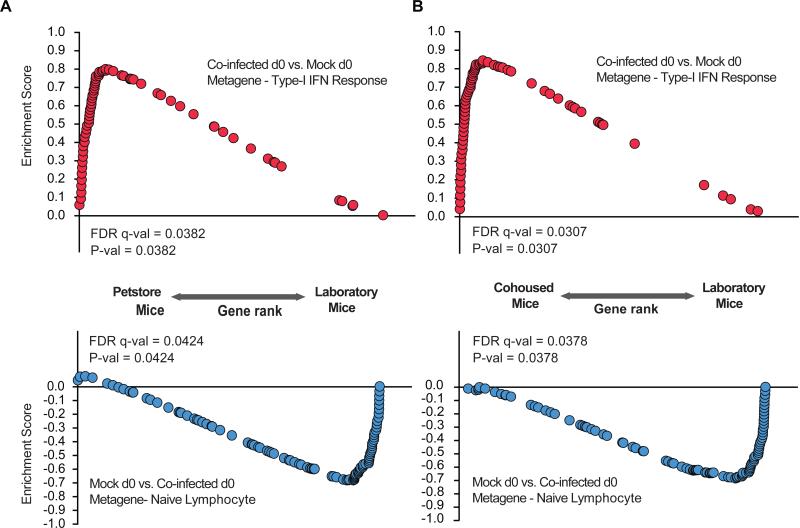
We next performed enrichment analysis of the co-infected day 0 versus mock day 0 type I interferon response metagene with gene expression observed in pet store mice compared to barrier-raised laboratory mice. In addition, we compared these metagenes to laboratory mice co-housed with pet store mice compared to barrier-raised laboratory mice (Beura et. al, in submission) (Figure 3A, B). The type I interferon metagene found in co-infected mice was significantly enriched in pet store mice as well as laboratory mice co-housed with pet store mice, while the naïve lymphocyte metagene found in mock mice was significantly enriched in laboratory mice (Figure 3). Thus co-infected mice express genes found in mice raised in a more “dirty” environment compared to a specific pathogen-free barrier.
Figure 3. Comparison of metagenes from co-infected and mock mice with pet store-raised and laboratory-raised mice indicates overlap of co-infected mice with pet store mice and mock mice with laboratory-raised mice.
(A) Enrichment analysis of a co-infected d0 vs. mock d0 metagene (type-I interferon response) and mock vs. co-infected d0 metagene (naive lymphocyte) in pet store vs. laboratory mice
(B) Enrichment analysis of a co-infected d0 vs. mock d0 metagene (type-I interferon response) and mock vs. co-infected d0 metagene (naive lymphocyte) in cohoused vs. laboratory array data.
Comparison of metagenes in mock and co-infected mice with neonate and adult blood gene expression signatures
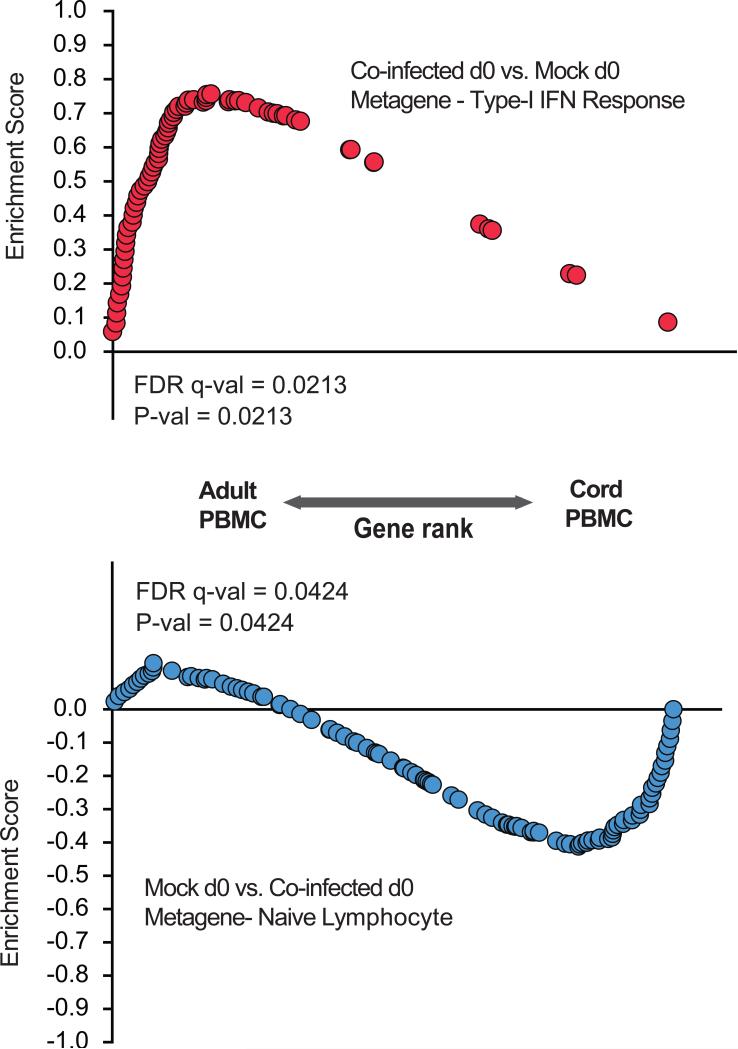
To understand whether these observations relate to species-specific gene expression patterns differing between barrier-raised mice and adult humans, we compared gene expression profiles prior to vaccination in mock and co-infected mice with human cord blood and adult blood (Votavova et al., 2011). We found that the type-I interferon metagene identified in co-infected mice was significantly enriched in maternal human blood while the naïve lymphocyte metagene in mock mice was significantly enriched in human cord blood (Figure 4). Taken together, these data suggest that mock mice from a barrier facility have a naïve or immature immune system that is more similar to human neonates than human adults. Co-infection of these barrier-raised mice promotes an immune system gene expression pattern more similar to pet store-raised mice and adult humans.
Figure 4. Co-infected mice have parallel gene expression with human maternal blood, whereas mock mice have parallel gene expression with human cord blood.
Enrichment analysis of co-infected d0 vs. mock d0 metagene (type-I interferon response) and mock vs. co-infected d0 metagene (naive lymphocyte) in human maternal and cord blood array data.
Discussion
These data indicate that studies of immune responses in mice should consider the virome, helminth infection, and, more broadly, infection history as potential determinants of species-specific immune responses. Chromosomal genes that differ between mice and humans do not solely determine inter-species differences in the immune system. Environmental factors that contribute to variation in immune response include nutritional status, age, socioeconomic status, geographic location, and the microbiome. While studies of the bacterial microbiome highlight the important contribution this variable makes to inflammatory responses (Honda and Littman, 2012; Hooper et al., 2012), there are fewer studies addressing the contribution of acute and chronic viruses and parasites to varying immune responses. Recent work showed that an acute infection compromised tissue-specific immunity even after clearance of the bacteria (Fonseca et al., 2015). Our work identifies chronic pathogens as important contributors to differences in response to vaccines, and likely other inflammatory conditions.
Potential mechanisms for how co-infections alter immunity are complex. Possible mechanisms include chronic stimulation of innate immune responses that regulate adaptive immunity, antigenic mimicry, cross-reactive immune responses, altered antigen presentation and changes in differentiation of memory and naïve lymphocyte responses (Hensley et al., 2007; Oldstone, 2005; Selin et al., 2006; Stelekati et al., 2014; Stelekati and Wherry, 2012; Virgin, 2014; Virgin et al., 2009). Latent infection with the γ-herpesvirus used in our studies, MHV68, alters the number and phenotype of memory CD8+ T cells (Barton et al., 2014). Latent MCMV infection also results in profound alteration in the T cell compartment, leading to impaired naïve T cell function (Cicin-Sain et al., 2012), despite no increase in susceptibility to secondary challenge with influenza virus, West Nile virus, or vesicular stomatitis (Marandu et al., 2015). Any or all of these mechanisms could be important contributors to co-infection associated changes in gene expression and altered antibody responses to YFV vaccination.
It is important to note that effects of co-infections and mechanisms that drive co-infection mediated changes in immune responses will likely differ depending on the co-infecting pathogens. Some pathogens will have more profound effects on T cells, for example, whereas others will have stronger effects on other aspects of immunity. Moreover, the effects driven by co-infections could be different for different vaccines. Some vaccine responses may be more affected by co-infections than others. More detailed studies will be required to determine which or what combination of our co-infecting pathogens are required for the effects we are seeing. Additionally, other studies examining different co-infecting pathogens and vaccines will be required to delineate mechanisms.
YFV vaccine is extremely efficacious, however immune responses in humans differ significantly. Evidence suggests that although vaccine provides protection in greater than 80% of vaccinees, levels of neutralizing antibody and T cell responses are significantly different (Querec et al., 2009). Additionally, B and T cell responses to YFV vaccination are measurably different in people from different parts of the world with variable levels of baseline immune activation prior to vaccination (Gaucher et al., 2008; Muyanja et al., 2014). Together, these data suggest that environmental factors, including possible co-infections may contribute to inter-individual variation.
In order to enhance reproducibility of mouse studies the biomedical research community rendered this important research animal specific-pathogen free. In doing so, researchers may have created an immune response that is less relevant to studies of humans or other mammals raised outside of barrier containment. Careful consideration of these issues may enhance the value of mouse models for experimental research relevant to human disease. As mechanisms for effects such as the co-infection-dependent changes in immunity identified here are defined, we speculate that it may become possible to manipulate the nature of mouse immune responses by reintroducing, under controlled circumstances, some of the natural infectious exposures that constitute the selective pressures that shaped the mouse immune system. This may partially humanize murine immune responses.
Experimental Procedures
Animals, infections, and sample collection
The individual experiments are outlined in Figure S1. In summary, at weaning mice were divided into two groups. The mock group was housed in the biohazard facility with the co-infected group. The co-infected mice were infected with 105 plaque forming units (PFU) intranasally of MHV68, followed by 105 PFU (in PBS) tissue culture passage MCMV intraperitoneally, 103 PFU influenza strain WSN (in PBS) intranasally, and 50 L3 larvae of H. polygyrus orally. Mice were then rested in the biohazard facility for 5-12 weeks. Mock and co-infected mice were challenged with 106 PFU of YFV-17D subcutaneously in the footpad. Mice were sacrificed on days 0, 3, 7, 21, and for experiments 1, 2, and 4 on days 34-40. Blood cells, serum, spleen, draining lymph node were collected. Blood cells were processed using RiboPure RNA purification kit from whole blood (Thermo Fisher Scientific). RNA from individual mice from each timepoint was pooled within an experiment for microarrays. Samples were hybridized to Affymetrix Mouse 430 2.0 arrays.
Analysis of microarray data
Prior to analysis, mouse microarray data were processed and normalized using the Affymetrix MAS5 algorithm and batch correction was performed using the ComBat algorithm. Genes exhibiting differential kinetics between co-infected and mock samples over the d0 to d21 time course were identified using maSigPro (Conesa et al., 2006) (default FDR <0.05). Gene set enrichment analysis (GSEA) was performed as described previously (Subramanian et al., 2005). LEM analysis was performed downstream of GSEA to yield groups of genes, termed metagenes, which are coordinately up-regulated in a given phenotypic comparison and common to multiple enriched gene sets. Metagenes were identified for d0 co-infected relative to d0 mock and vice versa. Enrichments of co-infected and mock metagenes in pet store vs. laboratory mouse (GSE78979), cohoused vs. laboratory mouse (GSE78979), and adult PBMC vs. cord PBMC array data (GSE27272) (Votavova et al., 2011) were determined using standard GSEA.
Supplementary Material
Acknowledgements
We acknowledge Darren Kreamalmeyer for mouse colony management; E. Pearce and S. Huang for H. polygyrus parasites; B. Parikh and W. Yokoyama for MCMV; J. Brien and M. Diamond for YFV-17D; the Genome Technology Access Center at Washington University for microarrays. A core funded by NIH P30AR048335 provided experimental support. NIH awards R24 OD019793, R01 OD011170, R01 AI111918, and R01 DK101354 supported HWV. Damon Runyon Postdoctoral Fellowship supported TAR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship Contributions
TAR and HWV designed the project and wrote the paper. TAR and AK performed experiments. KB and AFM performed bioinformatic analyses. WNH, BP and RS provided advice on bioinformatic analyses. All authors reviewed the manuscript.
Accession numbers
GEO accession number for microarray data is GSE79466.
Author Information:
The authors declare no competing financial interests statement.
References
- Barton ES, Rajkarnikar S, Langston PK, Price MJ, Grayson JM. Gammaherpesvirus Latency Differentially Impacts The Generation Of Primary Versus Secondary Memory CD8+T Cells During Subsequent Infection. J. Virol. 2014 doi: 10.1128/JVI.02106-14. doi:10.1128/JVI.02106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. doi:10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJL, Furman D, Shen-Orr S, Dekker CL, Swan GE, Butte AJ, Maecker HT, Davis MM. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. doi:10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny SP, Goel G, Reese TA, Zhang X, Xavier R, Virgin HW. Latent gammaherpesvirus 68 infection induces distinct transcriptional changes in different organs. J. Virol. 2014;88:730–738. doi: 10.1128/JVI.02708-13. doi:10.1128/JVI.02708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, Hoffner M, Streeck H, Ackerman ME, McElrath MJ, Schuitemaker H, Pau MG, Baden LR, Kim JH, Michael NL, Barouch DH, Lauffenburger DA, Alter G. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 2015;163:988–998. doi: 10.1016/j.cell.2015.10.027. doi:10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicin-Sain L, Brien JD, Uhrlaub JL, Drabig A, Marandu TF, Nikolich-Žugich J. Cytomegalovirus infection impairs immune responses and accentuates T-cell pool changes observed in mice with aging. PLoS Pathog. 2012;8:e1002849. doi: 10.1371/journal.ppat.1002849. doi:10.1371/journal.ppat.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Nueda MJ, Ferrer A, Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. doi:10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- Cooper PJ, Chico M, Sandoval C, Espinel I, Guevara A, Levine MM, Griffin GE, Nutman TB. Human infection with Ascaris lumbricoides is associated with suppression of the interleukin-2 response to recombinant cholera toxin B subunit following vaccination with the live oral cholera vaccine CVD 103-HgR. Infect. Immun. 2001;69:1574–1580. doi: 10.1128/IAI.69.3.1574-1580.2001. doi:10.1128/IAI.69.3.1574-1580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias D, Wolday D, Akuffo H, Petros B, Bronner U, Britton S. Effect of deworming on human T cell responses to mycobacterial antigens in helminth-exposed individuals before and after bacille Calmette-Guérin (BCG) vaccination. Clin. Exp. Immunol. 2001;123:219–225. doi: 10.1046/j.1365-2249.2001.01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca DMD, Hand TW, Han S-J, Gerner MY, Glatman Zaretsky A, Byrd AL, Harrison OJ, Ortiz AM, Quinones M, Trinchieri G, Brenchley JM, Brodsky IE, Germain RN, Randolph GJ, Belkaid Y. Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell. 2015;163:354–366. doi: 10.1016/j.cell.2015.08.030. doi:10.1016/j.cell.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman EF, Iwasaki A. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat. Rev. Microbiol. 2011;9:254–264. doi: 10.1038/nrmicro2541. doi:10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D, Jojic V, Sharma S, Shen-Orr SS, L Angel CJ, Onengut-Gumuscu S, Kidd BA, Maecker HT, Concannon P, Dekker CL, Thomas PG, Davis MM. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. 2015;7:281ra43–281ra43. doi: 10.1126/scitranslmed.aaa2293. doi:10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sékaly R-P. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. doi:10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, Mesirov JP, Haining WN. Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. doi:10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley SE, Cun AS, Giles-Davis W, Li Y, Xiang Z, Lasaro MO, Williams BRG, Silverman RH, Ertl HCJ. Type I interferon inhibits antibody responses induced by a chimpanzee adenovirus vector. Mol. Ther. 2007;15:393–403. doi: 10.1038/sj.mt.6300024. doi:10.1038/sj.mt.6300024. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. doi:10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. doi:10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff DA, Reese TA, Kimmey JM, Weiss LA, Song C, Zhang X, Kambal A, Duan E, Carrero JA, Boisson B, Laplantine E, Israël A, Picard C, Colonna M, Edelson BT, Sibley LD, Stallings CL, Casanova J-L, Iwai K, Virgin HW. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. Elife. 2015;4:e04494. doi: 10.7554/eLife.04494. doi:10.7554/eLife.04494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N, Aguirre M, Gulia S, Girerd-Chambaz Y, Colombani S, Moste C, Barban V. Standardized quantitative RT-PCR assays for quantitation of yellow fever and chimeric yellow fever-dengue vaccines. J. Virol. Methods. 2008;151:40–46. doi: 10.1016/j.jviromet.2008.03.026. doi:10.1016/j.jviromet.2008.03.026. [DOI] [PubMed] [Google Scholar]
- Marandu TF, Oduro JD, Borkner L, Dekhtiarenko I, Uhrlaub J, Drabig A, Kröger A, Nikolich-Žugich J, Cicin-Sain L. Immune protection against virus challenge in aging mice is not affected by latent herpesviral infections. J. Virol. 2015:JVI.01989–15. doi: 10.1128/JVI.01989-15. doi:10.1128/JVI.01989-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RA, Tauraso NM, Spertzel RO, Ginn RK. Yellow fever vaccine: direct challenge of monkeys given graded doses of 17D vaccine. Appl Microbiol. 1973;25:539–544. doi: 10.1128/am.25.4.539-544.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier KC, Gardner CL, Khoretonenko MV, Klimstra WB, Ryman KD. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS Pathog. 2009;5:e1000614. doi: 10.1371/journal.ppat.1000614. doi:10.1371/journal.ppat.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Muyanja E, Ssemaganda A, Ngauv P, Cubas R, Perrin H, Srinivasan D, Canderan G, Lawson B, Kopycinski J, Graham AS, Rowe DK, Smith MJ, Isern S, Michael S, Silvestri G, Vanderford TH, Castro E, Pantaleo G, Singer J, Gillmour J, Kiwanuka N, Nanvubya A, Schmidt C, Birungi J, Cox J, Haddad EK, Kaleebu P, Fast P, Sékaly R-P, Trautmann L. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. J. Clin. Invest. 2014;124:3147–3158. doi: 10.1172/JCI75429. doi:10.1172/JCI75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookala S, Srinivasan S, Kaliraj P, Narayanan RB, Nutman TB. Impairment of tetanus-specific cellular and humoral responses following tetanus vaccination in human lymphatic filariasis. Infect. Immun. 2004;72:2598–2604. doi: 10.1128/IAI.72.5.2598-2604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MBA. Molecular mimicry, microbial infection, and autoimmune disease: evolution of the concept. Curr. Top. Microbiol. Immunol. 2005;296:1–17. doi: 10.1007/3-540-30791-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, Tomov VT, Kobuley D, Tran SV, Bittinger K, Bailey AG, Laughlin AL, Boucher J-L, Wherry EJ, Bushman FD, Allen JE, Virgin HW, Artis D. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science. 2014;345:578–582. doi: 10.1126/science.1256942. doi:10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. doi:10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Wakeman BS, Choi HS, Hufford MM, Huang SC, Zhang X, Buck MD, Jezewski A, Kambal A, Liu CY, Goel G, Murray PJ, Xavier RJ, Kaplan MH, Renne R, Speck SH, Artyomov MN, Pearce EJ, Virgin HW. Helminth infection reactivates latent γ-herpesvirus via cytokine competition at a viral promoter. Science. 2014;345:573–577. doi: 10.1126/science.1254517. doi:10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Quaye L, Mangino M, Beddall MH, Mahnke Y, Chattopadhyay P, Tosi I, Napolitano L, Terranova Barberio M, Menni C, Villanova F, Di Meglio P, Spector TD, Nestle FO. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. doi: 10.1016/j.cell.2015.02.046. doi:10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin EA, Araujo MI, Carvalho EM, Pearce EJ. Impairment of tetanus toxoid-specific Th1-like immune responses in humans infected with Schistosoma mansoni. J. Infect. Dis. 1996;173:269–272. doi: 10.1093/infdis/173.1.269. [DOI] [PubMed] [Google Scholar]
- Salgame P, Yap GS, Gause WC. Effect of helminth-induced immunity on infections with microbial pathogens. Nat. Immunol. 2013;14:1118–1126. doi: 10.1038/ni.2736. doi:10.1038/ni.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim S-K, Clute SC, Welsh RM. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol. Rev. 2006;211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. doi:10.1111/j.0105- 2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG, Inflammation and Host Response to Injury, Large Scale Collaborative Research Program Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. doi:10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka MK, Homann D, Tishon A, Pagarigan R, Oldstone MBA. Measles virus infection results in suppression of both innate and adaptive immune responses to secondary bacterial infection. J. Clin. Invest. 2003;111:805–810. doi: 10.1172/JCI13603. doi:10.1172/JCI13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati E, Shin H, Doering TA, Dolfi DV, Ziegler CG, Beiting DP, Dawson L, Liboon J, Wolski D, Ali M-AA, Katsikis PD, Shen H, Roos DS, Haining WN, Lauer GM, Wherry EJ. Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity. 2014;40:801–813. doi: 10.1016/j.immuni.2014.04.010. doi:10.1016/j.immuni.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati E, Wherry EJ. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe. 2012;12:458–469. doi: 10.1016/j.chom.2012.10.001. doi:10.1016/j.chom.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. doi:10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. doi:10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW. The Virome in Mammalian Physiology and Disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. doi:10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. doi:10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Votavova H, Dostalova Merkerova M, Fejglova K, Vasikova A, Krejcik Z, Pastorkova A, Tabashidze N, Topinka J, Veleminsky M, Sram RJ, Brdicka R. Transcriptome alterations in maternal and fetal cells induced by tobacco smoke. Placenta. 2011;32:763–770. doi: 10.1016/j.placenta.2011.06.022. doi:10.1016/j.placenta.2011.06.022. [DOI] [PubMed] [Google Scholar]
- White DW, Keppel CR, Schneider SE, Reese TA, Coder J, Payton JE, Ley TJ, Virgin HW, Fehniger TA. Latent herpesvirus infection arms NK cells. Blood. 2010;115:4377–4383. doi: 10.1182/blood-2009-09-245464. doi:10.1182/blood-2009-09-245464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.