Summary
Depression is a complex, heterogeneous disorder and a leading contributor to the global burden of disease. Most previous research has focused on individual brain regions and genes contributing to depression. However, emerging evidence in humans and animal models suggests that dysregulated circuit function and gene expression across multiple brain regions drive depressive phenotypes. Here we performed RNA-sequencing on 4 brain regions from control animals and those susceptible or resilient to chronic social defeat stress at multiple time points. We employed an integrative network biology approach to identify transcriptional networks and key driver genes that regulate susceptibility to depressive-like symptoms. Further, we validated in vivo several key drivers and their associated transcriptional networks that regulate depression susceptibility and confirmed their functional significance at the levels of gene transcription, synaptic regulation and behavior. Our study reveals novel transcriptional networks that control stress susceptibility and offers fundamentally new leads for antidepressant drug discovery.
Introduction
Depression is a complex and heterogeneous disorder and a major contributor to the global burden of disease, yet current therapeutics have serious limitations (Greenberg et al., 2015; Steel et al., 2014). Virtually all drugs used to treat depression target the same basic mechanisms identified serendipitously more than 60 years ago and these existing pharmacotherapies induce full remission in fewer than 50% of people (Block and Nemeroff, 2014). Depression is thought to arise from a complex interaction of biological, psychological and social factors and, consequently, finding a single target that causes depression in all individuals is unlikely. Rather, depression may be better understood as a multi-gene syndrome, in which pathology arises from compounded small changes affecting many genes rather than large changes in a small subset of genes (Gaiteri et al., 2014). Genome-wide transcriptional profiling may shed new light on the molecular mechanisms of the illness and help identify transcriptional regulators that better account for the complexity of depression. This in turn will facilitate the development of truly novel antidepressant treatments that target defined transcriptional networks.
We aimed to gain novel insight into the molecular basis of depression by leveraging an unbiased, systems approach focused on transcriptional regulation. Weighted gene coexpression network analysis (WGCNA) (Zhang and Horvath, 2005) is one such approach that has been utilized successfully to provide new biological insight into gene networks involved in several CNS disorders, including autism (Parikshak et al., 2013), Alzheimer's disease (Miller et al., 2013; Zhang et al., 2013), schizophrenia (Maschietto et al., 2015) and alcoholism (Vanderlinden et al., 2013). Previous studies have utilized coexpression analyses in depressed human post-mortem tissue or mouse stress models to describe interesting network level changes in single brain regions, but the mechanistic role of such changes has not been examined (Chang et al., 2014; Gaiteri and Sibille, 2011; Malki et al., 2015; Malki et al., 2013).
Neuroimaging studies of depressed patients and related findings from animal models suggest that depression may be a circuit-level disorder in which several functionally inter-connected brain regions are affected (Bagot et al., 2015; Christoffel et al., 2015; Ressler and Mayberg, 2007). Accordingly, while studies of individual brain regions have yielded important information, a more global interrogation of transcriptional profiles within the several brain regions that comprise this broader circuitry may offer a fundamentally better understanding of the pathophysiology of the disorder. The nucleus accumbens (NAC) lies at the center of one such circuit implicated in depression (Epstein et al., 2006; Nestler and Carlezon, 2006; Schlaepfer et al., 2008). The NAC integrates information from diverse glutamatergic inputs from prefrontal cortex (PFC), amygdala (AMY) and ventral hippocampus (VHIP), among other regions (Goto and Grace, 2008). Structural, functional and transcriptional changes in each of these brain regions have been reported in both rodent depression models and depressed humans (Bagot et al., 2015; Chang et al., 2014; Covington et al., 2010; Ding et al., 2015; Guilloux et al., 2012; Jaworska et al., 2014; Kennedy et al., 2001; Mayberg et al., 2000; Sequeira et al., 2009; Vialou et al., 2014). We recently identified pathway-specific functional alterations in this circuitry (Bagot et al., 2015) in a highly validated mouse model of depression, chronic social defeat stress (CSDS) (Berton et al., 2006; Krishnan et al., 2007). Specifically, activity of VHIP projections to NAC mediated susceptibility to CSDS, while PFC and AMY projections to NAC mediated resilience; however, the molecular mechanisms of these changes are unknown.
To understand the transcriptional mechanisms of dysregulated circuit function in depression, we used CSDS and RNA-sequencing (RNA-seq) to generate transcriptional profiles in NAC, VHIP, PFC and AMY of control, susceptible and resilient mice at both early and late time-points post-CSDS. Utilizing these transcriptional profiles, we identified networks of co-regulated genes associated with susceptibility or resilience, Interestingly, some of the networks associated with susceptibility show opposite regulation in VHIP vs. PFC and we validate the ability of novel hub genes within these networks to drive functional abnormalities at molecular, synaptic and behavioral levels. Together these data provide important new insights into transcriptional mechanisms of stress susceptibility.
Results
Differential expression signatures of susceptibility and resilience to CSDS
To generate circuit-wide transcriptional profiles we used RNA-seq to analyze NAC, VHIP, PFC and AMY from control, susceptible and resilient mice at 3 time-points post-CSDS (Figure 1A). Previous work established that CSDS induces 2 phenotypes: mice that are susceptible to stress (~67%) and exhibit profound and enduring social avoidance, and those that are resilient to stress (~33%) and continue to show a preference for social interaction similar to control mice (Krishnan et al., 2007).
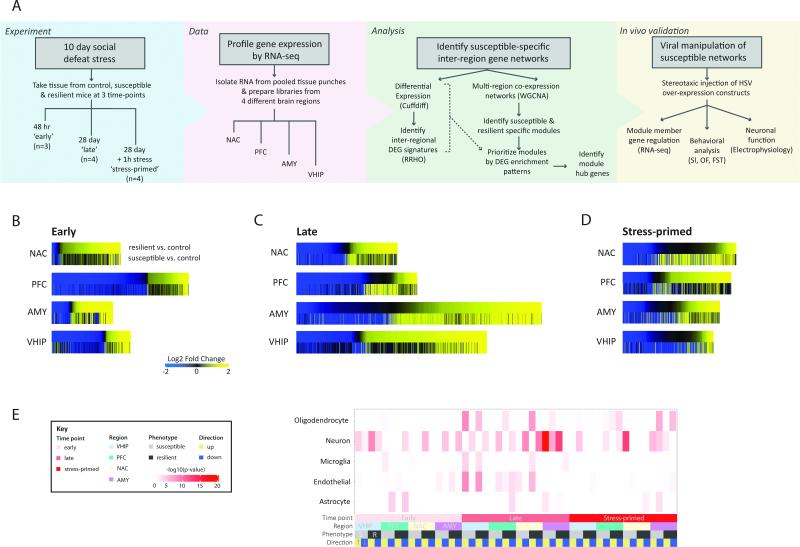
Figure 1. Overview of Experimental Design and Differential Gene Expression in Susceptible and Resilient Mice after CSDS.
(A) After CSDS 4 brain regions were collected at 3 post-defeat time-points (‘early’: 48h; ‘late’: 28d; ‘stress-primed’: 28d + 1 h post stress) for transcriptional profiling to identify expression networks underlying susceptible and resilient phenotypic adaptations to stress. Schematic diagram of experimental approach. (B-D) Union heatmaps show FC of all genes significantly differentially expressed (FC>1.3, p<0.05) in either comparison for resilient vs. control (R vs. C; top panel) or susceptible vs. control (S vs. C; lower panel) rank ordered by fold change in the R vs. C comparison in NAC, PFC, AMY and VHIP early (B), late, and stress-primed (D) scaled by number of DEGs. (E) Matrix summarizes enrichment of oligodendrocyte, neuron, microglia, endothelial or astrocyte genes (Zhang et al., 2014) in DEGs upregulated (yellow) and downregulated (blue) in R vs. C (dark grey) and S vs. C (light grey) conditions, early (light pink), late (dark pink) and stress-primed (red) in AMY (purple), NAC (cream), PFC (green), and VHIP (lightblue). Darker color indicates increasing –log10(p-value). See also Figure S1 and Table S1.
Profiling gene expression changes across brain region and time
Previous work has established that functional and transcriptional alterations associated with susceptibility vs. resilience to CSDS represent distinct processes, with resilience not simply being the absence of susceptibility (Dias et al., 2014; Friedman et al., 2014; Krishnan et al., 2007; Wilkinson et al., 2009). Thus, we aimed to identify the transcriptional alterations induced by CSDS in each population relative to the same non-stressed control animals to independently characterize stress-induced transcriptional profiles in susceptible and resilient states. We first profiled patterns of differential gene expression (susceptible vs. control [S vs. C] and resilient vs. control [R vs. C]) in each brain region at early (48h), late (28d) and stress-primed (28d, 5 min acute aggression, followed by 55 min housed adjacent to aggressor) time-points post-CSDS (Figure 1, Table S1). We selected the 48h and 28d time points to profile both the emergence and persistence of susceptible and resilient phenotypes as previous work has shown that the phenotypes emerge by 48h and are stable at 28 days post-CSDS (Krishnan et al., 2007). Broadly, at 48h, we identified more differentially expressed genes (DEGs; p<0.05, FC >1.3) in R vs. C than in S vs. C mice in all brain regions (Figure 1B). By contrast, at 28d, the largest changes were observed in S vs. C in AMY and in R vs. C in VHIP and PFC, with NAC exhibiting similar numbers of DEGs in the 2 conditions (Figure 1C).
Since enduring changes in brain function in depression can affect not only basal neural circuit function, but also how the same circuits respond to subsequent stressors (Admon et al., 2015; Hooley et al., 2009), we re-exposed a group of animals, 28d post-CSDS, to an acute defeat stress for 1h. Under this stress-primed condition, more DEGs were detected in S vs. C than in R vs. C in every brain region except PFC, where more DEGs were still detected in R vs. C (Figure 1D). Together, these results suggest that resilient animals show a greater initial response to stress, which may reflect active adaptation. Further, long after the stress, neural circuits in resilient animals may be less responsive to stress, with the important exception of PFC. The unique degree of gene regulation in the resilient PFC might indicate a homeostatic adaptation that serves to constrain excessive stress-induced activation in other brain regions (Buijs and Van Eden, 2000).
To further characterize the observed DEG patterns, we examined enrichment of cell-type associated genes and gene ontologies (GOs). Although patterns of cell-type enrichment varied across time, neuronal genes were enriched in all brain regions at each time point (Figure 1E). Early post-defeat (48h), DEGs were predominantly enriched for neuronal genes. Late post-defeat (28d), a diversity of cell-type associated genes was present, including neurons (Figure 1E). Furthermore, DEGs were enriched for several GOs including translation and biosynthetic processes, GPCR signaling, extracellular matrix, plasma membrane and protein metabolic processes (Figure S1). We also identified DEGs in each brain region regulated across time (28d vs. 48h) in control mice and observed that some of these age-related genes overlapped with DEGs regulated in susceptible or resilient mice at each time point (Table S2).
Regional comparisons of differential gene expression patterns after CSDS
Depression involves circuit-level functional alterations in multiple brain structures, which may be reflected in altered transcriptional synchrony (Chen et al., 2015; Gaiteri et al., 2010; Posner et al., 2013). To explore such transcriptional synchrony after CSDS, we compared differential gene expression patterns in susceptibility and resilience between pairs of brain regions 48h post-defeat, an early time point when such phenotypic alterations emerge. We used a rank rank hypergeometric overlap test (RRHO) to identify the patterns and significance of overlap between gene expression profiles (Figure 2) (Plaisier et al., 2010). In resilience (R vs. C), we identified a robust overlap between PFC and NAC (max –log10(p-value)=752) in genes upregulated in both brain regions (Figure 2A). We also observed a weaker overlap in co-upregulated genes between PFC and VHIP (max –log10(p-value)=320). In susceptibility (S vs. C), we identified weaker overlaps between PFC and VHIP (max –log10(p-value)=432), as well as between PFC and NAC (max –log10(p-value)=360), in genes upregulated in each pair of brain regions (Figure 2A). Further RRHO analysis confirmed significantly different strengths of the overlap between PFC and NAC (max-log10(p-value)=19) and between PFC and VHIP (max-log10(p-value)=3) in resilient vs. susceptible conditions (Figure S2).
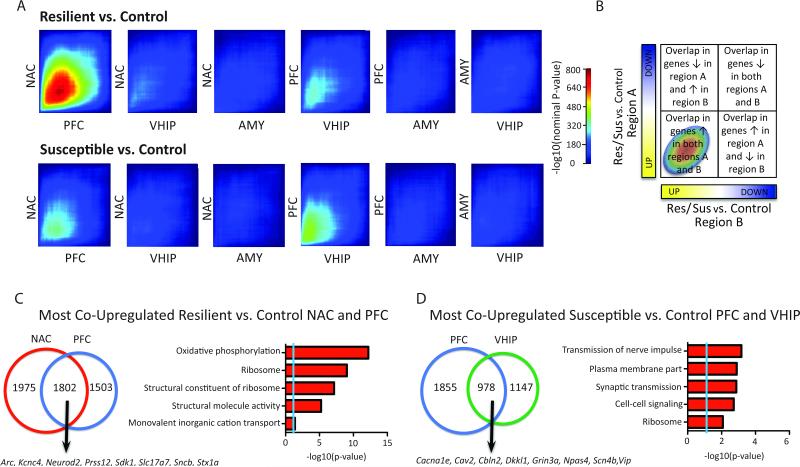
Figure 2. Inter-Regional Differential Expression Patterns Reveal Resilient- and Susceptible-Specific Co-Upregulation Signatures.
(A) RRHO maps compare threshold-free differential expression between pairs of brain regions in the resilient (R vs. C; upper panel) or susceptible (S vs. C; lower panel) transcriptome 48h post-CSDS. Each pixel represents the overlap between the resilient/ susceptible transcriptome of 2 brain regions (NAC, PFC, AMY, VHIP) with the significance of overlap (−log10(p-value) of a hypergeometric test; step size 200) color coded. The extent of overlap of upregulated genes is displayed in the bottom left corner, and in the top right the overlap of downregulated genes, illustrated in (B). Venn diagrams display the extent of overlap between genes upregulated in (C) NAC and PFC in resilient mice and (D) PFC and VHIP in susceptible mice, enriched gene ontology terms and examples of co-upregulated genes. See also Figure S2.
We next performed GO analysis on genes coordinately upregulated in PFC and NAC of resilient mice (R vs. C) (Figure 2C). Such genes enriched for the GOs oxidative phosphorylation (p=6.32×10−13, 3.56x), ribosome (p=8.96×10−10, 3.86x) and structural molecule activity (p=5.76×10−6, 2.41x). Likewise, genes coordinately upregulated in PFC and VHIP of susceptible mice (S vs. C) (Figure 2D) enriched for the GOs synaptic transmission (p=1.3×10−4, 2.92x) and ribosome (p=8.4×10−3, 3.43x).
Together, these analyses identified increased synchrony of transcriptional regulation between PFC and NAC in resilience, and between PFC and VHIP in susceptibility. The findings suggest that increased similarity of transcriptional profiles in PFC and NAC may drive resilience, whereas increased similarity in PFC and VHIP may drive susceptibility.
Coexpression analysis identifies susceptible- and resilient-specific gene networks
Having identified the broad pattern of transcriptome-wide changes across brain regions we then sought to resolve specific gene coexpression networks that could be critical in determining susceptiblity or resilience to CSDS. Previous findings suggest that gene coexpression analysis is especially useful in identifying transcriptional alterations in multi-gene diseases, where the phenotypic state emerges from the convergence of numerous small changes rather than from isolated single-gene effects (Gaiteri et al., 2014). We constructed 2 independent gene coexpression networks integrating expression data across brain regions (PFC, NAC, AMY, VHIP) and time-points (early, late, stress-primed) separately for susceptible and resilient mice (Figure 3A) to identify clusters (modules) of coexpressed genes (Langfelder et al., 2008; Zhang and Horvath, 2005). The susceptible network consisted of 52 modules and the resilient network consisted of 30 modules, with each module assigned an arbitrary color name (Table S3). Note that the 2 networks are completely independent and, while module names are reused across networks, there is no implied similarity of gene members.
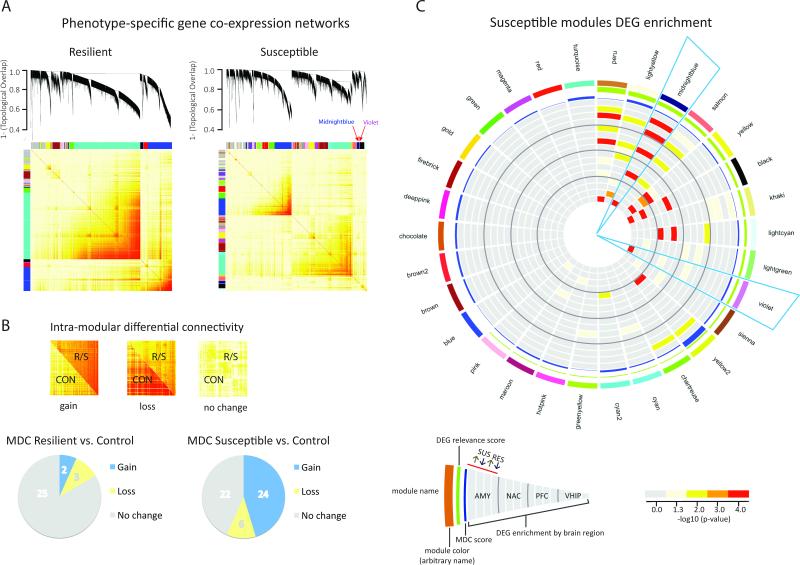
Figure 3. Identification of Resilient- and Susceptible-Specific Coexpression Networks and Key Modules.
(A) Multi-region coexpression network analysis identified coexpressed modules in resilient (left panel) and susceptible (right panel) mice across brain regions (NAC, PFC, AMY, VHIP) and time (early, late, stress-primed). Each module is arbitrarily assigned a unique color identifier, in bars on the left and top of each topological overlap matrix (TOM; lower panel). Increasing color intensity from white to dark red in TOM corresponds to increasing coexpression-based topological overlap. Dendograms (upper panel) show average linkage hierarchical clustering of genes. (B) Intra-modular connectivity of resilient and susceptible network modules was compared to that of corresponding genes in control mice to identify gain, loss or no change (upper panel). Pie charts (lower panel) summarize module differential connectivity (MDC) analysis. Proportionally, very few resilient modules (left panel) had significant MDC compared to more than half of all susceptible modules, which predominantly showed gain of connectivity. See also Figure S3. (C) Circos plot shows module name (ring 1), color (ring 2), differential expression relevance score (ring 3), and MDC score; increasing bar height shows increasing score (ring 4). Bar color indicates significance of enrichment for genes significantly up- or downregulated 48h post-defeat in R vs. C (RES) and S vs. C (SUS) with increasingly warm colors indicating increasing –log10(p-value). MB and V modules show gain of connectivity relative to controls and are enriched for genes that show opposing patterns of differential expression in PFC and NAC vs. VHIP. AMY SUS up (ring 5), AMY SUS down (ring 6), AMY RES up (ring 7), AMY RES down (ring 8), NAC SUS up (ring 9), NAC SUS down (ring 10), NAC RES up (ring 11), NAC RES down (ring 12), PFC SUS up (ring 13), PFC SUS down (ring 14), PFC RES up (ring 15), PFC RES down (ring 16), VHIP SUS up (ring 17), VHIP SUS down (ring 18), VHIP RES up (ring 19), VHIP RES down (ring 20). See also Figure S3 and Table S3.
To begin to probe the biological relevance of the identified coexpression modules we examined enrichment of GO terms (Table S3). Many of the identified biological processes have previously been implicated in depression (Arloth et al., 2015; Chang et al., 2014; Ding et al., 2015; Fabbri and Serretti, 2015; Kang et al., 2012; Philip et al., 2010; Sequeira et al., 2009; Tham et al., 2011; NPAS of PGI, 2015). GOs enriched in the susceptible network modules included microtubule cytoskeleton (Lightcyan; p=8.8×10−20 9.85x), extracellular matrix (Lightyellow; p=2.9×10−15, 6.36x), synaptic transmission (Yellow; p=4.8 × 10−14 3.7x), cell-cell signaling (Midnightblue; p=9.0×10−9, 3.16x), transcriptional activation (Purple; p=1.1×10−6, 3.1x) and NF-κB signaling (Red; p=6.3×10−4, 7.13x). Those enriched in the resilience network modules included synaptic transmission (Brown; p=2.80×10−18, 3.79x), ribosome (Blue; p=2.60×10−13, 7.15x), GABAA receptor activity (Cyan; p=3.0×10−9, 74.13x), extracellular matrix (Green; p=4.6×10−11, 4.6x), nerve ensheathement (Yellow; p=2.4×10−6, 14.56x) and acetylcholine metabolism (Greenyellow; p=3.3×10−3, 148.85x).
Greater coexpression differences in susceptibility vs. resilience
We next assessed the robustness of the coexpression modules identified in susceptible and resilient networks by determining whether their “connectivity”—the strength of coexpression—differs from control conditions. This is based on the hypothesis that gene modules that show altered connectivity in susceptibility or resilience as compared to the control state are the most functionally relevant (Figure 3B, S3A-B). A gene module can show either increased or decreased connectivity. Loss of connectivity describes a group of genes whose expression is highly coordinated in control mice but becomes less coordinated in susceptible or resilient mice, whereas gain of connectivity describes a group of genes more coordinately regulated in susceptible or resilient mice than in control mice. Loss of connectivity suggests a disruption or weakening of a basal transcriptional network, whereas gain of connectivity suggests strengthening or even emergence of a novel transcriptional network. Within the susceptible network, 24 modules (46.2%) showed gain of connectivity and 6 (11.5%) showed loss of connectivity, with the remaining modules (22; 42.3%) showing no change in connectivity compared to control. In stark contrast, within the resilient network, only 2 (6.7%) modules showed a gain of connectivity and 3 (10%) showed a loss of connectivity compared to control, with the majority (25; 83.3%) showing no differential connectivity (Table S3). This analysis indicates that susceptibility associates with much larger changes in network connectivity than resilience, which suggests that coexpression changes may be most relevant in susceptibility. We thus focused our subsequent analyses on susceptible modules with significant “module differential connectivity” or MDC. The robustness of the susceptible modules was further validated by additional statistical assessments (Table S4).
Probing susceptible-specific modules
To gain insight into the biology of the susceptible-specific modules, we identified the most interesting modules for further study. We first examined the enrichment of cell-type associated genes in susceptible modules and identified 9 enriched for cell-type signatures (Figure S3C). Neuronal genes were significantly enriched in 5 modules: Midnightblue (p=1.27×10−14, 4.80x), Salmon (p=2.67×10-9, 3.60x), Yellow (p=2.03 ×10−11, 2.83x), Cyan (p=0.002, 2 .48x) and Seashell (p=0.003, 3.78x). The Lightyellow module is enriched for oligodendrocyte (p=3.15×10−10, 13.94x), microglial (p=5.75×10−11, 4.06x) and endothelial (p=4.04×10−23, 6.62x) genes. Endothelial genes are also enriched in Peru (p=1.74×10−5, 4.05x), Purple (p=7.8×10−4, 2.47x), Chartreuse (p=0.004, 5.73x), Green (p=0.01, 1.75x) and Yellow2 (p=0.03, 7.55x), and Lightcyan enriched for astrocytic genes (p=5.50×10−14, 6.76x).
To further probe the biological significance of susceptible modules in the emergence of susceptibility, we examined the enrichment of S vs. C and R vs. C DEGs at the 48h time-point. Of the 30 significantly differentially connected modules, 19 were also enriched for DEGs in at least 1 brain region (Figure 3C). The Midnightblue (MB) module was of particular interest as it was highly enriched for DEGs across brain regions in a way that was consistent with regional RRHO patterns (Figure 2) and with our previous functional findings (Bagot et al., 2015). Specifically, this module robustly enriched for upregulated DEGs in PFC and NAC as well as downregulated DEGs in VHIP in R vs. C. MB also enriched for DEGs downregulated in AMY in both R vs. C and S vs. C, suggesting a lack of specificity within AMY that may relate to findings of elevated anxiety in both susceptible and resilient mice (Krishnan et al., 2007). One other module, Violet (V), was similarly enriched for DEGs upregulated in PFC and NAC and DEGs downregulated in VHIP in R vs. C. This region-specific pattern of DEG enrichment within the MB and V modules, along with the earlier identified regional RRHO analysis (Figure 2A), suggests that opposing PFC-VHIP regulation might be particularly important for determining susceptibility vs. resilience. Indeed, several of the most co-upregulated genes in the RRHO analysis were also found within either the MB or V module (e.g., Cbln2, Dkkl1, Neurod2, Prss12, Sdk1, Stx1a), and the synaptic transmission GO term is common to MB and to genes co-upregulated in PFC and VHIP (Figure 2C,D; Table S3). These shared patterns of gene regulation also relate to our recent findings of opposing functional adaptations between VHIP and PFC projections to NAC after CSDS (Bagot et al., 2015). Together, these analyses suggested that the MB and V modules may be particularly important in governing the emergence of susceptibility vs. resilience to CSDS.
Resolving the network structure of key susceptible-specific modules
Both the MB and V susceptible-specific modules show a gain of connectivity in susceptibility, suggesting that genes in these modules become coordinated in susceptible mice in a unique architecture or to a degree that does not occur in control mice (Figure S3B; Table S3). Further analysis revealed that the MB and V modules also exhibit a significant gain of connectivity in susceptibility relative to resilience (Table S4). As well, MB and V are neighboring modules within the hierarchical clustering network dendrogram and topological overlap matrix and as such highly correlated (Figure 3A), indicating that they are closely related. MB, a module of 433 genes, is significantly enriched for neuronal genes and GOs including cell-cell signaling and synaptic transmission, whereas V, a much smaller module of 77 genes, is not significantly enriched for specific cell-types or GOs.
As a prelude to testing the biological relevance of these networks in vivo, we reconstructed the network structure of genes within each of these modules based solely on their coexpression based connectivity and identified so-called ‘hub genes’ and ‘susceptible-specific hub genes’. Hub genes (or key drivers) are highly connected genes within a module that are predicted to control the expression of many other module members, although it is important to note that this prediction is derived from non-directed correlational analyses. Susceptible-specific hub genes are key drivers in susceptibility, but not under control conditions, and as such may be especially important in generating gene coexpression networks unique to susceptibility. Of the 60 hub genes in MB, 31 were susceptible-specific (Figure 4A), and of the 14 hub genes in V, 7 were susceptible-specific (Figure 4B; Table S5). We hypothesized that directed manipulation of susceptible-specific hub genes would regulate the expression of these key networks and subsequently direct the outcomes of CSDS towards susceptibility or resilience.
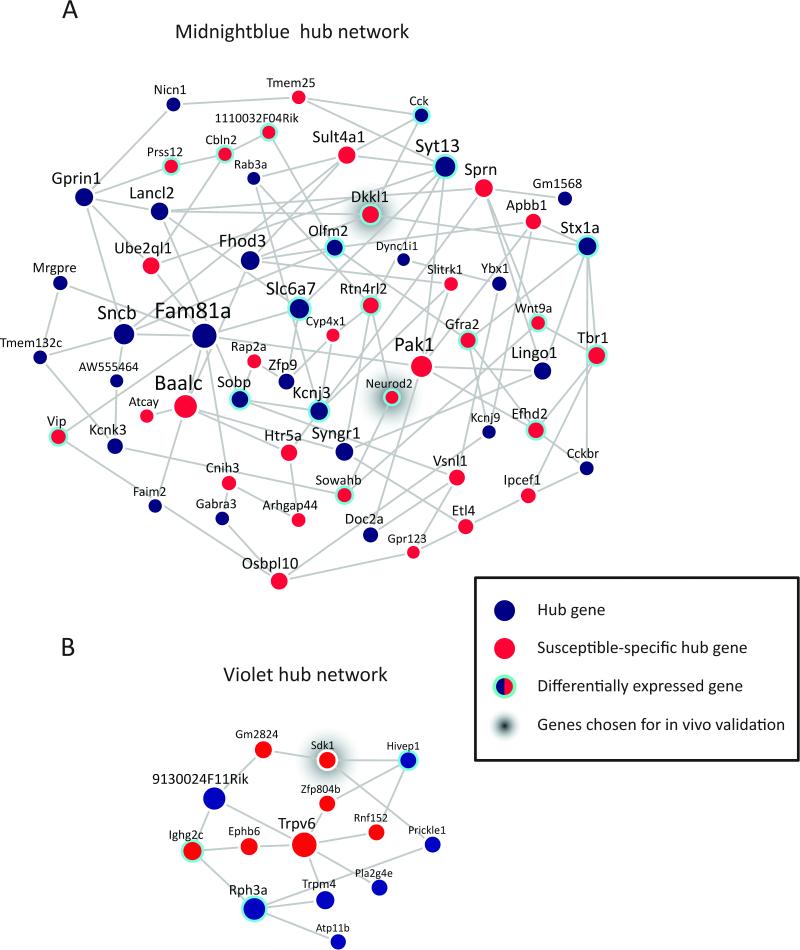
Figure 4. Hub Gene Coexpression Networks of MB and V Modules in Susceptible Mice.
(A) Network plot of hub genes within MB module. (B) Network plot of hub genes identified within V module. Node size is proportional to node's network centrality. Blue nodes indicate hub genes, red nodes indicate susceptible-specific hub genes and cyan halos indicate differential expression of a gene early post-defeat in at least 1 brain region. Edges reflect significant interactions between genes based on mutual information. Early post-defeat, Dkkl1 was differentially expressed in PFC (increased in R vs. C) and AMY (decreased in both S vs. C and R vs. C), and Neurod2 was differentially expressed in NAC (increased in R vs. C) early post-defeat, whereas Sdk1 was not differentially expressed in any region. See also Figure S4 and Table S5.
Over-expression of the hub gene Dkkl1 regulates the MB network
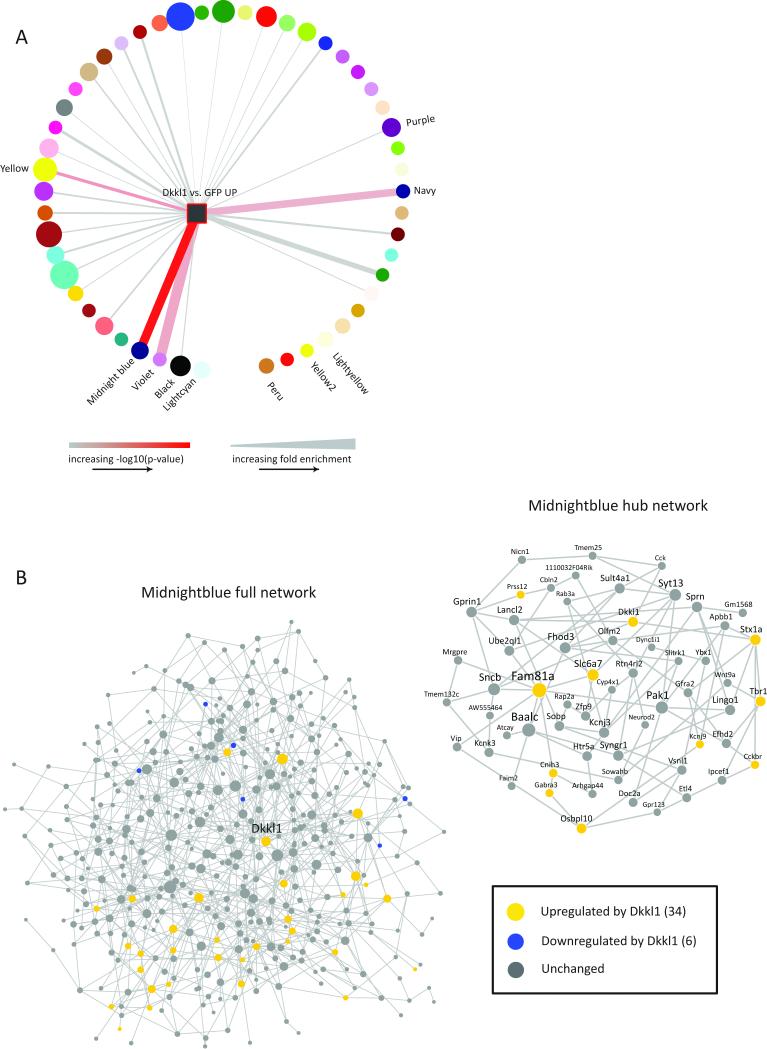
We first sought to validate our network analysis in vivo by testing the prediction that regulation of a hub gene would preferentially induce expression its module genes as opposed to genes in other modules. We focused on the larger MB module and the susceptible-specific hub gene Dkkl1 in VHIP, as our analyses predicted a regulatory role for this gene in VHIP even though it was not differentially expressed in this brain region. Using HSV vectors to infect neurons (Heller et al., 2014), thus targeting our manipulation to the cell-type for which the MB module was enriched, we over-expressed Dkkl1 plus GFP or GFP alone in VHIP of adult mice. We then subjected the mice to accelerated social defeat (coinciding with timing of maximal HSV expression) and performed RNA-seq on the virally infected tissue from VHIP. Differential expression analysis identified 184 genes upregulated by Dkkl1 over-expression and 1099 genes downregulated compared to GFP alone (p<0.05, FC>1.3; Figure S5A). We reasoned that, if Dkkl1 is a highly connected hub gene in the MB module, Dkkl1 over-expression should induce other MB module genes. Indeed, the genes upregulated in VHIP by Dkkl1 over-expression are highly significantly enriched in the MB module (p=1.46×10−21, 9.42x; Figure 5B). Notably, many other hub genes were regulated by Dkkl1 (Figure 5B). In addition, the genes upregulated by Dkkl1 over-expression were enriched for neuronal genes (p=1.96×10-13, 3.72x; Figure S5B), as was also observed for the MB module (Figure S4). We observed less significant enrichment in only a limited number of other susceptible modules (Figure 5A): V (p=5.46×10−5, 10.91x), a module closely related to MB as noted earlier; Yellow (p=1.51×10−6, 3.13x), a large module (1075 genes) that is enriched for similar GO terms as MB and shows a similar gain of connectivity and similar DEG enrichment (opposing PFC and VHIP regulation); and Navy (p=4.27×10−4, 7.85x), a smaller module (108 genes) that is enriched for the DEGs downregulated in VHIP of susceptible mice, suggesting its function may overlap with VHIP mechanisms of susceptibility. While the MB module was specifically enriched for DEGs upregulated by Dkkl1 over-expression, the DEGs downregulated by Dkkl1 overexpression were not enriched (data not shown). These observations provide critical validation of the hub gene status of Dkkl1 in regulating gene expression within the MB module in brain and thereby provide crucial in vivo support for our bioinformatic predictions.
Figure 5. In Vivo Over-Expression of Susceptible-Specific Hub Gene, Dkkl1, Upregulates MB Module Members.
Differential expression analysis identified 108 genes upregulated and 1075 genes downregulated in VHIP in HSV-Dkkl1-GFP vs. HSV-GFP at p<0.05, FC>1.3. (A) DEGs upregulated by Dkkl1 over-expression (yellow square) were significantly enriched in MB. Upregulated DEGs also enriched in V, Yellow and Navy. Increasing edge width indicates increasing fold enrichment (min=0, max=10). Increasing color gradient (grey to red) indicates increasing –log10(p-value) (min=0, max=21). Circle/square size indicates \ module size. (B) DEGs upregulated by Dkkl1 over-expression enriched in the MB network; left panel full MB network, right panel MB hub genes (see also Figure 5A). Yellow circles: genes upregulated by Dkkl1 (34).Blue circles: genes downregulated by Dkkl1; not significantly enriched (6). Grey circles: genes not regulated by Dkkl1. See also Figure S5.
Next, given that the manipulation of the MB module and Dkkl1 is predicted to induce behavioral susceptibility, we reasoned that it might reverse “resilient-like” transcriptional profiles in this brain region. As noted earlier, the MB module is enriched for the downregulated DEGs in R vs. C in VHIP. Thus, we asked whether the DEGs upregulated by Dkkl1 over-expression, which are enriched in the MB module and similarly enriched for neuronal genes, might overlap with the DEGs downregulated in R vs. C early post-defeat. Indeed, this enrichment was highly significant (p=1.5 × 10−8, 8.20x), indicating that over-expression of this susceptible-specific hub gene in VHIP induces a pattern of gene expression opposing that associated with resilience.
Over-expression of susceptible-specific hub genes in VHIP induces susceptibility
Having confirmed the potential of our coexpression network analysis to identify key drivers of the susceptible network, we examined the functional significance of predicted hub genes within the MB and V modules in directing behavioral outcomes of defeat stress. We selected 2 susceptible-specific hub genes, Dkkl1 and Neurod2, from the larger MB network and 1, Sdk1, from V for over-expression. Note that, while MB and V modules contain genes previously implicated in depression (see Discussion), we explicitly focused on genes not previously implicated in depression to test the utility of our unbiased approach to identify novel targets. Moreover, we selected susceptible-specific hub genes that were identified even though they did not show consistent differential expression changes in the targeted regions, a fact that would have excluded them from conventional DEG analyses. The selected hub genes mediate diverse biological processes. Little is known about the function of Dkkl1 in brain except that it bears sequence similarity to Dkk1, an antagonist of canonical Wnt signaling, implicated broadly in neuronal development and survival (Oliva et al., 2013; Sibbe and Jarowyj, 2013). Neurod2 encodes a transcription factor that is important in neuronal differentiation and synapse maturation (Messmer et al., 2012; Olson et al., 2001; Wilke et al., 2012). Sdk1 is a cell-adhesion molecule that guides synapse formation in retina (Yamagata and Sanes, 2008; Yamagata et al., 2002), and recent work from our group has implicated Sdk1 in cocaine-induced spinogenesis in NAC (Scobie et al., 2014). Based on the pattern of DEGs with the MB and V modules (Figure 3C), we predicted that over-expressing the selected hub genes in VHIP vs. PFC would induce different effects on susceptibility to social defeat.
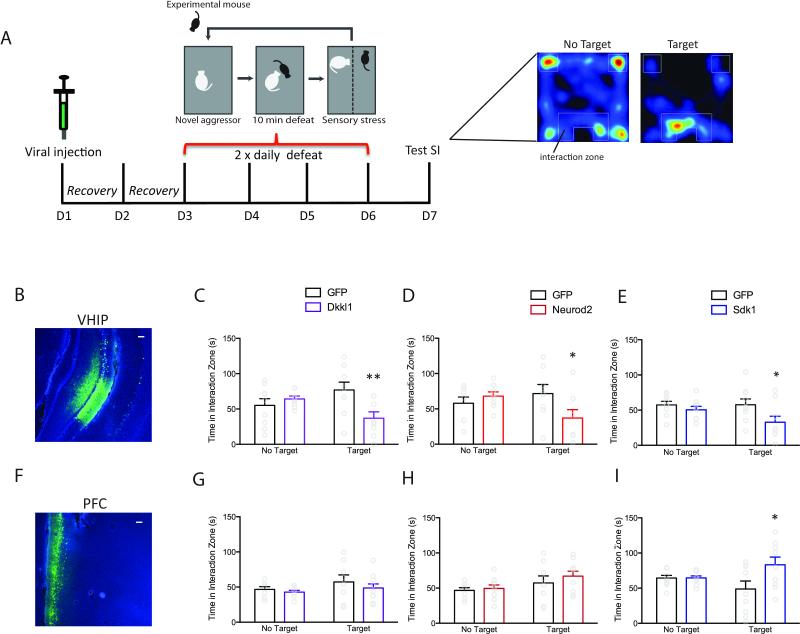
We over-expressed the gene of interest (Dkkl1, Neurod2 or Sdk1) plus GFP, or GFP alone, in either VHIP (Figure 6B) or PFC (Figure 6F), subjected mice to accelerated social defeat, and tested social interaction (Figure 6A). Mice in which Dkkl1 or Neurod2 (MB hub genes) was over-expressed in VHIP spent similar amounts of time investigating an empty interaction zone, but significantly less time interacting with a social target than HSV-GFP injected mice (F1,15=8.625, p=0.01, Bonferroni post-hoc p<0.01, n=9,8, Figure 6C; F1,14=5.087, p=0.04, Bonferroni post-hoc p<0.05,n=8,8, Figure 6D), indicating increased susceptibility to social defeat stress. In contrast, over-expression of Dkkl1 or Neurod2 in PFC did not alter the time mice spent investigating an empty interaction zone or interacting with a social target (F1,15=1.103, p=0.31, n=8,9; Figure 6G;F1,16=1.29, p=0.38, n=8,10; Figure 6H). Similar to manipulations of the MB hub genes, mice in which Sdk1 (a V hub gene) was over-expressed in VHIP also spent less time interacting with a social target (F1,16=4.959, p=0.04, Bonferroni post-hoc p<.05,n=9,9; Figure 6E), indicating increased stress susceptibility. Strikingly, over-expression of Sdk1 in PFC showed the opposite effect: mice spent more time interacting with a social target (F1,16=4.256, p=0.04, Bonferroni post-hoc, p<.05, n=9,9; Figure 6I), indicating increased resilience.
Figure 6. In Vivo Over-Expression of Susceptible-Specific Hub Genes Induces a Susceptible Behavioral Profile.
(A) Schematic of in vivo behavioral validation of susceptible-specific hub genes. Representative images of HSV-GFP infection in VHIP (B) and PFC (F). Scale bar=100μm. Mice injected with (C) HSV-Dkkl1-GFP, (D) HSV-Neurod2-GFP or (E) HSV-Sdk1-GFP in VHIP spent significantly less time in proximity to the wire mesh enclosure (Interaction Zone) compared to mice injected with HSV-GFP indicating increased susceptibility. Mice injected with the same viral constructs in PFC spent more time in the interaction zone (I; HSV-Sdk1-GFP) or an equivalent amount of time (G; HSV-Dkkl1-GFP, H; HSV-Neurod2-GFP) compared to HSV-GFP injected mice indicating increased resilience or lack of susceptibility. * p<0.05, **p<0.01. Bar graphs show mean ±SEM. See also Figure S6.
We also examined whether over-expression of these susceptible-specific hub genes in VHIP or PFC altered other measures of depression- and anxiety-like behaviors (Figure S6). Mice in which Neurod2 was over-expressed in PFC (Figure S6H) spent less time immobile in a forced swim test (t=2.512, p<0.05), an antidepressant-like effect, whereas over-expression of Sdk1 in VHIP (Figure S6K) trended towards increased immobility (t=1.767, p=0.099). Mice in which Sdk1 was over-expressed in VHIP (Figure S6I) also spent less time exploring the center of an open field, indicating an anxiogenic-like effect (t=2.370, p<0.05).
Since Neurod2 exhibited increased expression in NAC in R vs. C, and upregulation of MB module genes in NAC is also predicted to increase resilience (Figure 3C), we examined the effect of over-expressing Neurod2 in this brain region. Neurod2 over-expression significantly increased time interacting with a social target (F1,17=5.142, p=0.005, Bonferroni post-hoc p<.01, n=9,10, Figure S6N), supporting the prediction of increased resilience.
Finally, as a negative control for the predictive validity of our coexpression network analysis, we over-expressed 3 genes that are predicted to not regulate susceptibility in VHIP based on their assignment to the non-clustering Grey module (Agtr1b and Parg) or to the non-differentially connected Grey60 module (Parp). Over-expression of each of these genes failed to alter behavior in the social interaction, open field or forced swim tests (Figure S6Q-S; one-way ANOVA; GFP n=8, Agtr1b n=6, Parg n=7, Parp n=9).
MB and V hub genes increase sEPSC frequency in VHIP neurons
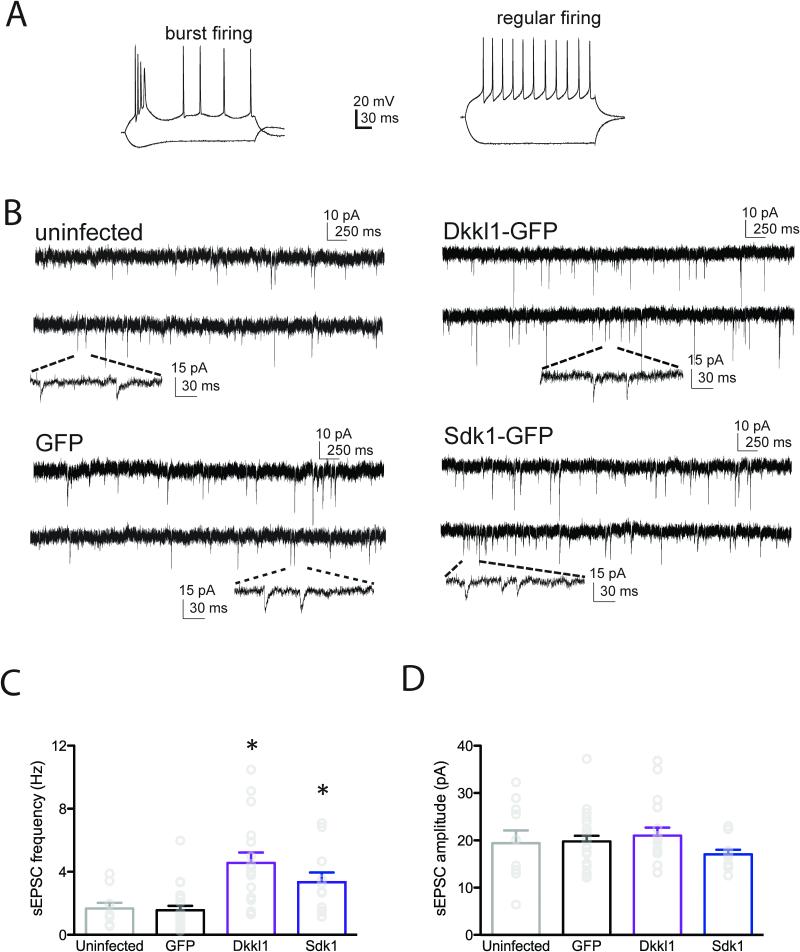
Having demonstrated that over-expression of susceptible-specific hub genes in VHIP induces a pro-susceptible phenotype, we next probed the cellular correlates of this increased susceptibility by examining the effects of over-expression prior to defeat. In a recent functional characterization of this circuitry, we found evidence of increased activity of VHIP neurons (projecting to NAC) in susceptible mice and optogenetically increasing activity of this pathway induced susceptibility (Bagot et al., 2015). Additionally, MB was significantly enriched for GOs cell-cell signaling and synaptic transmission. We thus reasoned that MB and V modules may be implicated in these synaptic alterations and that susceptible-specific hub genes may enhance VHIP excitability. We focused on Dkkl1, which robustly induced behavioral susceptibility in VHIP, and Sdk1, given its role in promoting spinogenesis in brain. Over-expression of either Dkkl1 or Sdk1 increased sEPSC frequency compared to cells infected with the control HSV-GFP vector, and importantly sEPSC frequency in GFP-overexpressing cells did not differ from non-infected cells (F3, 57=9.185, p < 0.0001; HSV-Dkkl1 vs. GFP p<0.0001; HSV-Sdk1 vs. GFP p<0.05, n cells/mice=10/5 non-infected, 23/5 HSV-GFP, 17/4 HSV-Dkkl1, 10/5 HSV-Sdk1; Figure 7B,C). In contrast, neither Dkkl1 or Sdk1 altered sEPSC amplitude (F3,57=0.8978, p=0.448; Figure 7B,D). These data suggest that early regulation of susceptible-specific hub genes in the MB and V modules establishes susceptibility by increasing activity at VHIP synapses.
Figure 7. In Vivo Over-Expression of Susceptible-Specific Hub Genes in VHIP Increases sEPSC Frequency.
Overexpression of Dkkl1 or Sdk1 increased the frequency of spontaneous EPSCs in VHIP neurons 24h after viral infection. (A) Among all 73 VHIP neurons recorded, 17 were burst firing (left) and 43 were regular firing (right). Subsequent synaptic analysis focused on regular firing neurons. (B) Representative sEPSCs from uninfected VHIP neurons (top left), neurons expressing GFP alone (bottom left), Dkkl1-GFP (top right) or Sdk1-GFP (bottom right). (C) sEPSC frequency was increased by either Dkkl1 or Sdk1 over-expression relative to uninfected or GFP infected neurons (the latter two did not differ). (D) sEPSC amplitude was not changed by either Dkkl1-GFP or Sdk1-GFP overexpression. * p<0.05, **p<0.01. Bar graphs show mean ±SEM.
Discussion
We developed molecular network models to significantly extend the understanding of transcriptional mechanisms of depression by performing a coexpression network analysis of the whole transcriptome in 4 inter-connected brain regions implicated in depression, over 3 time points after CSDS, in both susceptible and resilient populations (total 36 groups). Prior studies of transcriptional mechanisms of depression have focused primarily on identifying individual candidate genes, or profiling whole transcriptomes within single brain regions in isolation. Here we successfully leveraged a systems biology approach to reveal inter-regional co-regulation gene signatures of susceptibility and resilience and to then identify novel transcriptional networks associated with susceptibility or resilience to chronic stress. While coexpression analyses have been previously applied to generate insight into other syndromes, our study is unique in systematically examining the multifaceted dysregulation of gene networks within several inter-connected brain regions in depression and is the first study to demonstrate in vivo validation of key regulators of transcriptional networks. Critically, we demonstrate the utility and validity of this approach by directly manipulating predicted susceptibility-specific hub genes in vivo. We show that overexpressing one such hub gene in VHIP significantly increases the expression of other genes in its module selectively, and that over-expression of each of 3 hub genes induces predicted, region-specific regulation of behavioral susceptibility. Importantly, over-expression of genes predicted to not regulate susceptibility had no effect. Finally, we reveal a synaptic mechanism through which hub gene manipulations alter circuit function to regulate responses to chronic stress. This study provides a powerful example of how a network approach can be applied to integrate large-scale transcriptomic data to reveal novel insight into transcriptional mechanisms of a pathological state such as depression and, in so doing, create a platform for future studies to pursue novel targets for drug development.
We adapted RRHO analysis to characterize inter-regional patterns of transcriptional regulation and identified an important similarity of transcriptional regulation between PFC and NAC in resilience and between PFC and VHIP in susceptibility. These patterns were also seen when examining the enrichment of DEGs within key susceptible coexpression modules. This, combined with knowledge of functional connectivity between VHIP and PFC, informed our decision to focus upon the MB and V modules to reveal novel insights into transcriptional mechanisms of susceptibility. We demonstrated that key hub genes from each module regulated sEPSC frequency in VHIP, demonstrating the role of these transcriptional networks in regulating synaptic transmission. Intersection of our modules with DEG lists suggested opposing regulation of the modules in VHIP and PFC. It is interesting to note opposing functional adaptations in these 2 brain regions have been described in the context of depression in humans and stress in animal models, with the PFC exerting a pro-resilience effect and the VHIP promoting susceptibility (Bagot et al., 2015; Covington et al., 2010; Jaworska et al., 2014; Mayberg et al., 2000; Vialou et al., 2014), and that functionally increasing synaptic transmission between PFC and NAC enhances stress resilience while reducing PFC and VHIP synchrony is associated with reversal of depression-like phenotypes (Insel et al., 2015). Thus, our circuit-level transcriptional analyses coincide with findings of functional studies, including those in human depression, and greatly extend these observations by identifying putative transcriptional networks underpinning the functional adaptations. It will be interesting in future studies to expand this analysis to several additional brain regions that are also implicated in depression.
Our coexpression network analysis led us to investigate novel targets that we demonstrate regulate region-specific effects on synaptic function and behavioral susceptibility. Sdk1 showed a particularly striking phenotype: its overexpression in PFC vs. VHIP induced opposite behavioral effects, consistent with our derived predictions, and also increased synaptic transmission in VHIP, a potential cellular mechanism by which Sdk1 regulates increased susceptibility in this brain region (Bagot et al., 2015). The example of Sdk1 is especially compelling in its support of a systems biology approach as Sdk1 was not itself differentially expressed in any brain region studied, although we identified it as a susceptible-specific hub gene within a key coexpression module (V) that was enriched for other differentially expressed genes within VHIP and PFC. Thus, standard transcriptional analyses would have failed to detect this important target. Likewise, we validated 2 other genes, heretofore not examined in depression, Dkkl1 and Neurod2, which we show control stress susceptibility when over-expressed in VHIP and, for Neurod2, in NAC as well. We propose that hub gene manipulations regulate behavioral susceptibility by driving the expression of other genes within that module. This hypothesis is supported by the observed enrichment of MB module genes among genes induced by Dkkl1 over-expression in VHIP. However, Dkkl1 over-expression also affected numerous other genes and we cannot exclude the possibility that such regulation may contribute to the observed behavioral effects. In any event, these findings indicate that the susceptibility network generated in this study likely identifies numerous other hub genes which are also important in controlling stress responses, and provide a mechanism by which transcriptional networks can be regulated to drive susceptibility vs. resilience after CSDS.
In addition to identifying novel targets, our analyses identified genes and biological functions previously implicated in depression. Analyzing transcriptional alterations in postmortem tissue from depressed humans is one source of mechanistic insight, although the limited availability of human tissue coupled with its inherent variability (treatment history, years since diagnosis, age, post-mortem indices) can complicate such efforts. Mouse models of psychiatric disorders are thus essential tools in furthering our understanding of transcriptional mechanisms in human disease by both facilitating identification of disease-relevant transcriptional alterations and allowing the testing of causality of such changes through detailed in vivo validation in the same system. The present results attest to the utility of this approach. Our data sets are now a valuable resource with which to filter evolving transcriptional and genome sequence studies of depressed humans, which have not revealed significant depression risk genes in heterogeneous populations and ongoing transcriptomic studies. Indeed, we see overlap between MB genes and those identified as showing altered expression in depressed humans (e.g. ADRBK2, DNER, NCALD, NRXN2, PAK1, RGS7, SCAMP5, SYNGR1) and between MB and V genes and those in proximity to potential depression-associated single nucleotide polymorphisms (e.g. CACNA1G, CNIH3, DSCAM, ELAVL4, GFRA2, PAK1, PTK2, RPH3A, SULT4A1) (Chang et al., 2014; Ding et al., 2015; Sequeira et al., 2009). It is important to note that the current analyses focus on male mice. In light of documented sex-differences in depression incidence and clinical presentation future work should extend these analyses to females to determine the generalizability and specificity of transcriptional mechanisms of depression.
We observed clear differences between analyses of DEGs (Figure 1) and coexpression networks (Figure 3). At 48h post-defeat, more genes were differentially expressed in R vs. C than in S vs. C (Figure 1B). In contrast, in coexpression networks constructed across multiple time-points, greater changes in module connectivity were observed in S vs. C (Figure 3B). The 2 analyses capture different dimensions of the highly complex data set, with differential expression data reflecting changes in a single dimension—a gene—between groups. In contrast, differential connectivity implies a 2-dimensional phenomenon where correlations between pairs of genes have been altered. Although differential expression and differential connectivity can co-exist, they are essentially independent metrics. The finding of increased differential connectivity in susceptible modules suggests that susceptibility is associated with altered coordination of more biological pathways than resilience, even though resilience is associated with more DEGs. In turn, differential expression may indicate coordinated up- or downregulation of existing pathways without fundamentally altering pathway organization. Susceptibility thus is characterized by fundamental changes in the architecture of transcriptional networks rather than simply altered expression of existing networks, an important insight that is obscured in studies analyzing DEGs alone. Ultimately, designing therapeutic interventions to target differential connectivity networks rather than simply modulating DEGs may be critical to generate more effective treatments (Schrattenholz et al., 2010; Schrattenholz and Soskic, 2008).
We applied a systems biology approach to reveal novel insight into the transcriptional mechanisms of susceptibility and resilience. In this pursuit we have gleaned important biological insights from an abundance of data. Extracting meaning from such large data sets necessitates an exercise in reduction and clearly much more remains to be explored. The data and analyses generated will provide a valuable resource for other researchers. For example, referencing novel data sets to our coexpression networks offers the possibility to contextualize findings from other animal models or human data by identifying overlaps that may point to the broader transcriptional networks at play. By examining differential coexpression networks in addition to differential gene expression it is possible to obtain a more complete understanding of how susceptible and resilient responses to chronic social stress occur within specific brain regions over time. We demonstrate an approach to understand how numerous genes operate within functional clusters, across brain areas, to control stress responses, a critical element lacking in previous transcriptional analyses of depression-related phenotypes. In so doing, we identify novel molecular mechanisms controlling stress susceptibility. This work thus provides a template with which to characterize the molecular, cellular and circuit basis of pathological changes that underlie depression—or adaptive changes that promote resilience, information which can now be used to fundamentally advance the search for more effective antidepressant medications.
Experimental Procedures
(See also Extended Experimental Procedures)
Experimental subjects and CSDS
Mice were maintained on a 12h light-dark cycle (lights on at 7 am) at 22-25°. All experiments conformed to Mount Sinai IACUC guidelines. An established CSDS protocol induced depressive-like behaviors in mice (Berton et al., 2006). Male 8 week-old C57BL/6J mice were subjected to 10 daily, 5-min defeats by a novel 6-month-old CD1 aggressor mouse, then housed across a plexiglass divider for continued sensory contact. Control mice were housed in cages separated from another control. In viral manipulation experiments, a validated accelerated defeat protocol (2x daily, 10 min defeats, 4 days, starting 2 days after surgery) induced defeat during peak viral expression (Dias et al., 2014).
RNA isolation
Mice were killed directly from their home-cage 48h (early) or 28d (late) post CSDS or 28d post CSDS, 1h post 5min aggressor re-exposure (stress-primed). VHIP, PFC, NAC and AMY were dissected and flash frozen. Tissue from 3-5 mice was pooled for n=3 (early) or n=4 (late, stress-primed) independent biological replicates per brain region, phenotype and time-point and RNA isolated as described (Bagot et al., 2015). For viral experiments, mice were killed 24h post-defeat.
Statistical and bioinformatic data analysis
Pairwise differential expression analysis were performed with Cuffdiff (Trapnell et al., 2012) using the negative binomial distribution and a nominal significance threshold of p<0.05 and Fold Change(FC) >1.3. Rank Rank Hypergeometric Overlap (RRHO) evaluated the overlap of differential expression lists between pairs of brain regions (Plaisier et al., 2010). Susceptible and resilient data sets were independently processed through weighted gene coexpression network analysis (WGCNA) (Langfelder et al., 2008; Zhang and Horvath, 2005). Each module was assigned a unique, arbitrary color identifier.
Enrichment of GOs, cell-types, and DEGs in modules, as well as enrichment of module genes upon viral over-expression of a hub gene, was assessed through Fisher's exact test corrected for multiple testing (Benjamini-Hochberg FDR 0.05). A module differential connectivity (MDC) metric quantified differences in coexpression networks to identify phenotype-specific modules (Zhang et al., 2013). Key driver analysis (Zhang et al., 2013) applied to module-based unweighted coexpression networks derived from ARACNe (Margolin et al., 2006) identified module key driver genes.
Stereotaxic surgery
We over-expressed genes of interest using standard stereotaxic surgery procedures (Bagot et al., 2015) to bilaterally infuse 0.5 μl HSV-GFP, HSV-GFP-Sdk1, HSV-GFP-Neurod2 or HSV-GFP-Dkkl1 into PFC, VHIP or NAC.
Electrophysiology
Coronal slices were prepared 24h post viral injection. In current clamp mode, neurons in the ventral subiculum sub-region of VHIP—where our dissections focused—through which VHIP sends efferent projections (Groenewegen et al., 1987) – we characterized firing pattern (burst vs. regular) following reports of both neuronal types in this area (Figure 7A) (Cooper et al., 2003). As the large majority of neurons were regular firing (73/90) we limited analysis to this population. Spontaneous EPSCs were recorded for 3-5min in voltage clamp mode.
Supplementary Material
Highlights.
A large-scale multi-brain region transcriptomic cohort to probe stress susceptibility
Reveals susceptible and resilient transcriptional networks across brain regions
Identifies many novel hub genes that emerge in susceptible mice
In vivo validation of key regulators at molecular, synaptic and behavioral levels
Acknowledgments
This work is supported by P50 MH096890, a 2014 NARSAD Young Investigator Award 22713 from the Brain & Behavior Research Foundation to R.C.B., R01AG046170 from the NIH/National Institute on Aging to B.Z. and M.W., and U01AI111598-01 from NIH/National Institute of Allergy and Infectious Diseases to B.Z. and W.S., and K99 MH10237 to J.L.S. We thank Erin S. Calipari for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
R.C.B., B.Z. & E.J.N. designed the study. R.C.B., B.Z, & E.J.N. wrote the manuscript. R.C.B., I.P., Z.S.L., M.W., W.S., X.L., J.L.S., D.G., L.S. & B.Z. advised on analysis approaches and analyzed data. R.C.B., H.M.C., & I.M. prepared sequencing libraries, R.C.B., H.M.C., D.M.W., C.J.P., & M.A.D. performed stereotaxic surgeries, and behavioral manipulations, E.A.H., O.I., H.S.S. K.N.S. & R.N., cloned and packaged viral constructs and J.W., X.W. O.S. & Y.D. performed and analyzed electrophysiology recordings.
Accession Numbers
Raw and processed RNA-seq gene expression data are available via the Gene Expression Omnibus data (accession number GEO:GSE72343).
Supplemental Information
Supplemental information includes Extended Experimental Procedures and 7 figures and 6 tables.
References
- Network & Pathway Analysis Subgroup of the Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nature neuroscience. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Admon R, Holsen LM, Aizley H, Remington A, Whitfield-Gabrieli S, Goldstein JM, Pizzagalli DA. Striatal Hypersensitivity During Stress in Remitted Individuals with Recurrent Depression. Biological psychiatry. 2015;78:67–76. doi: 10.1016/j.biopsych.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arloth J, Bogdan R, Weber P, Frishman G, Menke A, Wagner KV, Balsevich G, Schmidt MV, Karbalai N, Czamara D, et al. Genetic Differences in the Immediate Transcriptome Response to Stress Predict Risk-Related Brain Function and Psychiatric Disorders. Neuron. 2015;86:1189–1202. doi: 10.1016/j.neuron.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Parise EM, Pena CJ, Zhang HX, Maze I, Chaudhury D, Persaud B, Cachope R, Bolanos-Guzman CA, Cheer J, et al. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nature communications. 2015;6:7062. doi: 10.1038/ncomms8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- Block SG, Nemeroff CB. Emerging antidepressants to treat major depressive disorder. Asian journal of psychiatry. 2014;12:7–16. doi: 10.1016/j.ajp.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Buijs RM, Van Eden CG. The integration of stress by the hypothalamus, amygdala and prefrontal cortex: balance between the autonomic nervous system and the neuroendocrine system. Progress in brain research. 2000;126:117–132. doi: 10.1016/S0079-6123(00)26011-1. [DOI] [PubMed] [Google Scholar]
- Chang LC, Jamain S, Lin CW, Rujescu D, Tseng GC, Sibille E. A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PloS one. 2014;9:e90980. doi: 10.1371/journal.pone.0090980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZQ, Du MY, Zhao YJ, Huang XQ, Li J, Lui S, Hu JM, Sun HQ, Liu J, Kemp GJ, et al. Voxel-wise meta-analyses of brain blood flow and local synchrony abnormalities in medication-free patients with major depressive disorder. Journal of psychiatry & neuroscience : JPN. 2015;40:140119. doi: 10.1503/jpn.140119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffel DJ, Golden SA, Walsh JJ, Guise KG, Heshmati M, Friedman AK, Dey A, Smith M, Rebusi N, Pfau M, et al. Excitatory transmission at thalamo-striatal synapses mediates susceptibility to social stress. Nature neuroscience. 2015;18:962–964. doi: 10.1038/nn.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DC, Moore SJ, Staff NP, Spruston N. Psychostimulant- induced plasticity of intrinsic neuronal excitability in ventral subiculum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:9937–9946. doi: 10.1523/JNEUROSCI.23-30-09937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, LaPlant Q, Mouzon E, Ghose S, Tamminga CA, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:16082–16090. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonte B, Ribeiro E, et al. beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516:51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H, French BJ, Lewis DA, Tseng GC, Sibille E. Molecular and Genetic Characterization of Depression: Overlap with other Psychiatric Disorders and Aging. Molecular neuropsychiatry. 2015;1:1–12. doi: 10.1159/000369974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J, Pan H, Kocsis JH, Yang Y, Butler T, Chusid J, Hochberg H, Murrough J, Strohmayer E, Stern E, et al. Lack of ventral striatal response to positive stimuli in depressed versus normal subjects. The American journal of psychiatry. 2006;163:1784–1790. doi: 10.1176/ajp.2006.163.10.1784. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Serretti A. Pharmacogenetics of major depressive disorder: top genes and pathways toward clinical applications. Current psychiatry reports. 2015;17:50. doi: 10.1007/s11920-015-0594-9. [DOI] [PubMed] [Google Scholar]
- Friedman AK, Walsh JJ, Juarez B, Ku SM, Chaudhury D, Wang J, Li X, Dietz DM, Pan N, Vialou VF, et al. Enhancing depression mechanisms in midbrain dopamine neurons achieves homeostatic resilience. Science. 2014;344:313–319. doi: 10.1126/science.1249240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes, brain, and behavior. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Guilloux JP, Lewis DA, Sibille E. Altered gene synchrony suggests a combined hormone-mediated dysregulated state in major depression. PloS one. 2010;5:e9970. doi: 10.1371/journal.pone.0009970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Sibille E. Differentially expressed genes in major depression reside on the periphery of resilient gene coexpression networks. Frontiers in neuroscience. 2011;5:95. doi: 10.3389/fnins.2011.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Grace AA. Limbic and cortical information processing in the nucleus accumbens. Trends in neurosciences. 2008;31:552–558. doi: 10.1016/j.tins.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg PE, Fournier AA, Sisitsky T, Pike CT, Kessler RC. The economic burden of adults with major depressive disorder in the United States (2005 and 2010). The Journal of clinical psychiatry. 2015;76:155–162. doi: 10.4088/JCP.14m09298. [DOI] [PubMed] [Google Scholar]
- Groenewegen HJ, Vermeulen-Van der Zee E, te Kortschot A, Witter MP. Organization of the projections from the subiculum to the ventral striatum in the rat. A study using anterograde transport of Phaseolus vulgaris leucoagglutinin. Neuroscience. 1987;23:103–120. doi: 10.1016/0306-4522(87)90275-2. [DOI] [PubMed] [Google Scholar]
- Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K, Tseng GC, Lewis DA, Sibille E. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Molecular psychiatry. 2012;17:1130–1142. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Pena CJ, Sun H, Shao N, Feng J, Golden SA, Herman JP, Walsh JJ, Mazei-Robison M, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nature neuroscience. 2014;17:1720–1727. doi: 10.1038/nn.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley JM, Gruber SA, Parker HA, Guillaumot J, Rogowska J, Yurgelun-Todd DA. Cortico-limbic response to personally challenging emotional stimuli after complete recovery from depression. Psychiatry research. 2009;172:83–91. doi: 10.1016/j.pscychresns.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Insel N, Pilkiw M, Nobrega JN, Hutchison WD, Takehara-Nishiuchi K, Hamani C. Chronic deep brain stimulation of the rat ventral medial prefrontal cortex disrupts hippocampal-prefrontal coherence. Experimental neurology. 2015;269:1–7. doi: 10.1016/j.expneurol.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska N, Yang XR, Knott V, Macqueen G. A review of fMRI studies during visual emotive processing in major depressive disorder. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry. 2014 doi: 10.3109/15622975.2014.885659. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, Lepack A, Majik MS, Jeong LS, Banasr M, et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nature medicine. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SH, Evans KR, Kruger S, Mayberg HS, Meyer JH, McCann S, Arifuzzman AI, Houle S, Vaccarino FJ. Changes in regional brain glucose metabolism measured with positron emission tomography after paroxetine treatment of major depression. The American journal of psychiatry. 2001;158:899–905. doi: 10.1176/appi.ajp.158.6.899. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, Laplant Q, Graham A, Lutter M, Lagace DC, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Malki K, Mineur YS, Tosto MG, Campbell J, Karia P, Jumabhoy I, Sluyter F, Crusio WE, Schalkwyk LC. Pervasive and opposing effects of Unpredictable Chronic Mild Stress (UCMS) on hippocampal gene expression in BALB/cJ and C57BL/6J mouse strains. BMC genomics. 2015;16:262. doi: 10.1186/s12864-015-1431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K, Tosto MG, Jumabhoy I, Lourdusamy A, Sluyter F, Craig I, Uher R, McGuffin P, Schalkwyk LC. Integrative mouse and human mRNA studies using WGCNA nominates novel candidate genes involved in the pathogenesis of major depressive disorder. Pharmacogenomics. 2013;14:1979–1990. doi: 10.2217/pgs.13.154. [DOI] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC bioinformatics 7 Suppl. 2006;1:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschietto M, Tahira AC, Puga R, Lima L, Mariani D, Paulsen Bda S, Belmonte de-Abreu P, Vieira H, Krepischi AC, Carraro DM, et al. Co-expression network of neural-differentiation genes shows specific pattern in schizophrenia. BMC medical genomics. 2015;8:23. doi: 10.1186/s12920-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS, Brannan SK, Tekell JL, Silva JA, Mahurin RK, McGinnis S, Jerabek PA. Regional metabolic effects of fluoxetine in major depression: serial changes and relationship to clinical response. Biological psychiatry. 2000;48:830–843. doi: 10.1016/s0006-3223(00)01036-2. [DOI] [PubMed] [Google Scholar]
- Messmer K, Shen WB, Remington M, Fishman PS. Induction of neural differentiation by the transcription factor neuroD2. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2012;30:105–112. doi: 10.1016/j.ijdevneu.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome medicine. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Carlezon WA., Jr. The mesolimbic dopamine reward circuit in depression. Biological psychiatry. 2006;59:1151–1159. doi: 10.1016/j.biopsych.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Oliva CA, Vargas JY, Inestrosa NC. Wnts in adult brain: from synaptic plasticity to cognitive deficiencies. Frontiers in cellular neuroscience. 2013;7:224. doi: 10.3389/fncel.2013.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JM, Asakura A, Snider L, Hawkes R, Strand A, Stoeck J, Hallahan A, Pritchard J, Tapscott SJ. NeuroD2 is necessary for development and survival of central nervous system neurons. Developmental biology. 2001;234:174–187. doi: 10.1006/dbio.2001.0245. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip NS, Carpenter LL, Tyrka AR, Price LH. Nicotinic acetylcholine receptors and depression: a review of the preclinical and clinical literature. Psychopharmacology. 2010;212:1–12. doi: 10.1007/s00213-010-1932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene- expression signatures. Nucleic acids research. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner J, Hellerstein DJ, Gat I, Mechling A, Klahr K, Wang Z, McGrath PJ, Stewart JW, Peterson BS. Antidepressants normalize the default mode network in patients with dysthymia. JAMA psychiatry. 2013;70:373–382. doi: 10.1001/jamapsychiatry.2013.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Mayberg HS. Targeting abnormal neural circuits in mood and anxiety disorders: from the laboratory to the clinic. Nature neuroscience. 2007;10:1116–1124. doi: 10.1038/nn1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer TE, Cohen MX, Frick C, Kosel M, Brodesser D, Axmacher N, Joe AY, Kreft M, Lenartz D, Sturm V. Deep brain stimulation to reward circuitry alleviates anhedonia in refractory major depression. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2008;33:368–377. doi: 10.1038/sj.npp.1301408. [DOI] [PubMed] [Google Scholar]
- Schrattenholz A, Groebe K, Soskic V. Systems biology approaches and tools for analysis of interactomes and multi-target drugs. Methods in molecular biology. 2010;662:29–58. doi: 10.1007/978-1-60761-800-3_2. [DOI] [PubMed] [Google Scholar]
- Schrattenholz A, Soskic V. What does systems biology mean for drug development? Current medicinal chemistry. 2008;15:1520–1528. doi: 10.2174/092986708784638843. [DOI] [PubMed] [Google Scholar]
- Scobie KN, Damez-Werno D, Sun H, Shao N, Gancarz A, Panganiban CH, Dias C, Koo J, Caiafa P, Kaufman L, et al. Essential role of poly(ADP- ribosyl)ation in cocaine action. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2005–2010. doi: 10.1073/pnas.1319703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PloS one. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbe M, Jarowyj J. Region-specific expression of Dickkopf-like1 in the adult brain. Abbreviated title: Dkkl1 in the adult brain. Neuroscience letters. 2013;535:84–89. doi: 10.1016/j.neulet.2012.12.040. [DOI] [PubMed] [Google Scholar]
- Steel Z, Marnane C, Iranpour C, Chey T, Jackson JW, Patel V, Silove D. The global prevalence of common mental disorders: a systematic review and meta-analysis 1980-2013. International journal of epidemiology. 2014;43:476–493. doi: 10.1093/ije/dyu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernandez IA, Marchetto MC, Baker DK, Lu D, Hinman CR, Lowe JK, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham MW, Woon PS, Sum MY, Lee TS, Sim K. White matter abnormalities in major depression: evidence from post-mortem, neuroimaging and genetic studies. Journal of affective disorders. 2011;132:26–36. doi: 10.1016/j.jad.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Kechris K, Miles MF, Hoffman PL, Tabakoff B. Whole brain and brain regional coexpression network interactions associated with predisposition to alcohol consumption. PloS one. 2013;8:e68878. doi: 10.1371/journal.pone.0068878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialou V, Bagot RC, Cahill ME, Ferguson D, Robison AJ, Dietz DM, Fallon B, Mazei-Robison M, Ku SM, Harrigan E, et al. Prefrontal cortical circuit for depression- and anxiety-related behaviors mediated by cholecystokinin: role of DeltaFosB. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:3878–3887. doi: 10.1523/JNEUROSCI.1787-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke SA, Hall BJ, Antonios JK, Denardo LA, Otto S, Yuan B, Chen F, Robbins EM, Tiglio K, Williams ME, et al. NeuroD2 regulates the development of hippocampal mossy fiber synapses. Neural development. 2012;7:9. doi: 10.1186/1749-8104-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MB, Xiao G, Kumar A, LaPlant Q, Renthal W, Sikder D, Kodadek TJ, Nestler EJ. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co- expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.