Abstract
Purpose
This study aimed to investigate the genetic effects underlying non-familial sporadic congenital cataract (SCC).
Methods
We collected DNA samples from 74 patients with SCC and 20 patients with traumatic cataract (TC) in an age-matched group and performed genomic sequencing of 61 lens-related genes with target region capture and next-generation sequencing (NGS). The suspected SCC variants were validated with MassARRAY and Sanger sequencing. DNA samples from 103 healthy subjects were used as additional controls in the confirmation examination.
Results
By filtering against common variants in public databases and those associated with TC cases, we identified 23 SCC-specific variants in 17 genes from 19 patients, which were predicted to be functional. These mutations were further confirmed by examination of the 103 healthy controls. Among the mutated genes, CRYBB3 had the highest mutation frequency with mutations detected four times in four patients, followed by EPHA2, NHS, and WDR36, the mutation of which were detected two times in two patients. We observed that the four patients with CRYBB3 mutations had three different cataract phenotypes.
Conclusions
From this study, we concluded the clinical and genetic heterogeneity of SCC. This is the first study to report broad spectrum genotyping for patients with SCC.
Introduction
Congenital cataract (CC) is defined as lens opacity that is present at birth or occurs within the first year after birth; this condition is also called infantile cataract. CC is one of the most common causes of childhood blindness (approximately 200,000 children worldwide are blind from cataracts) [1] and might result in permanent blindness due to the irreversible damage to the responsible brain zone during visual development. Previous studies indicated that about one-third of CC cases are hereditary [2] and, in most cases, represent an autosomal dominant trait, although autosomal recessive and X-linked cases are also observed [3,4]. Meanwhile, the majority of the unilateral cases and approximately half of the bilateral cases are not due to familial history [5]; these cases are called sporadic cases and mostly occur due to unknown causes. Little attention has been paid to these non-familial cases in terms of broad spectrum genetic identification.
With an extensive genetic heterogeneity, cataract has been linked to more than 20 genes, the majority of which are located in crystalline and connexin (gap junction) genes [6,7], but the pool is continuously expanding. The most acknowledged cataract genes encode crystallins (CRYAA-Gene ID 1409, OMIM 123580, CRYAB-Gene ID 1410, OMIM 123590, CRYBA1-Gene ID 1411, OMIM 123610, CRYBB1-Gene ID 1414, OMIM 600929, CRYBB2-gene ID 1415, OMIM 123620, CRYBB3-Gene ID 1417, OMIM 123630, CRYGC-Gene ID 1420, OMIM 123680, and CRYGD-gene ID 1421, OMIM 123690), membrane transport proteins (GJA1-Gene ID 2697, OMIM 121014, GJA3 -Gene ID 2700, OMIM 121015, GJA8-Gene ID 2703, OMIM 600897, and MIP-Gene ID 4284, OMIM 154050), a cytoskeletal protein (BFSP2-Gene ID 8419, OMIM 603212), and developmental regulators (PITX3-Gene ID 5309, OMIM 602669, MAF-Gene ID 4094, OMIM 177075, and HSF4-Gene ID 3299, OMIM 602438) [8]. In the present study, we examined 61 lens-related genes in 74 SCC cases using target capture and next-generation sequencing (NGS). In addition to the identified CC genes described, we included other lens disease-related genes: microphthalmia (SOX2-Gene ID 6657, OMIM 184429, CYP1B1-Gene ID 1545, OMIM 601771, RAX-Gene ID 30062, OMIM 601881, FOXE3-Gene ID 2301, OMIM 601094, VSX2-Gene ID 338917, OMIM 142993, and CRYBA4-Gene ID 1413, OMIM 123631) [9,10], aniridia (FOXC1-Gene ID 2296, OMIM 601090, PITX2-Gene ID 5308, OMIM 601542, and PITX3) [11-13], Marfan syndrome (FBN1-Gene ID 2200, OMIM 134797) [14], ectopia lentis (ADAMTSL4-Gene ID 54507, OMIM 610113) and ADAMTS17-Gene ID 170691, OMIM 607511) [15], Alport syndrome (COL4A5-Gene ID 1287, OMIM 303630) [16], glaucoma (CYP1B1-Gene ID 1545, OMIM 601771, MYOC-Gene ID 4653, OMIM 601652, OPTN-Gene ID 10133, OMIM 602432, and WDR36-Gene ID 134430, OMIM 609669) [17], Nance-Horan syndrome (NHS-Gene ID 4810, OMIM 300457) [18], and exfoliation syndrome (LOXL1-Gene ID 4016, OMIM 153456) [19]. The total gene list is provided in Appendix 1. The aim of this study was to investigate what proportion of the SCC cases had a genetic basis and the distribution of these tested genes with functional mutations. We showed that in 19 out of 74 patients with SCC, genetic mutations were detected in the lens-related genes, and CRYBB3 appeared to have the highest mutation frequency. Here, we provide the first report on the mutation distribution and frequency of known genes from SCC cases in a Han Chinese population.
Methods
Patients
Informed consent in written form to collect the blood sample and to publish case details was obtained from the parents of the patients. All procedures were reviewed and approved by the ethics committee of Eye & ENT Hospital of Fudan University. The study was adhered to ARVO statement on human subjects and the tenets of the Declaration of Helsinki. A total of 74 patients from the pediatric congenital cataract clinic were diagnosed with SCC with no familial history (i.e., lens opacity occurred at birth or within the first year after birth and no family member was diagnosed with congenital cataract within three generations; in addition, the lens of the parents were diagnosed as normal at the time of surgery) between September 2012 and April 2013. During the same period, 20 patients from the pediatric congenital cataract clinic were diagnosed with traumatic cataract (TC). Meticulous clinical examinations were performed by experienced specialists. All patients were Han Chinese from 11 provinces in China. To confirm the identified SCC variants, DNA samples from 103 healthy controls were obtained from Shanghai Children’s Hospital of Shanghai Jiao Tong University and used as additional controls.
Molecular genetic analysis
A total of 1 to 2 ml peripheral blood was collected from patients and controls in EDTA blood collection tubes and stored in -80 °C prior to use. Genomic DNA was extracted from the whole blood cells using a genomic DNA extraction and purification kit (FlexiGene DNA kit #51206, QiagenChina, Shenzhen, China) according to the manufacturer’s protocol. The DNA concentration was measured using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA), and 100 ng DNA was used for molecular analysis.
Capture of the target DNA region was completed using the Agilent SureSelect DNA Design (Agilent Technologies, Santa Clara, CA). Each captured library was loaded on the HiSeq 2000 platform (Illumina, Santa Clara, CA); high-throughput sequencing for each captured library was performed independently to ensure that each sample met the desired average fold-coverage. The paired raw data were deposited in the NCBI Sequencing Read Archive under accession number SRP062799. The bioinformatics analysis began from the sequencing data (raw data) that had been generated from the Illumina pipeline. First, the adaptor sequence in the raw data was removed, and low-quality reads that had too many Ns or low base quality were discarded. This step produced clean data. Second, Burrows-Wheeler Aligner (BWA) was used to do the alignment. BWA gives results in BAM format files. Single nucleotide polymorphisms (SNPs) were investigated with SOAPsnp SAMtools or GATK. Then the software AnnoDB was used in-house to annotate the confident variant results. The SNPs with 5X or more coverage and insertions or deletions were annotated to genes using Ensembl release version 73. The minimum thresholds were set at 50X/45 mqV. Additional annotation was provided from Online NCBI dbSNP, as well as population frequencies from the 1000 Genomes Project (including the 1000 Genomes All, Asian, African, American, and European categories). Then we filtered the resulting SNPs in the following steps: 1) exclude database SNPs with minor allele frequency (MAF) equal to or greater than 0.01 in the 1000 Genome database, 2) exclude the variants that existed in the TC controls, 3) exclude the silent or synonymous variants without altering splicing sites in genes, and 4) exclude the missense variants with low effects in SIFT and Polyphen-2 predictions. We obtained 32 SCC-specific variants, 27 of which were novel (not deposited in dbSNP and not described in published reports). All of the novel SCC SNPs were examined in each patient and healthy controls with Sequenom MassARRAY genotyping [20]; Sanger sequencing was performed instead for the samples that failed in the MassARRAY. (The primers are listed in Appendix 2) The variants listed in the dbSNP were confirmed with the TaqMan SNP genotyping assay kit (Thermo Fisher Scientific, Carlsbad, CA; the product number of each SNP is listed in Appendix 2). SIFT [21] and PolyPhen [22] analysis were performed to predict whether the identified SNPs would affect the protein structure and function.
Results
Clinical observations
A total of 52.7% of the 74 patients with SCC were male and 47.3% were female; the average age was 4.6 years (range 0 to 11 years). A total of 43 patients were diagnosed with bilateral congenital cataract (58.1%), and 31 patients were diagnosed with unilateral congenital cataract (41.9%); this incidence was considerably higher than the rate observed in family hereditary cataract cases (only approximately 6% were unilateral cases in our database; data not shown). The 20 control subjects diagnosed with traumatic lens opacity included 13 boys and 7 girls, with an average age of 6.1 years (range from 3 to 11 years); all of the control subjects were diagnosed with unilateral cataract. Detailed clinical observations are presented in Table 1.
Table 1. Clinical observations of the investigated subjects.
| Clinical characteristics | N (range) | Percentage (%) |
|---|---|---|
| SCC patients |
74 |
|
| Age (years) |
4.6 (0–11) |
|
| Male |
39 |
52.7 |
| Female |
35 |
47.3 |
| Double eye |
44 |
59.5 |
| Single eye |
30 |
40.5 |
| TC patients (control) |
20 |
|
| Age (years) |
6.9 (3–11) |
|
| Male |
13 |
65 |
| Female |
7 |
35 |
| Double eye |
0 |
0 |
| Single eye | 20 | 100 |
Analysis of the genetic variants in SCC
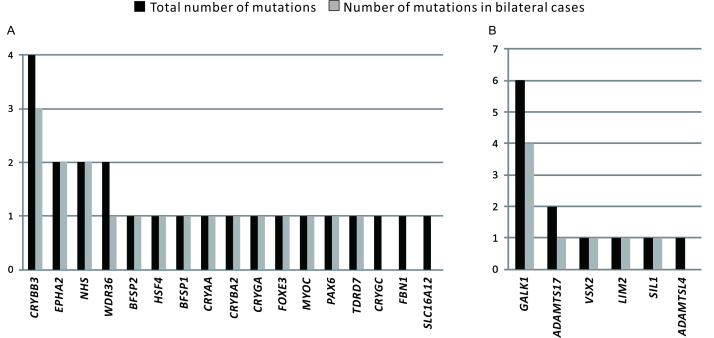
The average mean read depth for the target region (whole sequences from the 61 genes, including introns and exons) was 152.7X; moreover, 93.7% of the target region achieved 10X coverage. By filtering the variants following the four steps described in the Methods section, we identified 32 SCC-specific variants, all of which were heterozygous except one homozygous nonsense mutation in the NHS gene. Additional sequencing analysis of the 103 healthy controls confirmed that the 32 variants did not exist in the controls. In addition, we searched each variant in the ExAC East Asian samples database. None of the 32 SCC variants had an allele frequency of more than 0.1%: 16 were not available (NA), four were 0, and 12 were <0.1%. However, nine of the heterozygous variants were from six genes (GALK1, SIL1, ADAMTSL4, ADAMTS17, VSX2, and LIM2) responsible for an autosomal recessive pattern of cataract, which should not be considered causative mutation. However, we listed these variants separately (Appendix 3) because they still could be risk sites for CC as none existed in the TC and healthy controls. As a result, 23 predicted functional SCC mutations remained in the main list (Table 2), including 16 missense, five nonsense, one spicing site acceptor, and one splicing site donor mutations. These mutations were located in 17 genes related to lens disease; the mutation frequencies (the number of detected times) of these genes were analyzed and are presented in Figure 1A. CRYBB3 was the most mutated gene in the samples, followed by EPHA2, NHS, and WDR36. The six recessive genes were also analyzed, as shown in Figure 1B. GALK1 appeared to be the most frequently mutated gene (six mutations in six patients) among the recessive genes.
Table 2. Summary of 23 SCC-specific mutations.
| Gene |
Exonic/ |
Transcript |
Codon |
dbSNP |
Status |
SIFT |
PolyPhen |
SCC |
EXAC |
|---|---|---|---|---|---|---|---|---|---|
| Biotype | patient (N) | (East Asia) | |||||||
| BFSP1 |
MISSENSE |
NM_001195:p.Glu468Lys |
Gag/Aag |
|
Het |
0.02 damaging |
0.531 possibly damaging |
1 |
0 |
| BFSP2 |
MISSENSE |
NM_003571:p.Gly68Asp |
gGc/gAc |
|
Het |
0.01 damaging |
0.981 probably damaging |
1 |
NA |
| CRYAA |
MISSENSE |
NM_000394:p.Val161Met |
Gtg/Atg |
|
Het |
0 damaging |
0.965 probably damaging |
1 |
0 |
| CRYBA2 |
MISSENSE |
NM_005209:p.Phe63Ser |
tTc/tCc |
|
Het |
0 damaging |
1.000 probably damaging |
1 |
NA |
| CRYBB3 |
MISSENSE |
NM_004076:p.Gly156Arg |
Ggg/Agg |
|
Het |
0 damaging |
1.000 probably damaging |
2 |
NA |
| CRYBB3 |
MISSENSE |
NM_004076:p.Gly76Arg |
Ggg/Agg |
|
Het |
0 damaging |
1.000 probably damaging |
1 |
0.000231 |
| CRYBB3 |
MISSENSE |
NM_004076:p.Arg105Gln |
cGg/cAg |
rs17670506 |
Het |
0.03 damaging |
1.000 probably damaging |
1 |
0.000694 |
| CRYGA |
SPLICE_SITE
ACCEPTOR |
NM_014617 |
|
|
Het |
|
N/A |
1 |
NA |
| CRYGC |
NONSENSE |
NM_020989:p.Gln113* |
Cag/Tag |
|
Het |
N/A |
|
1 |
NA |
| CRYGC |
MISSENSE |
NM_020989:p.Glu94Lys |
Gag/Aag |
rs200572745 |
Het |
0 damaging |
0.999 probably damaging |
1 |
0.000116 |
| EPHA2 |
NONSENSE |
NM_004431:p.Lys935* |
Aag/Tag |
|
Het |
N/A |
|
1 |
NA |
| EPHA2 |
MISSENSE |
NM_004431:p.Glu934Lys |
Gag/Aag |
|
Het |
0.01 damaging |
0.985 probably damaging |
1 |
0 |
| FBN1 |
MISSENSE |
NM_000138:p.Asn497Lys |
aaC/aaG |
|
Het |
0.04 damaging |
0.856 possibly damaging |
1 |
NA |
| FOXE3 |
MISSENSE |
NM_012186:p.Pro149Ser |
Ccc/Tcc |
|
Het |
0 damaging |
0.999 probably damaging |
1 |
NA |
| HSF4 |
MISSENSE |
NM_001040667:p.Pro60His |
cCc/cAc |
|
Het |
0 damaging |
1.000 probably damaging |
1 |
NA |
| MYOC |
MISSENSE |
NM_000261:p.Leu215Pro |
cTg/cCg |
|
Het |
0 damaging |
1.000 probably damaging |
1 |
0.000578 |
| NHS |
NONSENSE |
NM_198270:p.Arg248* |
Cga/Tga |
|
Het |
N/A |
|
1 |
NA |
| NHS |
NONSENSE |
NM_198270:p.Arg565* |
Cga/Tga |
|
Hom |
N/A |
|
1 |
NA |
| PAX6 |
NONSENSE |
NM_001604:p.Glu123* |
Gag/Tag |
|
Het |
N/A |
|
1 |
NA |
| SLC16A12 |
MISSENSE |
NM_213606:p.Asn333Asp |
Aat/Gat |
|
Het |
0 damaging |
1.000 probably damaging |
1 |
NA |
| TDRD7 |
MISSENSE |
NM_014290:p.Gln28Pro |
cAa/cCa |
|
Het |
0 damaging |
0.999 probably damaging |
1 |
0.000116 |
| WDR36 |
SPLICE_SITE
_DONOR |
NM_139281 |
|
|
Het |
N/A |
|
1 |
NA |
| WDR36 | MISSENSE | NM_139281:p.Arg703Gln | cGg/cAg | Het | N/A | 0.987 probably damaging | 1 | NA |
Figure 1.
The mutation frequencies of the selected genes. A: The mutations in genes responsible for autosomal dominant or X-linked lens disease. B: The mutations in genes responsible for autosomal recessive lens disease. Black bar, total number of mutations; gray bar, number of mutations in the patients with bilateral cataract.
The genetic heterogeneity in SCCs
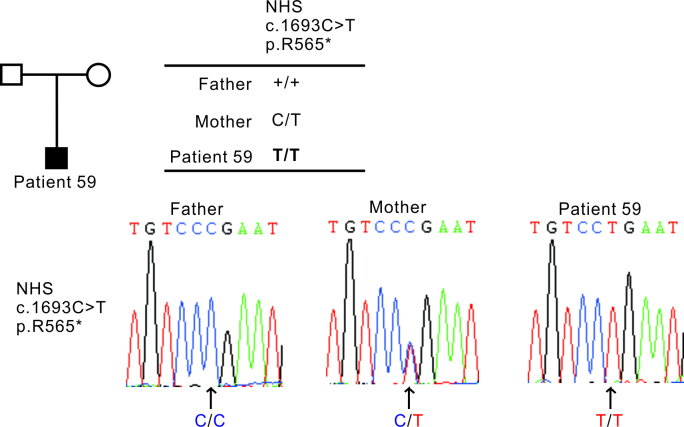
Compared to familial CC, the genetic effect is ambiguous in SCC cases. As displayed in Table 3, in the 19 patients with identified SCC variants, five were found to have mutations in more than one gene. These complexities suggested combined effects of multiple variants acting together. For example, patient 3, who presented with bilateral total CC, was found to have one missense mutation in EPHA2 and one splice site acceptor mutation in CRYGC; patient 51, who presented with bilateral nuclear CC, had missense mutations in CRYBB3 and BFSP2. Notably, with the only detected homozygous mutation, patient 59, who presented with bilateral nuclear CC, was found to have a homozygous mutation c.1693C>T, p.R565* in NHS located in chromosome X. Based on this result, we suspected this patient had Nance-Horan syndrome, a rare X-linked syndrome characterized by congenital cataract and dental anomalies [18]. Although we found dental anomalies in this patient (Appendix 4), we were not confident to attribute this feature to the syndrome because the patient was growing permanent teeth when examined. The patient was intellectually normal and did not appear with other obvious dysmorphic features. By comparing the patient’s DNA sequences with the parents’ DNA sequences at the corresponding sites, we found that the homozygous c.1693C>T, p.R565* in X-linked NHS was inherited from the mother (Figure 2). Both parents had normal transparent lenses. The mother’s cornea diameter was 11 mm and the ocular axis was 24.69 mm (D), 24.50 mm (S) on an ultrasound A-scan, both of which were within normal range. This case is an example indicating that genetic effects could be underestimated in sporadic cases when the patient presents a recessively inherited mutation (X-linked recessive in this case). Segregation analysis could not be performed on the other patients because the parents’ DNA samples were not available.
Table 3. Predicted functional mutations identified in 19 SCC patients.
| Patient # | Identified variant/variants SIFT Polyphen-2 | Uni/Bi- lateral | Phenotype |
|---|---|---|---|
| 1 |
CRYAA, p.Val161Met 0 0.965 |
B |
Nuclear |
| 3 |
EPHA2, p.Lys935* 0.56 N/A |
B |
Total |
|
CRYGA, Splice site N/A N/A acceptor |
|||
| 4 |
WDR36, Splice site N/A N/A donor |
U |
Posterior polar |
| 5 |
CRYBA2, p.Phe63Ser, 0 1.000 |
B |
Posterior polar |
| 10 |
CRYBB3, p.Gly156Arg 0 1 |
B |
Nuclear |
| 24 |
FBN1, p.Asn497Lys 0.04 0.856 |
U |
Nuclear |
| 31 |
PAX6, p.Glu123* N/A N/A |
B |
Total |
|
FOXE3, p.Pro149Ser 0 0.999 |
|||
| 32 |
NHS, p.Arg248* (X-link) N/A N/A |
B |
Cortical |
|
EPHA2, p.Glu934Lys 0.01 0.985 |
|||
| 35 |
MYOC, p.Leu215Pro 0 1.000 |
B |
Nuclear |
| 37 |
CRYGC, p.Glu94Lys 0 0.999 |
U |
Total |
| 50 |
BFSP1, p.Glu468Lys 0.02 0.531 |
B |
Nucelar |
| 51 |
CRYBB3, p.Gly156Arg 0 1.000 |
B |
Nuclear |
|
BFSP2, p.Gly68Asp 0.01 0.981 |
|||
| 56 |
WDR36, p.Arg703Gln N/A 0.987 |
B |
Nuclear |
| 57 |
CRYGC, p.Gln113* 0.2 N/A |
B |
Nuclear |
|
HSF4, p.Pro60His 0 1.000 |
|||
| 59 |
NHS, p.Arg565*,(X-link) N/A N/A (Hom) |
B |
Nuclear |
| 60 |
CRYBB3, p.Gly76Arg 0 1.000 |
U |
Cortical |
| 62 |
TDRD7, p.Gln28Pro 0 0.999 |
B |
Posterior polar |
| 68 |
CRYBB3, p.Arg105Gln 0.03 1.000 |
B |
Total |
| 70 | SLC16A12, p.Asn333Asp 0 1.000 | U | Nuclear |
Figure 2.
The NHS homozygous mutation identified in patient 59. The sequence chromatography and pedigree of patient 59 showed that the homozygous mutation of NHS was inherited from the patient’s mother.
Association between SCC-SNPs and the cataract phenotype
Six cataract subtypes (classified as total, punctate, nuclear, posterior polar, anterior polar, and cortical subtypes) associated with genes that have been mutated at least two times were listed in Table 4. The four patients with CRYBB3 mutations presented with three cataract subtypes, indicating the phenotype heterogeneity of SCC. Of note, patient 10 and patient 51 shared a mutation site, c.466G>A, p.G156R; both presented with bilateral nuclear cataract.
Table 4. The number of the cases of specific cataract subtypes associated with the gene mutations.
| Gene | Total | Punctate | Nuclear | posterior polar | Anterior polar | Cortical |
|---|---|---|---|---|---|---|
|
CRYBB3 |
1 |
|
2 |
|
|
1 |
|
EPHA2 |
1 |
|
|
|
|
1 |
|
NHS |
|
|
1 |
|
|
1 |
| WDR36 | 1 | 1 |
Discussion
In the pediatric cataract clinic in our tertiary care hospital, we found that SCC cases account for 71% of total congenital cataract cases (data not shown), which presented with an unclear etiology without familial history. Genetic studies have revealed numerous mutations in diverse genes associated with congenital cataracts; these studies most frequently used whole exome sequencing followed by linkage analysis within family members [23-28] or screened only one specific gene in a larger group [29]. In a previous study conducted in an Indian population (100 congenital cataract cases), the exon sequencing of 14 genes (ten of which were crystalline genes) identified 18 variants, 14 of which were from crystalline genes [6]. In the present study, we extended the genetic analysis to 61 genes in 74 cases (the familial factor was excluded) using target capture and NGS. We found 32 SCC-specific SNPs in 29 patients, 31 of which were heterozygous. After nine variants located in genes responsible for recessive cataracts were removed, 23 mutations in 17 genes from 19 patients were predicted to be functional. Among the mutated genes, CRYBB3 appeared with the highest mutation frequency, followed by 16 genes that have been mutated at least once, suggesting a broad spectrum of genes could contribute to SCC (Figure 1A). The mutation pick-up rate in this study was 26% (19 out of 74), which was much lower than in a recent study of general CC cases in the United Kingdom (not only SCC) that was 75% (27 out of 35 patients) using a similar method [30]. This result suggested that a genetic factor has much less of an effect in sporadic cases. Notably, putative functional mutations in more than one gene were found in five patients, suggesting a combined genetic effect for SCC. However, we did not obtain the DNA samples from the parents of each patient (except patient 59), and thus, we could not confirm through segregation study whether these SNPs are disease-causing.
As the most mutated gene, CRYBB3 (encoding crystalline beta B3) was mutated four times in patients with SCC in the present results. The mutation in CRYBB3 has been reported in autosomal dominant [28] and autosomal recessive cataract [31]. The four mutations in CRYBB3 presented no significant association with any specific subtype of cataract phenotype (Table 4).
We propose that examining the whole genomic sequence of the selected genes rather than screening known SNPs represents an efficient method for discovering susceptible SNPs involved in SCC, because the genomic variants in most non-familial cases differ from previously described variants. The majority of the identified SCC-specific SNPs in the present results have not been reported. Compared to whole exome sequencing, this target region sequencing method is a better option for exploring the SNPs in sporadic cases because it is more economical and the results are easier to analyze (considering the sample size and the fact that these subjects are not related). Additionally, this method successfully identified cataract patient-specific mutations in three families when applied to genomic studies of familial cataracts (data not shown). However, novel genes cannot be revealed using this sequencing strategy. Taken together, our study may help enrich the knowledge of the etiology of congenital cataract, especially SCC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China, grant number 81200668 and 81270989. The samples used for the analyses described in this manuscript were obtained from the EENT Biobank; we would like to thank all the participants and the staffs for their valuable contribution to this research. We thank Dr. Jihong Wu for her precious advice. Dr. Jin Yang (jin_er76@hotmail.com) and Yi Lu (luyieent@126.com) are co-corresponding authors of this work.
Appendix 1. List of the 61 selected genes and related lens diseases.
To access these data, click or select the words "Appendix 1".
Appendix 2. Primers used for Sanger sequencing and the product number of Taqman SNP genotyping.
To access these data, click or select the words "Appendix 2".
Appendix 3. Summary of 9 SCC-specific mutations in recessive genes.
To access these data, click or select the words "Appendix 3".
Appendix 4. The dental feature of patient 59.
To access these data, click or select the words "Appendix 4".
References
- 1.Chan WH, Biswas S, Ashworth JL, Lloyd IC. Congenital and infantile cataract: aetiology and management. Eur J Pediatr. 2012;171:625–30. doi: 10.1007/s00431-012-1700-1. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22383071&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 2.Lambert SR, Drack AV. Infantile cataracts. Surv Ophthalmol. 1996;40:427–58. doi: 10.1016/s0039-6257(96)82011-x. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8724637&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Scott MH, Hejtmancik JF, Wozencraft LA, Reuter LM, Parks MM, Kaiser-Kupfer MI. Autosomal dominant congenital cataract. Interocular phenotypic variability. Ophthalmology. 1994;101:866–71. doi: 10.1016/s0161-6420(94)31246-2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8190472&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 4.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–49. doi: 10.1016/j.semcdb.2007.10.003. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18035564&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ionides A, Francis P, Berry V, Mackay D, Bhattacharya S, Shiels A, Moore A. Clinical and genetic heterogeneity in autosomal dominant cataract. Br J Ophthalmol. 1999;83:802–8. doi: 10.1136/bjo.83.7.802. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10381667&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar M, Agarwal T, Kaur P, Khokhar S, Dada R. Molecular and structural analysis of genetic variations in congenital cataract. Mol Vis. 2013;19:2436–50. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24319337&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 7.Hejtmancik JF, Smaoui N. Molecular genetics of cataract. Dev Ophthalmol. 2003;37:67–82. doi: 10.1159/000072039. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12876830&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 8.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–15. doi: 10.1016/j.survophthal.2004.02.013. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15110667&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 9.Mauri L, Franzoni A, Scarcello M, Sala S, Garavelli L, Modugno A, Grammatico P, Patrosso MC, Piozzi E, Del Longo A, Gesu GP, Manfredini E, Primignani P, Damante G, Penco S. SOX2, OTX2 and PAX6 analysis in subjects with anophthalmia and microphthalmia. Eur J Med Genet. 2015;58:66–70. doi: 10.1016/j.ejmg.2014.12.005. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25542770&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Williamson KA, FitzPatrick DR. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet. 2014;57:369–80. doi: 10.1016/j.ejmg.2014.05.002. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24859618&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 11.Ito YA, Footz TK, Berry FB, Mirzayans F, Yu M, Khan AO, Walter MA. Severe molecular defects of a novel FOXC1 W152G mutation result in aniridia. Invest Ophthalmol Vis Sci. 2009;50:3573–9. doi: 10.1167/iovs.08-3032. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19279310&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 12.Law SK, Sami M, Piri N, Coleman AL, Caprioli J. Asymmetric phenotype of Axenfeld-Rieger anomaly and aniridia associated with a novel PITX2 mutation. Mol Vis. 2011;17:1231–8. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21617748&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 13.Khan AO, Aldahmesh MA, Alkuraya FS. Genetic and genomic analysis of classic aniridia in Saudi Arabia. Mol Vis. 2011;17:708–14. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21423868&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 14.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, Stetten G, Meyers DA, Francomano CA. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–9. doi: 10.1038/352337a0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=1852208&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 15.Zhou XM, Wang Y, Zhao L, Yu WH, Fan N, Yan NH, Su Q, Liang YQ, Li LP, Cai SP, Jonas JB, Liu XY. Novel compound heterozygous mutations identified in ADAMTSL4 gene in a Chinese family with isolated ectopia lentis. Acta Ophthalmol. 2015;93:e91–2. doi: 10.1111/aos.12399. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24802351&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 16.Barker DF, Hostikka SL, Zhou J, Chow LT, Oliphant AR, Gerken SC, Gregory MC, Skolnick MH, Atkin CL, Tryggvason K. Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science. 1990;248:1224–7. doi: 10.1126/science.2349482. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2349482&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 17.Abu-Amero KK, Bosley TM, Morales J. Analysis of nuclear and mitochondrial genes in patients with pseudoexfoliation glaucoma. Mol Vis. 2008;14:29–36. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18246027&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 18.Walpole IR, Hockey A, Nicoll A. The Nance-Horan syndrome. J Med Genet. 1990;27:632–4. doi: 10.1136/jmg.27.10.632. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=2246772&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Stefansdottir G, Masson G, Hardarson GA, Petursson H, Arnarsson A, Motallebipour M, Wallerman O, Wadelius C, Gulcher JR, Thorsteinsdottir U, Kong A, Jonasson F, Stefansson K. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–400. doi: 10.1126/science.1146554. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17690259&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 20.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet 2009; Chapter 2:Unit 2 12. [DOI] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11337480&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12202775&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun W, Xiao X, Li S, Guo X, Zhang Q. Exome sequencing of 18 Chinese families with congenital cataracts: a new sight of the NHS gene. PLoS One. 2014;9:e100455. doi: 10.1371/journal.pone.0100455. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24968223&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narumi Y, Nishina S, Tokimitsu M, Aoki Y, Kosaki R, Wakui K, Azuma N, Murata T, Takada F, Fukushima Y, Kosho T. Identification of a novel missense mutation of MAF in a Japanese family with congenital cataract by whole exome sequencing: a clinical report and review of literature. Am J Med Genet A. 2014;164A:1272–6. doi: 10.1002/ajmg.a.36433. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24664492&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Huerta LM, Messina-Baas O, Urueta H, Toral-Lopez J, Cuevas-Covarrubias SA. A CRYGC gene mutation associated with autosomal dominant pulverulent cataract. Gene. 2013;529:181–5. doi: 10.1016/j.gene.2013.07.044. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23954869&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, Chen P, Li J, Zhu Y, Zhai Y, Yao K. A novel MIP gene mutation associated with autosomal dominant congenital cataracts in a Chinese family. BMC Med Genet. 2014;15:6. doi: 10.1186/1471-2350-15-6. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24405844&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Zhang T, Wu D, Zhang J. A novel beaded filament structural protein 1 (BFSP1) gene mutation associated with autosomal dominant congenital cataract in a Chinese family. Mol Vis. 2013;19:2590–5. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24379646&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 28.Reis LM, Tyler RC, Muheisen S, Raggio V, Salviati L, Han DP, Costakos D, Yonath H, Hall S, Power P, Semina EV. Whole exome sequencing in dominant cataract identifies a new causative factor, CRYBA2, and a variety of novel alleles in known genes. Hum Genet. 2013;132:761–70. doi: 10.1007/s00439-013-1289-0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23508780&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dave A, Laurie K, Staffieri SE, Taranath D, Mackey DA, Mitchell P, Wang JJ, Craig JE, Burdon KP, Sharma S. Mutations in the EPHA2 gene are a major contributor to inherited cataracts in South-Eastern Australia. PLoS One. 2013;8:e72518. doi: 10.1371/journal.pone.0072518. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24014202&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillespie RL, O’Sullivan J, Ashworth J, Bhaskar S, Williams S, Biswas S, Kehdi E, Ramsden SC, Clayton-Smith J, Black GC, Lloyd IC. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology. 2014;121:2124–37. doi: 10.1016/j.ophtha.2014.06.006. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25148791&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 31.Riazuddin SA, Yasmeen A, Yao W, Sergeev YV, Zhang Q, Zulfiqar F, Riaz A, Riazuddin S, Hejtmancik JF. Mutations in betaB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2100–6. doi: 10.1167/iovs.04-1481. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15914629&dopt=Abstract [DOI] [PubMed] [Google Scholar]