Abstract
OBJECTIVES:
Celiac disease (CD)-associated duodenal dysbiosis has not yet been clearly defined, and the mechanisms by which CD-associated dysbiosis could concur to CD development or exacerbation are unknown. In this study, we analyzed the duodenal microbiome of CD patients.
METHODS:
The microbiome was evaluated in duodenal biopsy samples of 20 adult patients with active CD, 6 CD patients on a gluten-free diet, and 15 controls by DNA sequencing of 16S ribosomal RNA libraries. Bacterial species were cultured, isolated and identified by mass spectrometry. Isolated bacterial species were used to infect CaCo-2 cells, and to stimulate normal duodenal explants and cultured human and murine dendritic cells (DCs). Inflammatory markers and cytokines were evaluated by immunofluorescence and ELISA, respectively.
RESULTS:
Proteobacteria was the most abundant and Firmicutes and Actinobacteria the least abundant phyla in the microbiome profiles of active CD patients. Members of the Neisseria genus (Betaproteobacteria class) were significantly more abundant in active CD patients than in the other two groups (P=0.03). Neisseria flavescens (CD-Nf) was the most abundant Neisseria species in active CD duodenum. Whole-genome sequencing of CD-Nf and control-Nf showed genetic diversity of the iron acquisition systems and of some hemoglobin-related genes. CD-Nf was able to escape the lysosomal compartment in CaCo-2 cells and to induce an inflammatory response in DCs and in ex-vivo mucosal explants.
CONCLUSIONS:
Marked dysbiosis and an abundance of a peculiar CD-Nf strain characterize the duodenal microbiome in active CD patients thus suggesting that the CD-associated microbiota could contribute to the many inflammatory signals in this disorder.
INTRODUCTION
Celiac disease (CD) is an autoimmune, inflammatory disorder of the small intestine that involves interactions between genetic and environmental factors (1). In genetically susceptible individuals (human leukocyte antigen (HLA)-DQ2/DQ8 carriers), ingestion of gluten leads to an abnormal intestinal immune response involving both the adaptive and the innate immunity, which is characterized by a failure to establish and/or maintain tolerance to dietary peptides present in wheat, barley, and rye (particularly wheat gliadin) (2) and by intestinal damage (3). Subsequent studies challenged the gluten-hypersensitivity dogma and suggested that exposure to gluten may not be the only trigger of CD (4, 5). In the case of gluten exposure, it remains a mystery why: (i) the frequency of DQ2/DQ8 molecules in the general population is about 30%, but only 1−3% of individuals develop CD (1); (ii) CD is increasingly being diagnosed in adulthood, many years after introduction of gluten in the diet (6); and (iii) the prevalence of CD is rapidly increasing in the western world (although consumption of gliadin-containing food has not increased) (7). In this context, other environmental factors such as the intestinal ecosystem have been implicated in the pathogenesis of CD (8, 9). The rationale for this concept is that the gut microbiota is involved in the following: (i) differentiation of the intestinal epithelium; (ii) regulation of gut permeability and immunity; (iii) development of adequate responses to microbial and dietary antigens; and (iv) avoidance of overreactions (8, 10, 11, 12). Consequently, there is growing interest in understanding whether interactions between intestinal microbes and innate immunity of the host could influence CD expression by promoting inflammation and/or impairing mucosal function.
The present study was prompted by our previous finding of increased proliferation and decreased differentiation of intestinal cells toward the secretory goblet cell lineage in children with active CD and in children on a gluten-free diet (GFD) with respect to controls (13). Notably, maintenance of a correct number of functional goblet cells is required for homeostasis of the intestinal mucosal environment, and deficiencies in mucin composition render the mucosa more susceptible to damaging agents (e.g., bacterial infections) in the lumen (14). In line with these observations, studies conducted on fecal samples revealed the differences between CD patients and healthy controls in terms of microbiota composition and related metabolites (8, 9, 15, 16, 17), whereas data about the mucosa-associated microbiota are inconclusive, mainly because they focused on specific groups of bacteria, used different methodologies and specimens, or investigated heterogeneous CD populations in terms of age, clinical conditions, and medications (18, 19, 20, 21, 22, 23, 24, 25). Thanks to the advent of next-generation sequencing, we can now study simultaneously all the genomes of a microbial community directly recovered from specific environments (26, 27, 28, 29). This technique can also reveal changes in the gut microbiome profile before and after therapy (25, 30).
The aim of this study was to use 16S ribosomal RNA (rRNA) metagenomics to determine the CD-associated duodenal dysbiosis in adult celiac patients and elucidate the mechanisms by which gut CD-associated dysbiosis could concur to CD development or exacerbation.
METHODS
Patients and samples
In this cohort study, 41 unrelated Caucasian individuals were recruited, over 2 years, from among patients attending the Departments of Gastroenterology of the Universities of Salerno and of Roma-Tor Vergata, Italy. Subjects were enrolled from among patients with symptoms suggestive of CD, from among patients with a diagnosis of gastroesophageal reflux disease (GERD), and from among GFD CD patients undergoing CD follow-up, all of whom consented to participate in the study. The exclusion criteria for enrollment were any known food intolerance apart from gluten, IgA deficiency, treatment with antibiotics, proton pump inhibitors, antiviral or corticosteroids, or assumption of probiotics in the 2 months before sampling. Twenty patients with active CD were on a gluten-containing diet. These patients were positive for CD-specific antibodies (IgA anti-endomysium (EMA) and/or IgA anti-tissue transglutaminase 2 (TG2)) and had villous atrophy on histological examination (31). Six patients had been on a GFD for at least 2 years, were EMA and anti-TG2 negative, and none of them had villous atrophy on histological examination. HLA-DQ2 and/or –DQ8 molecules were present in 100% of CD and GFD patients. The control group was constituted by seven patients with irritable bowel syndrome and/or dyspepsia, five patients with GERD, two patients with chronic gastritis, and one patient with iron loss caused by a fibromatous uterus.
We collected blood samples (to obtain DNA and serum) and duodenal biopsies, under sterile conditions to avoid contamination, from all enrolled individuals. Biopsies not used for histological examination were immediately cooled in dry ice and stored at −80 °C until genetic analysis, and/or stored in a transport medium for microbiological assays, and/or immediately cultured for in vitro experiments. The study was approved by the University of Naples Federico II Ethics Committee (Prot. N. 36/13). Clinical and laboratory features of the 3 study groups are shown in Table 1.
Table 1. Clinical and laboratory features of the three groups enrolled in the study.
|
Group |
|||
|---|---|---|---|
| Clinical features | Controls | Active CD patients | GFD patients |
| No. of subjects | 15 | 20 | 6 |
| Age in years (Mean±s.d.) | 42±16 | 38±12 | 39±11 |
| Sex F/M (%F) | 13F/2M (85%) | 18F/2M (89%) | 5F/1M (83%) |
| Clinical data (%) | |||
| Anemiaa | 6/15 (40%) | 9/20 (45%) | 1/6 (17%) |
| Weight loss | 1/15 (7%) | 2/20 (10%) | 0/6 (0%) |
| Diarrhea | 1/15 (7%) | 5/20 (25%) | 2/6 (33%) |
| Dyspepsia and/or IBS | 7/15 (47%) | 10/20 (50%) | 2/6 (33%) |
| Chronic gastritis | 2/15 (13%) | 0/20 (0%) | 0/6 (0%) |
| GERD | 5/15 (33%) | 0/20 (0%) | 0/6 (0%) |
| Presence of CD antibodies (EMA and/or TG2 IgA) | 10(+), 5b/15 | 20 (+)/20 | 0 (+)/6 |
| Positive family history of CD (%) | 1/15 (7%) | 5/20 (25%) | 1/6 (17%) |
| Duodenal biopsy (Marsh Index) (%) | 15/15 M0 (100%) | 4/20 M3A (20%) 8/20 M3B (40%) 8/20 M3C (40%) | 4/6 M0 (67%) 2c/6 M1 (33%) |
CD, celiac disease; EMA, antiendomysium IgA; F, female; GERD, gastroesophageal reflux disease; GFD, gluten-free diet; IBS, irritable bowel syndrome; M, male; TG2, anti tissue-transglutaminase 2 IgA.
Iron-deficiency anemia.
Not assayed in GERD.
Partial villous atrophy and intra-epithelial lymphocyte count =25/100.
DNA extraction and 16S rRNA sequencing
Total DNA was extracted from duodenal biopsies (3 mg/sample) using the QIAamp DNA Mini Kit (Qiagen,Venlo, The Netherlands) according to the manufacturer's instructions. A 548 bp amplicon, spanning from V4 to V6 variable regions of the 16S rRNA gene, was amplified using modified fusion primers (Supplementary Table S1 and Supplementary Material online) (32). Next-generation sequencing reactions were carried out on a 454 Genome Sequencer instrument (Roche, Penzberg, Germany) according to the manufacturer's instructions (33). Sequences were obtained in all enrolled subjects and analyzed using QIIME v. 1.9.1 (Supplementary Material) (34).
Microbiological analysis
Culture-dependent microbiological analysis and isolation of aerobic bacterial species (both adherent and intracellular microbial flora) were performed in individuals (9/20 active CD patients and 8/15 controls) for whom duodenal biopsies were available. In this study, we did not consider anaerobic species. Bacterial species were identified by mass spectrometry using the Matrix Assisted Laser Desorption/Ionization (MALDI) mass spectrometer (Bruker Daltonics MALDI Biotyper, Fremont, CA; Supplementary Material). We also cultured and isolated aerobic bacterial species from the oropharynx of 2 control subjects.
Neisseria flavescens whole-genome sequencing
Genomic DNA from Neisseria flavescens (Nf)-isolated strains was prepared as previously reported (35). Their whole-genome shotgun sequencing was carried out using the Genome Sequencer FLX System (454 Life Sciences and Roche, Basel, Switzerland), as previously described (Supplementary Material) (36). Sequencing reads were subjected to quality trimming, error correction, and assembly using the SPAdes software (37); default parameters and genome annotation were performed with the RAST Genome Annotation Server (38). Custom scripts were used to automate and parse the results of all-against-all tBlastx searches (incorporating 23 representative Neisseria genomes) and to identify clusters of orthologous genes (COGs).
Phylogenetic analyses
The sequences of 904 unambiguous orthologous single copy genes present in all the considered Neisseria genomes were aligned at the amino acid level using the muscle software (39), and alignments were subjected to exclusion of low-quality regions using the GBlocks software (40), before concatenation to leave 93,642 amino acid positions. Maximum likelihood bootstrap phylogenetic analysis of protein sequences was performed using the RAxML software (41) with invariable and 4 variable gamma-distributed site rate categories under the WAG amino acid substitution model (42).
Iron acquisition systems: Genetic and phenotypic characterization
The coding regions of the hemoglobin receptor (hmbR) gene, of the haptoglobin-hemoglobin (hpuA/B), and transferrin (tbpA/B)-binding protein genes, all involved in iron uptake from different sources, were amplified from the genomic DNA of the isolated Nf strains and sequenced by Sanger methods (Supplementary Table S2 and Supplementary Material). The ability to utilize different iron sources was assessed by growing CD-Nf and control-Nf isolates on iron-restricted media supplemented with hemoglobin (Hb), transferrin, and ferric nitrate (FeN) nonahydrate impregnated discs. N. meningitidis (Nm) (MC58 serogroup B) was used as pathogenic control bacterium (Supplementary Material).
Human and mouse dendritic cells and ex vivo mucosal explants in vitro experiments
Splenic and bone marrow-derived dendritic cells (DCs) were isolated from six C57BL/6 mice diluting 1 ml of a collagenase type IV solution (Sigma-Aldrich, St Lois, MO) in 9 mL RMPI IX and using a mechanical razor blade to disrupt the organs. After tissue disruption, red blood cells were lysed and CD11c+ DCs were isolated using magnetic MicroBeads (Miltenyi Biotec, Cambridge, MA) according to the manufacturer's instructions.
Monocyte-derived DCs were derived from peripheral blood monocytes of three healthy volunteers. CD14+ monocytes were isolated from total peripheral blood mononuclear cells using magnetic MicroBeads (Miltenyi Biotec) according to the manufacturer's instructions and cultured for 6 days in the presence of interleukin-4 (IL-4) and granulocyte macrophage colony-stimulating factor (GM-CSF) as previously described (43).
Both human and murine DCs were cultured in complete medium in the presence of 0.1 nM retinoic acid and 2 ng/ml transforming grow factor-β, to mimic the small intestinal micro-environment, as previously described (44). DCs were stimulated for 48 h with 50 μg/ml of filtered bacterial lysates (Supplementary Materials and Methods) from Nf strains isolated from five active CD patients (Nf1,2,3,7,8). Bacterial lipopolysaccharide (LPS; 100ng/ml) was used as a positive control. In addition, for murine DCs, lysates from Serratia marcescens (Sm, Gammaproteobacteria) isolated from a septic patient were used as pro-inflammatory controls, whereas Lactobacillus GG (LGG, Bacilli) was used as a negative non-inflammatory control as described previously (44). Forty-eight hours later, supernatants were harvested and tested by ELISA to check the levels of IL-12p40 (BD OptEIA, Becton Dickinson, Milano, Italia) and tumor necrosis factor-α (TNF-α eBioscience, San Diego, CA). All experiments were performed in duplicate. Mice were housed in the animal facility of the University of Chicago, Knapp Center for the Biomedical Discovery. All experiments were performed in accordance with the Institutional Biosafety Committee and the Institutional Care and Use Committee.
Duodenal fragments sampled from three control subjects were collected, oriented, and used for in vitro challenge with CD-Nf to evaluate the inflammatory properties of this bacterium. We chose to perform ex-vivo challenge with CD-Nf on duodenal mucosa from controls and not from CD patients (active and treated) in order to evaluate whether the inflammatory effect of the bacterium occurred regardless of other contributing factors likely present in CD patients. Mucosal explants were collected in ice-chilled tissue culture medium and cultured for 20 min. Fragments were placed on a stainless-steel mesh positioned over the central well of an organ-culture dish containing culture medium (37 °C), with the epithelium facing upward. One fragment per subject remained untreated and used as reference, whereas the other two fragments were treated with the CD-Nf by using two multiplicities of infection (MOI)—namely, 1:10 and 1:50. Briefly, fragments were cultured for 6 h, and incubation was stopped by washing, embedding tissues in optimal cutting temperature (OCT, Tissue TEK; Milews Laboratories, Elkhart, IN), and snap freezing them in cooled isopenthane. Samples and supernatants were then stored in liquid nitrogen until analysis. Cryostat sections (5 μm) of each sample were air dried and then processed. We performed immunofluorescence analysis of cells positive for the induced markers of inflammation HLA-DR (the major histocompatibility complex class II molecules) and COX-2 (enzyme rapidly induced by pro-inflammatory signals in monocytes or macrophages) (Supplementary Material). Using ELISA, we also evaluated the levels of interferon-γ, TNF-α, IL-6, IL-23, IL-17A, IL-17F, and IL-12p70 (Procarta Plex multiplex immunoassay; eBioscience) in supernatants of untreated and treated mucosal fragments according to the manufacturer's instructions. All experiments were performed in duplicate.
Internalization and intracellular localization of N. flavescens in epithelial cells
CaCo-2 epithelial cells were obtained from a Bank of Human and Animal Continuous Cell Lines (CEINGE-Biotecnologie Avanzate, S.C.a R.L. Naples, Italy). The cells were grown in Eagle's minimal essential medium (Sigma-Aldrich) and supplemented with 10% fetal bovine serum (Lonza, Basel, Switzerland), 1% l-glutamine (Sigma-Aldrich), and 1% non essential amino acids in the presence of penicillin (50 U/ml) and streptomycin (50 μg/ml). To distinguish between extracellular and intracellular bacteria, Nf internalization was evaluated by adapting the infection protocol to the immunofluorescence analysis (Supplementary Material) (45, 46). Specimens were viewed with the Leica TCS SP5 confocal laser-scanning microscope equipped with a 63x oil immersion objective. Images were processed using Adobe Photoshop CS5.
Statistical analysis
Differences in abundance of specific taxa across the study groups were identified with analysis of variance together with a Bonferroni correction within QIIME. Differences in Operational taxonomic units with P≤0.05 after Bonferroni adjustment were considered statistically significant. To compare the alpha diversity across sample groups, a non-parametric two sample t-test was run using 999 Monte Carlo permutations. Non-parametric multivariate analysis of variance (ADONIS) (47) using Unifrac distance matrices was used to test the significance of differences in beta diversity. The mean fold change value (untreated/treated) and the s.e.m. were calculated for each investigated cytokine in supernatants of ex vivo mucosal explants. Between-group comparisons were tested by the Friedman test for paired data with Bonferroni correction and the differences considered significant at a P-level <0.05. A two-way analysis of variance test was used to compare means between different groups of human and murine DCs treated with different bacterial lysates.
RESULTS
Microbiome profiles
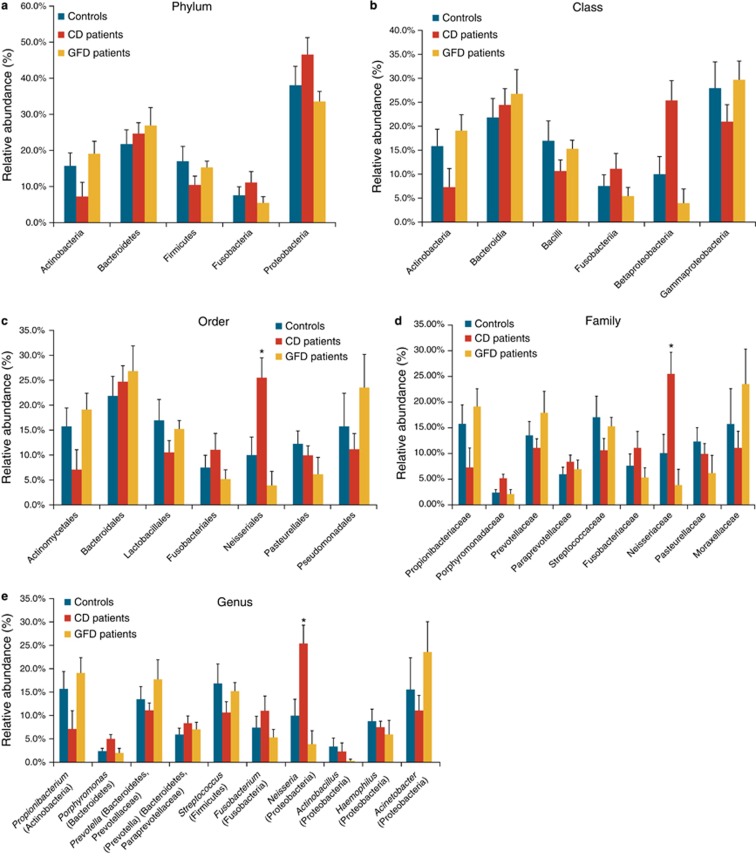
Sample metadata obtained from 16S bacterial rRNA sequencing of duodenal samples from all enrolled subjects are summarized in Supplementary Table 3. Twenty distinct phyla were identified (Supplementary Figure S1), of which Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Fusobacteria were the only ones present at a relative abundance >1%. Proteobacteria was the most abundant phylum in the three study groups, whereas the Actinobacteria and Firmicutes phyla were less abundant in the microbiome profiles of active CD patients than in the other two groups (Figure 1a). Class, order, family, and genus in each phylum were also identified (Figures 1b–e). There were no significant differences between the three groups within the Firmicutes phylum. Within the Proteobacteria phylum, the Betaproteobacteria class was highly represented in the gut microbiome of active CD patients, although its levels did not directly correlate with disease severity. In addition, within the Betaproteobacteria class, the Neisseriales order (P=0.009), the Neisseriaceae family (P=0.01), and the Neisseria genus (P=0.03) were significantly more abundant in active CD patients than in the other 2 study groups, and the Neisseria genus was the most abundant genus (99.8%) within the Neisseriaceae family (P<0.05) (Figure 1e). The Gammaproteobacteria class was less abundant in active CD patients than in the other two study groups (Figure 1b), whereas the Acinetobacter genus was more abundant in GFD patients and controls than in active CD patients, although the difference was not significant (Figure 1e).
Figure 1.
Duodenal microbiome taxonomic composition in controls, in active and in gluten-free diet (GFD) celiac disease (CD) patients. (a) Relative abundance at phylum level. Proteobacteria was the most abundant phylum in all 3 groups with an average abundance of 39.3%. (b) Class level classification in the 3 groups showed a decreasing trend for the Betaproteobacteria class in GFD patients (4%) and controls (10%) versus CD patients (26%). The Gammaproteobacteria class was less abundant in CD patients (21%) than in controls (28%) and GFD patients (30%). (c) Order level classification revealed a significant difference (P=0.009) in the Neisseriales order among controls (~10%), GFD patients (~4%), and CD patients (~26%). (d) Also at family level, the Neisseriaceae family was significantly less abundant (P=0.01) in controls (~10%) and GFD patients (4%) than in CD patients (~32%). (e) At genus level, Neisseria was significantly more abundant (P=0.03) in active CD patients (26%) than in controls (10%) and GFD patients (4%). Taxa (in parentheses) refer to the phylum to which the genus belongs. The data reported are filtered by a frequency higher than 1%. Error bars indicate standard error. Asterisks refer to taxa that differed significantly among the three groups (P<0.05, analysis of variance).
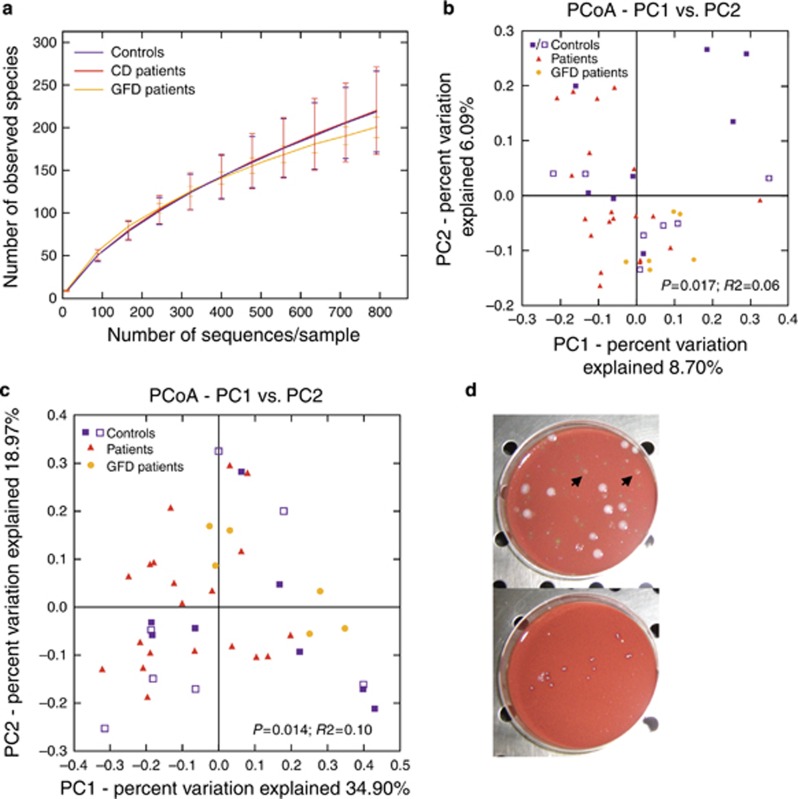
Alpha rarefaction curves (obtained with the QIIME pipeline, Supplementary Material) showed that bacterial community richness did not differ between CD patients and controls (Figure 2a). The Beta diversity of bacterial communities was statistically significant within the three groups computed with both the unweighted (P=0.017, R2=0.06; ADONIS) and the weighted Unifrac (P=0.014, R2=0.10; ADONIS) method (Figures 2b, c). This analysis suggests that active and GFD CD microbiomes are more similar to each other than control microbiomes are to the active or GFD CD microbiomes. This finding is supported by the tighter clustering of active and GFD CD points compared with control points (see Figures 2b, c). The high heterogeneity of the volunteers in the control group (which was inevitable given the difficulty in collecting biopsy samples) caused a slight variability in the microbiome composition, although no significant differences were observed within the control group in terms of the presence/absence of dyspepsia or irritable bowel syndrome (Figure 2b, c, Supplementary Figure S1 and Supplementary Tables S4 and S5). Furthermore, we did not find any correlation between the differences in intestinal microbiome composition and the histological features among the controls enrolled in the study.
Figure 2.
Alpha rarefaction curves and beta diversity analysis in controls, in active and in gluten-free diet (GFD) celiac disease (CD) patients. (a) Alpha rarefaction curves at normalization by depth of 792 sequences per sample (obtained with the QIIME pipeline, Supplementary Materials and Methods) showed that bacterial community richness did not differ between CD patients and controls. (b) Principal-coordinates analysis applied to the unweighted and (c) weighted UniFrac distances shows a difference between CD patients and controls. In fact, both active CD and GFD patients are distinct from controls, which moreover had a random distribution (P=0.017, R2=0.06; ADONIS and P=0.014, R2=0.10; ADONIS, respectively). Filled blue squares refer to a subgroup of control subjects affected by gastroesophageal reflux disease, chronic gastritis, or fibromatous uterus, whereas opened blue squares refer to a subgroup of controls affected by dyspepsia or irritable bowel syndrome; both these control groups are CD negative. (d) This figure shows the viability of N. flavescens species (arrows) in duodenal mucosa cultures from active CD patients (top panel), whereas the bacterium did not grow in duodenal cultures from control subjects (bottom panel).
Microbiological assays
To verify the viability of the aerobic species identified by 16S sequencing, which include Neisseria, we performed standard microbiological cultures of duodenal biopsy specimens. Differential analysis (differential culture media and mass spectrometry analysis) revealed a cultivable aerobic biodiversity in duodenal microflora of the samples tested (Supplementary Table S6). In particular, neither Lactobacillus nor Bacillus cereus (which belong to Firmicutes) was found in the microbiome of the active CD patients, although both are commensal bacteria in the healthy gut. In contrast, Nf was viable in all duodenal mucosa samples from active CD patients (CD-Nf), with a MALDI-TOF score above 2 (data not shown), but was undetectable in the duodenal mucosa samples from controls (Figure 2d). However, we were able to culture and isolate Nf from the oropharynx of two control subjects (control-Nf), in whom this bacterium is commensal (data not shown).
Genotypic and phenotypic features of the isolated CD-Neisseria flavescens
Nfs isolated from three active CD patients (namely CD-Nf1, CD-Nf2, and CD-Nf3), whose mucosal intestinal biopsies were available, and from the oropharynx of one healthy individual (control-Nf) were subjected to whole-genome shotgun sequencing as reported in the Methods section. Genome assembly and annotation were performed using standard approaches (37, 38) and resulted in assemblies ranging in size from 2.22 to 2.30 Mb (Supplementary Table S7). Phylogenetic analyses of the tested Nf genomes confirmed the close phylogenetic relationships between all the strains recovered in the current study and those annotated in GenBank (http://www.ncbi.nlm.nih.gov/genome/genomes), namely, the N. flavescens strains SK114 and NRL 30031, N. mucosa strain C102, and N. subflava strain NJ9703 (Supplementary Figure S2).
We used reciprocal best blast analyses to identify genes present in all CD-Nf genomes, but absent in at least one of the other studied Nf genomes (see above), and in control-Nf. Notably, the isolates from our control-Nf and the two commensal Nf genomes (SK114 and NRL 30031) annotated in GenBank possess neisserial haptoglobin-hemoglobin A/B (hpuA/B) and transferrin A/B (tbpA/B) binding protein genes but lack the hmbR gene. On the contrary, all isolates from active CD patients (CD-Nf1, CD-Nf2, and CD-Nf3) lack the hpuA/B and tbpA/B genes but possess the hmbR gene (data not shown). This result was confirmed by PCR analysis in 5 CD-Nf samples (Supplementary Table S8). The hmbR gene encodes a Hb receptor that has more frequently been associated with disease isolates of N. meningitidis (Nm) than with carriage isolates, suggesting that this receptor contributes to Nm-related disease (48, 49, 50). As the process of iron scavenging from the host is an important virulence attribute of bacterial pathogens, we evaluated the ability of Nf isolated from the duodenal mucosa of CD patients to scavenge iron from Hb and from other sources compared with control-Nf. We observed that, on Mueller-Hinton (MH) agar plus desferal, strains isolated from CD patients (CD-Nf1, CD-Nf2, CD-Nf3, CD-Nf7, and CD-Nf8) and Nm (pathogenic positive control) grew well with Hb as the sole iron source, which indicates that these strains express at least one Hb receptor, in agreement with our sequence data. In contrast, the control-Nf strain grew well also using transferrin as an iron source, which indicates that this strain utilizes iron through the hpuA/B and tbpA/B gene systems— a finding that is in accordance with our control-Nf genomic sequence and PCR results (Supplementary Table 8).
Internalization of N. flavescens in epithelial cells
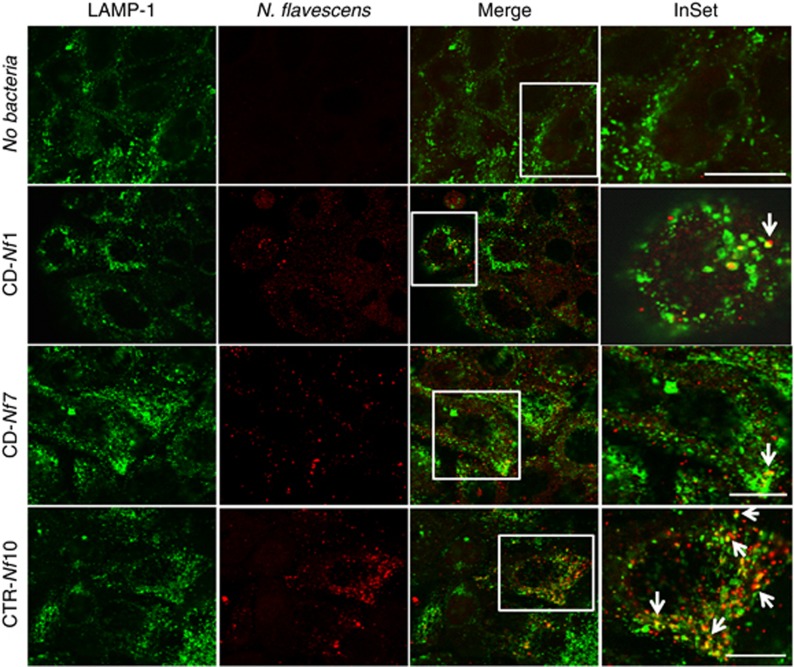
To evaluate how CD-Nf and control-Nf are internalized in the host epithelial cells, we infected CaCo-2 epithelial cells with Nf from two active CD patients (CD-Nf1 and CD-Nf7) and used confocal microscopy to compare their intracellular localization with that of control-Nf up to 6 h after infection (Figure 3). At the end of the experiment, although CD-Nfs (CD-Nf1and CD-Nf7) were internalized (red signals) and seldom colocalized with the marker of the late/endosome-lysosomal pathway (LAMP-1, green signals), control-Nf (CTR-Nf10) clearly colocalized with LAMP1, indicating that most CD-Nfs escape this intracellular compartment.
Figure 3.
N. flavescens internalization in epithelial cells. N. flavescens isolated from the gut mucosa of active celiac disease (CD) patients (CD-Nf) seldom colocalized with lysosomes (LAMP-1) in CaCo-2 cells, whereas N. flavescens isolated from the oropharynx of control subjects (CTR-Nf) clearly colocalized with LAMP-1. The figure (which is representative of three independent experiments) shows the results obtained 6 h after infection of CaCo-2 cells with N. flavescens (multiplicity of infection 1:50) isolated from 2 active CD patients (CD-Nf1/Nf7) and from a control subject (CTR-Nf10). An antibody against N. flavescens was used before permeabilization followed by a secondary Cy5-conjugated antibody and, after permeabilization, with an Alexafluor-546-conjugated secondary antibody (red). We used LAMP-1 and an Alexafluor-488 (green)-conjugated secondary antibody to detect endo/lysosomes. Merge and InSet images show colocalization of the bacteria with lysosomes (white arrows). Uninfected CaCo-2 cells (NO bacteria), stained with anti N. meningitidis antibody and processed for immunofluorescence as the other samples, served as control of the procedure. Samples were analyzed by confocal microscopy with × 63 oil immersion objective. Bars = 10 μm.
In vitro inflammation effect of CD-Nf
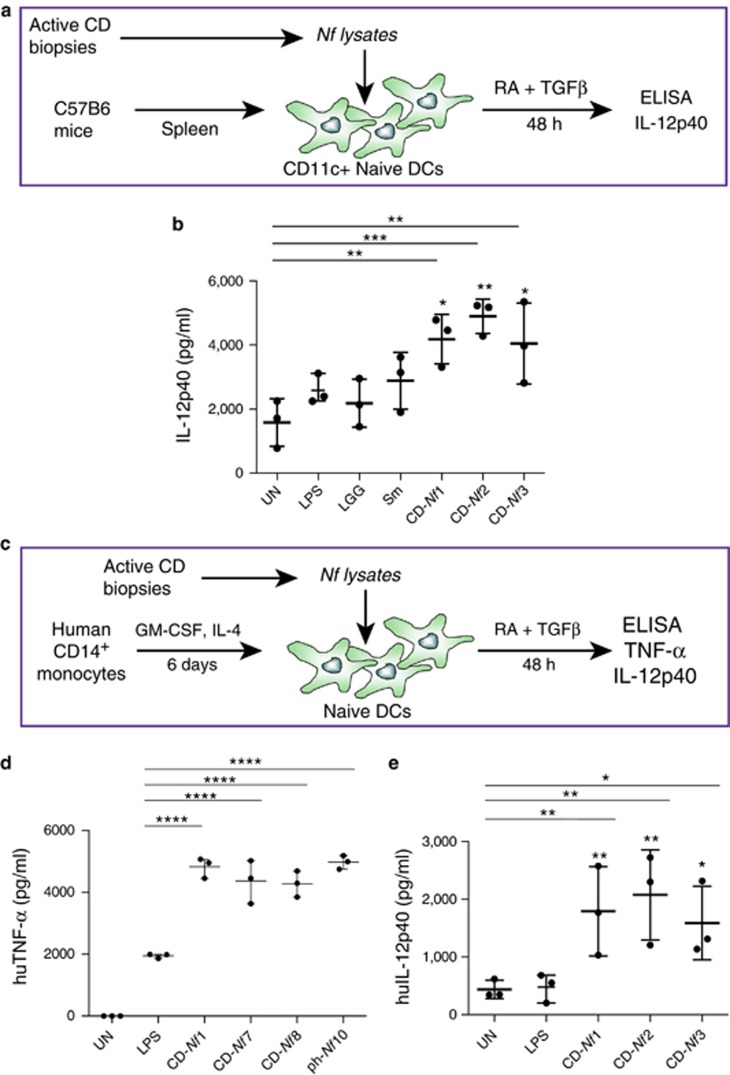
We investigated the pro-inflammatory properties of CD-Nf bacterial strains in vitro using both murine (Figure 4a) and human monocyte-derived (Figure 4c) DCs challenged for 48 h with bacterial lysates obtained from CD-Nf1, CD-Nf2, CD-Nf3, CD-Nf7, and CD-Nf8 (Figures 4b, d and e). DCs were cultured in the presence of retinoic acid and transforming growth factor-β, both of which are abundant in the small intestinal microenvironment, to mimic physiological intestinal conditions. After 48 h of stimulation, the level of IL-12p40, a cytokine produced by DCs under inflammatory conditions, was higher in murine splenic DCs treated with CD-Nf strains than in untreated DCs (untreated vs. Nf1 P=0.0021, vs. Nf2 P=0.0003, vs. Nf3 P=0.0031) (Figures 4a and b) and in murine splenic DCs challenged with LGG (LGG vs. Nf1 P=0.01, vs. Nf2 P=0.0016, vs. Nf3 P=0.0161), a bacterium that colonizes the gut of healthy subjects and that in normal concentrations does not exert an inflammatory effect (Figure 4b). Levels of IL-12p40 were also higher in supernatants of DCs treated with the three strains of CD-Nf than in DC supernatants after stimulation with bacterial LPS (LPS vs. Nf1 P=0.0339, vs. Nf2 P=0.0047, vs. Nf3 P=0.05) and comparable to or even higher than (as assessed for Nf2 (P=0.01) those detected upon challenge with lysates of Sm (Gammaproteobacteria), a bacterium isolated from patients affected by sepsis and known to exert pro-inflammatory effects on DCs (Figure 4b) (51). The results of this set of experiments indicate that each of the 3 strains isolated from the small intestinal mucosa of CD patients was able to imprint a pro-inflammatory phenotype on murine splenic DCs in vitro.
Figure 4.
N. flavescens (Nf) isolated from celiac disease (CD) patients induce a pro-inflammatory phenotype on dendritic cells. Filtered bacterial lysates from Neisseria flavescens (Nf), isolated from 5 active celiac patients (CD-Nf1, 2, 3, 7, and 8), promoted the production of pro-inflammatory cytokines by dendritic cells (DCs). CD11c+ DCs isolated from the spleens of C57B6 mice were cultured for 48 h in the presence of retinoic acid (RA) and transforming growth factor-β (TGF-β) with filtered Nf lysates from three active CD patients (a). All three tested bacterial lysates promoted the production of interleukin (IL)-12p40 by murine DCs, whereas untreated (stars upon horizontal bars), lipopolysaccharide (LPS)-treated (stars), and LGG-treated DCs did not (b). Human monocyte-derived DCs were cultured for 48 h in the presence of RA and TGF-β with filtered Nf lysates isolated from active CD patients (c). Tumor necrosis factor-α (TNF-α) levels were higher in supernatants of human DCs treated with Nf 1, Nf 7, and Nf 8 bacterial strains isolated from small intestinal biopsies of CD patients, and in those challenged with Nf10 isolated from a pharyngeal swab of a control subject (d), than in those treated with bacterial LPS, used as a pro-inflammatory positive control (stars upon horizontal bars) and in untreated (UN) cells (P<0.0001 for each of the treatments). IL-12p40 levels were higher in supernatants of human DCs challenged with Nf1,2,3 filtered lysates than in those treated with bacterial LPS (stars) used as a pro-inflammatory positive control and in untreated (UN) cells (stars upon horizontal bars) (e). Dot plots represent values from three independent experiments. Each experiment was performed in duplicate. A two-way analysis of variance was performed to check for inter-group differences. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
We confirmed these results in bone marrow-derived murine CD11c+ DCs (data not shown). Furthermore, we tested the pro-inflammatory properties of the lysates of Nf strains isolated from active CD subjects on human monocyte-derived DCs (Figure 4c) and found that the production of both TNF-α (Figure 4d) and IL12p40 (Figure 4e) was higher in Nf-stimulated DCs than in either untreated or LPS-treated DCs (P<0.0001, one-way analysis of variance and multiple comparison analysis with Bonferroni correction). This finding suggests that Nf strains exert a pro-inflammatory effect on human monocyte-derived DCs. Of note, the Nf10 strain isolated from a pharyngeal swab of a control subject enhanced the production of the pro-inflammatory cytokines TNF-α when cultured in conditions mimicking the small intestinal environment (medium containing retinoic acid and transforming growth factor-β). This may suggest that not only the presence of a specific bacterial strain but also particularly the specific microenvironment and milieu may determine which effect is exerted on immune cells.
To verify the inflammatory in vitro effect of CD-Nf, we exposed ex-vivo mucosal explants from control subjects to CD-Nf bacteria for up to 6 h, and using immunofluorescence we found increased epithelial expression of the inflammation markers HLA-DR and COX-2. In particular, HLA-DR expression (Supplementary Figure S3) was enhanced in villus enterocytes, in the basal cytoplasmatic compartment, as well as on the brush border and basolateral membranes. HLA-DR was overexpressed also in the lamina propria, particularly near the surface epithelium. In addition, in experiments with COX-2 (data not shown), the mean number of COX-2-positive epithelial cells increased from 16/100 (untreated mucosa) up to 25/100 after CD-Nf challenge.
CD-Nf also promoted the release of inflammatory cytokines (interferon-γ, TNF-α, IL-6, IL-23, IL-17A, IL-17F, and IL-12p70). In fact, the levels of these cytokines were higher in the supernatants of treated than in untreated biopsies as assessed in organ-culture experiments (Table 2), and the increase was significant for interferon-γ (P=0.001), TNF-α (P<0.011), and IL-23 (P=0.015). We show that IL-12p70 and IL-23 share the subunit IL-12p40 thus suggesting that there might be an increase in IL-12p40 subunit production.
Table 2. CD-Nf challengea induces duodenal mucosa from control subjects to secrete pro-inflammatory cytokines in supernatants.
| Treatment |
Levels of cytokines (pg/ml) measured in supernatants mean (s.e.m.) |
||||||
|---|---|---|---|---|---|---|---|
| IFN-γ | TNF-α | IL-6 | IL-23 | IL-17A | IL-17F | IL-12p70 | |
| No treatment | 23.44 (9.34) | 10.10 (0.48) | 135.29 (72.08) | 38.69 (8.86) | 8.42 (5.98) | 5.17 (4.18) | 6.43 (1.22) |
| Challenge with CD-Nf (MOI 1:10) | 59.23 (2.40) | 14.83 (2.12) | 253.34 (147.57) | 50.39 (11.84) | 11.16 (7.92) | 7.12 (5.01) | 10.61 (2.15) |
| Challenge with CD-Nf (MOI 1:50) | 107.94 (2.34) | 23.24 (2.49) | 452.66 (19.74) | 106.01 (3.68) | 25.01 (3.80) | 21.06 (14.15) | 14.71 (2.43) |
| P valueb | 0.001 | 0.011 | 0.249 | 0.015 | 0.320 | 0.353 | 0.082 |
IFN-γ, interferon-γ IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
Ex- vivo mucosal explants from three control subjects were treated for 6 h with CD-Nf at a multiplicity of infection (MOI) 1:10 and 1:50, and then levels of inflammatory cytokines were measured in supernatants of those treated with respect to untreated biopsies. Differences between mean levels were tested by the Friedman test with Bonferroni correction, and the p value is reported under each column.
Boldface type indicates statistically significant differences among groups (P<0.05).
DISCUSSION
CD-associated duodenal dysbiosis is not yet clearly defined, and the mechanisms by which gut CD-associated dysbiosis could concur to CD development or exacerbation remain largely unknown. In the attempt to shed light on this issue, we used 16S rRNA metagenomics to determine whether the duodenum microbiome differed among three groups of adult individuals: active CD patients, GFD CD patients, and controls.
Of the five major phyla present with a frequency above 1% in the gut microbiome of our three study groups (Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Fusobacteria), members of the phylum Proteobacteria were more abundant and those of the Firmicutes and Actinobacteria phyla less abundant in active CD than in either GFD patients or controls. A similar dysbiosis was previously reported in the gut microbiome of four adult CD patients with gastrointestinal symptoms studied by 16S rRNA gene sequencing (23). The same group of investigators confirmed this dysbiosis in the gut microbiome of 17 GFD CD patients with persistent symptoms but not in 17 asymptomatic GFD patients (52). Similar results were obtained in biopsy specimens from active CD children when compared with GFD and control children (25). All the above data support the concept that the gut dysbiosis associated with the active stage of CD is characterized by an increase in Proteobacteria and a decrease in Firmicutes phyla. However, no other group has previously explored how specific bacterial species detected in the duodenal mucosa of CD patients could contribute to the CD-associated gut malfunction.
Within the bacterial genera detected in the gut microbiome of the 3 study groups by 16S rRNA gene sequencing, the relative abundance of the Neisseria genus was significantly higher (P=0.03) in active CD patients (26%), than in either GFD patients (4%) or controls (10%). We confirmed the viability of the CD-associated Neisseria identified with next-generation sequencing by culturing 17 biopsies (9 from active CD subjects and 8 from controls) under selective conditions for aerobic Neisseria growth. Subsequently, mass spectrometry revealed that Nf was the major species among those present in the Neisseria isolates from the gut microbiome of active CD patients but absent from the gut of controls. Other relevant differences between the microbiomes of active CD patients and controls were the reduced levels of Actinobacteria and Firmicutes in the active CD microbiome. Notably, two cultivable bacteria belonging to the Firmicutes phylum (Lactobacillus and Bacillus cereus), which are involved in gluten metabolism, were not found in the active CD microbiome. Low levels of Lactobacilli are one of the most consistent findings in both adults and children with active CD (16, 18, 21); however, the relevance of this information is unclear. The fecal levels of Firmicutes (Lactobacillus) and Actinobacteria were recently reported to be lower in active CD patients than in healthy controls, and this result was associated with differences in gluten metabolism among the two groups of individuals (53).
We next asked the following questions: (1) Are there genetic and/or phenotypic differences between CD-Nf and commensal control-Nf?; and (2) If there is a difference, does that difference enable CD-Nf to promote an abnormal response in the intestinal epithelium? To address these questions, we used next-generation sequencing to sequence the whole Nf genome of strains isolated from the gut of active CD patients (CD-Nf1, CD-Nf2, and CD-Nf3) and from the oropharynx of a control subject (control-Nf). Then we compared these sequences to those of other pathogenic and commensal Nf genomes present in GenBank. Notably, all CD-Nf and Nm genomes showed genetic diversity of the iron acquisition systems with respect to the control-Nf and the other tested commensal Nf. In fact, the CD-Nf genomes lacked the hpuA/B and tbpA/B genes but possessed the hmbR gene. This finding was supported by an in vitro test of iron uptake showing that CD-Nf and control-Nf acquired iron from different sources. However, although the hmbR gene has been widely associated with pathogenicity in Nm (48, 49, 50), the significance of its presence in our CD-Nf genomes remains to be further investigated. Notably, our data show that gene structure and function differ between CD-Nf and control-Nf.
To answer the second question, we explored the inflammatory effect of CD-Nf because overgrowth of pathogenic bacteria could lead to aberrant activation of the immune system. Interestingly, in vitro, all the Nf strains isolated from 5 adults affected by active CD induced an inflammatory phenotype in human and murine DCs, as shown also for other bacterial strains (44). In detail, IL-12p40 and TNF-α levels were higher in pure bacterial lysates from CD-Nf than in lysates stimulated with a control bacterium (LGG) or with LPS. These findings support the notion that Nf may promote a pro-inflammatory phenotype in DCs residing in the intestinal mucosa of celiac subjects and thus contribute to the CD immune response characterized by high levels of inflammatory cytokines. Furthermore, interferon-γ, TNF-α, and IL-23 levels were higher in the culture supernatants of biopsies treated with CD-Nf than in untreated biopsies from controls. These data support our hypothesis that Nf contributes to promoting inflammation in the small intestine. Whether this inflammation precedes or follows the onset of full-blown CD remains, however, to be studied. It is feasible that the inflammatory conditions occurring in the gut of CD patients may favor the colonization of Nf strains thus promoting the maintenance of a pro-inflammatory immune response; on the other hand, under certain conditions (environmental factors, viral infections, genetic predisposing factors, etc.), Nf colonization of the duodenal tract is favored, and this event could contribute to triggering CD development in genetically susceptible individuals.
Beside modulating the immune response, the microbiota may also play a role in CD by degrading toxic gluten peptides, thereby aiding gluten detoxification in vivo (54). In this context, it is noteworthy that members of the Neisseria genus are poorly active against intact and toxic gliadin peptides (55). Moreover, CD-Nf could affect intracellular trafficking in two ways: (a) CD-Nf escape from lysosomal degradation could trigger a stress pathway independent of the gliadin peptide pathways; (b) CD-Nf could interfere with the pathway involving the toxic gliadin peptides by following the intracellular pathway previously identified in CaCo-2 cells (56). Both these hypotheses deserve further investigation. However, it remains to be established whether the CD-associated bacterial species contributes to the pathogenesis of the disease or whether it is a consequence of it. At present, the induction of inflammatory responses in DCs and in ex-vivo mucosal explant supports the first possibility.
In conclusion, marked dysbiosis and an abundance of a peculiar CD-Nf strain characterize the duodenal microbiome of active CD patients, which suggests that the CD-associated microbiota could contribute to the many inflammatory signals in this disorder.
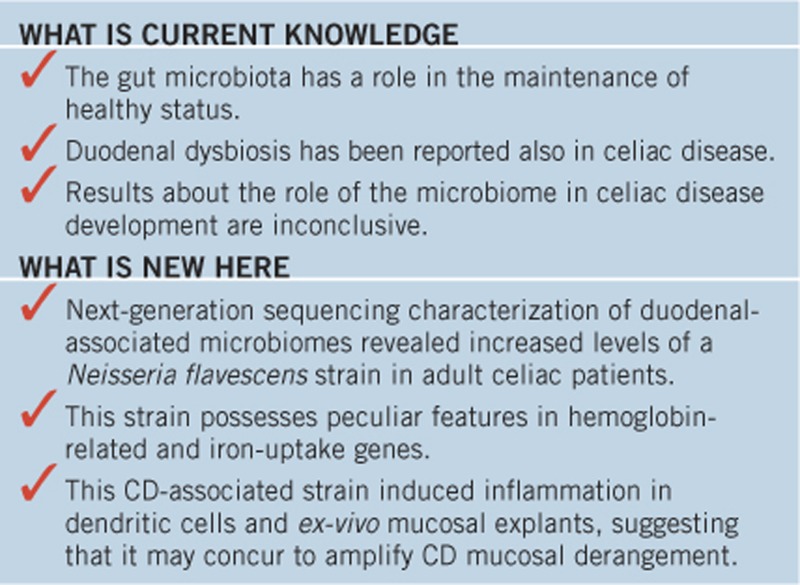
Study Highlights
Acknowledgments
The authors thank Jean Ann Gilder (Scientific Communication srl., Naples) for editing the text and Vittorio Lucignano, CEINGE–Biotecnologie Avanzate, for technical assistance related to graphics.
Guarantor of the article: Francesco Salvatore, MD, PhD.
Specific author contributions: Conceived and designed the study: Valeria D'Argenio, Francesco Salvatore, and Lucia Sacchetti; enrolled the study subjects and collected the biological samples: Carolina Ciacci, Giovanni Monteleone, and Giovanna Del Vecchio Blanco; performed challenging experiments on ex vivo mucosal explants: Ilaria Russo; performed the sequencing experiments: Valeria D'Argenio, and Vincenza Precone; conceived and carried out the bioinformatic analysis of metagenomic data: Giorgio Casaburi and Gregory J. Caporaso; conceived and carried out the genome assembly and annotation and performed phylogenetic analysis: Graziano Pesole, David S. Horner, and Matteo Chiara; conceived and microbiologically characterized and functionally evaluated N. flavescens features: Chiara Pagliuca, Roberta Colicchio, and Paola Salvatore; conceived and performed confocal microscopy assays: Daniela Sarnataro; conceived the experiments to evaluate the immune properties of N. flavescens: Valentina Discepolo, Sangman M. Kim, and Bana Jabrì performed the experiments to evaluate the immune properties of N. flavescens: Valentina Discepolo and Sangman M. Kim; analyzed the experiments to evaluate the immune properties of N. flavescens: Valentina Discepolo; analyzed the final data and wrote the manuscript: Valeria D'Argenio, Francesco Salvatore, and Lucia Sacchetti. All authors have read, contributed to the writing, and approved the final manuscript.
Financial support: This work was supported by grant 007_FC_2014 from the “Fondazione Italiana Celiachia” (to L.S.), by DIAINTECH-Regione Campania (to F.S.), by PRIN 2012 (n. 2012WJSX8K) and by POR Campania FSE 2007-2013, Project CREME (to P.S.), grant PS 35-126/Ind and grant PON01_02589 (MICROMAP) 2012 from the Ministry of University and Research (to F.S.), and grant RF-2010-2318372 from the Ministry of Health (to F.S.).
Potential competing interests: None.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/ajg
Supplementary Material
References
- Green PH, Cellier C. Celiac disease. N Engl J Med 2007;357:1731–1743. [DOI] [PubMed] [Google Scholar]
- Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology 2009;137:1912–1933. [DOI] [PubMed] [Google Scholar]
- Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol 2002;2:647–655. [DOI] [PubMed] [Google Scholar]
- Stene LC, Honeyman MC, Hoffenberg EJ et al. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am J Gastroenterol 2006;101:2333–2340. [DOI] [PubMed] [Google Scholar]
- Akobeng AK, Ramanan AV, Buchan I et al. Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies. Arch Dis Child 2006;91:39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas S, Ruiz de Morales JM, Fernandez M et al. Age-related clinical, serological, and histopathological features of celiac disease. Am J Gastroenterol 2008;103:2360–2365. [DOI] [PubMed] [Google Scholar]
- Lohi S, Mustalahti K, Kaukinen K et al. Increasing prevalence of coeliac disease over time. Aliment Pharmacol Ther 2007;26:1217–1225. [DOI] [PubMed] [Google Scholar]
- Sanz Y, Sánchez E, Marzotto M et al. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol Med Microbiol 2007;51:562–568. [DOI] [PubMed] [Google Scholar]
- Collado MC, Calabuig M, Sanz Y. Differences between the fecal microbiota of celiac infants and healthy controls. Curr Issues Intest Microbiol 2007;8:9–14. [PubMed] [Google Scholar]
- Conte MP, Schippa S, Zamboni I et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006;55:1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard CL, Elson CO, Hatton RD et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012;489:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Oelschlaeger TA, Wullaert A et al. Bacteria regulate intestinal epithelial cell differentiation factors both in vitro and in vivo. PLoS One 2013;8:e55620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano M, Iaffaldano L, Tinto N et al. MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients. PLoS One 2011;6:e29094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Linden SK, Png CW et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest 2007;117:2313–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G, Nadal I, Medina M et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol 2010;10:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistal E, Caminero A, Vivas S et al. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie 2012;94:1724–1729. [DOI] [PubMed] [Google Scholar]
- Sellitto M, Bai G, Serena G et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS One 2012;7:e33387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal I, Donat E, Ribes-Koninckx C et al. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol 2007;56:1669–1674. [DOI] [PubMed] [Google Scholar]
- Collado MC, Donat E, Ribes-Koninckx C et al. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol 2009;62:264–269. [DOI] [PubMed] [Google Scholar]
- Ou G, Hedberg M, Hörstedt P et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol 2009;104:3058–3067. [DOI] [PubMed] [Google Scholar]
- Di Cagno R, De Angelis M, De Pasquale I et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol 2011;11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistal E, Caminero A, Herrán AR et al. Differences of small intestinal bacteria populations in adults and children with/without celiac disease: effect of age, gluten diet, and disease. Inflamm Bowel Dis 2012;18:649–656. [DOI] [PubMed] [Google Scholar]
- Wacklin P, Kaukinen K, Tuovinen E et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis 2013;19:934–941. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kalliomäki M, Heilig HG et al. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol 2013;13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez E, Donat E, Ribes-Koninckx C et al. Duodenal-mucosal bacteria associated with celiac disease in children. Appl Environ Microbiol 2013;79:5472–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier DM, Riehle K, Mistretta TA et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011;141:1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 2013;9:e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebinger RM, Liu MM, Dowd SE et al. Examination with next-generation sequencing technology of the bacterial microbiota in bronchoalveolar lavage samples after traumatic injury. Surg Infect 2013;14:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzberg K, Le R, Bharti B et al. The personal human oral microbiome obscures the effects of treatment on periodontal disease. PLoS One 2014;9:e86708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio V, Precone V, Casaburi G et al. An altered gut microbiome profile in a child affected by Crohn's disease normalized after nutritional therapy. Am J Gastroenterol 2013;108:851–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P, Austin A, Forsyth J et al. British Society of Gastroenterology guidelines on the diagnosis and management of coeliac disease. Gut 2015;64 (4): 691–692. [DOI] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 2003;55:541–555. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Lavitola A, Salvatore P et al. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol Cell 1999;3:435–445. [DOI] [PubMed] [Google Scholar]
- D'Argenio V, Petrillo M, Cantiello P et al. De novo sequencing and assembly of the whole genome of Novosphingobium sp. strain PP1Y. J Bacteriol 2011;193:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004;5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000;17:540–552. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 2005;21:456–463. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 2001;18:691–699. [DOI] [PubMed] [Google Scholar]
- Ardavin C, Martinez del Hoyo G, Martin P et al. Origin and differentiation of dendritic cells. Trends Immunol 2001;22:691–700. [DOI] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012;487:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinosa MR, Progida C, Talà A et al. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect Immun 2007;75:3594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talà A, De Stefano M, Bucci C et al. Reverse transcriptase-PCR differential display analysis of meningococcal transcripts during infection of human cells: Up-regulation of priA and its role in intracellular replication. BMC Microbiol 2008;8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol 2001;26:32–46. [Google Scholar]
- Evans NJ, Harrison OB, Clow K et al. Variation and molecular evolution of HmbR, the Neisseria meningitidis haemoglobin receptor. Microbiology 2010;156:1384–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauseef I, Harrison OB, Wooldridge KG et al. Influence of the combination and phase variation status of the haemoglobin receptors HmbR and HpuAB on meningococcal virulence. Microbiology 2011;157:1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OB, Bennett JS. Derrick JP et al. Distribution and diversity of the haemoglobin-haptoglobin iron-acquisition systems in pathogenic and non-pathogenic Neisseria. Microbiology 2013;159:1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlen SD. Serratia Infections: from military experiments to current practice. In:. Clin Microbiol Rev 2011;24:755–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P, Laurikka P, Lindfors K et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am J Gastroenterol 2014;109:1933–1941. [DOI] [PubMed] [Google Scholar]
- Caminero A, Nistal E, Herrán AR et al. Differences in gluten metabolism among healthy volunteers, coeliac disease patients and first-degree relatives. Br J Nutr 2015;114:1157–1167. [DOI] [PubMed] [Google Scholar]
- Caminero A, Herrán AR, Nistal E et al. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol 2014;88:309–319. [DOI] [PubMed] [Google Scholar]
- Fernandez-Feo M, Wei G, Blumenkranz G et al. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin Microbiol Infect 2013;19:E386–E394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone MV, Zanzi D, Maglio M et al. Gliadin-mediated proliferation and innate immune activation in celiac disease are due to alterations in vesicular trafficking. PLoS One 2011;6:e17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.