Dear Editor,
Haploid embryonic stem cells (haESCs) have been recently generated from parthenogenetic (PG) or androgenetic (AG) blastocysts of different mammals, including mouse, rat and monkey, enabling genetic screening at both cellular and organism levels1,2,3,4,5,6. However, whether haESCs can be generated from human remains unknown. In this study, we establish protocols for the derivation of stable haESCs from human PG haploid blastocysts obtained by microsugical removal of male pronucleus from fertilized eggs. Human haESCs are pluripotent and sustain typical maternal imprinting status.
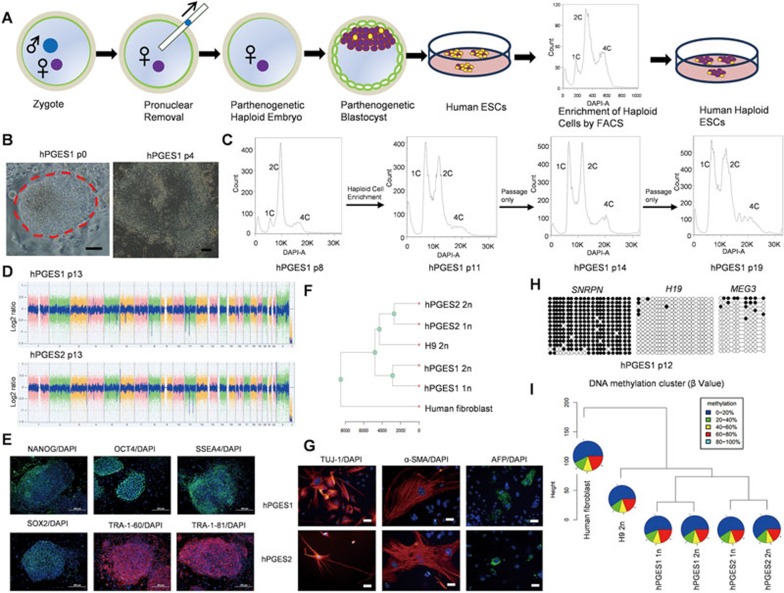
To generate human PG haploid embryos, we adopted two different strategies. In the first set of experiments, we artificially activated human mature metaphase II (MII) oocytes according to a previously reported protocol7 that has been successfully used for generation of monkey PG blastocysts5 and human PG blastocysts from reconstructed oocytes8. A total of 25 cumulus-free MII oocytes from human were treated sequentially with ionomycin, followed by cycloheximide (CHX), resulting in extrusion of the second polar body and development to the blastocyst stage. A total of 10 PG blastocysts were obtained and used for ESC derivation, leading to generation of 3 ESC lines (Supplementary information, Table S1). However, no haploid cells could be enriched by viable fluorescence-activated cell sorting (FACS) with Hoechst 33342 staining in the initial cell sorting at passages 4-8 (Supplementary information, Figure S1). In the second set of experiments, PG haploid blastocysts were generated by removal of the male pronucleus from fertilized oocytes (Figure 1A and Supplementary information, Figure S2A). From 23 manipulated fertilized oocytes, we obtained 6 blastocysts and derived 4 ESC lines. Strikingly, two lines contained substantial haploid cells in the initial cell sorting that could be enriched for haploid cell derivation (Supplementary information, Table S1). After multiple rounds of FACS enrichment of haploid cells following passaging in vitro, two individual PG-haESC lines were successfully obtained (referred to as hPGES1 and hPGES2; Figure 1B, 1C and Supplementary information, Figure S2B and S2C). These cells were cultured in standard human ESC culture conditions for over 30 passages and the haploidy could be well maintained by regular FACS in both cell lines. Karyotyping of hPGES1 and hPGES2 cells revealed that both of them had a haploid set of 23 chromosomes (Supplementary information, Figure S2D). Copy number variation (CNV) analysis of hPGES1 and hPGES2 cells confirmed that both human haploid cell lines sustained genome integrity during haploid cell derivation and maintenance (Figure 1D).
Figure 1.
Derivation of human haploid ESCs from parthenogenetic embryos. (A) Schematic of the generation of human haploid ESCs from parthenogenetic blastocysts obtained after removing male pronucleus from zygotes. (B) Outgrowth (passage 0) and ESC colony (passage 4) of hPGES1 cells. Scale bar, 100 μm. (C) Establishment of hPGES1 cell line after one round of FACS enrichment of haploid cells. A DAPI filter was used to detect signal of Hoechst-stained DNA. High ratio of haploid cells could be detected at passages 11, 14 and 19 after one round of FACS enrichment. (D) CNV analysis of hPGES1 (passage 13) and hPGES2 (passage 13) indicated no major genomic alternations in human haploid cells. (E) Expression of human ESC markers in hPGES1 cells, including NANOG, OCT4, SSEA4, SOX2, TRA-1-60 and TRA-1–81. Shown are the representative immunostaining images of human haploid ESC colonies. Scale bar, 200 μm. (F) Gene expression profiles of human haploid ESCs using RNA-seq analysis. Gene expression profiles were clustered using all expressed genes. (G) Expression of markers of various cells from different germ layers in EBs formed from human haploid ESCs, including TUJ1 (ectoderm), α-SMA (mesoderm) and AFP (endoderm). Scale bar, 50 μm. (H) Methylation state of the DMRs of SNRPN, H19 and MEG3 in hPGES1 (passage 12). Open and filled circles represent unmethylated and methylated CpG sites, respectively. (I) Pie chart of DNA methylation profiles of haploid ESCs based on RRBS. The pies show different methylation levels of CpG sites in different colors.
Human haploid ESCs exhibited typical colony morphology and expressed classical human ESC markers, including alkaline phosphatase, OCT4, NANOG, SSEA4, SOX2, TRA-1-81 and TRA-1-60 (Figure 1E and Supplementary information, Figure S3A-S3C). We next investigated the gene expression profiles of haploid cells by performing RNA sequencing (RNA-seq) analysis. To avoid the influence of different cell cycle stages on the expression profile, we collected samples after FACS of cells in the G1/G0 phase from haploid cells, recently diploidized cells and normal human ESCs from the H9 cell line. Meanwhile, human fibroblasts were used as control. Clustering of these cells showed a high correlation between hPGES cells and the diploid ESCs, but not human somatic cells (Figure 1F and Supplementary information, Figure S3D). As expected, pluripotency genes are highly expressed in both haploid and diploid ESCs but not in somatic cells, while fibroblast-specific genes are expressed only in human fibroblasts (Supplementary information, Figure S3E). To test the differentiation potential of hPGES1 and hPGES2 cells, we suspended cells for the formation of embryoid body (EB) in vitro (Supplementary information, Figure S3F). Immuostaining analysis of EBs formed from hPGES1 and hPGES2 cells showed that both cell lines could differentiate into various cell types originating from all three embryonic germ layers, demonstrated by the expression of AFP (endoderm marker), α-SMA (mesoderm marker) and TUJ1 (ectoderm marker) (Figure 1G). Moreover, teratoma analysis indicated that human haploid ESCs could differentiate into a variety of tissues from all three embryonic germ layers in vivo (Supplementary information, Figure S3G). To determine whether haploidy can be maintained in differentiated cells, we enriched haploid cells from EBs and found that both the undifferentiated and differentiated populations contained haploid cells (Supplementary information, Figure S4). Taken together, these results demonstrate that human haploid ESCs derived from PG blastocysts are pluripotent as normal diploid ESCs.
As hPGES1 and hPGES2 were derived from two individual zygotes, in which, male pronucleus has been removed, their genomes are expected to originate from oocyte donors but not sperm. To test this, we performed short tandem repeat (STR) genotyping using genomic DNA extracted from haploid cells and zygote donor parents. As expected, both hPGES1 and hPGES2 cells were precisely inherited from the corresponding female pronucleus donors (Supplementary information, Table S2). Having confirmed their oocyte origin, we next examined whether maternal imprints are maintained in human haploid ESCs. We first compared the expression of imprinted genes in hPGES1 and hPGES2 cells and diploid cells from RNA-seq results. As expected, both cell lines sustained a typical maternal imprinting status, demonstrated by upregulation of paternally imprinted genes and downregulation of maternally imprinted genes in haploid cells (Supplementary information, Figure S5A). We further assessed epigenetic inheritance by performing bisulfite-sequencing analysis to determine the DNA methylation profiles of differentially methylated regions (DMRs) of one maternally imprinted gene, Snrpn, and two paternally imprinted genes, H19 and Meg3. The results showed that, as expected, the DMRs of H19 and Meg3 were free of methylation while DMR of Snrpn largely retained methylation in both cell lines (Figure 1H). Interestingly, different from mouse PG-haESCs, in which, DNA methylation was quickly lost at maternally imprinted loci during haploid ESC derivation and in vitro culture9, hPGES1 and hPGES2 cells maintained a stable maternal imprinting state in early and late passages (Supplementary information, Figure S5B), consistent with previous observations in monkey PG-haESCs5. Finally, we investigated the methylation profile of all genes in haploid cells and control diploid cells by performing whole-genome bisulfite sequencing (WGBS) analysis using DNA samples extracted from the cells that we prepared for RNA-seq analysis. The results, based on the methylation patterns of all detected CpGs and the DMRs of imprinted genes, indicated that hPGES1 and hPGES2 cells sustained a typical maternal imprinting state (Figure 1I and Supplementary information, Figure S5C and S5D).
In summary, we have demonstrated that human haploid ESCs can be generated from PG embryos obtained after removal of male pronucleus from zygotes. During the preparation of this work for publication, an independent study also reported the generation of human PG haploid ESCs following activation of oocytes10. Interestingly, similar to our observations when we used the same strategy to obtain haploid embryos, the authors observed no obvious 1c peak at the first sorting, and therefore sorted sub-2c cells for further maintenance and haploid cell line derivation. By contrast, our method by removal of male pronucleus can result in a large amount of haploid cells from the first round of FACS enrichment, leading to efficient derivation of haploid cell lines (2 of 4 derived cell lines). One potential reason could be that sperm-mediated oocyte activation resulted in better embryonic development with high ratio of haploid inner cell mass than chemically induced activation. Comprehensive comparative analysis is needed in future to reveal the underlying differences between human haploid ESCs generated by these two approaches. The establishment of stable human haploid ESC lines thus provides a novel tool for genetic analysis in human.
Acknowledgments
We thank W Bian, X Wang and F Zhang from Cell Biology Facility for support with cell sorting, Y Chen and X Ding from the Stem Cell Core Facility at SIBCB for support with cell cultures, H Li, M Li, H Deng, R Hou and H Xiao from Shanghai Biochip Company for sequencing data analysis, and Y Jin and Y Zeng from SIBS for providing antibodies. This study was supported by the Ministry of Science and Technology of China (2014CB964803, 2012CB944700, 2015AA020307 and 2013CB967103) and the National Natural Science Foundation of China (91319310, 31225017, 31530048, 81430029, 81490743, 31371453 and 31571548).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary information
References
- Leeb M, Wutz A. Nature 2011; 479:131–134. [DOI] [PMC free article] [PubMed]
- Elling U, Taubenschmid J, Wirnsberger G, et al. Cell Stem Cell 2011; 9:563–574. [DOI] [PMC free article] [PubMed]
- Yang H, Shi L, Wang BA, et al. Cell 2012; 149:605–617. [DOI] [PubMed]
- Li W, Li X, Li T, et al. Cell Stem Cell 2014; 14:404–414. [DOI] [PubMed]
- Yang H, Liu Z, Ma Y, et al. Cell Res 2013; 23:1187–1200. [DOI] [PMC free article] [PubMed]
- Zhong C, Yin Q, Xie Z, et al. Cell Stem Cell 2015; 17:221–232. [DOI] [PubMed]
- Mitalipov SM, Nusser KD, Wolf DP. Biol Reprod 2001; 65:253–259. [DOI] [PubMed]
- Paull D, Emmanuele V, Weiss KA, et al. Nature 2013; 493:632–637. [DOI] [PMC free article] [PubMed]
- Zhong C, Xie Z, Yin Q, et al. Cell Res 2016; 26:131–134. [DOI] [PMC free article] [PubMed]
- Sagi I, Chia G, Golan-Lev T, et al. Nature 2016; 532:107–111. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information