Abstract
Tenomodulin (TNMD, Tnmd) is a gene highly expressed in tendon known to be important for tendon maturation with key implications for the residing tendon stem/progenitor cells as well as for the regulation of endothelial cell migration in chordae tendineae cordis in the heart and in experimental tumour models. This review aims at providing an encompassing overview of this gene and its protein. In addition, its known expression pattern as well as putative signalling pathways will be described. A chronological overview of the discovered functions of this gene in tendon and other tissues and cells is provided as well as its use as a tendon and ligament lineage marker is assessed in detail and discussed. Last, information about the possible connections between TNMD genomic mutations and mRNA expression to various diseases is delivered. Taken together this review offers a solid synopsis on the up-to-date information available about TNMD and aids at directing and focusing the future research to fully uncover the roles and implications of this interesting gene.
Keywords: tenomodulin, BRICHOS, tendon, eye, chordae tendineae cordis, vasculature, periodontal ligament, knockout mice, tendon stem/progenitor cells, cell differentiation, gene marker, single nucleotide polymorphism, obesity, metabolic syndrome
2. Introduction
Tendons are dense connective tissues arranged in a hierarchical manner. Mature tendons are normally characterized by low cellular density and this is one of the most obvious features when looking at tendon histological preparations. About 90-95% of the cellular content of tendon comprises tendon-specific cell types described in the literature as tenoblasts and tenocytes, the latter being the terminally differentiated form (Kannus 1997, Chuen et al. 2004). During development new-born tendons have a very high cell-to-matrix ratio with tenoblasts of various shapes and sizes aligned in long parallel chains. Following tendon maturation, the ratio between cells and matrix gradually decreases, with tenoblasts transforming from an ovoid to a spindle and elongated shape, specific for their differentiated counterparts, tenocytes (Ippolito et al. 1980).
Although the general knowledge about the differentiated cells residing in tendon tissue has been developing in the latest years, still little is known about their precursors. Stem/progenitor cells of mesenchymal origin, such as the tendon lineage, are of great interest in order to understand tendon development and healing processes. In 2007 (Bi et al. 2007) demonstrated that human and mouse tendons harbour a unique cell population which has universal stem cell characteristics such as clonogenicity, self-renewal capacity and multipotency. Compared to bone marrow stromal stem cells, these tendon-derived cells expressed high levels of Scleraxis (SCX), cartilage oligomeric matrix protein (COMP), Tenascin-C and tenomodulin (TNMD), all tendon-related factors, thus identifying their origin. Additionally, these isolated cells showed the ability to regenerate tendon-like tissue after extended expansion in vitro and transplantation in vivo. However, the fact that the cells of this population showed heterogeneity in their stem properties and the possibility of containing tendon progenitor cells as well, made the authors name them-tendon stem/progenitor cells (TSPCs).
The repair of musculoskeletal tissues often recapitulates the cellular and molecular events of development. Thus, understanding the process of tendon development can significantly help to develop novel repair strategies. So far, the knowledge on the ontogeny of the tendon lags far behind other mesodermal tissues due to both the lack of specific markers exclusive to the tendon lineage. However, the identification of the basic helix-loop-helix (bHLH) transcription factor Scx as a specific and early marker of tendon progenitors during embryonic development (Cserjesi et al. 1995, Schweitzer et al. 2001) and the study of mice harbouring genetic mutations leading to tendon phenotypes, reviewed in (Tozer and Duprez 2005, Liu et al. 2011), have provided some insights into the onset of the tendon lineage and the molecules involved in this process. Using knockout mouse models, further transcription factors were identified to be essential for tendon differentiation and development such as Mohawk (Mkx), Egr1 and Egr2 (Ito et al. 2010, Lejard et al. 2011, Guerquin et al. 2013).
Tenomodulin was discovered in 2001 by (Brandau et al. 2001) and (Shukunami et al. 2001) as a gene sharing high homology with chondromodulin-1 (CHM1, alternative names chondromodulin-1 and alternative abbreviations, CHM-I, LECT1, BRICD3 and MYETS1) (Hiraki et al. 1991). Both research groups described high expression in tendons, explaining the rationale behind its name. It was hinted to be useful as a tendon specific marker, which later on became an established marker for the mature tendon and ligamentous lineage.
Despite great progress in deciphering tendon development, the exact molecular pathways orchestrating tendon progenitor specification and differentiation still remain largely unknown. Therefore, further studies are required to identify novel tendon-specific markers, to understand their roles and to elucidate the molecular cascades occurring during tendon development and maintenance.
In view of the above, the focus in this review is on TNMD one of the best so far tendon-specific marker genes. We carried out a systematic review analysis on all available articles in the PubMed databank. The examination was performed by searching tenomodulin under its full name, but also by its alternative names and abbreviations (tendin and myodulin, TNMD and TeM). A summary of the selection of articles for this review is shown in Fig. 1.
Fig. 1.
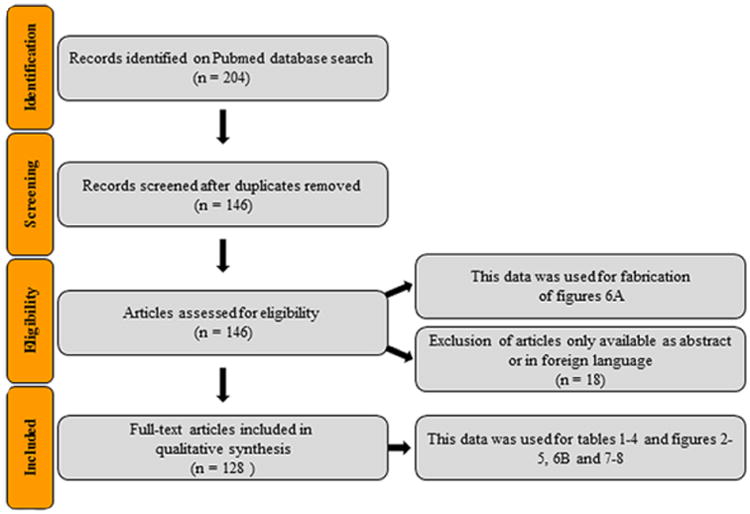
Flow chart of the search strategy and study selection used in this systematic review.
3. Tenomodulin
3.1. Gene discovery and nomenclature
TNMD was found independently by (Brandau et al. 2001) and (Shukunami et al. 2001). Brandau's team named it tendin with the justification that it is highly expressed in tendons and ligaments. Shukunami's team, on the other hand named it tenomodulin, since it shares high homology to chondromodulin-I, but is expressed in tendons, hence postulating a similar modulatory function in these tissues. Tenomodulin also circulated around the literature very briefly under the name of myodulin (Pisani et al. 2004, Pisani et al. 2005). The authors gave the alternative name myodulin based on detecting Tnmd mRNA in Northern blot analysis of rat gastrocnemius muscles. As previously mentioned, its abbreviations in the literature are found under TNMD and TeM.
3.2. Gene and protein structure
TNMD, Tnmd belongs to the new family of type II transmembrane glycoproteins with a highly conserved cleavable C-terminal cysteine-rich domain (Brandau et al. 2001, Shukunami et al. 2001). The TNMD, Tnmd gene consists of seven exons localized on the X chromosome (Fig. 2) and accounts for an approximately 1.4 kb transcript and a predicted protein consisting of 317 amino acids (Brandau et al. 2001, Shukunami et al. 2001). Analysis of primary amino acid sequences reveals several structural features in the putative TNMD, Tnmd protein (Fig. 3).
Fig. 2.
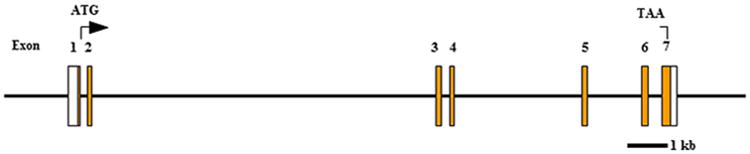
Schematic representation of the human TNMD gene.
White boxes represent 5′ and 3′ UTR sites, while orange boxes represent exons. Abbreviations: ATG, start codon; kb, kilo base; TAA, stop codon.
Fig. 3.
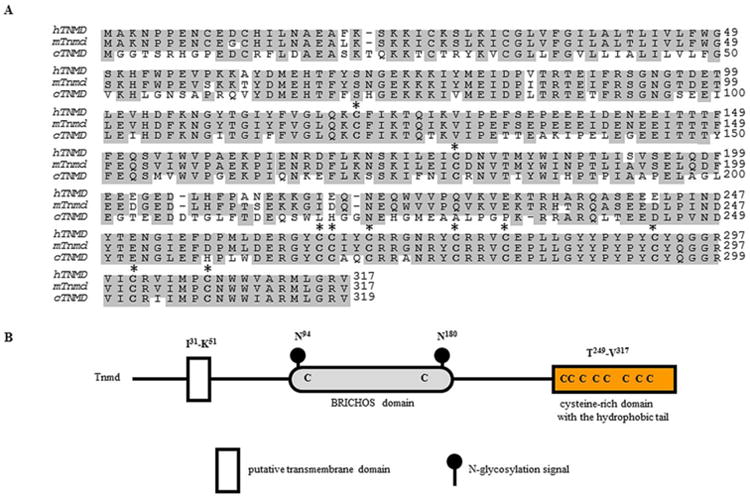
A comparison of the deduced amino acid sequences of tenomodulin proteins and structural features of the human TNMD. (A) Predicted amino acid sequences of human (Shukunami et al. 2001, and Yamana et al. 2001), mouse (Brandau et al. 2001, Shukunami et al. 2001, and Yamana et al. 2001), and chick (Shukunami et al. 2006) tenomodulin. The conserved amino acid residues are shaded and gaps were introduced for optimal alignment. Conserved cysteine residues are indicated in bold with an asterisk. The chicken sequence shares 62% homology with the mouse and human orthologs. The conserved amino.GenBank accession numbers for the aligned sequences are as follows: human TNMD, AF234259.1; mouse Tnmd, AF219993.1; chick TNMD, AY156693. (B) Human TNMD protein includes a type II transmembrane domain at the N terminus, a BRICHOS domain (Sanchez-Pulido et al. 2002) and a C-terminal cysteine-rich domain. The TNMD protein contains two N-glycosylation sites within the BRICHOS domain. Abbreviations: C, cysteine; cTnmd; chicken tenomodulin; hTNMD, human tenomodulin; I, isoleucine; K, lysine; mTnmd, mouse tenomodulin; N, asparagine; Q, glutamine; V, valine; TNMD, Tnmd, tenomodulin.
The gene is composed of seven exons among which the second encodes for the transmembrane domain and the last for the C-terminal cysteine-rich domain that is conserved across species (Fig. 3A). Unlike the Chm1 precursor, the Tnmd protein does not include a furin cleavage site, although a putative protease recognition sequence (Arg-Xxx-Xxx-Arg) was identified at the position 233-236 (Barr 1991, Shukunami et al. 2005). The extracellular part, prior the cleavage site, contains a BRICHOS extracellular domain (Fig. 3B). This domain consists of a homologous sequence of approximately 100 amino acids containing a pair of conserved cysteine residues (Sanchez-Pulido et al. 2002). BRICHOS has been found in several un-related to Tnmd genes involved in dementia, respiratory distress and cancer. It has been suggested that it participates in post-translational processing, however the function remains unclear (Sanchez-Pulido et al. 2002). In contrast to Chm1, which has two N-glycosylation sties and one O-glycosylation site (Neame et al. 1990), Tnmd is absent of these three sites, while it contains two N-glycosylation sites at position 94 and 180 (Brandau et al. 2001). Protein analyses in eye and periodontal ligament revealed full length Tnmd protein as a double band of 40 and 45 kDa (Barr 1991, Shukunami et al. 2005, Komiyama et al. 2013). It has been experimentally proven that the 45 kDa band corresponds to the glycosylated Tnmd, while the 40 kDa band refers to the non-glycosylated Tnmd (Barr 1991, Shukunami et al. 2005).
Tnmd contains a highly conserved C-terminal cysteine-rich domain (Shukunami et al. 2001), making up the part of the protein sharing the highest resemblance to Chm1 (77% similarity/66% identity) (Brandau et al. 2001). This domain contains C-terminal hydrophobic tail with eight Cys residues forming four disulphide-bridges, which are well conserved across vertebrate species (Shukunami et al. 2005, Shukunami et al. 2006, Kondo et al. 2011) as shown in Fig. 3. A smaller cyclic structure forming by single Cys280-Cys292 disulphide bridge in tenomodulin has been shown to exert an anti-angiogenic function, while the other three disulphide-bridges are speculated to hold this cyclic structure and the C-terminal hydrophobic tail separated from each other to avoid the formation of intramolecular aggregates (Miura et al. 2012). In certain tendon tissues, such as Achilles tendon and chordae tendineae cordis (CTCs), 16 kDa cleaved C-terminal part of Tnmd was detected in the collagenous extracellular matrix, which suggested tissue-specific Tnmd secretion (Docheva et al. 2005, Kimura et al. 2008).
Since TNMD and its homolog CHM1 are a novel class of proteins it is very relevant to clarify the existence of isoforms as well as their exact protein structure and function. A study dealing with human tendon cells and tissues, and using the I-TASSER molecular modelling program, has suggested that the TNMD gene has 3 possible isoforms I, II and III with molecular weights of 37.1, 20.3 and 25.4 kDa, respectively (Qi et al. 2012). However, further validation by carrying out Northern blot analyses as well as specific PCR designed to clearly discriminate and demonstrate individual isoforms is necessary. With regards to mouse, (Brandau et al. 2001) (Northern blot analyses on multiple mouse tissues with Tnmd cDNA probe for exons 4-7) and (Shukunami et al. 2001) (Northern blot with 8 different mouse tissues and cDNA probe encoding two thirds of the entire coding region) did not detect Tnmd isoforms. Similar findings were observed in Northern blot investigations on samples from wild type and knockout mice with full cDNA probe and western blotting analyses of mouse tissues and anti-Tnmd antibodies (Docheva et al. 2005, Shukunami et al. 2006, Takimoto et al. 2012, Komiyama et al. 2013). This data suggests that TNMD isoforms might be species-specific. Interestingly, the program-based study predicted for each isoform a very different protein function: isoform I to be a cytosine methyltransferase; isoform II a SUMO-1-like SENP-1 protease; and isoform III to be an α-syntrophin, pleckstrin homology domain scaffolding protein. A possible explanation behind the suggestion that each isoform functions entirely different could be that the novel and unstudied protein domain structures of TNMD and CHM1 causes the available prediction platforms to search for minimal homology scores, hence identifying totally different proteins. Despite of the many open questions left, the study of Qi et al. 2012 strongly urges for further analyses on the TNMD isoforms and exact protein domain structure and function.
3.3. Expression pattern
TNMD is a protein predominantly expressed in tendons and ligaments. The first northern blot analysis on new-born mouse tissue showed the highest expression of Tnmd in skeletal muscle (Shukunami et al. 2001), diaphragm and eye, even though an overall weak signal was visible in almost all other screened tissues (Brandau et al. 2001). Following in situ hybridization and western blotting studies revealed that the signal from muscle tissue is mainly due to expression of Tnmd in tendons and ligaments as well as in the muscle sheet epimysium (Brandau et al. 2001, Docheva et al. 2005, Kimura et al. 2008, Komiyama et al. 2013, Sato et al. 2014). In a temporal expression analysis in mice the presence of the Tnmd transcript was weekly detected already at embryonic day E9.5 in developing limb buds (Brandau et al. 2001); however a recent paper has firmly reported very strong Tnmd mRNA expression at day E14.5 corresponding to the differentiation stage of tendon progenitors (Havis et al. 2014). Furthermore, also tendon-like structures in the heart, namely chordae tendineae cordis, demonstrated Tnmd messenger and protein existence in these areas (Kimura et al. 2008). Interestingly, several papers have found Tnmd expression in the maxilla-facial region. Komiyama et al. 2013 reported high Tnmd expression, by immunohistochemical analysis, in periodontal ligaments at 3 and 4 postnatal weeks, marking the molar eruptive and post-eruptive phases when occlusal forces are transferred to the teeth and the surrounding tissues. Watahiki et al. 2008 discovered a specific expression of Tnmd in mandibular condylar cartilage at 1 week in rats. The masseter muscle is compartmentalized by a laminar structure, which was shown to express by in situ hybridization Tnmd in mouse embryos at E12.5 to E17.5 ((Sato et al. 2014)). Furthermore, Oshima et al. 2003 revealed Tnmd expression at E15.5 in skin and eye using Northern blotting. The exact localization of Tnmd mRNA in the eye was shown by in situ hybridization in the sclerocornea, tendon of the extraocular muscle, ganglion cell layer, lens fibre cells, inner nuclear layer cells and pigment epithelium of the retina (Oshima et al. 2003). TNMD expression was also found by PCR in human subcutaneous adipose tissue and adipocytes (Saiki et al. 2009). Last, in situ hybridization showed Tnmd expression in various parts of the adult mouse brain such as the dentate gyrus, CA regions of the hippocampus, neurons in the cerebral nuclei, cerebellum, Purkinje cells and neuronal cells in the cerebellar nucleus (Brandau et al. 2001). However, a general notion should be taken that Tnmd expression outside dense connective tissues, such as tendons and ligaments, is very low. The main expression sites of TNMD, Tnmd are summarised in Table. 1.
Table 1.
Expression pattern of TNMD, Tnmd.
| Species | Experimental source | Methods | Expression domains | References |
|---|---|---|---|---|
| Human | Musculoskeletal tissues | Real-time PCR | Tendon, fat, muscle in | Jelinsky et al., 2010 |
| CTC from patients | IHC | Elastin-rich mid layer of CTC | Kimura et al., 2008 | |
| Adipose tissue | DNA-MA, Real-time PCR | Lean and obese adipose subcutaneous tissue, more specifically mammary glands, omental fat, tongue superior part and main corpus, but also urethra, coronary artery, oral mucosa, ovary, skeletal muscle, bone marrow, colon, substantia nigra and pituitary gland | Saiki et al., 2009 | |
| Cultured cells | ICC | Bone marrow-derived MSCs expressing Scx | Alberton et al., 2012 | |
| PCR | TSPCs | Bi et al., 2007 | ||
| Porcine | Pig heart and eyes | RT-PCR, WB | Chordae and eye | Kimura et al., 2008 |
| Rabbit | 1-week-old Japanese white rabbits | ISH | Achilles tendon | Yukata et al., 2010 |
| 12-week-old Japanese white rabbits | PCR | CTC interstitial cells | Kimura et al., 2008 | |
| Rat | Rotator cuff healing model in 19-21-week-old rats | Real-time PCR ISH |
Healing tissues at 2-, 4-, 6-, 8-, 12-weeks, but significantly upregulated in animals treated with FGF-2 at weeks 4, 6, 8 and 12 compared to normally healing tendons Midsubstance of tendon at 2- and 8-weeks |
Tokunaga et al., 2015 |
| Achilles tendon healing model with 9-10-week-old rats | RT-PCR | Upregulation in the normally loaded Achilles tendon compared to the animal where normal loading of the muscle-tendon unit was prevented by intramuscular Botox injection | Eliasson et al., 2009 | |
| Mouse | Embryos | NB | Whole embryos at E15.5 and E17.5 | Oshima et al., 2003 |
| High expression in skeletal muscle, diaphragm and eye, weak signals in brain, lung, liver, heart, kidney, skin, thymus and perichondrium- and periosteum-free rib cartilage | Brandau et al., 2001 | |||
| IHC | Secondary lens fibres and retina at E16.5 | Oshima et al., 2003 | ||
| ISH | Tendon of extrinsic ocular muscle, sclera, equatorial lens, epithelial cells at E16.5 Tendons at E14.5, E15.5, and E16.5 | Oshima et al., 2003 | ||
| CD31- negative regions of tendons at E16.5. Strong signals in limb tendons and cruciate ligaments, tendons of the masculus flexor digitorum, ligament intercostal, diaphragm, cortex, medulla of the thymus at E17.5 | Shukunami et al., 2008 | |||
| PCR | Heart from E14.5 to E18.5 | Kimura et al., 2008 | ||
| New-born | IHC | Patellar ligament, anterior and posterior cruciate ligament, Achilles tendon of Sox9Cre/+;R26R | Sugimoto et al., 2013 | |
| RT-PCR | Heart | Kimura et al., 2008 | ||
| 2- and 3-week-old mice | IHC | Peridontal ligaments | Komiyama et al, 2013 | |
| 4-week-old mice | NB/WB | Whole eye, skin | Oshima et al., 2003 | |
| PCR | Whole eye, cornea, retina, sclera with ocular muscles | |||
| ISH | Cornea, sclera, retina | |||
| Tendon, epimysium of skeletal muscle | Shukunami et al., 2001 | |||
| WB | Skeletal muscle | Oshima et al., 2004 | ||
| IHC | Achilles tendon | Docheva et al., 2005 | ||
| NB | High in tendon, low in eye and muscle | |||
| Cultured cells | PCR/IHC | TSPC | Bi et al., 2007; Alberton et al., 2015 | |
| Chick | Embryos (from HH stage 23-41) | ISH | Tendon primordia and tendons | Shukunami et al., 2006 |
| Tendon primordia and tendons, ligaments and perichondrium | Shukunami et al., 2008 | |||
| Hind limb tendons at HH stage 30 | Takimoto et al, 2012 | |||
| NB | Whole embryos from HH stage 25 to stage 35, tendon, skeletal muscle, heart, skin and eye at HH stage 45 | |||
| Cultured cells | NB | Tenocytes isolated from leg tendons at HH stage 41 | Takimoto et al, 2012 |
Abbreviations: CTC, chordae tendineae cordis; DNA, deoxyribonucleic acid; E, embryonic day; HC, immunohistochemistry; HH, Hamburger Hamilton; ICC, immunocytochemistry; ISH, in situ hybridization; MA, microarray; MSC, mesenchymal stem cell; NB, Northern blot; PCR, polymerase chain reaction; RT, reverse transcriptase; Scx, scleraxis; TSPC, tendon stem/progenitor cell; WB, western blot.
3.4. Putative signalling pathway
TNMD is an established marker for the mature tendon and ligament lineage (Shukunami et al. 2006) due to its high expression in these tissues, segregating uncommitted from committed cells of this lineage. In order to decipher upstream and downstream effectors of Tnmd many knockout mouse models with tendon phenotypes have helped in understanding which factors or pathways affect Tnmd. Similarly, the loss of Tnmd allows suggestions on which molecules might be downstream effectors. It is important to emphasise that the most of the following studies show correlations between Tnmd expression or function to other genes and not a direct link in a common signalling cascade. In Fig. 4, we have summarised the so far reported, mostly indirect, links between Tnmd and putative upstream and downstream factors.
Fig. 4.
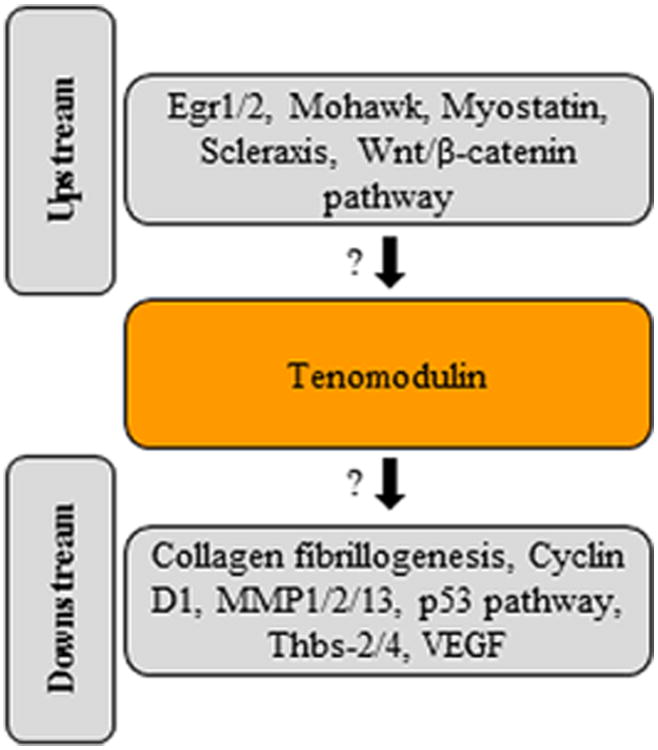
Summary of putative upstream and downstream factors in Tnmd-related signalling. The figure is based on studies showing mostly correlations, indicated by the question marks, between TNMD expression or function to other genes and not a direct link in a common signalling cascade. Based on Alberton et al. 2012; Alberton et al. 2015; Brent et al. 2005; Docheva et al. 2005; Frolova et al. 2014; Kimura et al. 2008; Kyriakides et al. 1998; Lejard et al. 2011; Liu et al. 2010; Mendias et al. 2008; Miyabara et al. 2014; Murchison et al. 2007; Shukunami et al. 2006. Abbreviations: Egr, early growth response protein; MMP, matrix metalloproteinase; Thbs, thrombospondin; VEGF, vascular endothelial growth factor.
Brent et al. 2005 reported that in Sox5 and Sox6 double knockout mice, where skeletal elements failed to undergo normal chondrogenic differentiation, the domain of Scx is broadened and accompanied by the expression of Tnmd. These observations proposed that Tnmd might be a direct target of Scx transcriptional activities. Towards the same suggestion led also an in vitro experiment where over-expression of Scx triggered the up-regulation of Tnmd in cultured chicken tenocytes (Shukunami et al. 2006). Additional evidence came from the study of Murchison et al. 2007 which reported the phenotype of the Scx knockout mice (Murchison et al. 2007). These mice are characterized by severe tendon defects, ranging from a dramatic failure in progenitor differentiation to the formation of small and poorly organized force transmitting tendons. Interestingly, in these mice the screening of the expression of several genes known to be expressed in tendons revealed the complete loss of expression of Tnmd and type XIV collagen at E16.5 (Murchison et al. 2007). Another study in 2012 validated that by overexpression of Scx in mesenchymal stem cells Tnmd expression is significantly upregulated in a cell type which normally does not express Scx nor Tnmd (Alberton et al. 2012).
Another factor participating in Tnmd signalling is the growth differentiation factor myostatin. The deletion of myostatin in mice resulted in small, brittle and hypocellular tendons (Mendias et al. 2008), a phenotype similar, in term of cell density, to the one found in the Tnmd-deficient mice (Docheva et al. 2005). Moreover, these mice showed a decrease in Scx and Tnmd expression (Mendias et al. 2008) and myostatin stimulation of fibroblasts led to Scx and Tnmd mRNA upregulation, suggesting myostatin as an upstream factor in the Tnmd pathway by first inducing Scx expression.
The expression of an Egr-like transcription factor (Frommer et al. 1996) in tendon precursor cells of Drosophila motivated the generation of Egr1 and Egr2 mutant mice and chicks in 2011 for investigation of their involvement in vertebrate tendon development (Lejard et al. 2011). This study concluded that Egr1 and Egr2 expression in tendon cells was associated with the upregulation of Col1a1 from E11.5 onwards by trans-activating its proximal promoter as well as other collagens in tendon cell differentiation in embryonic limbs. Furthermore, the authors established that Egr genes were activated by muscle-derived FGF4 and are able to induce de novo expression of Scx. Moreover, both Egr1 and Egr2 knockouts were characterized by reduced Col1a1 transcripts together with decreased number of collagen fibrils (Lejard et al. 2011). It would be interesting to investigate if loss of Egr1/2 also affects Tnmd expression.
The absence of the Mohwak (Mkx) homebox gene in mouse led to an abnormal tendon phenotype including an effect on Tnmd expression (Liu et al. 2010). These mice exhibited tendons with inferior size compared to normal mice. On a molecular level, Mkx mutants revealed significantly lower Tnmd levels as well as collagen I and fibromodulin (Liu et al. 2010). Tnmd mRNA was downregulated in Scx and Mkx mutant mice around E16.5 (Murchison et al. 2007, Liu et al. 2010). Interestingly, Scx mutant mice show tendon defects as early as E13.5, while the tendon phenotype of Mkx mutants is first visible at E16.5 despite that Scx expression is sustained in these animals (Murchison et al. 2007, Liu et al. 2010). This clearly suggests that in tendon development Scx acts prior to Mkx, but with regards to regulation of Tnmd both Scx and Mkx can perform independently from each other as upstream factors.
The use of gene silencing experiments was valuable for understanding why bone marrow-derived stem cells (BMDSCs) from horse, which in culture have very low amounts of TNMD compared to tendons, showed similar TNMD expression when cultured in collagen gels containing a glycogen synthase kinase-3 (GSK-3) inhibitor (Miyabara et al. 2014). Application of inhibitor or small interference RNA corresponding to GSK-3α/β into BMDSCs resulted in nuclear translocation of β-catenin and this drove the expression of TNMD. Since the levels of Scx and Mkx were unaffected in the above conditions, it points out that the Wnt/β-catenin signalling works independent from these transcription factors (Miyabara et al. 2014).
Lastly, a Tnmd knockout mouse model study revealed that loss of Tnmd results in reduced proliferation and earlier onset of senescence in TSPCs (Alberton et al. 2015). The study compared tenogenic markers in both genotypes of which no differences were observed, except for the absence of Tnmd transcript in mutant mice. However, analysis of TSPCs self-renewal showed that Tnmd knockout TSPCs had significantly lower clonogenic ability as well as proliferated less after passage 3 and reached an earlier plateau compared to wildtype TSPCs. This was further accompanied with significant downregulation of the proliferative marker Cyclin D1 in Tnmd knockout TSPCs. Another striking feature of the Tnmd knockout TSPCs is that at an earlier passage they show signs of senescence, revealed by β-galatosidase staining, and the number of senescent cells was always larger in the knockouts. In addition a profound upregulation of p53 was detected in the knockout cells. Taken together, these findings suggest that loss of Tnmd results in reduced proliferation and premature ageing of the TSPCs (Alberton et al. 2015) and secondary to this there is a distorted gene expression balance (Cyclin D1 and p53). This finding is in line with the analyses of the Tnmd knockout mouse model, which revealed reduced cellular density and proliferation in vivo (Docheva et al. 2005). In addition, Tnmd mutants showed an abnormal collagen fibril phenotype with pathologically thicker fibrils, resembling signs of premature tendon matrix aging. In sum, it can be suggested that collagen type I, proliferation and senescent-related factors are belonging to the putative downstream effectors. Still, at this stage of the research it is not possible to clarify how exactly Tnmd is regulating the above factors. Since Tnmd shares high homology with Chm1, it was obvious to actually consider Chm1 as a possible compensatory factor. However, the study of Tnmd expression in Chm1 null mice and vice versa of Chm1 expression in Tnmd-deficient mice, showed no upregulation of neither of the genes, suggesting that the loss of these genes is likely to be compensated by other factors (Brandau et al. 2002, Docheva et al. 2005).
Interestingly, both Thrombospondin-2 (Thbs-2) and Thbs-4 knockout mice both display tendon phenotypes consisting of abnormally large collagen fibrils (Kyriakides et al. 1998, Frolova et al. 2014), a similar outcome to the collagen fibril phenotype of Tnmd deficient mice. These observations are open invitations for exploring if Thbs members and Tnmd are participating or connected in the same molecular cascade.
Last, the study analysing ruptures of human CTCs revealed that in the affected area TNMD expression is downregulated, but VEGF-A and several MMP (MMP1, 2 and 13) expressions are upregulated. Furthermore, tube formation and mobilization of human coronary artery endothelial cells were dramatically inhibited when treated with conditioned media from CTC or NIH3T3 cells that were transfected with the Tnmd C-terminal domain. On the contrary, when cultured with conditioned media from CTC cells treated with Tnmd siRNA their capability forming ability was recovered. Hence these experimental findings confirm an antiangiogenic role of Tnmd in CTCs (Kimura et al. 2008).
We strongly stress that in future research it would be essential to decipher what are the exact binding partners of Tnmd. This may help us to explain, for example, opposing effects such as stimulating the proliferation of tendon-derived cells, but inhibiting vascular cells. At present, we speculate that Tnmd might be a co-factor regulating the function of a growth factor or growth factor receptor and hence, depending on the availability, act as a stimulator or inhibitor.
3.5. Tenogenic differentiation cascade
The up-to-date paradigm underlining the putative tendon cell commitment and differentiation process is still very controversial and not fully understood; however the involvement of Tnmd in several steps of this process became very apparent in recent studies. Fig. 5 represents the molecular orchestra of tendon cell differentiation and importantly, it marks the participation of Tnmd in specific commitment steps. In early tendon development, embryonic mesenchymal progenitors at E10.5 commit into the tendon lineage by first elevating the Scx transcription factor in developing sclerotomes of the somites, mesenchymal cells in the body wall and limb buds in mouse embryo (Brent et al. 2003). Furthermore, follow up studies suggested that the initial Scx expression is mediated by transcription factors Pea3 and Erm, which are activated by FGF signalling (Brent and Tabin 2004, Eloy-Trinquet et al. 2009). Among the FGF family the critical members in mice are FGFs 4 and 6, and in chick FGFs 4 and 8. Pryce et al. 2009 reported that in addition to FGFs, TGFβ (Tgfβ) signalling is also critical in tendon progenitor differentiation via direct induction of Scx expression. In this paper, the tendon phenotypes of Tgfb2–/–;Tgfb3–/– double mutant, or the corresponding receptors, revealed that TGFβ signalling acts later than FGF signalling since the tendon phenotype was first manifested at E12.5, while the tendon condensation and Scx-positive tendon cells were normal at E11.5.
Fig. 5.
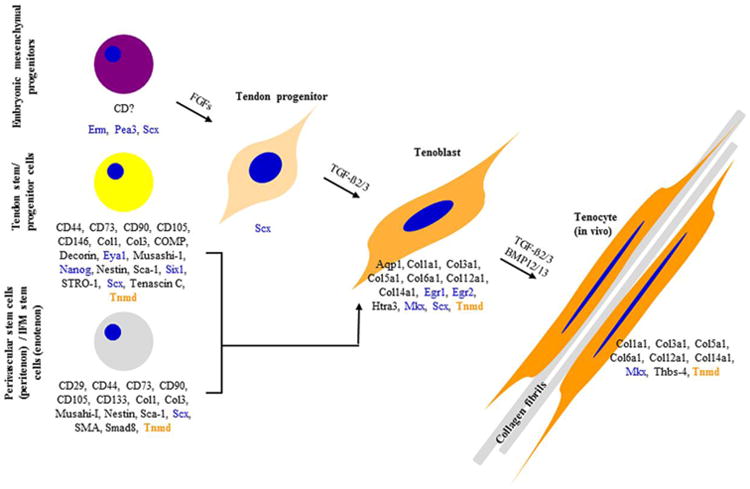
Current model of the tenogenic cascade and TNMD involvement. TNMD is marked in orange, transcription factors in blue and other genes in black. Based on Alberton et al. 2015; Berasi et al. 2011; Eloy-Trinquet et al. 2009; Havis et al. 2014; Huang et al. 2015; Lee et al. 2015; Liu et al. 2014; Mienaltowski et al. 2013; Pryce et al. 2009; Shen et al. 2013; Tempfer et al. 2009. Abbreviations: Aqp1, aquaporin 1; BMP, bone morphogenetic protein; Col, collagen; COMP, cartilage oligomeric matrix protein; Egr, early growth response protein; Eya, eyes absent transcription factor; FGF, fibroblast growth factor; Htra3, HtrA serine peptidase 3; IFM, interfascicular matrix; Mkx, Mohawk; Sca-1, stem cells antigen-1; Scx, scleraxis; SMA, smooth muscle actin; TGF, transforming growth factor; Thbs, thrombospondin; Tnmd, tenomodulin.
Next, a breakthrough paper based on transcriptome analysis comparing expression profiles of a variety of genes in ScxGFP-positive tendon cells derived from different embryonic tendon development stages, resulted in more clear picture of the molecular events during tendon cell differentiation (Havis et al. 2014). By comparing cells from E11.5, corresponding to tendon progenitors, to cells from E14.5, corresponding to tendon differentiated cells (tenoblast), the authors found a significant upregulation of Tnmd, Col1a1, Col3a1, Col5a1, Col6a1, Col12a1 and Col14a1 and two novel and unrelated so far to tendon genes, namely aquaporin 1 (Aqp1) and HtrA serine peptidase 3 (Htra3). With regards to the further maturation of tenoblasts towards tenocytes the following studies suggested the essential contribution of the Tnmd, Mkx, Erg 1/2, and Thbs-4 (Docheva et al. 2005, Liu et al. 2010, Lejard et al. 2011, Alberton et al. 2012, Barsby et al. 2014, Onizuka et al. 2014). The Scx expression in tenocytes remains at present is debatable. Scx signals are visible in the mature cells in ScxGFP reporter mice (Sugimoto et al., 2013). However, from this transgenic model is not clear how strong the levels of endogenous Scx in these cells are.
With regards to tendon differentiation during tendon postnatal homeostasis/maintenance or at times of tendon healing, the commitment cascade is still very elusive. Cumulatively the few articles providing some information on this topic are suggesting that the above process might be mediated by the adult TSPCs or perivascular stem cells or tendon intra-fascicular matrix stem cells, or even a mixture of all these cell types depending on the circumstances (Bi et al. 2007, Tempfer et al. 2009, Rui et al. 2010, Mienaltowski et al. 2013, Liu et al. 2014, Alberton et al. 2015, Docheva et al. 2015, Lee et al. 2015). Mienaltowski et al., 2013 actually proposed that in tendons there is a regional distribution of different stem/progenitor cells. Specifically, they analysed two subpopulations; one originating from the peritenon and the other from the tendon proper of mouse Achilles tendons. Comparison between these subpopulations revealed that the cells from the tendon proper are more proliferative and exhibit higher levels of tendon-related markers, such as Tnmd and Scx, while peritenon-derived cells showed increased vascular and pericyte markers. As illustrated in Fig. 5 these adult cell types share some marker gene expression; however their identity, density, exact location and distribution are still not fully understood. Importantly, Tnmd expression has been strongly connected with the adult TSPC cell type (Bi et al. 2007, Alberton et al. 2015).
Taken together, even though our knowledge is being gradually enriched about what genes are expressed in tendon precursor cells, still very little is known whether they originate from embryonic-, TSPC- or the perivascular cell lineages as well as how exactly the various molecular factor are interwoven in the progression of the tendon differentiation program. For these reasons it is highly important to identify further markers exclusive to tendons and their residing stem, progenitor and mature cells. Specifically for the Tnmd signalling pathway a detailed analysis on the expression of other genes in loss-of-function experimental models may clarify the compensatory mechanisms. Chromatin immunoprecipitation can rule out which transcription factors directly interact with the Tnmd promoter, while pool-down assays with Tnmd antibody might help identifying possible directly binding molecules. Last, since tendons are mechano-sensitive tissues, follow up studies can aim at testing if and how Tnmd is regulated in vitro and in vivo by mechanical stimuli.
3.6. Tenomodulin functional analyses
Ever since the discovery in 2001, TNMD gained gradual attention in the tendon research field with an immense rise of publications just in the recent years as depicted in Fig. 6A. Altogether 146 articles and abstracts on Pubmed have been published covering TNMD until the end of 2015. After exclusion of articles only available in abstract form and foreign language articles, we can group the remaining 128 full-text publications into four categories; namely into studies looking into functions of TNMD, articles using TNMD as a tendon marker, research observing correlations between TNMD mutations and a variety of diseases, and lastly reviews Fig. 6B.
Fig. 6.
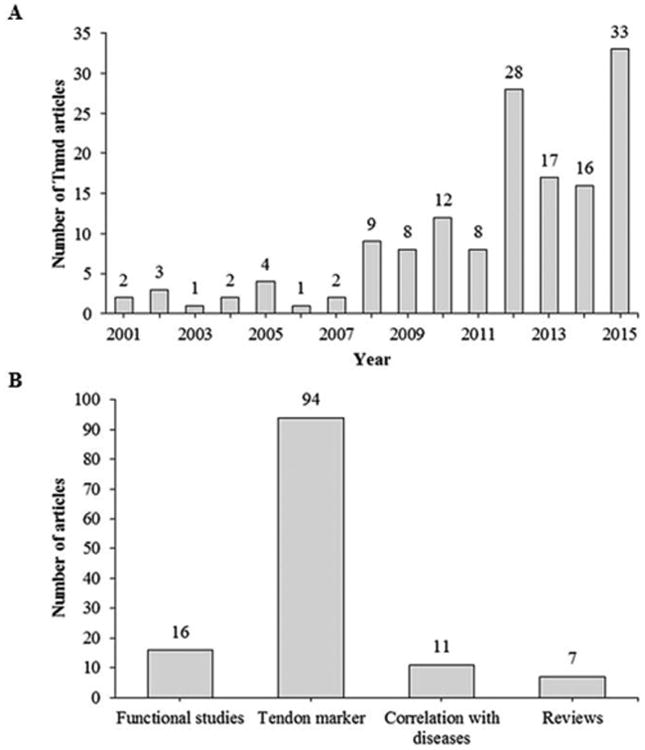
Research articles published annually including tenomodulin. (A) All articles published on Pubmed covering tenomodulin and its alternative names tendin and myodulin as well as its abbreviations TNMD and TeM. (B) Distribution of the published research on tenomodulin into four main categories. This figure only includes research published in English and full-text research articles.
From this analysis it becomes obvious that most of the studies utilize TNMD in research as a tendon marker. In contrast, very little research, 16 full-text studies, were conducted on determining the functional role of TNMD in tendons and other tissues. A summary and detail comments on the so far known TNMD functions are shown in Table 2, which is organized in chronological appearance of the articles from the discovery of TNMD in 2001 to the end of 2015. From this table we see major breakthroughs in the functions of TNMD not just in tendons, but also in other tissues and cells. In tendons it proves to have beneficial functions for the maintenance of healthy and functional tissue, because its loss results in distorted collagen fibrillogenesis, reduced cell density and overall premature tendon ageing. Tnmd exerts a positive effect on TSPCs by sustaining their stem cell like features such as promoting self-renewal and delaying senescence, and the proliferative effect can be carried out exclusively by the Tnmd C-terminal cysteine-rich domain (Alberton et al. 2015). Another discovered function of Tnmd is the contribution to proper adhesion of periodontal fibroblasts which have high gene expression (Komiyama et al. 2013).
Table 2.
Summary of the discovered functions of Tnmd.
| Reference | Experimental model | Study type | Findings and functions of Tnmd |
|---|---|---|---|
| Brandau et al., 2001 | Northern blot analysis of new born mouse RNA; ISH on E17.5 mouse embryos and adult mouse brain; cloning of human and mouse Tnmd cDNA; genomic analysis of human Tnmd organization | Genomic study and in vivo | - Strong expression of 1.5 kb Tnmd transcript in diaphragm, skeletal muscle and eye; low expression in brain, liver, lung, kidney, heart, thymus and perichondrium- and periosteum-free ribcage; Tnmd transcript found in tendons and ligaments but also in the brain, spinal cord, liver, lungs, bowel, thymus and eye of E17.5 mouse embryos; high expression of Tnmd mRNA in the dentate gyrus, CA regions of the hippocampus, neurones in the cerebral nuclei, Purkinje cells and neuronal cells in the cerebellar nucleus, neurons in the anterior and posterior horn of the spinal cord and neurons from all cell layers in the cerebral cortex of adult mice. - Mouse and human Tnmd share 89% homology, similarity (54%) and identity (31%) to Chm1, especially with the C-terminal (77% and 66%); 7 exons found. |
| Shukunami et al., 2001 | Cloning of mouse Tnmd; Northern blot analysis of mouse tissues; ISH of mouse skeletal muscle | Genomic study and in vivo | - Cloning of Tnmd full-length reveals novel protein of 317 amino acid residues; predicted amino acid sequence revealed 33% overall identity with mouse Chm1 precursor; structural features include singly transmembrane domain at the N-terminal region and the putative angiogenic domain with eight cysteine residues; Tnmd lacked a hormone-processing signal compared to Chm1, suggesting it may function as a type II transmembrane protein on cell surface. - Tnmd transcript of 1.4 kb detected in skeletal muscle; Tnmd expression not associated with muscle fibres, but rather with epimysium and tendon. |
| Oshima et al., 2003 | Northern blot analysis on mouse embryos; ISH on eyes of mouse embryos; human retinal endothelial cells (HRECs) transduced with Tnmd; human umbilical vein endothelial cells (HUVECs) co-cultured with HRECs transduced with Tnmd | In vivo and in vitro | -Tnmd mRNA expression detectable from day E15.5 and present in eye and skin; localization on Tnmd transcript on the extraocular muscle, sclerocornea, lens fibre cells, ganglion cell layer, inner nuclear layer cells and pigment epithelium of the retina. - Effective autocrine suppression of cell proliferation and capillary-like morphogenesis of retina vascular endothelial cells; conditioned media from soluble Tnmd-overexpressing cells also showed marked inhibitory effect on angiogenesis. |
| Oshima et al., 2004 | Adenoviral expression system to force expression of Tnmd C-Terminal in HUVECs; C57BL/6 mouse model subcutaneously injected with melanoma tumour cells; histological analysis of melanoma tumour cells injected into mice; transduction of Tnmd into BL-6 melanoma cells | In vivo and in vitro | - Transduction of Tnmd into HUVECS impaired tube formation of HUVECs cultured in Matrigel; conditioned media from COS7 cells transfected with Tnmd impaired tube formation of HUVECs; transformation of HUVECS with Tnmd downregulated VEGF synthesis to 4-50% of normal levels; migration of HUVECs in response to VEGF was significantly affected after Tnmd transduction. - Formed tumours were 46% smaller compared to Ad-EGFP-transduced melanoma cells; visible inhibition of angiogenesis in implanted tumours and decreased microvessel density; no effect on growth rate. |
| Pisani et al., 2004 | Northern blot analysis on gastrocnemius muscle of rats; RT-PCR analysis of tendons separated from soleus muscle and tendon-free soleus muscle of rats and Tnmd in primary human muscle cells derived from single satellite cells; quantitative Northern blot analysis of C2C12 mouse cell line; overexpression of Tnmd fused to FLAG peptide and immunofluorescence to FLAG; Western blot analysis of myodulin-FLAG fusion protein; co-culture experiment of C2C12 mouse myoblast with H5V cells; proliferation of WT muscle cells and Tnmd-FLAG C2C12 cells compared; Tnmd expression in hind limb suspension model in rats | In vivo and in vitro | -Tnmd mRNA found in muscle fibres and tendons; Tnmd transcript found at the myoblast proliferating stage and myotube differiated stage; Tnmd transcript found in C2C12 cells without any significant difference in expression levels between proliferating and differentiated stages; evidence of a muscle cell surface protein. - Tnmd-FLAG fusion protein = 44 kDa, FLAG = 1kDa so mass of Tnmd = 43 kDa.Calculated mass = 37.047. - Tnmd has an active role in invasive action of endothelial cells, without evidence of extracellular Tnmd secretion; no differences observed in proliferative capacity between WT muscle cells and Tnmd-FLAG C2C12 cells, even with the addition of FGF-2; soleus muscle mass decrease resulted in capillary loss resulting in three-fold Tnmd downregulation and 2-fold upregulation the in opposite condition. |
| Docheva et al., 2005 | Generation of the Tnmd KO mouse model; Northern blot analysis of Tnmd in new-born thymus, eye, tendon, muscle and heart tissues; oxygen-induced retinopathy model (OIR); electron microscopy of Achilles tendons; immunohistochemical analysis of collagens, decorin, lumican, aggrecan and matrilin-2 | In vivo | - Successful generation of Tnmd KO mouse line validated by lack of Tnmd transcript; high Tnmd expression in tendons and at lower levels in eye and muscle. - Probing of tendon extracts with an antibody raised against the C-terminal domain of Tnmd, detected a protein signal of 16 kDa corresponding to the cleavable C-terminal cysteine-rich extracellular domain; suggestion that C-terminal cysteine rich domain of Tnmd is rapidly cleaved in vivo - No changes in retinal vascularization seen in both genotypes after OIR. - Loss of Tnmd expression resulted in decreased tenocyte proliferation and reduced tenocyte density in vivo. - No changes in extracellular matrix protein production like collagen type I, II, III and VI and decorin, lumican, aggrecan and matrilin-2; collagen fibrils showed significantly increased average diameters |
| Pisani et al., 2005 | Yeast transformation with mouse Tnmd vector including C-terminal region and Western blotting; purification of Tnmd in yeast by column purification; co-culturing C2C12 and H5V cells together with modified yeast | In vitro | - Mouse Tnmd could be expressed at the plasma membrane of Saccharomyces cervisiae in an N-glycosylated state. - Murine Tnmd can be purified from yeast; detection of three different bands of which one shows Tnmd in a glycosylated state (65 kDa). - Mouse Tnmd expressed in yeast fully functional by increasing invasive potential of C2C12 cells in H5V co-culture system. |
| Shukunami et al., 2006 | ISH on developing chick embryos; retroviral expression of Scx in chick tenocytes and chondrocytes; misexpression of RCAS-cScx by electroporation into hind limb | In vivo and in vitro | - At stage 23, Scx expression in the syndetome has extended to the tail region and Tnmd detectable in anterior eight somites; at stage 25, Tnmd and Scx detectable in regions adjacent to the myotome; at stage 32 and later, Scx and Tnmd displayed similar expression profiles in developing tendons. - Upregulation of Tnmd in tenocytes but not chondrocytes; no induction of generation of additional tendons but resulted in upregulation of Tnmd in tendons at stage 33 and later. |
| Watahiki et al., 2007 | Quantitative PCR of mandibular condylar cartilage and tibial growth plate cartilage of 1-and 5 week old rats; IHC analysis on mandibular-, condylar- and tibial cartilage of 1-and 5- week old rats | In vivo | - Mandibular condylar cartilage showed higher mRNA levels of Tnmd than tibial growth plate cartilage; mandibular condylar cartilage 1-week-old rats presented higher Tnmd mRNA levels than 5-weeks-old rats; Tnmd was found on condylar cartilage except on fibrous layer; Tnmd only detected in the hypertrophic layer and part of articular cartilage. |
| Shukunami et al., 2008 | In situ hybridization on developing mouse at E14.5; immunohistochemical analysis on developing mouse forelimb; ISH on avascular mesenchyme and vertebrae of developing chick | In vivo | - mRNA localized in developing tendons which were co-localized to Scx expression; Tnmd staining spotted in cartilage and localization restricted to the PECAM-1 negative region of the developing tendons; first identified in posterior regions of limb mesenchyme at HH stage 23 and later expressed in attachment sites connecting cartilage and muscle; at HH stage 30, distinct expression pattern adjacent to MyoD-positive domain marking the myotome and partially overlapping with Scx expression; Tnmd-positive domains were lacking blood vessels; Tnmd expression exclusive to connective tissues including intervertebral region, perichondrium of vertebrae and ligaments; Tnmd only identified in peripheral region where thick bundles of collagen fibers were present. |
| Wang et al., 2012 | OIR in mice; injection of commercially produced recombinant Tnmd into eye for analysis of blood vessel patterns | In vivo | - In OIR model, fewer central non-perfused areas were observed in Tnmd-injected eyes than in PBS-injected eyes; significantly less nuclei in new blood vessels breaking in each retinal cross-section of Tnmd-treated- compared to PBS-treated mice; suggestion of potential role of Tnmd in prevention and treatment of ocular neovascularization. |
| Qi et al., 2012 | I-TASSER protein three-dimensional conformation modeling prediction; human and porcine tenocytes; knock- down of human FCR cells; Tnmd Western blotting of pig, horse and human tendon cells; ICC on Tnmd isoforms overexpressed in COS-7 cells | Computer analysis and in vitro | - Three TNMD transcripts with molecular weights of 37.1, 20.3 and 25.4 kDa for isoforms I, II and II respectively proposed and verified by Western blot from human cells; isoforms I and II localized perinuclear and isoform III on cytoplasm; predicted function of TNMD I = cytosine methyltransferase, II= SUMO--1-like SENP-1 protease, III= α-syntrophin, pleckstrin homology domain scaffolding protein. - Knockdown of all isoforms resulted in reduced cell proliferation and downregulation of myostatin and scleraxis. |
| Komiyama et al., 2013 | Tnmd IHC on WT mouse molars; FL-, CTD-, CS-, BRICHOS-, and EC Tnmd overexpression in NIH3T3 and hPDL cells; cleavage of N-glycans and inhibition of glycosylation in vitro analysis by adding tunicamycin and digesting cell lysates with PNGaseF respectively; analysis of adhesion capacity of Tnmd KO mouse fibroblasts | In vivo and in vitro | -Tnmd expression is related to time of tooth eruption at 2-3 weeks of age; Tnmd localized in Golgi apparatus, microtubules and plasma membrane of PDL cells. - 45-kDa Tnmd protein is likely to be glycosylated form; 40-kDa protein the non-glycosylated Tnmd protein. - Tnmd overexpression resulted in in enhanced adhesion of PDL and NIH3T3 cells to collagen I and increased surface area; Tnmd KO fibroblasts showed decreased cell adhesion; BRICHOS domain or CS region seem responsible for Tnmd-mediated improvement of cell adhesion in Tnmd overexpressing NIH3T3 cells. |
| Miyabara et al., 2014 | Wnt in vitro model; equine BMSCs cultured in collagen gel; nuclear translocation of ß-catenin in glycogen synthase kinase-3 inhibitor-treated BMSCs | In vitro | - mRNA expression of Tnmd in equine BMSCs increased when Wnt pathway was activated, comparable to levels in tendon. - Concluded regulation of tenomodulin expression via Wnt/ß-catenin signaling. |
| Sato et al., 2014 | Tnmd messenger analysis using qPCR of masseter muscle (MM) from C57/BL6 mice; ISH of Tnmd transcript MM | In vivo | - Level of Tnmd RNA increased from E12.5-E17.5 and decreased after birth; CD31 and VEGF mRNA levels in the MM remained constant from E12.5-E18.5 and low after birth; Tnmd mRNA found in the middle region of the MM at E12.5 as well as in the muscle-bone junction from E14.5 onwards. |
| Alberton et al. 2015 | Self-renewal comparison between WT and Tnmd KO TSPC; transfection of full-length and C-terminal domain of Tnmd in Tnmd KO mouse TSPCs; multipotential comparison on TSPCs | In vitro | - Tnmd KO TSPCs show reduced proliferative capacity. - C-terminal alone restores proliferation in Tnmd KO TSPCs. - Loss of Tnmd results in premature and increased senescence of TSPCs compared to WT. - Lack of Tnmd does not affect multipotential in TSPC. |
Abbreviations: Ad-EGFP, adenovirus vector encoding enhanced green fluorescent protein; BMSCs, bone marrow stromal cells; C2C12, mouse myoblast cell line; C57BL/6, C57 black 6; CA region, Cornu Ammonis region; cDNA, complementary deoxyribonucleic acid; Chm1; chondromodulin-1; COS-7, African green monkey fibroblast-like kidney cell line; CS, mutant with deleted cleavage site; CTD, C-terminal domain deletion mutant; E, embryonic day; EC domain, mutant with entire extracellular portion of Tnmd deleted; FCR, flexor carpi radialis; FGF-2, fibroblast growth factor-2; FL, full length Tnmd; H5V, mouse embryonic heart endothelial cells; HH, Hamburger-Hamilton stage; hPDL, human periodontal ligament; ICC, immunocytochemistry; IHC, immunohistochemistry; ISH, in situ hybridization; I-TASSER, iterative, threading assembly refinement; kb, kilo base; kDa, kilodalton; KO, knockout; mRNA, messenger ribonucleic acid; NIH3T3, mouse embryonic fibroblasts; OIR, oxygen-induced retinopathy; PBS, phosphate-buffered saline; PDL, periodontal ligament; PNGaseF, peptide-N-glycosidase F; qPCR, quantitative PCR; RCAS-cScx, replication-competent avian sarcoma-leukosis virus-copy Scleraxis; RNA, ribonucleic acid; RT-PCR, reverse transcriptase-polymerase chain reaction; Scx, Scleraxis; SUMO, small ubiquitin-like modifier; Tnmd, tenomodulin; VEGF, vascular endothelial growth factor; WT, wildtype.
Intriguingly, the proliferative influence of Tnmd was shown to be dependent on the cell type. For example, human retinal endothelial cells transduced with Tnmd exhibited a reduction in proliferation. In addition, Tnmd lowered the angiogenic migration of human umbilical vein endothelial cells (HUVECs) as reported by (Oshima et al. 2003, Oshima et al. 2004). Transduction of Tnmd in human melanoma cells resulted in suppression of tumour growth in ectopic in vivo model, due to decreased vessel density most likely because of reduced endothelial cell migration (Oshima et al. 2004). With respect to organismal vessel formation in vivo it is clear that lack of Tnmd does not affect angiogenesis in tendon and retina development since the knockout model revealed no major differences to wilt type mice (Docheva et al. 2005). Still, the latter finding is open for discussion because a study with recombinant Tnmd has shown an obliterating effect on retinal vessels when Tnmd was injected in the vitreous body in vivo (Wang et al. 2012). In this study, however, information and validation of the produced Tnmd were not included and at present the protein is not available for purchase from the producer company. Last, the study on CTCs demonstrated that in this tissue type Tnmd prevents vascularization (Kimura et al. 2008).
Now that some Tnmd functions have been exposed it is also equally important to decipher in follow up studies how exactly Tnmd exerts these. As already discussed previously, the way to deliver the necessary explanations is via narrowing down the exact signalling pathway of Tnmd. This will not only allow a complete understanding of the role of Tnmd in the domains where is expressed, but will also enable concrete manipulation of its effect in possible clinical applications.
3.7. Tenomodulin as lineage marker
As observed in Fig. 6 most of the full-text research papers containing TNMD have used it as a gene marker for the tendon and ligament lineage. Table 3 summarises these studies as all findings were clustered into 6 categories. Table 3 makes it evident that most research looks at TNMD on a messenger level with the use of PCR to demonstrate that the used cell types are or have differentiated into tendon-like cells. Protein data on the other hand is shown very rarely. Based on our experience, it is the protein data that is most difficult to gain, but it is an essential proof of the true involvement of TNMD in the investigated model. We and our Japanese collaborators have produced self-made antibodies and examined their specificity using Tnmd knockdown or knockout samples (Docheva et al. 2005, Shukunami et al. 2006, Komiyama et al. 2013). It is therefore crucial for researchers to carefully validate the antibody specificity.
Table 3.
The use of TNMD as a lineage marker.
Abbreviations: 3D, three dimensional; ICC, immunocytochemistry; IHC, immunohistochemistry; ISH, in situ hybridization; MA, microarray; NB, Northern blot; PCR, polymerase chain reaction; WB, western blot.
Interestingly, several wound healing models, namely in skin, rotator cuff and patella tendons, revealed differential Tnmd expression. In skin, Tnmd expression significantly downregulated between day 1 and 3 after skin incision, while in the tendon defect studies Tnmd expression significantly upregulated in the period between 1-2 weeks in the patella and between 4-12 weeks in the rotator cuff that was FGF-2 treated (Kameyama et al. 2015, Omachi et al. 2015, Tokunaga et al. 2015). This data strongly motivates further investigations on the exact roles of Tnmd in tissue repair.
3.8. Tenomodulin correlations to various diseases
In recent years, research mostly conducted on a genomic level by single nucleotide polymorphism (SNP), has presented very interesting correlations between TNMD and a variety of diseases. Specifically, TNMD was selected as a candidate gene for obesity, type 2 diabetes, metabolic syndrome, Alzheimer's disease and age-related macular degeneration, etc. The stated association studies are summarized in Table 4, and the concrete TNMD SNPs are shown in Fig. 7. Associations with obesity, type 2 diabetes and metabolic syndrome were investigated in two study populations; the Finnish diabetes prevention study and the metabolic syndrome in men (Tolppanen et al. 2007, Tolppanen et al. 2008a, Tolppanen et al. 2008b). In these studies several gender-specific SNPs were identified; however it is still not exactly known how these SNPs affect TNMD transcription, splicing or protein amino acid sequence. Intriguingly most of the identified SNPs are found in introns and not in exons or near exon-intron splice sites. Only SNP rs2073162 is found on exon 3, suggesting that the key SNPs might have a long-distance effect on transcriptional control. A follow up study has revealed that TNMD expression is increased in obesity and down-regulated during weight-reducing diet (Saiki et al. 2009). A similar finding was also reported by Kolehmainen et al. 2008, the authors found a strong downregulation of the TNMD gene along with weight reduction. Moreover, the expression of TNMD was associated with several characteristics of the metabolic syndrome, but the mechanisms by which TNMD may be involved in insulin sensitivity remain elusive. Other three studies have looked into correlations between TNMD SNPs, or expression, to metabolic syndrome and reported certain links to serum lipoproteins levels and inflammatory factors (Tolppanen et al. 2008a, Tolppanen et al. 2008b, Gonzalez-Muniesa et al. 2013). One circulated idea is that an increase in TNMD expression during obesity might exert a protective mechanism aimed at limiting growth of new blood vessels in periods of adipose tissue expansion. There are still many open questions to these studies: for example, which cell type expresses TNMD mRNA and how abundant is the protein; if and how TNMD regulates the vascular formation and can it even directly affect adipose cells; how Tnmd might be involved in cholesterol metabolism or can affect systemic immune mediators.
Table 4.
Summary of studies correlating TNMD with various diseases.
| Diseases | Study model | TNMD analysis | Main conclusions | References |
|---|---|---|---|---|
| Obesity | Longitudinal study, 166 men and 341 women with overweight and impaired glucose tolerance | TNMD SNPs | rs5966709 and rs4828037 were associated with adiposity in women; rs11798018 was related with adiposity in men. | Tolppanen et al., 2007 |
| Longitudinal study, 68 obese cases, subcutaneous adipose tissue, omental adipose tissue | TNMD mRNA | TNMD was highly expressed in adipose tissue and increased in obesity and was downregulated during diet-induced weight loss in men and women. | Saiki et al., 2009 | |
| Longitudinal study, 28 weight reduction cases and 18 controls, fat tissue | TNMD mRNA | Strong downregulation of TNMD expression after long-term weight reduction in men and women. | Kolehmainen et al., 2008 | |
| Type 2 diabetes | Longitudinal study, 166 men and 341 women with overweight and impaired glucose tolerance | TNMD SNPs | rs2073163 and rs1155974 were linked with 2-hour plasma glucose in men; rs2073163, rs1155974, and rs2073162 were associated with type 2 diabetes in men; only rs2073162 associated with 2-hour plasma glucose in women. | Tolppanen et al., 2007 |
| Metabolic syndrome | Cross-sectional study, random sample of 5298 men | TNMD SNPs | rs2073162 was related with serum lipoproteins in a BMI-dependent manner in men; no women were involved in the research. | Tolppanen et al., 2008a |
| Longitudinal study, 9 lean and 9 obese male high fat cases, subcutaneous abdominal adipose tissue | TNMD mRNA | Expression of TNMD was linked with some relevant metabolic syndrome features in men; no women were involved in the research. | González-Muniesa et al., 2013 | |
| Cross-sectional study, 166 men and 341 women with overweight and impaired glucose tolerance | TNMD SNPs | rs2073163, rs1155974, and rs2073162 were connected with acute phase reactants in men; rs5966709 and rs4828037 were associated with inflammatory factors in men and women; rs2073163, rs4828038, and rs1155974 were related with inflammatory factors in women. | Tolppanen et al., 2008b | |
| Alzheimer's disease | Cross-sectional study, 526 Alzheimer's disease cases and 672 controls | TNMD SNPs | rs5966709 was connected with risk of Alzheimer's disease among women with AOPE ε4-allele; none of the SNPs associated with Alzheimer's disease in men. | Tolppanen et al., 2011 |
| Age-related macular degeneration | Cross-sectional study, 307 age-related macular degeneration cases and 168 controls | TNMD SNPs | rs1155974 and rs2073163 were associated with a risk of age-related macular degeneration in women; none of the SNPs associated with prevalence of age-related macular degeneration among men. | Tolppanen et al., 2009 |
| Rupture of the chordae tendineae cordis | Cross-sectional study, 12 normal and 16 ruptured chordae tendineae cordis (CTC), specimens obtained from autopsy or surgery, immunohisto-chemical analysis | TNMD protein | TNMD was locally absent in the rupture area, where abnormal vessel formation, strong expression of vascular endothelial growth factor-A and matrix metalloproteinases, and infiltration of inflammatory cells were observed, but not in the normal or non-ruptured CTC. | Kimura et al., 2008 |
| Juvenile dermatomyositis | Cross-sectional study, 31 girls with Juvenile dermatomyositis and 4 healthy controls, muscle biopsies | TNMD mRNA | TNMD was up-regulated during the chronic inflammatory state of the disease, and downregulated when disease duration was short in girls, no boys were involved in the research. | Chen et al., 2008 |
| Intellectual disability and seizures | Case report, two female patients, deletions spanning PCDH19 | TNMD gene | Genomic deletions spanning the TNMD gene in female-restricted epilepsy with mental retardation. | Vincent et al., 2012 |
Abbreviations: APOE, apolipoprotein E; CTC, chordae tendineae cordis; mRNA, messenger ribonucleic acid; PCDH19, Protocadherin 19; rs, reference SNP; SNP, single nucleotide polymorphism; TNMD, tenomodulin.
Fig. 7.
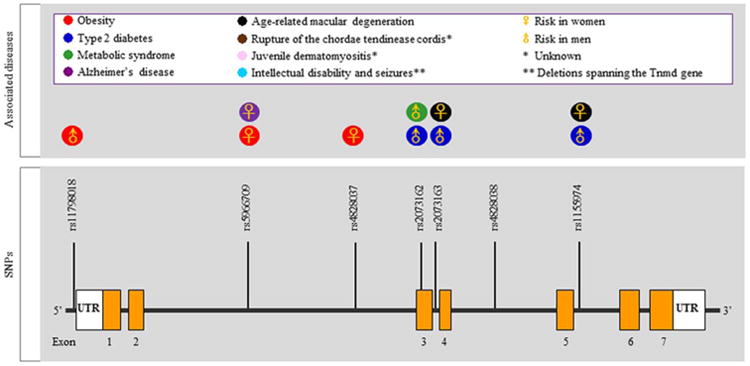
Single nucleotide polymorphism (SNPs) and other putative mutations in the TNMD gene locus correlating with various diseases. Orange boxes represent exons, while the thick black lines connecting them are introns. Abbreviations: rs, reference SNP; SNPs, small nucleotide polymorphisms; UTR, untranslated region.
(Tolppanen et al. 2011) suggested TNMD as an interesting candidate gene for Alzheimer's disease since they detected associations between TNMD SNPs and known risk factors for this disease. The sequence variation of TNMD was not connected with the prevalence of Alzheimer's disease when the results were adjusted to the APOE genotype, but the marker rs5966709 was linked with disease risk among women with APOE ε4-allele. These results suggest that the effect of APOE may be modified by TNMD. (Brandau et al. 2001) has shown by in situ hybridization a Tnmd messenger in multiple areas of the adult mouse brain, however it is not clear if the protein is expressed, hence it would be very interesting to follow up on the above studies and explain possible functions of TNMD in the brain and brain diseases.
Age-related macular degeneration can be divided into atrophic and exudative forms, the latter being more common and accounting for approximately 80% of age-related macular degeneration cases. Since dysregulated neovascularization is involved in the pathogenesis of age-related macular degeneration, Tolppanen et al. (Tolppanen et al. 2009) also investigated the associations of TNMD SNPs with this condition and found rs1155974, rs2073163 and rs7890586 were correlating with higher risk in women. This study further supported the theory of a disrupted balance between stimulators and inhibitors of neovascularization in the pathogenesis of exudative age-related macular degeneration. However, functional studies are needed to reveal the exact mechanisms and involvement of TNMD in these associations.
Due to the presence of TNMD in tendinous structures TNMD was in the spotlight also in heart-related studies. In fact, the anatomical organization of the CTCs, which connect the papillary muscle to the atrioventricular valves, is very similar to tendons but of much smaller size. Their rupture is a well-known cause of mitral regurgitation and cardiac valvular syndromes. At the base of these failures is the abrogated avascularity of the cordis (Kimura et al. 2008, Hakuno et al. 2011, Kusumoto and Fukuda 2013). The study by Kimura et al. 2008 strongly confirmed that the local absence of TNMD leads to enhanced angiogenesis, VEGF-A and MMPs activation following the rupture of the heart chordae tendineae. In the above study TNMD knockdown experiments have suggested that TNMD can directly inhibit endothelial cell migration, but the receptor or molecular agent through which Tnmd acts has not been found.
The associations with juvenile dermatomyositis were studied by Chen et al. 2008 revealing that TNMD mRNA was upregulated during the chronic inflammatory phase. The authors suggested that TNMD might regulate contractility of vascular smooth muscle cells and speculated that interventions that diminish the anti-angiogenic remodeling may be beneficial for children with longer duration of untreated juvenile dermatomyositis. However, there is little information describing how exactly TNMD is involved in vasculature loss and tissue remodeling.
Last, a case report study of two female patients with intellectual disability and seizures related to female-restricted epilepsy with mental retardation found a genomic deletion at PCDH19, spanning the TNMD gene, (Vincent et al. 2012). Despite being a case study, it would be very interesting, after validation of TNMD protein expression in the brain, to examine if Tnmd knockout animals exhibit a brain phenotype and altered behavior.
In summary, now that it is apparent that certain SNPs linked to several health conditions are located in the TNMD locus and that changes in the TNMD levels are associated with at least three different disease it becomes almost obligatory in future research to prove whether or not these associations have a critical role in the establishment of these diseases and especially how the SNPs affect TNMD gene expression or protein production and function. One way to continue is to challenge the Tnmd knockout strain by crossing it with mouse strains harbouring a concrete disease phenotype. We believe that the uncovering of the exact TNMD functions will be essential for improving our understanding of the mentioned clinical conditions, which subsequently will permit the development of appropriate counterstrategies.
4. Conclusion and future perspectives
Since its discovery in 2001, TNMD has gained significantly more attention as demonstrated in the increasing number of articles in the last 15 years. The alternative names have been condensed to the single name tenomodulin with its corresponding abbreviation as TNMD. It is a gene bearing high and only homology to Chm1, but still exhibits important differences, such as the absence of a furin cleavage signal, different glycosylation sites and expression pattern. Although the predominant expression of TNMD is found in tendons and ligaments, it is also shown to be expressed in other tissues such as parts of the eyes, cordae tendineae cordis, muscle sheaths, brain, and skin. The presence of isoforms needs to be carefully verified in different species. Another very important future goal should be the better understanding of the TNMD protein domain structure. The TNMD signalling pathway is still very elusive as up to date only few upstream factors, such as Scx and Mkx, have been identified. With regards to downstream effectors, there is a complete lack of knowledge on the direct protein binding partners of TNMD. The full clarification of Tnmd signalling must become central in follow up research, because by only elucidating TNMD modes of action we would be able to specifically manipulate its effect in tendons and other tissues. We have summarized the current discovered functions of Tnmd in Fig. 8. Firstly, TNMD has been strongly justified as the best tendon and ligament marker in more than 90 different studies. Next, a combination of in vivo and in vitro investigations has revealed its positive role on tendon/ligament cell and tissue functions as well as is negative effect on vessels in specific regions of the heart and in tumour models. Last, we are just starting to comprehend the potential involvement of TNMD in various diseases such as obesity and metabolic syndrome, where TNMD expression is positively correlated to an advanced disease state. However, since most of the disease-associated studies are based on identification of SNPs in TNMD gene locus it is very important to find out how exactly these mutations translate into these diseases. In sum, we believe that this review not only summarises the current state of affairs of our knowledge of TNMD gene, protein, expression and functions, but also will excite international researchers to further study this mysterious tenogenic modulating insider factor in terms of complete deciphering of its signalling pathway, contribution to certain pathologies as well as possible development of therapeutic strategies.
Fig. 8.
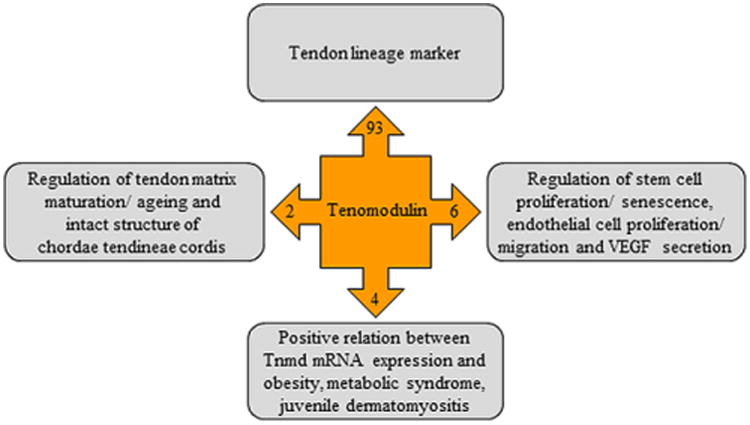
Schematic summary of TNMD known functions. Abbreviations: mRNA, messenger ribonucleic acid; Tnmd, tenomodulin; VEGF, vascular endothelial growth factor.
Highlights.
Tenomodulin gene and protein structure are elucidated in text and figure form.
The expression pattern of tenomodulin is described and complemented with detailed table.
Putative signalling pathway and tenogenic cascade are critically reviewed and presented graphically.
Chronological and thorough description of the discovered tenomodulin functions is provided.
Correlations between tenomodulin mutations and various diseases are newly summarized.
Acknowledgments
This review and the corresponding Gene Wiki article, https://en.wikipedia.org/wiki/Tenomodulin, are written as part of the Gene Wiki Review series – a series resulting from collaboration between the journal Gene and the Gene Wiki initiative. The Gene Wiki Initiative is supported by the National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. D.D. acknowledges the German Research Foundation (DFG grant DO1414/3-1) and the Bavarian Research Foundation (BFS grant DOK-100-08) for the support on TNMD-related research. We thank Angela Chapman for final proof reading.
Abbreviations
- 3D
three dimensional
- Ad-EGFP
adenovirus vector encoding enhanced green fluorescent protein
- APOE
apolipoprotein E
- Aqp1
aquaporin 1
- Arg
arginine
- bHLH
basic helix-loop-helix
- BMDSC
bone marrow-derived stem cell
- BMP
bone morphogenetic protein
- BMSCs
bone marrow stromal cells
- C
cysteine
- C2C12
mouse myoblast cell line
- C57BL/6
C57 black 6
- CA region
Cornu Ammonis region
- cDNA
complementary deoxyribonucleic acid
- Chm1
chondromodulin-1
- Col
collagen
- COMP
cartilage oligomeric matrix protein
- COS-7
African green monkey fibroblast-like kidney cell line
- CS
mutant with deleted cleavage site
- CTC
chordae tendineae cordis
- CTD
C-terminal domain deletion mutant
- C-terminal
carboxyl-terminal
- cTnmd
chicken tenomodulin
- Cys
cysteine
- DNA
deoxyribonucleic acid
- E
embryonic day
- EC domain
mutant with entire extracellular portion of Tnmd deleted
- Egr
early growth response protein
- Eya
eyes absent transcription factor
- FCR
flexor carpi radialis
- FGF
fibroblast growth factor
- FL
full length tenomodulin
- GSK-3
glycogen synthase-3
- H5V
mouse embryonic heart endothelial cells
- HH
Hamburger-Hamilton stage
- hPDL
human periodontal ligament
- hTNMD
human tenomodulin
- Htra3
HtrA serine peptidase 3
- HUVEC
human umbilical vein endothelial cell
- I
isoleucine
- ICC
immunocytochemistry
- ICC
immunocytochemistry
- IFM
interfascicular matrix
- IHC
immunohistochemistry
- ISH
in situ hybridization
- I-TASSER
iterative, threading assembly refinement
- K
lysine
- kb
kilo base
- kDa
kilodalton
- KO
knockout
- MA
microarray
- Mkx
Mohawk
- MMP
matrix metalloproteinase
- mRNA
messenger ribonucleic acid
- MSC
mesenchymal stem cell
- mTnmd
mouse tenomodulin
- N
asparagine
- NB
Northern blot
- NIH3T3
mouse embryonic fibroblasts
- OIR
oxygen-induced retinopathy
- p53
tumour protein 53
- PBS
phosphate-buffered saline
- PCDH19
Protocadherin 19
- PCR
polymerase chain reaction
- PDL
periodontal ligament
- PNGaseF
peptide-N-glycosidase F
- Q
glutamine
- qPCR
quantitative PCR
- RCAS-cScx
replication-competent avian sarcoma-leukosis virus-copy Scleraxis
- RNA
ribonucleic acid
- rs
reference small nucleotide polymorphism
- RT-PCR
reverse transcriptase-polymerase chain reaction
- Sca-1
stem cells antigen-1
- Scx
Scleraxis
- SMA
smooth muscle actin
- SNP
small nucleotide polymorphism
- Sox
sex determining region Y
- SUMO
small ubiquitin-like modifier
- TGF
transforming growth factor
- Thbs
thrombospondin
- TNMD
Tnmd, tenomodulin
- TSPC
tendon stem/progenitor cell
- UTR
untranslated region
- V
valine
- VEGF
vascular endothelial growth factor
- WB
western blot
- WT
wildtype
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberton P, Dex S, Popov C, Shukunami C, Schieker M, Docheva D. Loss of tenomodulin results in reduced self-renewal and augmented senescence of tendon stem/progenitor cells. Stem Cells Dev. 2015;24(5):597–609. doi: 10.1089/scd.2014.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberton P, Popov C, Pragert M, Kohler J, Shukunami C, Schieker M, Docheva D. Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev. 2012;21(6):846–858. doi: 10.1089/scd.2011.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman LJ, Fong G, Andersson G, Scott A, Danielson P. Substance P is a mechanoresponsive, autocrine regulator of human tenocyte proliferation. PLoS One. 2011;6(11):e27209. doi: 10.1371/journal.pone.0027209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr PJ. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Barsby T, Bavin EP, Guest DJ. Three-dimensional culture and transforming growth factor beta3 synergistically promote tenogenic differentiation of equine embryo-derived stem cells. Tissue Eng Part A. 2014;20(19-20):2604–2613. doi: 10.1089/ten.tea.2013.0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashur CA, Shaffer RD, Dahlgren LA, Guelcher SA, Goldstein AS. Effect of fiber diameter and alignment of electrospun polyurethane meshes on mesenchymal progenitor cells. Tissue Eng Part A. 2009;15(9):2435–2445. doi: 10.1089/ten.tea.2008.0295. [DOI] [PubMed] [Google Scholar]
- Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, Zhang L, Shi S, Young MF. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13(10):1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- Brandau O, Aszodi A, Hunziker EB, Neame PJ, Vestweber D, Fassler R. Chondromodulin I is dispensable during enchondral ossification and eye development. Mol Cell Biol. 2002;22(18):6627–6635. doi: 10.1128/MCB.22.18.6627-6635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandau O, Meindl A, Fassler R, Aszodi A. A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Dev Dyn. 2001;221(1):72–80. doi: 10.1002/dvdy.1126. [DOI] [PubMed] [Google Scholar]
- Brent AE, Braun T, Tabin CJ. Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development. 2005;132(3):515–528. doi: 10.1242/dev.01605. [DOI] [PubMed] [Google Scholar]
- Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113(2):235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development. 2004;131(16):3885–3896. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- Brown JP, Galassi TV, Stoppato M, Schiele NR, Kuo CK. Comparative analysis of mesenchymal stem cell and embryonic tendon progenitor cell response to embryonic tendon biochemical and mechanical factors. Stem Cell Res Ther. 2015;6:89. doi: 10.1186/s13287-015-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch F, Mobasheri A, Shayan P, Lueders C, Stahlmann R, Shakibaei M. Resveratrol modulates interleukin-1beta-induced phosphatidylinositol 3-kinase and nuclear factor kappaB signaling pathways in human tenocytes. J Biol Chem. 2012;287(45):38050–38063. doi: 10.1074/jbc.M112.377028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Ade M, Badial PR, Alvarez LE, Yamada AL, Borges AS, Deffune E, Hussni CA, Garcia Alves AL. Equine tendonitis therapy using mesenchymal stem cells and platelet concentrates: a randomized controlled trial. Stem Cell Res Ther. 2013;4(4):85. doi: 10.1186/scrt236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang B, Zhang WJ, Zhou G, Cao Y, Liu W. In vivo tendon engineering with skeletal muscle derived cells in a mouse model. Biomaterials. 2012;33(26):6086–6097. doi: 10.1016/j.biomaterials.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Chen JW, Galloway JL. The development of zebrafish tendon and ligament progenitors. Development. 2014;141(10):2035–2045. doi: 10.1242/dev.104067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Tang H, Zhou M, Hu C, Zhang J, Tang K. Dexamethasone inhibits the differentiation of rat tendon stem cells into tenocytes by targeting the scleraxis gene. J Steroid Biochem Mol Biol. 2015;152:16–24. doi: 10.1016/j.jsbmb.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Chen YW, Shi R, Geraci N, Shrestha S, Gordish-Dressman H, Pachman LM. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 2008;9:43. doi: 10.1186/1471-2172-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuen FS, Chuk CY, Ping WY, Nar WW, Kim HL, Ming CK. Immunohistochemical characterization of cells in adult human patellar tendons. J Histochem Cytochem. 2004;52(9):1151–1157. doi: 10.1369/jhc.3A6232.2004. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Ligon KL, Lyons GE, Copeland NG, Gilbert DJ, Jenkins NA, Olson EN. Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121(4):1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- Docheva D, Hunziker EB, Fassler R, Brandau O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol Cell Biol. 2005;25(2):699–705. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docheva D, Muller SA, Majewski M, Evans CH. Biologics for tendon repair. Adv Drug Deliv Rev. 2015;84:222–239. doi: 10.1016/j.addr.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson P, Andersson T, Aspenberg P. Rat Achilles tendon healing: mechanical loading and gene expression. J Appl Physiol (1985) 2009;107(2):399–407. doi: 10.1152/japplphysiol.91563.2008. [DOI] [PubMed] [Google Scholar]
- Eloy-Trinquet S, Wang H, Edom-Vovard F, Duprez D. Fgf signaling components are associated with muscles and tendons during limb development. Dev Dyn. 2009;238(5):1195–1206. doi: 10.1002/dvdy.21946. [DOI] [PubMed] [Google Scholar]
- Erisken C, Zhang X, Moffat KL, Levine WN, Lu HH. Scaffold fiber diameter regulates human tendon fibroblast growth and differentiation. Tissue Eng Part A. 2013;19(3-4):519–528. doi: 10.1089/ten.tea.2012.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong G, Backman LJ, Andersson G, Scott A, Danielson P. Human tenocytes are stimulated to proliferate by acetylcholine through an EGFR signalling pathway. Cell Tissue Res. 2013;351(3):465–475. doi: 10.1007/s00441-012-1530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova EG, Drazba J, Krukovets I, Kostenko V, Blech L, Harry C, Vasanji A, Drumm C, Sul P, Jenniskens GJ, Plow EF, Stenina-Adognravi O. Control of organization and function of muscle and tendon by thrombospondin-4. Matrix Biol. 2014;37:35–48. doi: 10.1016/j.matbio.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer G, Vorbruggen G, Pasca G, Jackle H, Volk T. Epidermal egr-like zinc finger protein of Drosophila participates in myotube guidance. EMBO J. 1996;15(7):1642–1649. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Muniesa P, Marrades MP, Martinez JA, Moreno-Aliaga MJ. Differential proinflammatory and oxidative stress response and vulnerability to metabolic syndrome in habitual high-fat young male consumers putatively predisposed by their genetic background. Int J Mol Sci. 2013;14(9):17238–17255. doi: 10.3390/ijms140917238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerquin MJ, Charvet B, Nourissat G, Havis E, Ronsin O, Bonnin MA, Ruggiu M, Olivera-Martinez I, Robert N, Lu Y, Kadler KE, Baumberger T, Doursounian L, Berenbaum F, Duprez D. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J Clin Invest. 2013;123(8):3564–3576. doi: 10.1172/JCI67521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati BR, Kumar R, Mohanty N, Kumar P, Somasundaram RK, Yadav PS. Bone morphogenetic protein-12 induces tenogenic differentiation of mesenchymal stem cells derived from equine amniotic fluid. Cells Tissues Organs. 2013;198(5):377–389. doi: 10.1159/000358231. [DOI] [PubMed] [Google Scholar]
- Haddad-Weber M, Prager P, Kunz M, Seefried L, Jakob F, Murray MM, Evans CH, Noth U, Steinert AF. BMP12 and BMP13 gene transfer induce ligamentogenic differentiation in mesenchymal progenitor and anterior cruciate ligament cells. Cytotherapy. 2010;12(4):505–513. doi: 10.3109/14653241003709652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakuno D, Kimura N, Yoshioka M, Fukuda K. Role of angiogenetic factors in cardiac valve homeostasis and disease. Journal of cardiovascular translational research. 2011;4(6):727–740. doi: 10.1007/s12265-011-9317-8. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Chosa N, Asakawa T, Yoshimura Y, Ishisaki A, Tanaka M. Establishment of immortalized human periodontal ligament cells derived from deciduous teeth. Int J Mol Med. 2010;26(5):701–705. doi: 10.3892/ijmm_00000516. [DOI] [PubMed] [Google Scholar]
- Havis E, Bonnin MA, Olivera-Martinez I, Nazaret N, Ruggiu M, Weibel J, Durand C, Guerquin MJ, Bonod-Bidaud C, Ruggiero F, Schweitzer R, Duprez D. Transcriptomic analysis of mouse limb tendon cells during development. Development. 2014;141(19):3683–3696. doi: 10.1242/dev.108654. [DOI] [PubMed] [Google Scholar]
- Herchenhan A, Bayer ML, Eliasson P, Magnusson SP, Kjaer M. Insulin-like growth factor I enhances collagen synthesis in engineered human tendon tissue. Growth Horm IGF Res. 2015;25(1):13–19. doi: 10.1016/j.ghir.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Hiraki Y, Tanaka H, Inoue H, Kondo J, Kamizono A, Suzuki F. Molecular cloning of a new class of cartilage-specific matrix, chondromodulin-I, which stimulates growth of cultured chondrocytes. Biochem Biophys Res Commun. 1991;175(3):971–977. doi: 10.1016/0006-291x(91)91660-5. [DOI] [PubMed] [Google Scholar]
- Inoue M, Ebisawa K, Itaya T, Sugito T, Yamawaki-Ogata A, Sumita Y, Wadagaki R, Narita Y, Agata H, Kagami H, Ueda M. Effect of GDF-5 and BMP-2 on the expression of tendo/ligamentogenesis-related markers in human PDL-derived cells. Oral Dis. 2012;18(2):206–212. doi: 10.1111/j.1601-0825.2011.01871.x. [DOI] [PubMed] [Google Scholar]
- Ippolito E, Natali PG, Postacchini F, Accinni L, De Martino C. Morphological, immunochemical, and biochemical study of rabbit achilles tendon at various ages. J Bone Joint Surg Am. 1980;62(4):583–598. [PubMed] [Google Scholar]
- Itaya T, Kagami H, Okada K, Yamawaki A, Narita Y, Inoue M, Sumita Y, Ueda M. Characteristic changes of periodontal ligament-derived cells during passage. J Periodontal Res. 2009;44(4):425–433. doi: 10.1111/j.1600-0765.2008.01137.x. [DOI] [PubMed] [Google Scholar]
- Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S, Nishida K, Akimoto T, Takahashi M, Miyaki S, Asahara H. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci U S A. 2010;107(23):10538–10542. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Archambault J, Li L, Seeherman H. Tendon-selective genes identified from rat and human musculoskeletal tissues. J Orthop Res. 2010;28(3):289–297. doi: 10.1002/jor.20999. [DOI] [PubMed] [Google Scholar]
- Johansson LE, Danielsson AP, Parikh H, Klintenberg M, Norstrom F, Groop L, Ridderstrale M. Differential gene expression in adipose tissue from obese human subjects during weight loss and weight maintenance. Am J Clin Nutr. 2012;96(1):196–207. doi: 10.3945/ajcn.111.020578. [DOI] [PubMed] [Google Scholar]
- Kameyama H, Udagawa O, Hoshi T, Toukairin Y, Arai T, Nogami M. The mRNA expressions and immunohistochemistry of factors involved in angiogenesis and lymphangiogenesis in the early stage of rat skin incision wounds. Leg Med (Tokyo) 2015;17(4):255–260. doi: 10.1016/j.legalmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Kannus P. Tendons--a source of major concern in competitive and recreational athletes. Scand J Med Sci Sports. 1997;7(2):53–54. [PubMed] [Google Scholar]
- Kato S, Saito M, Funasaki H, Marumo K. Distinctive collagen maturation process in fibroblasts derived from rabbit anterior cruciate ligament, medial collateral ligament, and patellar tendon in vitro. Knee Surg Sports Traumatol Arthrosc. 2015;23(5):1384–1392. doi: 10.1007/s00167-013-2773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaux JF, Drion PV, Colige A, Pascon F, Libertiaux V, Hoffmann A, Janssen L, Heyers A, Nusgens BV, Le Goff C, Gothot A, Cescotto S, Defraigne JO, Rickert M, Crielaard JM. Effects of platelet-rich plasma (PRP) on the healing of Achilles tendons of rats. Wound Repair Regen. 2012;20(5):748–756. doi: 10.1111/j.1524-475X.2012.00826.x. [DOI] [PubMed] [Google Scholar]
- Kaux JF, Janssen L, Drion P, Nusgens B, Libertiaux V, Pascon F, Heyeres A, Hoffmann A, Lambert C, Le Goff C, Denoel V, Defraigne JO, Rickert M, Crielaard JM, Colige A. Vascular Endothelial Growth Factor-111 (VEGF-111) and tendon healing: preliminary results in a rat model of tendon injury. Muscles Ligaments Tendons J. 2014;4(1):24–28. [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Shukunami C, Hakuno D, Yoshioka M, Miura S, Docheva D, Kimura T, Okada Y, Matsumura G, Shin'oka T, Yozu R, Kobayashi J, Ishibashi-Ueda H, Hiraki Y, Fukuda K. Local tenomodulin absence, angiogenesis, and matrix metalloproteinase activation are associated with the rupture of the chordae tendineae cordis. Circulation. 2008;118(17):1737–1747. doi: 10.1161/CIRCULATIONAHA.108.780031. [DOI] [PubMed] [Google Scholar]
- Kishore V, Bullock W, Sun X, Van Dyke WS, Akkus O. Tenogenic differentiation of human MSCs induced by the topography of electrochemically aligned collagen threads. Biomaterials. 2012;33(7):2137–2144. doi: 10.1016/j.biomaterials.2011.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte-Schulz F, Gerhardt C, Scheibel M, Wildemann B, Pauly S. Relationship between muscle fatty infiltration and the biological characteristics and stimulation potential of tenocytes from rotator cuff tears. J Orthop Res. 2014;32(1):129–137. doi: 10.1002/jor.22481. [DOI] [PubMed] [Google Scholar]
- Kolehmainen M, Salopuro T, Schwab US, Kekalainen J, Kallio P, Laaksonen DE, Pulkkinen L, Lindi VI, Sivenius K, Mager U, Siitonen N, Niskanen L, Gylling H, Rauramaa R, Uusitupa M. Weight reduction modulates expression of genes involved in extracellular matrix and cell death: the GENOBIN study. International journal of obesity. 2008;32(2):292–303. doi: 10.1038/sj.ijo.0803718. [DOI] [PubMed] [Google Scholar]
- Komiyama Y, Ohba S, Shimohata N, Nakajima K, Hojo H, Yano F, Takato T, Docheva D, Shukunami C, Hiraki Y, Chung UI. Tenomodulin expression in the periodontal ligament enhances cellular adhesion. PLoS One. 2013;8(4):e60203. doi: 10.1371/journal.pone.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo J, Shibata H, Miura S, Yamakawa A, Sato K, Higuchi Y, Shukunami C, Hiraki Y. A functional role of the glycosylated N-terminal domain of chondromodulin-I. J Bone Miner Metab. 2011;29(1):23–30. doi: 10.1007/s00774-010-0193-0. [DOI] [PubMed] [Google Scholar]
- Kusumoto D, Fukuda K. The role of angiogenetic factors in the pathogenesis and the progression of cardiac valve disease. Clin Calcium. 2013;23(4):481–488. [PubMed] [Google Scholar]
- Kyriakides TR, Zhu YH, Smith LT, Bain SD, Yang Z, Lin MT, Danielson KG, Iozzo RV, LaMarca M, McKinney CE, Ginns EI, Bornstein P. Mice that lack thrombospondin 2 display connective tissue abnormalities that are associated with disordered collagen fibrillogenesis, an increased vascular density, and a bleeding diathesis. J Cell Biol. 1998;140(2):419–430. doi: 10.1083/jcb.140.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Zhou Z, Taub PJ, Ramcharan M, Li Y, Akinbiyi T, Maharam ER, Leong DJ, Laudier DM, Ruike T, Torina PJ, Zaidi M, Majeska RJ, Schaffler MB, Flatow EL, Sun HB. BMP-12 treatment of adult mesenchymal stem cells in vitro augments tendon-like tissue formation and defect repair in vivo. PLoS One. 2011;6(3):e17531. doi: 10.1371/journal.pone.0017531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Clegg P, Comerford E, Canty-Laird E. The stem cell niche in tendon and ligament: Investigating alterations with ageing and disease. Orthopaedic Proceedings. 2015;97-B(SUPP 11):23–23. [Google Scholar]
- Lee WY, Lui PP, Rui YF. Hypoxia-mediated efficient expansion of human tendon-derived stem cells in vitro. Tissue Eng Part A. 2012;18(5-6):484–498. doi: 10.1089/ten.tea.2011.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejard V, Blais F, Guerquin MJ, Bonnet A, Bonnin MA, Havis E, Malbouyres M, Bidaud CB, Maro G, Gilardi-Hebenstreit P, Rossert J, Ruggiero F, Duprez D. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J Biol Chem. 2011;286(7):5855–5867. doi: 10.1074/jbc.M110.153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone L, Vetrano M, Ranieri D, Raffa S, Vulpiani MC, Ferretti A, Torrisi MR, Visco V. Extracorporeal Shock Wave Treatment (ESWT) improves in vitro functional activities of ruptured human tendon-derived tenocytes. PLoS One. 2012;7(11):e49759. doi: 10.1371/journal.pone.0049759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ramcharan M, Zhou Z, Leong DJ, Akinbiyi T, Majeska RJ, Sun HB. The Role of Scleraxis in Fate Determination of Mesenchymal Stem Cells for Tenocyte Differentiation. Sci Rep. 2015;5:13149. doi: 10.1038/srep13149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu CF, Aschbacher-Smith L, Barthelery NJ, Dyment N, Butler D, Wylie C. Spatial and temporal expression of molecular markers and cell signals during normal development of the mouse patellar tendon. Tissue Eng Part A. 2012;18(5-6):598–608. doi: 10.1089/ten.tea.2011.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CF, Aschbacher-Smith L, Barthelery NJ, Dyment N, Butler D, Wylie C. What we should know before using tissue engineering techniques to repair injured tendons: a developmental biology perspective. Tissue Eng Part B Rev. 2011;17(3):165–176. doi: 10.1089/ten.teb.2010.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu S, Zhang C, Lu P, Hu J, Yin Z, Ma Y, Chen X, OuYang H. Crucial transcription factors in tendon development and differentiation: their potential for tendon regeneration. Cell Tissue Res. 2014;356(2):287–298. doi: 10.1007/s00441-014-1834-8. [DOI] [PubMed] [Google Scholar]
- Liu J, Tao X, Chen L, Han W, Zhou Y, Tang K. CTGF positively regulates BMP12 induced tenogenic differentiation of tendon stem cells and signaling. Cell Physiol Biochem. 2015;35(5):1831–1845. doi: 10.1159/000373994. [DOI] [PubMed] [Google Scholar]
- Liu W, Watson SS, Lan Y, Keene DR, Ovitt CE, Liu H, Schweitzer R, Jiang R. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol Cell Biol. 2010;30(20):4797–4807. doi: 10.1128/MCB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorda-Diez CI, Montero JA, Martinez-Cue C, Garcia-Porrero JA, Hurle JM. Transforming growth factors beta coordinate cartilage and tendon differentiation in the developing limb mesenchyme. J Biol Chem. 2009;284(43):29988–29996. doi: 10.1074/jbc.M109.014811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovati AB, Corradetti B, Cremonesi F, Bizzaro D, Consiglio AL. Tenogenic differentiation of equine mesenchymal progenitor cells under indirect co-culture. Int J Artif Organs. 2012;35(11):996–1005. doi: 10.5301/ijao.5000129. [DOI] [PubMed] [Google Scholar]
- Mazzocca AD, Chowaniec D, McCarthy MB, Beitzel K, Cote MP, McKinnon W, Arciero R. In vitro changes in human tenocyte cultures obtained from proximal biceps tendon: multiple passages result in changes in routine cell markers. Knee Surg Sports Traumatol Arthrosc. 2011 doi: 10.1007/s00167-011-1711-x. [DOI] [PubMed] [Google Scholar]
- Mendias CL, Gumucio JP, Bakhurin KI, Lynch EB, Brooks SV. Physiological loading of tendons induces scleraxis expression in epitenon fibroblasts. J Orthop Res. 2012;30(4):606–612. doi: 10.1002/jor.21550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendias CL, Bakhurin KI, Faulkner JA. Tendons of myostatin-deficient mice are small, brittle, and hypocellular. Proc Natl Acad Sci U S A. 2008;105(1):388–393. doi: 10.1073/pnas.0707069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mienaltowski MJ, Adams SM, Birk DE. Regional differences in stem cell/progenitor cell populations from the mouse achilles tendon. Tissue Eng Part A. 2013;19(1-2):199–210. doi: 10.1089/ten.tea.2012.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minogue BM, Richardson SM, Zeef LA, Freemont AJ, Hoyland JA. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther. 2010;12(1):R22. doi: 10.1186/ar2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani G, Sato M, Yamato M, Kokubo M, Takagaki T, Ebihara G, Okano T, Mochida J. Potential utility of cell sheets derived from the anterior cruciate ligament and synovium fabricated in temperature-responsive culture dishes. J Biomed Mater Res A. 2014;102(9):2927–2933. doi: 10.1002/jbm.a.34962. [DOI] [PubMed] [Google Scholar]
- Miura S, Kondo J, Kawakami T, Shukunami C, Aimoto S, Tanaka H, Hiraki Y. Synthetic disulfide-bridged cyclic peptides mimic the anti-angiogenic actions of chondromodulin-I. Cancer Sci. 2012;103(7):1311–1318. doi: 10.1111/j.1349-7006.2012.02276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabara S, Yuda Y, Kasashima Y, Kuwano A, Arai K. Regulation of Tenomodulin Expression Via Wnt/beta-catenin Signaling in Equine Bone Marrow-derived Mesenchymal Stem Cells. J Equine Sci. 2014;25(1):7–13. doi: 10.1294/jes.25.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani N, Kageyama S, Yamada M, Hasegawa M, Miyamoto K, Horiuchi T. The behavior of ligament cells cultured on elastin and collagen scaffolds. J Artif Organs. 2014;17(1):50–59. doi: 10.1007/s10047-013-0736-y. [DOI] [PubMed] [Google Scholar]
- Mohanty N, Gulati BR, Kumar R, Gera S, Kumar P, Somasundaram RK, Kumar S. Immunophenotypic characterization and tenogenic differentiation of mesenchymal stromal cells isolated from equine umbilical cord blood. In Vitro Cell Dev Biol Anim. 2014;50(6):538–548. doi: 10.1007/s11626-013-9729-7. [DOI] [PubMed] [Google Scholar]
- Moshaverinia A, Xu X, Chen C, Ansari S, Zadeh HH, Snead ML, Shi S. Application of stem cells derived from the periodontal ligament or gingival tissue sources for tendon tissue regeneration. Biomaterials. 2014;35(9):2642–2650. doi: 10.1016/j.biomaterials.2013.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Sunohara M, Sato I. Expression of DMP1 in the developing mouse tongue embryo. Ann Anat. 2015;200:136–148. doi: 10.1016/j.aanat.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134(14):2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- Musson DS, Naot D, Chhana A, Matthews BG, McIntosh JD, Lin ST, Choi AJ, Callon KE, Dunbar PR, Lesage S, Coleman B, Cornish J. In vitro evaluation of a novel non-mulberry silk scaffold for use in tendon regeneration. Tissue Eng Part A. 2015;21(9-10):1539–1551. doi: 10.1089/ten.tea.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neame PJ, Treep JT, Young CN. An 18-kDa glycoprotein from bovine nasal cartilage. Isolation and primary structure of small, cartilage-derived glycoprotein. J Biol Chem. 1990;265(17):9628–9633. [PubMed] [Google Scholar]
- Omachi T, Sakai T, Hiraiwa H, Hamada T, Ono Y, Nakashima M, Ishizuka S, Matsukawa T, Oda T, Takamatsu A, Yamashita S, Ishiguro N. Expression of tenocyte lineage-related factors in regenerated tissue at sites of tendon defect. J Orthop Sci. 2015;20(2):380–389. doi: 10.1007/s00776-014-0684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omae H, Sun YL, An KN, Amadio PC, Zhao C. Engineered tendon with decellularized xenotendon slices and bone marrow stromal cells: an in vivo animal study. J Tissue Eng Regen Med. 2012;6(3):238–244. doi: 10.1002/term.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omae H, Zhao C, Sun YL, An KN, Amadio PC. Multilayer tendon slices seeded with bone marrow stromal cells: a novel composite for tendon engineering. J Orthop Res. 2009;27(7):937–942. doi: 10.1002/jor.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizuka N, Ito Y, Inagawa M, Nakahara H, Takada S, Lotz M, Toyama Y, Asahara H. The Mohawk homeobox transcription factor regulates the differentiation of tendons and volar plates. J Orthop Sci. 2014;19(1):172–180. doi: 10.1007/s00776-013-0485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Sato K, Tashiro F, Miyazaki J, Nishida K, Hiraki Y, Tano Y, Shukunami C. Anti-angiogenic action of the C-terminal domain of tenomodulin that shares homology with chondromodulin-I. J Cell Sci. 2004;117(Pt 13):2731–2744. doi: 10.1242/jcs.01112. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Shukunami C, Honda J, Nishida K, Tashiro F, Miyazaki J, Hiraki Y, Tano Y. Expression and localization of tenomodulin, a transmembrane type chondromodulin-I-related angiogenesis inhibitor, in mouse eyes. Invest Ophthalmol Vis Sci. 2003;44(5):1814–1823. doi: 10.1167/iovs.02-0664. [DOI] [PubMed] [Google Scholar]
- Otabe K, Nakahara H, Hasegawa A, Matsukawa T, Ayabe F, Onizuka N, Inui M, Takada S, Ito Y, Sekiya I, Muneta T, Lotz M, Asahara H. Transcription factor Mohawk controls tenogenic differentiation of bone marrow mesenchymal stem cells in vitro and in vivo. J Orthop Res. 2015;33(1):1–8. doi: 10.1002/jor.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A, Hogan MV, Kesturu GS, James R, Balian G, Chhabra AB. Adipose-derived mesenchymal stem cells treated with growth differentiation factor-5 express tendon-specific markers. Tissue Eng Part A. 2010;16(9):2941–2951. doi: 10.1089/ten.tea.2009.0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly S, Klatte F, Strobel C, Schmidmaier G, Greiner S, Scheibel M, Wildemann B. Characterization of tendon cell cultures of the human rotator cuff. Eur Cell Mater. 2010;20:84–97. doi: 10.22203/ecm.v020a08. [DOI] [PubMed] [Google Scholar]
- Pawelec KM, Wardale RJ, Best SM, Cameron RE. The effects of scaffold architecture and fibrin gel addition on tendon cell phenotype. J Mater Sci Mater Med. 2015;26(1):5349. doi: 10.1007/s10856-014-5349-3. [DOI] [PubMed] [Google Scholar]
- Peach MS, James R, Toti US, Deng M, Morozowich NL, Allcock HR, Laurencin CT, Kumbar SG. Polyphosphazene functionalized polyester fiber matrices for tendon tissue engineering: in vitro evaluation with human mesenchymal stem cells. Biomed Mater. 2012;7(4):045016. doi: 10.1088/1748-6041/7/4/045016. [DOI] [PubMed] [Google Scholar]
- Pisani DF, Pierson PM, Massoudi A, Leclerc L, Chopard A, Marini JF, Dechesne CA. Myodulin is a novel potential angiogenic factor in skeletal muscle. Exp Cell Res. 2004;292(1):40–50. doi: 10.1016/j.yexcr.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Pisani DF, Rivoyre MD, Ruel L, Bonino F, Bidet M, Dechesne CA, Mus-Veteau I. Mouse myodulin, a new potential angiogenic factor, functionally expressed in yeast. Biochem Biophys Res Commun. 2005;331(2):552–556. doi: 10.1016/j.bbrc.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Pryce BA, Watson SS, Murchison ND, Staverosky JA, Dunker N, Schweitzer R. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development. 2009;136(8):1351–1361. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Dmochowski JM, Banes AN, Tsuzaki M, Bynum D, Patterson M, Creighton A, Gomez S, Tech K, Cederlund A, Banes AJ. Differential expression and cellular localization of novel isoforms of the tendon biomarker tenomodulin. J Appl Physiol (1985) 2012;113(6):861–871. doi: 10.1152/japplphysiol.00198.2012. [DOI] [PubMed] [Google Scholar]
- Qin TW, Sun YL, Thoreson AR, Steinmann SP, Amadio PC, An KN, Zhao C. Effect of mechanical stimulation on bone marrow stromal cell-seeded tendon slice constructs: a potential engineered tendon patch for rotator cuff repair. Biomaterials. 2015;51:43–50. doi: 10.1016/j.biomaterials.2015.01.070. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Wang X, Zhang Y, Rout R, Carr AJ, Zhu L, Xia Z, Sabokbar A. Development of a refined tenocyte differentiation culture technique for tendon tissue engineering. Cells Tissues Organs. 2013;197(1):27–36. doi: 10.1159/000341426. [DOI] [PubMed] [Google Scholar]
- Rothan HA, Suhaeb AM, Kamarul T. Recombinant human adiponectin as a potential protein for treating diabetic tendinopathy promotes tenocyte progenitor cells proliferation and tenogenic differentiation in vitro. Int J Med Sci. 2013;10(13):1899–1906. doi: 10.7150/ijms.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Azpeitia E, Sanchez P, Delgado D, Andia I. Three-Dimensional Platelet-Rich Plasma Hydrogel Model to Study Early Tendon Healing. Cells Tissues Organs. 2015;200(6):394–404. doi: 10.1159/000441053. [DOI] [PubMed] [Google Scholar]
- Rui YF, Lui PP, Li G, Fu SC, Lee YW, Chan KM. Isolation and characterization of multipotent rat tendon-derived stem cells. Tissue Eng Part A. 2010;16(5):1549–1558. doi: 10.1089/ten.TEA.2009.0529. [DOI] [PubMed] [Google Scholar]
- Rui YF, Lui PP, Wong YM, Tan Q, Chan KM. Altered fate of tendon-derived stem cells isolated from a failed tendon-healing animal model of tendinopathy. Stem Cells Dev. 2013;22(7):1076–1085. doi: 10.1089/scd.2012.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V, Mauro A, Martelli A, Di Giacinto O, Di Marcantonio L, Nardinocchi D, Berardinelli P, Barboni B. Cellular and molecular maturation in fetal and adult ovine calcaneal tendons. J Anat. 2015;226(2):126–142. doi: 10.1111/joa.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki A, Olsson M, Jernas M, Gummesson A, McTernan PG, Andersson J, Jacobson P, Sjoholm K, Olsson B, Yamamura S, Walley A, Froguel P, Carlsson B, Sjostrom L, Svensson PA, Carlsson LM. Tenomodulin is highly expressed in adipose tissue, increased in obesity, and down-regulated during diet-induced weight loss. The Journal of clinical endocrinology and metabolism. 2009;94(10):3987–3994. doi: 10.1210/jc.2009-0292. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Valencia A. BRICHOS: a conserved domain in proteins associated with dementia, respiratory distress and cancer. Trends Biochem Sci. 2002;27(7):329–332. doi: 10.1016/s0968-0004(02)02134-5. [DOI] [PubMed] [Google Scholar]
- Sassoon AA, Ozasa Y, Chikenji T, Sun YL, Larson DR, Maas ML, Zhao C, Jen J, Amadio PC. Skeletal muscle and bone marrow derived stromal cells: a comparison of tenocyte differentiation capabilities. J Orthop Res. 2012;30(11):1710–1718. doi: 10.1002/jor.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato I, Miwa Y, Hara S, Fukuyama Y, Sunohara M. Tenomodulin regulated the compartments of embryonic and early postnatal mouse masseter muscle. Ann Anat. 2014;196(6):410–415. doi: 10.1016/j.aanat.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Schneider PR, Buhrmann C, Mobasheri A, Matis U, Shakibaei M. Three-dimensional high-density co-culture with primary tenocytes induces tenogenic differentiation in mesenchymal stem cells. J Orthop Res. 2011;29(9):1351–1360. doi: 10.1002/jor.21400. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128(19):3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- Scott A, Sampaio A, Abraham T, Duronio C, Underhill TM. Scleraxis expression is coordinately regulated in a murine model of patellar tendon injury. J Orthop Res. 2011;29(2):289–296. doi: 10.1002/jor.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Gelberman RH, Silva MJ, Sakiyama-Elbert SE, Thomopoulos S. BMP12 induces tenogenic differentiation of adipose-derived stromal cells. PLoS One. 2013;8(10):e77613. doi: 10.1371/journal.pone.0077613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Wada S, Inoue K, Ideno H, Kamiunten T, Komatsu K, Kudo A, Nakamura Y, Sato T, Nakashima K, Nifuji A. Efficient expansion of mouse primary tenocytes using a novel collagen gel culture method. Histochem Cell Biol. 2014;142(2):205–215. doi: 10.1007/s00418-014-1191-4. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Oshima Y, Hiraki Y. Molecular cloning of tenomodulin, a novel chondromodulin-I related gene. Biochem Biophys Res Commun. 2001;280(5):1323–1327. doi: 10.1006/bbrc.2001.4271. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Oshima Y, Hiraki Y. Chondromodulin-I and tenomodulin: a new class of tissue-specific angiogenesis inhibitors found in hypovascular connective tissues. Biochem Biophys Res Commun. 2005;333(2):299–307. doi: 10.1016/j.bbrc.2005.05.133. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol. 2006;298(1):234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Takimoto A, Miura S, Nishizaki Y, Hiraki Y. Chondromodulin-I and tenomodulin are differentially expressed in the avascular mesenchyme during mouse and chick development. Cell Tissue Res. 2008;332(1):111–122. doi: 10.1007/s00441-007-0570-8. [DOI] [PubMed] [Google Scholar]
- Subramony SD, Su A, Yeager K, Lu HH. Combined effects of chemical priming and mechanical stimulation on mesenchymal stem cell differentiation on nanofiber scaffolds. J Biomech. 2014;47(9):2189–2196. doi: 10.1016/j.jbiomech.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Takimoto A, Akiyama H, Kist R, Scherer G, Nakamura T, Hiraki Y, Shukunami C. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013;140(11):2280–2288. doi: 10.1242/dev.096354. [DOI] [PubMed] [Google Scholar]
- Takimoto A, Oro M, Hiraki Y, Shukunami C. Direct conversion of tenocytes into chondrocytes by Sox9. Exp Cell Res. 2012;318(13):1492–1507. doi: 10.1016/j.yexcr.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Tan Q, Lui PP, Rui YF. Effect of in vitro passaging on the stem cell-related properties of tendon-derived stem cells-implications in tissue engineering. Stem Cells Dev. 2012a;21(5):790–800. doi: 10.1089/scd.2011.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Lui PP, Rui YF, Wong YM. Comparison of potentials of stem cells isolated from tendon and bone marrow for musculoskeletal tissue engineering. Tissue Eng Part A. 2012b;18(7-8):840–851. doi: 10.1089/ten.tea.2011.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SW, Tong WY, Shen W, Yeung KW, Lam YW. Stringent requirement for spatial arrangement of extracellular matrix in supporting cell morphogenesis and differentiation. BMC Cell Biol. 2014;15:10. doi: 10.1186/1471-2121-15-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempfer H, Wagner A, Gehwolf R, Lehner C, Tauber M, Resch H, Bauer HC. Perivascular cells of the supraspinatus tendon express both tendon- and stem cell-related markers. Histochem Cell Biol. 2009;131(6):733–741. doi: 10.1007/s00418-009-0581-5. [DOI] [PubMed] [Google Scholar]
- Theiss F, Mirsaidi A, Mhanna R, Kummerle J, Glanz S, Bahrenberg G, Tiaden AN, Richards PJ. Use of biomimetic microtissue spheroids and specific growth factor supplementation to improve tenocyte differentiation and adaptation to a collagen-based scaffold in vitro. Biomaterials. 2015;69:99–109. doi: 10.1016/j.biomaterials.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Tokunaga T, Shukunami C, Okamoto N, Taniwaki T, Oka K, Sakamoto H, Ide J, Mizuta H, Hiraki Y. FGF-2 Stimulates the Growth of Tenogenic Progenitor Cells to Facilitate the Generation of Tenomodulin-Positive Tenocytes in a Rat Rotator Cuff Healing Model. Am J Sports Med. 2015;43(10):2411–2422. doi: 10.1177/0363546515597488. [DOI] [PubMed] [Google Scholar]
- Tolppanen AM, Helisalmi S, Hiltunen M, Kolehmainen M, Schwab U, Pirttila T, Pulkkinen L, Uusitupa M, Soininen H. Tenomodulin variants, APOE and Alzheimer's disease in a Finnish case-control cohort. Neurobiology of aging. 2011;32(3):546 e547–549. doi: 10.1016/j.neurobiolaging.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Tolppanen AM, Nevalainen T, Kolehmainen M, Seitsonen S, Immonen I, Uusitupa M, Kaarniranta K, Pulkkinen L. Single nucleotide polymorphisms of the tenomodulin gene (TNMD) in age-related macular degeneration. Molecular vision. 2009;15:762–770. [PMC free article] [PubMed] [Google Scholar]
- Tolppanen AM, Pulkkinen L, Herder C, Koenig W, Kolehmainen M, Lindstrom J, Tuomilehto J, Uusitupa M. The genetic variation of the tenomodulin gene (TNMD) is associated with serum levels of systemic immune mediators--the Finnish Diabetes Prevention Study. Genetics in medicine : official journal of the American College of Medical Genetics. 2008a;10(7):536–544. doi: 10.1097/gim.0b013e3181772129. [DOI] [PubMed] [Google Scholar]
- Tolppanen AM, Pulkkinen L, Kolehmainen M, Schwab U, Lindstrom J, Tuomilehto J, Uusitupa M. Tenomodulin is associated with obesity and diabetes risk: the Finnish diabetes prevention study. Obesity. 2007;15(5):1082–1088. doi: 10.1038/oby.2007.613. [DOI] [PubMed] [Google Scholar]
- Tolppanen AM, Pulkkinen L, Kuulasmaa T, Kolehmainen M, Schwab U, Lindstrom J, Tuomilehto J, Uusitupa M, Kuusisto J. The genetic variation in the tenomodulin gene is associated with serum total and LDL cholesterol in a body size-dependent manner. International journal of obesity. 2008b;32(12):1868–1872. doi: 10.1038/ijo.2008.217. [DOI] [PubMed] [Google Scholar]
- Tong WY, Shen W, Yeung CW, Zhao Y, Cheng SH, Chu PK, Chan D, Chan GC, Cheung KM, Yeung KW, Lam YW. Functional replication of the tendon tissue microenvironment by a bioimprinted substrate and the support of tenocytic differentiation of mesenchymal stem cells. Biomaterials. 2012;33(31):7686–7698. doi: 10.1016/j.biomaterials.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Tozer S, Duprez D. Tendon and ligament: development, repair and disease. Birth Defects Res C Embryo Today. 2005;75(3):226–236. doi: 10.1002/bdrc.20049. [DOI] [PubMed] [Google Scholar]
- Vincent AK, Noor A, Janson A, Minassian BA, Ayub M, Vincent JB, Morel CF. Identification of genomic deletions spanning the PCDH19 gene in two unrelated girls with intellectual disability and seizures. Clin Genet. 2012;82(6):540–545. doi: 10.1111/j.1399-0004.2011.01812.x. [DOI] [PubMed] [Google Scholar]
- Violini S, Ramelli P, Pisani LF, Gorni C, Mariani P. Horse bone marrow mesenchymal stem cells express embryo stem cell markers and show the ability for tenogenic differentiation by in vitro exposure to BMP-12. BMC Cell Biol. 2009;10:29. doi: 10.1186/1471-2121-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ideno H, Shimada A, Kamiunten T, Nakamura Y, Nakashima K, Kimura H, Shinkai Y, Tachibana M, Nifuji A. H3K9MTase G9a is essential for the differentiation and growth of tenocytes in vitro. Histochem Cell Biol. 2015;144(1):13–20. doi: 10.1007/s00418-015-1318-2. [DOI] [PubMed] [Google Scholar]
- Wagenhauser MU, Pietschmann MF, Docheva D, Gulecyuz MF, Jansson V, Muller PE. Assessment of essential characteristics of two different scaffolds for tendon in situ regeneration. Knee Surg Sports Traumatol Arthrosc. 2015;23(4):1239–1246. doi: 10.1007/s00167-013-2820-5. [DOI] [PubMed] [Google Scholar]
- Wagenhauser MU, Pietschmann MF, Sievers B, Docheva D, Schieker M, Jansson V, Muller PE. Collagen type I and decorin expression in tenocytes depend on the cell isolation method. BMC Musculoskelet Disord. 2012;13:140. doi: 10.1186/1471-2474-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li J, Wang K, Zhang Z, Zhang W, Zhou G, Cao Y, Ye M, Zou H, Liu W. Induction of predominant tenogenic phenotype in human dermal fibroblasts via synergistic effect of TGF-beta and elongated cell shape. Am J Physiol Cell Physiol. 2015 doi: 10.1152/ajpcell.00300.2015. ajpcell 00300 02015. [DOI] [PubMed] [Google Scholar]
- Wang W, Li Z, Sato T, Oshima Y. Tenomodulin inhibits retinal neovascularization in a mouse model of oxygen-induced retinopathy. Int J Mol Sci. 2012;13(11):15373–15386. doi: 10.3390/ijms131115373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki J, Yamaguchi T, Enomoto A, Irie T, Yoshie K, Tachikawa T, Maki K. Identification of differentially expressed genes in mandibular condylar and tibial growth cartilages using laser microdissection and fluorescent differential display: chondromodulin-I (ChM-1) and tenomodulin (TeM) are differentially expressed in mandibular condylar and other growth cartilages. Bone. 2008;42(6):1053–1060. doi: 10.1016/j.bone.2007.09.048. [DOI] [PubMed] [Google Scholar]
- Watts AE, Nixon AJ, Yeager AE, Mohammed HO. A collagenase gel/physical defect model for controlled induction of superficial digital flexor tendonitis. Equine Vet J. 2012;44(5):576–586. doi: 10.1111/j.2042-3306.2011.00471.x. [DOI] [PubMed] [Google Scholar]
- Watts AE, Yeager AE, Kopyov OV, Nixon AJ. Fetal derived embryonic-like stem cells improve healing in a large animal flexor tendonitis model. Stem Cell Res Ther. 2011;2(1):4. doi: 10.1186/scrt45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Sumita Y, Liu Y, Tai Y, Wang J, Uehara M, Agata H, Kagami H, Fan Z, Asahina I, Wang S, Tran SD. GDFs promote tenogenic characteristics on human periodontal ligament-derived cells in culture at late passages. Growth Factors. 2013;31(5):165–173. doi: 10.3109/08977194.2013.830611. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang Q, Li Y, Gan Y, Li P, Li S, Zhou Y, Zhou Q. Cyclic Tensile Strain Induces Tenogenic Differentiation of Tendon-Derived Stem Cells in Bioreactor Culture. Biomed Res Int. 2015;2015:790804. doi: 10.1155/2015/790804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh LC, Tsai AD, Lee JC. Cartilage-derived morphogenetic proteins induce osteogenic gene expression in the C2C12 mesenchymal cell line. J Cell Biochem. 2005;95(1):173–188. doi: 10.1002/jcb.20402. [DOI] [PubMed] [Google Scholar]
- Yeh LC, Tsai AD, Lee JC. Bone morphogenetic protein-7 regulates differentially the mRNA expression of bone morphogenetic proteins and their receptors in rat achilles and patellar tendon cell cultures. J Cell Biochem. 2008;104(6):2107–2122. doi: 10.1002/jcb.21768. [DOI] [PubMed] [Google Scholar]
- Younesi M, Islam A, Kishore V, Anderson JM, Akkus O. Tenogenic Induction of Human MSCs by Anisotropically Aligned Collagen Biotextiles. Adv Funct Mater. 2014;24(36):5762–5770. doi: 10.1002/adfm.201400828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukata K, Matsui Y, Shukunami C, Takimoto A, Hirohashi N, Ohtani O, Kimura T, Hiraki Y, Yasui N. Differential expression of Tenomodulin and Chondromodulin-1 at the insertion site of the tendon reflects a phenotypic transition of the resident cells. Tissue Cell. 2010;42(2):116–120. doi: 10.1016/j.tice.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang JH. The effects of mechanical loading on tendons--an in vivo and in vitro model study. PLoS One. 2013;8(8):e71740. doi: 10.1371/journal.pone.0071740. [DOI] [PMC free article] [PubMed] [Google Scholar]


