Abstract
Streptococcus suis serotype 2 is an emerging zoonotic pathogen that triggered two outbreaks of streptococcal toxic shock syndrome (STSS) in China. Our previous research demonstrated that a type IV secretion system (T4SS) harbored in the 89K pathogenicity island contributes to the pathogenicity of S. suis 2. In the present study, a shotgun proteomics approach was employed to identify the effectors secreted by T4SS in S. suis 2, and surface-associated subtilisin-like protease-1 (SspA-1) was identified as a potential virulence effector. Western blot analysis and pull-down assay revealed that SspA-1 secretion depends on T4SS. Knockout mutations affecting sspA-1 attenuated S. suis 2 and impaired the pathogen’s ability to trigger inflammatory response in mice. And purified SspA-1 induced the secretion of IL-6, TNF-α, and IL-12p70 in THP-1 cells directly. SspA-1 is the first T4SS virulence effector reported in Gram-positive bacteria. Overall, these findings allow us to gain further insights into the pathogenesis of T4SS and STSS.
Bacteria are equipped with various mechanisms to secrete proteins essential for their pathogenicity and survival. In addition to the typical general secretion (Sec) and twin-arginine translocation (Tat) systems1, at least seven specialized protein-secretion systems have been reported in bacteria2,3,4. Among the known secretion systems, type IV secretion systems (T4SSs) are the most versatile and widespread in archaea and bacteria5. T4SSs are also unique because of their ability to transport DNA substrates, and pathogenic effectors across the cell envelope. However, studies on T4SSs have mainly focused on Gram-negative bacteria, such as the VirB/D4 system from Agrobacterium tumefaciens and closely related systems from Escherichia coli encoded by conjugative plasmids6. Hence, limited information is available regarding Gram-positive T4SSs and their substrates.
Streptococcus suis serotype 2 (S. suis 2) is a Gram-positive zoonotic pathogen responsible for a variety of life-threatening infections in humans and pigs, such as meningitis, pneumonia, arthritis, and septicaemia7,8. In the two human outbreaks caused by S. suis 2 in China (1998 and 2005), a high proportion of patients manifested the typical symptoms of streptococcal toxic shock syndrome (STSS) characterized by a very short disease course and high mortality9,10. As such, the emergence of highly pathogenic S. suis 2 poses a serious threat to public health.
However, the pathogenetic mechanisms employed by the highly pathogenic S. suis 2 have yet to be clarified. Chen et al.11 conducted a comparative genomics research and found that a unique pathogenicity island (PAI) designated as 89K is specific to the two epidemic strains, namely, 98HAH12 and 05ZYH33, which caused the STSS outbreaks. We demonstrated that a functional VirB/D4 T4SS located at the 5′-end of 89 K mediates the conjugal transfer of 89 K, and this T4SS may secrete some unknown effectors triggering an excessive host inflammatory response and inducing STSS12,13. Therefore, the repertoire of T4SS effectors should be identified to elucidate the pathogenic mechanisms of STSS caused by S. suis 2.
In the current study, a shotgun proteomics strategy14,15 was applied to analyze the secretome of S. suis 2 and a T4SS-deficient mutant. A T4SS effector termed surface-associated subtilisin-like protease-1 (SspA-1), which belongs to the subtilase family, was identified using this strategy. SspA-1 can trigger an excessive inflammatory response in a mouse infection model and further promote STSS development. To our knowledge, SspA-1 is the first T4SS effector identified in Gram-positive bacteria. Overall, our research provided significant insights into the pathogenesis of the highly virulent S. suis 2 strain.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1. S. suis 2 strains were cultured in Todd–Hewitt broth containing 2% yeast extract. E. coli strains were grown in Luria-Bertani medium. If necessary, antibiotics were added to the media with the following concentrations: 100 mg/L spectinomycin, 100 mg/L ampicillin, 50 mg/L kanamycin, 1 mg/L erythromycin for S. suis, and 250 mg/L erythromycin for E. coli.
Preparation of culture supernatants and whole-cell proteins
Cultures of different S. suis 2 strains harvested in the late exponential growth phase were centrifuged at 10,000 × g for 10 min at 4 °C. Supernatants and cell pellets were prepared as follows. The supernatants were precipitated with acetone–trichloroacetic acid in accordance with previously described methods16. The cell pellets were washed with PBS, resuspended in a lysis buffer (50 mM Tris–HCl, 2 mM EDTA, 100 mM NaCl, 0.5% Triton X-100, 10 mg/ml lysozyme, and protease inhibitor cocktail at pH 8.5–9.0), and incubated at 37 °C for 4 h. After disruption was performed with three cycles of alternating ultrasound and freezing/thawing, the lysates were centrifuged at 2,000 × g for 5 min to remove debris. The resulting supernatants were collected as whole-cell proteins.
LC-MS/MS analysis
The precipitated proteins from the culture supernatants of wild-type S. suis 2 05ZYH33 and T4SS-deficient mutant strain (ΔvirD4-89K) were analyzed through LC-MS/MS to identify the putative proteins secreted via T4SS. Protein samples were separated through sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), pretreated with trypsin, and analyzed through LC-MS/MS by using UltiMate3000 RSLCnano liquid chromatography/Bruker maxis 4G Q-TOF. The resulting peptide mass fingerprints were compared against the ORFs of the genome of 05ZYH33 by using Mascot and Mascot Daemon software (Matrix Science); matches with P < 0.05 were considered with high confidence. Signal peptides were predicted using SignalP 4.1 server.
Construction of sspA-1 knockout mutant and complemented CΔsspA-1 strain
The sspA-1 mutant was generated through allelic replacement with a spectinomycin (spc) resistance gene cassette as previously described17. The sspA-1 upstream flanking sequence (left arm) was cloned as an EcoR I/BamH I fragment and the downstream flanking sequence (right arm) as a Pst I/Hind III fragment at both sides of the spectinomycin resistance gene in the pUC18 plasmid. The primers used to amplify the left and right arms are listed in Table S2. The recombinant plasmid was electrotransformed into competent cells of 05ZYH33. Positive transformants, which were resistant to spectinomycin, were further confirmed through PCR and DNA sequencing. A double-crossover mutant of sspA-1 was isolated and designated as ΔsspA-1.
A 5049 bp fragment containing sspA-1 and the promotor sequence was divided into two sequential fragments and amplified from the 05ZYH33 chromosome by using the PCR primers CsspA-1-F/CsspA-1-MR and CsspA-1-MF/CsspA-1-R (Table S2). The resulting PCR products were digested with BamH I/Sph I and Sph I/EcoR I and then cloned into E. coli–S. suis pVA838 shuttle vector successively, as a result, pVA838-sspA-1 was generated. After the result was verified through DNA sequencing, the resulting plasmid was electrotransformed into ΔsspA-1 and designated as the complemented strain CΔsspA-1.
Quantitative real-time PCR (qRT-PCR)
Total RNA of various S. suis 2 strains was extracted from cultures grown to the late exponential phase by using an SV total RNA isolation system (Promega). RNA was reverse transcribed to cDNA by using a Transcriptor first-strand cDNA synthesis kit (Roche). qRT-PCR was conducted using SYBR premix Ex TaqTM (TaKaRa) in an Eco Real-Time PCR System (Illumina). Levels of 16S rRNA were used as internal control18. The primers used for qRT-PCR are shown in Table S2. The fold changes of the sspA-1 transcripts were quantified using a comparative threshold cycle (ΔΔCT) program in the Eco software package. The assays were performed in triplicate.
Cloning, overexpression, and purification of recombinant proteins
The possible B-cell epitopes of SspA-1 were analyzed and a 2349 bp DNA fragment encoding all the predicted functional domains of SspA-1 was selected and cloned into the pET-28a expression vector by using the primers listed in Table S2. E. coli BL21 harboring the SspA-1-expressing plasmid was induced with 1 mM IPTG at 30 °C for 6 h. Cells were harvested and resuspended in PBS containing 1 mM PMSF. After disruption was performed through ultrasound in an ice bath, cell lysates were centrifuged, and supernatant was collected and filtered through a 0.45 μm membrane. His–SspA-1 in the cleared supernatant was purified using a His GraviTrap column (Bio-Rad) in accordance with the manufacturer’s instructions.
VirD4–89 K was overexpressed with the GST fusion vector pGEX-6P-1 in E. coli BL21, and this process was similar to that applied to induce His–SspA-1. GST–VirD4 and GST alone were purified using gravity columns (Bio-Rad) with a Uniflow glutathione resin in accordance with the manufacturer’s recommended protocol.
Preparation of polyclonal antibodies against SspA-1
Five six-week-old BALB/c mice were immunized subcutaneously and intraperitoneally on days 0, 14, and 28 with 40 μg of recombinant SspA-1 formulated with a complete Freund adjuvant (Sigma) for the first dose, incomplete Freund adjuvant (Sigma) for the second dose, and SspA-1 alone for the third inoculation per mouse. Sera were collected from the mice at 7 days after the last immunization19. Western blot analysis and enzyme-linked immunosorbent assay (ELISA) were performed to determine the specificity and titers of the antibodies against SspA-1. The polyclonal antibodies against SspA-2 were prepared in the same manner. The animal experiments were performed in accordance with the International Guiding Principles for Biomedical Research involving Animals-1985 and approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University.
Western blot analysis
The protein concentrations in whole-cell lysates and culture supernatants were calculated using a Bradford protein assay kit (Beyotime) or a Bio-Rad DC protein assay kit. An equal amount of total protein from each sample was loaded and separated in 10% SDS–PAGE. The proteins were transferred to a polyvinylidene fluoride membrane through electrophoresis. The membrane was blocked in 5% milk for 1 h at room temperature and incubated with primary antibodies at 4 °C overnight. After the membrane was washed thrice in PBST buffer, the membrane was incubated with a horseradish-peroxidase-conjugated secondary antibody (ZSGB-BIO) for 1 h at room temperature. The signals were detected using SuperSignal West Pico (Pierce) and Image Station 4000MM PRO Digital Imaging System (Kodak). The primary antibodies used in this study were polyclonal mouse anti-SspA-1 (prepared in our laboratory), monoclonal mouse anti-RNA polymerase subunit RpoB (Abcam), and monoclonal mouse anti-GST antibody (ZSGB-BIO).
GST pull-down assay
The purified GST–VirD4 or GST (negative control) was mixed with glutathione beads equilibrated with TBST (20 mM Tris–HCl, 200 mM NaCl, and 1% Tween 20 at pH 8.0) at 4 °C overnight. After the beads were washed and resuspended in TBST, the purified His–SspA-1 or total protein from ΔvirD4–89K lysates, which contained the abundant native SspA-1, was added. Bovine serum albumin (BSA) and ΔsspA-1 lysates were used as negative controls of the prey. The mixture was incubated with rotation at 4 °C overnight; the beads were then washed thoroughly with TBST. The final samples and the input samples were subjected to SDS–PAGE and Western blot to detect His–SspA-1, native SspA-1, GST, and GST–VirD420,21.
Virulence studies
A total of 40 four-week-old female BALB/c mice were randomly allocated to four groups (10 animals per group), and each mouse was challenged by intraperitoneally injecting wild-type S. suis 2, T4SS-deficient mutant (ΔvirD4-89K), ΔsspA-1, or CΔsspA-1 strains at a dose of 2.5 × 107 CFU/mice. Clinical signs and survival times were observed and recorded for 7 days after infection.
Measurement of inflammatory cytokines in infected mice
A total of 60 four-week-old BALB/c mice were randomly assigned to three groups, and each mouse was injected intraperitoneally with wild-type S. suis 2, T4SS-deficient mutant (ΔvirD4-89K), or ΔsspA-1 at a dose of 106 CFU/mouse. At 4, 8, 12, and 16 h after infection, five mice per group were euthanized, and serum samples were collected in accordance with previously described methods13. Cytokines were quantified using a Quantikine ELISA kit (R&D Systems) in accordance with the manufacturer’s recommendations. In addition, the bacterial loads in the blood samples of infected mice were determined by the drop plate method.
Induction of inflammatory cytokines in THP-1 cells
The human leukemia monocytic cell line THP-1 was cultivated in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C in a 5% CO2 atmosphere and induced by PMA (200 ng/ml). When appropriate, cells were seeded in a 24-well plate (5 × 106 cells/well) and treated separately with purified SspA-1 (100 μg/ml), Trypsin (100 μg/ml), SspA-1 (100 μg/ml) with polymyxin B (1 μg/ml), LPS (5 μg/ml), and PBS for 24 hours. Trypsin and LPS were negative control and positive control respectively. Polymyxin B was used to block the effect of any contaminating LPS in the purified SspA-1. After these treatments, the cell culture supernatants were collected and the cytokines were quantified as previously described.
Immunization and challenge of mice
A total of 20 six-week-old female BALB/c mice were randomly assigned to two groups. Group 1 was immunized as described in the preparation of polyclonal antibodies against SspA-1. Group 2 was immunized in the same way except that the recombinant SspA-1 was replaced with PBS. Blood samples were collected before each vaccination to determine the antibody response. Mouse serum titers were determined through ELISA in accordance with previously described methods22. One week after the third vaccination, the mice were challenged intraperitoneally with wild-type S. suis 2 at a dose of 108 CFU/mouse. Survival times were recorded for 7 days after infection.
Statistical analysis
Data were analyzed using Wilcoxon rank sum test, Kruskal–Wallis test, or Nemenyi test as appropriate. A value of P < 0.05 was considered significant.
Results
Screening of putative effectors of T4SS
A shotgun proteomics strategy was applied to identify the T4SS-dependent effectors secreted into the extracellular matrix and compare the secretomes of wild-type S. suis 2 (05ZYH33) and a T4SS-deficient mutant (ΔvirD4-89K), which was attenuated in virulence and defective in T4SS-dependent horizontal gene transfer12,13. The extracellular proteins from the wild-type and T4SS-deficient mutant cultures in the late exponential phase were identified through LC-MS/MS. Three independent experiments were performed. Only the proteins detected at least twice in the wild-type supernatant and absent in the T4SS-deficient mutant supernatants were considered as putative T4SS effectors. Seven proteins satisfied these criteria (Table 1). The six other candidates, except prolyl–tRNA synthetase, exhibited putative signal peptides; these candidates are also predicted as extracellularly secreted. Among these proteins, two subtilisin-like serine proteases designated as SspA-1 and SspA-2 which were both detected three times in the wild-type supernatants were examined. Some members of the subtilase family are virulence determinants of S. suis 2 and can trigger a proinflammatory response in macrophages23,24,25,26. Therefore, these two putative effectors may be correlated with the development of STSS caused by S. suis 2. As such, the two putative effectors were further examined.
Table 1. Putative effectors of T4SS by shotgun proteomic analysis.
| Locus | GI | Mass | Score | No. of peptides matched (% coverage) | Product | Signal peptide position |
|---|---|---|---|---|---|---|
| 05SSU1982 | gi|146319636 | 186968 | 105 | 8(6) | Subtilisin-like serine protease (SspA-1) | 1–40 |
| 05SSU0552 | gi|146318206 | 31367 | 50 | 3(2) | Amino acid ABC transporter substrate-binding protein | 1–30 |
| 05SSU0811 | gi|146318465 | 31665 | 41 | 4(3) | Subtilisin-like serine protease (SspA-2) | 1–19 |
| 05SSU2133 | gi|146319787 | 44302 | 27 | 1(1) | Sugar ABC transporter substrate-binding protein | 1–34 |
| 05SSU1311 | gi|146318965 | 117795 | 22 | 19(1) | Hypothetical protein | 1–32 |
| 05SSU1961 | gi|146319615 | 68516 | 16 | 9(1) | Prolyl-tRNA synthetase | None |
| 05SSU0214 | gi|146317870 | 137100 | 14 | 2(1) | Sugar ABC transporter periplasmic protein | 1–25 |
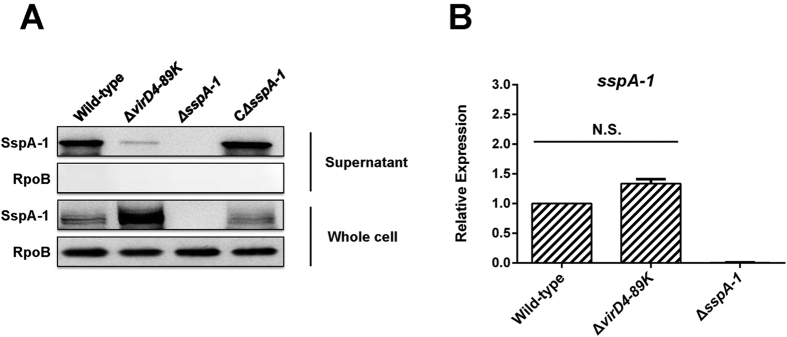
SspA-1 is secreted in a T4SS-dependent manner
The culture supernatants and pellets from a wild-type strain, T4SS-deficient mutant (ΔvirD4-89K), ΔsspA-1, or CΔsspA-1 were harvested and analyzed through Western blot with polyclonal antibodies against the two putative protein effectors. SspA-1 is abundant in the culture supernatants from the wild-type strain but is barely detected in those of ΔvirD4–89K (Fig. 1A). By contrast, the reactivity bands of SspA-1 were significantly stronger in the cell pellets of ΔvirD4–89K than in the wild-type strain. The results indicate that the deficiency of T4SS blocks the secretion of SspA-1 and leads to the accumulation of SspA-1 in the mutant cells. However, Western blot analysis revealed that the SspA-2 levels did not show any significant difference between the culture supernatants and pellets from S. suis 2 wild-type strain and T4SS-deficient mutant (data not shown). Thus, we focused on SspA-1.
Figure 1. SspA-1 is secreted in a T4SS-dependent manner.
(A) Western blot analysis comparing the levels of SspA-1 in the supernatants and cell pellets of the designated S. suis 2 strains. RNA polymerase subunit RpoB was used as loading control. (B) sspA-1 transcripts quantified through real-time PCR. N. S. indicates no significant difference (P > 0.05) between S. suis 2 wild-type stain 05ZYH33 and T4SS-deficient mutant (ΔvirD4–89K).
RT-PCR assay was conducted to evaluate sspA-1 transcription, as well as to exclude the possibility that T4SS may affect this transcription. As shown in Fig. 1B, the sspA-1 transcription in the wild-type strain did not significantly differ from that of the T4SS-deficient mutant; therefore, various SspA-1 levels in the supernatants are not attributed to transcriptional differences.
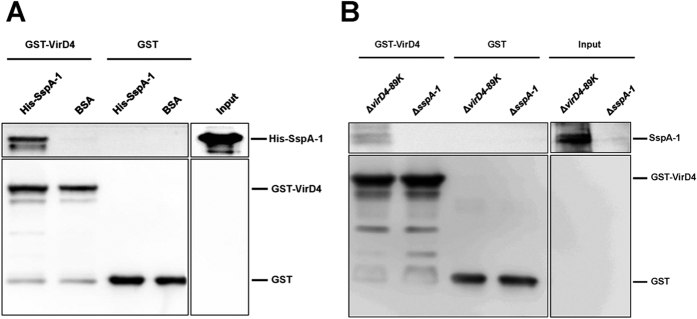
Interaction between SspA-1 and VirD4–89K
In T4SS, VirD4 acts as a type 4 coupling protein, which binds T4SS substrates directly before these substrates are delivered into the protein channel. As a result, the interaction between SspA-1 and VirD4–89K was investigated through pull-down assays. In one assay, GST–VirD4 was incubated with a purified truncated poly-histidine-tagged SspA-1 (His–SspA-1), which contained all the predicted functional domains of SspA-1. In the other assay, native SspA-1, which exists in ΔvirD4–89K lysates, was used instead of His–SspA-1. GST–VirD4 was co-precipitated with purified (Fig. 2A) or native (Fig. 2B) SspA-1; by contrast, GST alone as a negative control was not co-precipitated with either of the SspA-1 types. This finding suggested that SspA-1 directly interacts with VirD4–89K.
Figure 2. Direct interaction between SspA-1 and VirD4–89K.
(A) Pull-down assay between GST–VirD4 and purified His–SspA-1. GST–VirD4 or GST alone was pre-absorbed with glutathione beads and mixed with purified His-SspA-1 or BSA. GST or BSA was used as a negative control for bait or prey, respectively. (B) Pull-down assay between GST–VirD4 and native SspA-1 in the ΔvirD4–89K mutant. GST–VirD4 or GST alone was pre-absorbed with glutathione beads and then mixed with the total proteins from ΔvirD4–89K or ΔsspA-1 lysates. GST or ΔsspA-1 lysates were used as negative controls for the bait or prey, respectively. Pull-down samples and input samples (0.1% input His-SspA-1 or 2% input ΔvirD4–89K lysates) were subjected to Western blot with anti-SspA-1 or anti-GST.
Role of SspA-1 in the virulence of S. suis2
A sspA-1 knockout mutant designated as ΔsspA-1 and a complementary strain termed CΔsspA-1 were constructed to examine the role of SspA-1 in the pathogenicity of the highly virulent S. suis 2. The growth rate, chain length, capsular material thickness, and hemolytic activity of ΔsspA-1 and CΔsspA-1 did not significantly differ from those of the wild-type strain (data not shown). BALB/c mice were used to assess the virulence of wild-type strain 05ZYH33, T4SS-deficient mutant (ΔvirD4-89K), ΔsspA-1, and CΔsspA-1. The mice inoculated with the wild-type strain manifested typical symptoms, such as rough hair coat, hypnesthesia, swollen eyes, and suppuration in the inner canthus but died within 24 h. By contrast, the mice infected with ΔsspA-1 or ΔvirD4-89K exhibited less serious symptoms and yielded 40% and 60% survival rates at the end of the experiment, respectively. When the mice were challenged with the complementary CΔsspA-1 strain, the results were similar to those obtained from the wild-type strain (Fig. 3). These results implied that SspA-1 contributes to the virulence of S. suis 2.
Figure 3. Survival curves of mice infected with the designated S. suis 2 strains.
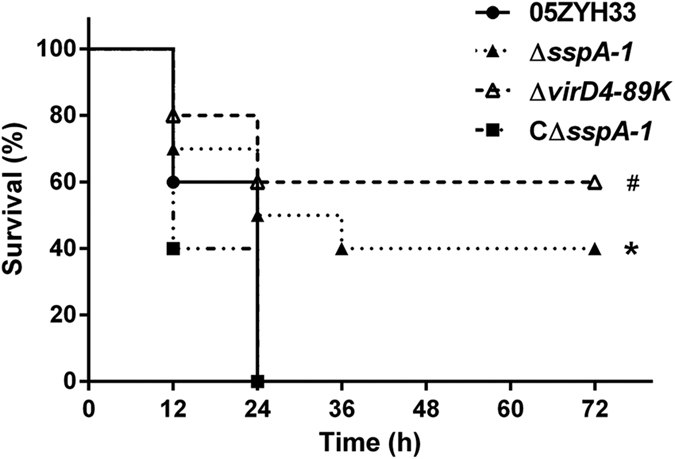
A total of 40 four-week-old female BALB/c mice were randomly allocated to four groups (10 animals per group) and challenged by intraperitoneally injecting a dose of 2.5 × 107 CFU/mice. The results are representative of three independent experiments. *P < 0.05 for comparison between the mutant ΔsspA-1 and the wild-type strain 05ZYH33 or CΔsspA-1 strain; #P < 0.05 for comparison between the mutant ΔvirD4-89K and the wild-type strain or CΔsspA-1.
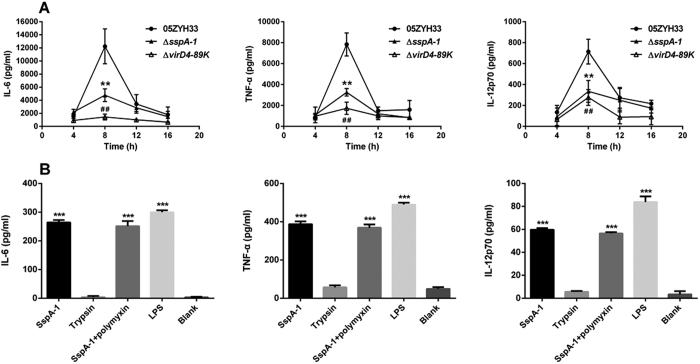
Proinflammatory effect of SspA-1 in vivo and in vitro
We assessed the kinetic profiles of cytokine secretion in the mice infected with the wild-type strain, T4SS-deficient mutant (ΔvirD4–89K), and ΔsspA-1 at 4, 8, 12, and 16 h after infection. The serum levels of interleukin 6 (IL-6), tumor necrosis factor-α (TNF-α), and interleukin 12p70 (IL-12p70) from the three mouse groups showed a sharp peak at 8 h after infection (Fig. 4A); the peak values returned to basal levels at 12 or 16 h after infection. However, the IL-6, TNF-α, and IL-12p70 levels induced by either ΔsspA-1 or ΔvirD4–89K were markedly lower than those induced by the wild-type strain, particularly at 8 h after infection. The production of IL-6, TNF-α, and IL-12p70 in the mice infected with ΔsspA-1 was slightly higher than that in mice infected with ΔvirD4–89K; nevertheless, the two groups did not significantly differ from each other. Meanwhile, the bacterial loads in the blood samples of the three groups showed no significant difference at any of the same time point (Fig. S1).
Figure 4. Cytokine levels in infected mice serum and culture supernatants of stimulated THP-1 cells.
(A) Cytokine production in the BALB/c mice infected with the designated strains at 4, 8, 12, and 16 h after infection. **P < 0.01 for the comparison between the wild-type strain 05ZYH33 and ΔsspA-1 mutant; ##P < 0.01 for the comparison between the wild-type strain 05ZYH33 and ΔvirD4–89K mutant. (B) Cytokine production by stimulated THP-1 cells. ***P < 0.001 indicates a significant difference in comparison with the non-stimulated cells (Blank)c. All these data were obtained from three independent experiments.
In vitro, the treatment of purified recombinant SspA-1 resulted in a significant increase of IL-6, TNF-α, and IL-12p70 secretion in THP-1 cells while trypsin did not (Fig. 4B). And the proinflammatory effect of SspA-1 could not be blocked by polymyxin B, so it excluded the possible contribution of contaminating LPS in the cell stimulation.
These data indicated that SspA-1, as the effector of the T4SS, triggered the excessive secretion of proinflammatory cytokines both in vivo and in vitro.
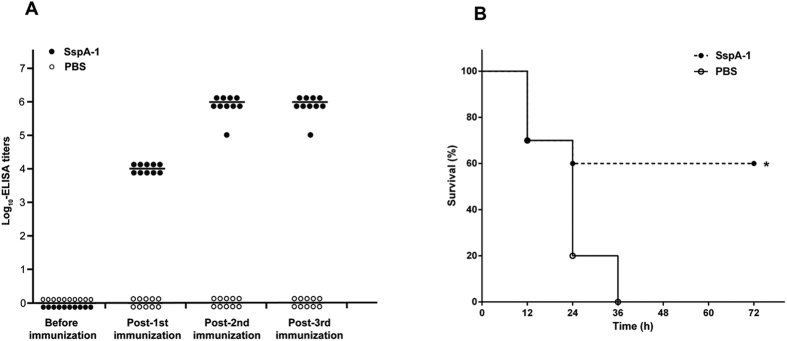
SspA-1-induced immune response and protection of mice against S. suis 2
The mice immunized with SspA-1 elicited a specific humoral IgG response. By contrast, SspA-1-specific antibody titers were below the limit of detection in the sera of the non-immunized mice (Fig. 5A). When the mice were infected with S. suis 2, the mice in the non-immunized control group exhibited typical clinical signs and died within 36 h; by contrast, 60% of the SspA-1-immunized mice manifested only mild clinical signs and survived the infection (Fig. 5B). These results indicated that the SspA-1-induced immune response protects the mice against S. suis 2 infection.
Figure 5. Immune response and survival curves of mice immunized with PBS or recombinant SspA-1.
(A) SspA-1 specific antibody titers for individual mice immunized with PBS or recombinant SspA-1; the average titer is represented as a bar. (B) Survival curves of mice immunized with PBS or recombinant SspA-1 challenged with the wild-type strain 05ZYH33, *P < 0.05.
Discussion
Although T4SS is implicated in the pathogenicity of the highly virulent S. suis 213,27, the exact effectors secreted by this system have yet to be identified. In our study, seven candidate T4SS effectors were identified through LC-MS/MS. Among these seven candidates, two subtilisin-like serine proteases (SspA-1 and SspA-2) were selected for further analysis because subtilisin-like serine proteases often act as virulence determinants in certain bacteria.
To verify whether SspA-1 and SspA-2 are secreted by the T4SS of S. suis 2, we separately determined the levels of the two proteins in the supernatants and whole cells of the wild-type strain and T4SS-deficient mutant (ΔvirD4–89K) through Western blot. The results revealed that the loss of T4SS function almost completely blocked the secretion of SspA-1 and led to abundant accumulation of SspA-1 in the cells of the mutant strain, indicating that the secretion of SspA-1 depends on the T4SS. This finding is consistent with the LC-MS/MS results. Similar to VirD4 of A. tumefaciens, the VirD4–89K of S. suis 2 is a key component of T4SS, which acts as a coupling factor that binds T4SS substrates or effectors directly prior to delivery into the VirB transmembrane channel28,29. In the present study, the interaction between SspA-1 and VirD4-89K was verified by pull-down assays. These tests further confirmed that SspA-1 is the effector of T4SS of the highly virulent S. suis 2. However, Western blot results also indicated that the secretion of SspA-2 was unaffected by T4SS (data not shown). Because Western blot is much more sensitive than LC-MS/MS, it could detect SspA-2 in the supernatant of ΔvirD4-89K which was ignored by LC-MS/MS, thereby leading to the inconsistent results of Western blot and LC-MS/MS on SspA-2. As such, we focused on SspA-1.
Moreover, a trace amount of SspA-1 can still be detected in the culture supernatant of the T4SS-deficient mutant (ΔvirD4–89K) (Fig. 1A), suggesting that SspA-1 secretion is not completely blocked when T4SS is inactivated. Hu et al.23 demonstrated that SspA is also a surface antigen anchored on the cell wall of S. suis 2 (SC19 strain). Thus, a trace amount of proteolytic fragments derived from the anchored SspA-1 may be detected inevitably in the supernatant of the VirD4–89K mutants, although the SspA-1 secretion through T4SS is inhibited. Hence, the SspA-1 of S. suis 2 may be secreted in two forms; one of these forms is anchored to the cell wall, and the other is released into the extracellular matrix via T4SS.
To our knowledge, SspA-1 is the first effector identified in Gram-positive bacteria that is transported by T4SS, a system generally thought to be used in conjugation to transfer DNA30. Gram-negative bacteria deploy dedicated secretion systems (type I to VI) to overcome the outer membrane barrier in the translocation of proteins. Although Gram-positive bacteria are generally considered lacking in outer membrane, their peptidoglycan layer is much thicker than that of Gram-negative microbes, and they can still block the diffusion of proteins greater than 25–50 kDa across the cell envelope31,32. It is favored by the evidence that boiling staphylococci in hot SDS does not release lipoproteins from the murein sacculus whereas advanced puncturing murein with specific hydrolases extracts such lipoproteins33. Therefore, Gram-positive bacteria may also require specialized secretion machinery or transport channels to translocate proteins. For instance, the most studied specialized protein export system in some Gram-positive bacteria (e.g. Mycobacterium tuberculosis and Staphylococcus aureus) is the type VII secretion system which mediates the secretion of some important virulence factors34,35. Moreover, in Streptococcus pyogenes, a microdomain designated as ExPortal is related to the secretion of precursor proteins across the thick peptidoglycan layer32,36. In our study, the acquisition of T4SS in 89K PAI during evolution enables the highly virulent S. suis 2 to secrete SspA-1 across the thick peptidoglycan barrier into the extracellular milieu.
A majority of patients in the two outbreaks of S. suis 2 infection in China developed STSS characterized by a severe systemic inflammatory response37. Our previous study suggested that T4SS in 89K PAI of S. suis 2 secretes unknown effectors that can trigger an excessive inflammatory response and lead to STSS13. Furthermore, a member of the subtilase family which shares 29% identities with SspA-1 can reportedly induce massive secretion of IL-1β, IL-6, TNF-α, CXCL8, and CCL5 in macrophages25. Therefore, we wondered whether SspA-1 is responsible for STSS caused by S. suis 2. The results from the experimental infection of mice showed that deletion of either SspA-1 or VirD4-89K attenuates the virulence of S. suis 2. Additionally, the proinflammatory cytokines released in the sera after infection with ΔsspA-1 or ΔvirD4-89K were found at much lower levels than those induced by a wild-type infection. And in vitro study also showed that purified SspA-1 could induce the secretion of IL-6, TNF-α, and IL-12p70 in THP-1 cells directly. These data provide further evidence that SspA-1 is an effector of T4SS and enhances the virulence of S. suis 2 by triggering an excessive inflammatory response, thereby contributing to the development of STSS. In addition, the protective role of SspA-1-induced immune response suggests that SspA-1 might be a potential vaccine candidate.
The ΔsspA-1 mutant seems slightly more virulent than the ΔvirD4-89K mutant on the basis of their survival rates and cytokine stimulation, although the difference is not significant. This finding implies that other T4SS effectors may exist in S. suis 2, or T4SS is independently pathogenic, as in the case of Helicobacter pylori38,39. Notably, the reliance on a single culture condition may restrict the discovery of effectors induced in alternative environments or in vivo. Considering that T4SS plays a role in effector transport, these possibilities should be further investigated to understand the pathogenic mechanisms of S. suis 2.
Additional Information
How to cite this article: Yin, S. et al. Subtilisin-like protease-1 secreted through type IV secretion system contributes to high virulence of Streptococcus suis 2. Sci. Rep. 6, 27369; doi: 10.1038/srep27369 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81301398); the Military Medical Science and Technique Youth training project (13QNP106) and the Military Medical Science and Technique Foundation during the 12th Five-Year Plan Period (CWS11J135).
Footnotes
Author Contributions Y.Z., F.H. and M.L. designed the study; S.Y., M.L., X.Y., Q.Z., M.W. and J.W.performed the experiments; Y.Z., J.T. and Y.P. analyzed the data; S.Y., Y.Z. and X.R. wrote the paper.
References
- Kudva R. et al. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 164, 505–534 (2013). [DOI] [PubMed] [Google Scholar]
- Preston G. M., Studholme D. J. & Caldelari I. Profiling the secretomes of plant pathogenic Proteobacteria. FEMS Microbiol Rev 29, 331–360 (2005). [DOI] [PubMed] [Google Scholar]
- Hood R. D. et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudl R. Leaving home ain’t easy: protein export systems in Gram-positive bacteria. Res Microbiol 164, 664–674 (2013). [DOI] [PubMed] [Google Scholar]
- Voth D. E., Broederdorf L. J. & Graham J. G. Bacterial Type IV secretion systems: versatile virulence machines. Future Microbiol 7, 241–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokter M., Felisberto-Rodrigues C., Christie P. J. & Waksman G. Recent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol 27, 16–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk M., Xu J., Calzas C. & Segura M. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 5, 371–391 (2010). [DOI] [PubMed] [Google Scholar]
- Segura M. Streptococcus suis: an emerging human threat. J Infect Dis 199, 4–6 (2009). [DOI] [PubMed] [Google Scholar]
- Tang J. et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLos Med 3, e151 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C. et al. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis 12, 1203–1208 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. et al. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLos One 2, e315 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol Microbiol 79, 1670–1683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. Role of a type IV-like secretion system of Streptococcus suis 2 in the development of streptococcal toxic shock syndrome. J Infect Dis 204, 274–281 (2011). [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Chung P. C., Shih H. W., Wen S. R. & Lai E. M. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190, 2841–2850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. L. et al. Strategic shotgun proteomics approach for efficient construction of an expression map of targeted protein families in hepatoma cell lines. Proteomics 3, 2472–2486 (2003). [DOI] [PubMed] [Google Scholar]
- Geng H. et al. Identification and characterization of novel immunogenic proteins of Streptococcus suis serotype 2. J Proteome Res 7, 4132–4142 (2008). [DOI] [PubMed] [Google Scholar]
- Li M. et al. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLos One 3, e2080 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. et al. Identification of three novel in vivo-induced expressed antigens during infection with Streptococcus suis serotype 2. FEMS Microbiol Lett 295, 17–22 (2009). [DOI] [PubMed] [Google Scholar]
- Liu F., Wu X., Li L., Liu Z. & Wang Z. Expression, purification and characterization of two truncated peste des petits ruminants virus matrix proteins in Escherichia coli, and production of polyclonal antibodies against this protein. Protein Expr Purif 91, 1–9 (2013). [DOI] [PubMed] [Google Scholar]
- de Barsy M. et al. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol 13, 1044–1058 (2011). [DOI] [PubMed] [Google Scholar]
- Akeda Y. et al. Identification of the Vibrio parahaemolyticus type III secretion system 2-associated chaperone VocC for the T3SS2-specific effector VopC. FEMS Microbiol Lett 324, 156–164 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y. Y. et al. Immunization with recombinant Sao protein confers protection against Streptococcus suis infection. Clinical and Vaccine Immunology 14, 937–943 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. et al. Identification of a cell wall-associated subtilisin-like serine protease involved in the pathogenesis of Streptococcus suis serotype 2. Microb Pathog 48, 103–109 (2010). [DOI] [PubMed] [Google Scholar]
- Bonifait L., Vaillancourt K., Gottschalk M., Frenette M. & Grenier D. Purification and characterization of the subtilisin-like protease of Streptococcus suis that contributes to its virulence. Vet Microbiol 148, 333–340 (2011). [DOI] [PubMed] [Google Scholar]
- Bonifait L. & Grenier D. The SspA subtilisin-like protease of Streptococcus suis triggers a pro-inflammatory response in macrophages through a non-proteolytic mechanism. BMC Microbiol 11, 47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifait L. et al. The cell envelope subtilisin-like proteinase is a virulence determinant for Streptococcus suis. BMC Microbiol 10, 42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q. et al. A functional peptidoglycan hydrolase characterized from T4SS in 89K pathogenicity island of epidemic Streptococcus suis serotype 2. BMC Microbiol 14, 73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R., Christie P. J. & Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol 7, 703–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Kozjak-Pavlovic V., Rudel T. & Rikihisa Y. Anaplasma phagocytophilum Ats-1 Is Imported into Host Cell Mitochondria and Interferes with Apoptosis Induction. PLos Pathogens 6, e1000774 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessweiner-Mohr N., Arends K., Keller W. & Grohmann E. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70, 289–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A. & Marquis H. Restricted translocation across the cell wall regulates secretion of the broad-range phospholipase C of Listetia monocytogenes. J Bacteriol 185, 5953–5958 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J. & Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science 304, 1513–1515 (2004). [DOI] [PubMed] [Google Scholar]
- Navarre W. W., Daefler S. & Schneewind O. Cell wall sorting of lipoproteins in Staphylococcus aureus. J Bacteriol 178, 441–446 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah A. M. et al. Type VII secretion - mycobacteria show the way. Nature Reviews Microbiology 5, 883–891 (2007). [DOI] [PubMed] [Google Scholar]
- Burts M. L., Williams W. A., DeBord K. & Missiakas D. M. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci USA 102, 1169–1174 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J. W. & Caparon M. G. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol 58, 959–968 (2005). [DOI] [PubMed] [Google Scholar]
- Ye C. et al. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J Infect Dis 199, 97–107 (2009). [DOI] [PubMed] [Google Scholar]
- Tegtmeyer N., Wessler S. & Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J 278, 1190–1202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M., Mollenkopf H. J., Klemm U. & Meyer T. F. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc Natl Acad Sci USA 109, E1153–1162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.