Abstract
Microbiological contamination in the food industry is often attributed to the presence of biofilms in processing plants. Bacterial biofilms are complex communities of bacteria attached to a surface and surrounded by an extracellular polymeric material. Their extreme resistance to cleaning and disinfecting processes is related to a unique organization, which implies a differential bacterial growth and gene expression inside the biofilm. The impact of biofilms on health, and the economic consequences, has promoted the development of different approaches to control or remove biofilm formation. Recently, successful results in phage therapy have boosted new research in bacteriophages and phage lytic proteins for biofilm eradication. In this regard, this review examines the environmental factors that determine biofilm development in food-processing equipment. In addition, future perspectives for the use of bacteriophage-derived tools as disinfectants are discussed.
Keywords: biofilm, bacteriophage, phage lytic proteins, food industry, disinfection
Introduction
Food safety is an important issue for health authorities and industries due to the health impact and economic losses caused by the contamination of foodstuffs. Despite the implementation of Good Manufacturing Practices (GMP) and Hazard Analysis Critical Control Point (HACCPs) in food industries, in 2014 the European Food Safety Authority (EFSA) reported a total of 5,251 foodborne outbreaks resulting in 6,438 hospitalizations (EFSA and ECDC, 2016). In the United States, 866 foodborne outbreaks were reported in 2014, resulting in 714 hospitalizations: (http://wwwn.cdc.gov/foodborneoutbreaks/; accessed: November 27, 2015).
Food is often contaminated during processing and packaging through contact with equipment surfaces. Of note, contamination with hemolytic bacteria (Staphylococcus aureus and Streptococcus agalactiae) was detected in hands, hand-contact and food-contact surfaces in foodservice settings (DeVita et al., 2007); the presence of coliforms in washing water and industrial facilities are involved in the low microbiological quality of tomatoes (van Dyk et al., 2016) or the notable incidence of S. aureus and other pathogenic bacteria on food industry surfaces in Spain (Gutiérrez et al., 2012a) are some of the great number of reported examples.
In fact, elimination of bacteria in food processing environments is greatly hindered by the presence of biofilms which provide a reservoir of foodborne pathogens. Usually most bacteria are organized in multispecies communities attached to a surface as biofilms, which confer ecological advantages that free-living bacteria in planktonic cultures do not have. Extracellular matrix, composed of a mixture of polymeric compounds such as polysaccharides, proteins, nucleic acids, and lipids, keeps the bacteria in close proximity each other and forms channels to distribute water, nutrients, oxygen, enzymes, and cell debris. This structure provides a microenvironment with physicochemical gradients, horizontal gene transfer, and inter-cell communication. In addition, biofilm matrix protects the involved bacteria from environmental damages, antimicrobial agents, and host immune defenses (Flemming and Wingender, 2010). The low diffusion of antimicrobial substances through the matrix, together with an altered growth rate of bacteria constitutes the main barrier in the fight against relevant microorganisms living in biofilms (Donlan and Costerton, 2002).
Biofilm formation has notable implications in industrial processes, in particular in food processing, with a negative impact on food safety and the subsequent economic losses (Van Houdt and Michiels, 2010). In this regard, further studies about biofilm development and disassembly have been performed for important pathogenic bacteria such as S. aureus (Boles and Horswill, 2011; Periasamy et al., 2012) and Listeria monocytogenes (da Silva and De Martinis, 2013). Numerous biofilm control strategies have been proposed but the problem remains unsolved, probably because of the complexity of these structures, which contain both cells and extracellular substances. Ideally, a biofilm removal system should be able to get inside the biofilm structure and eliminate efficiently all the matrix components and the bacteria.
New approaches are focused on preventing biofilm formation by the development of anti-adhesive surfaces (Gao et al., 2011; Kesel et al., 2014; Salwiczek et al., 2014) or by the inhibition or reduction of bacterial adhesion (Cegelski et al., 2009; Pimentel-Filho et al., 2014). Moreover, removal strategies like physical and chemical treatments (Van Houdt and Michiels, 2010), antimicrobial photodynamic therapy (Sharma et al., 2011), induction of biofilm detachment (Cerca et al., 2013), blocking of biofilm regulation (Romling and Balsalobre, 2012), matrix degradation (Ramasubbu et al., 2005; Alkawash et al., 2006), and quorum sensing inhibitors (Hentzer et al., 2003) have been explored.
Another promising approach to control and eradicate biofilms is the use of bacteriophages. These viruses are harmless to humans, animals, and plants because they specifically target and kill bacteria. Virulent phages follow a lytic cycle where they multiply within bacteria to finally release the phage progeny by lysis of the cell. This process confers phages their antimicrobial activity. Phages have been used as treatment against human infections in countries from Eastern Europe, but the increase in antibiotic resistance has boosted new research and a notable interest worldwide for the use of phages to fight against pathogenic bacteria in clinical, veterinary, food safety, and environment (O’Flaherty et al., 2009; García et al., 2010). Phage-encoded lytic proteins such as endolysins and virion-associated peptidoglycan hydrolases (VAPGHs) have also been assessed as antimicrobial agents against pathogens (Schmelcher et al., 2012; Rodríguez-Rubio et al., 2013) and other phage-encoded proteins with polysaccharide depolymerase activity can be used as anti-biofilm agents (Cornelissen et al., 2011; Gutiérrez et al., 2012b, 2015a). Therefore, bacteriophages are not only bacterial killers but also a source of antimicrobial phage-derived proteins that can be exploited to fight against pathogenic bacteria.
Overall, the aim of this review is to assess both bacteriophages and bacteriophage-derived proteins as potential compounds to be applied as part of the cleaning and disinfecting processes of food-contact surfaces in the food industry.
Relevance of Biofilms in the Food Industry and Disinfection Hurdles
Biofilm formation is a major concern in industrial settings, since it is one of the causes of operating troubles by decreasing heat transfer, blocking tubes, plugging filters, and causing damage to surfaces (Myszka and Czaczyk, 2011). Specifically in the food industry, the ability of bacteria to attach to food-contact surfaces provides a reservoir of contamination for pathogens with the consequent risks to human health. Analysis of the microbial composition of biofilms formed on food industrial surfaces revealed the presence of mixed biofilms including pathogenic and spoilage bacteria (Gounadaki et al., 2008; Gutiérrez et al., 2012a). These microorganisms can reach the food industry through several sources such as water, raw foods, animals, and can persist in the equipment for long periods of time. Therefore, food products can be contaminated at any stage of the food chain, even though all required cleaning protocols have been applied, because disinfecting and cleaning processes in the food industry are often ineffective. For instance, some microorganisms are able to survive after cleaning-in-place procedures, like in the case of dairy industries (Anand and Singh, 2013).
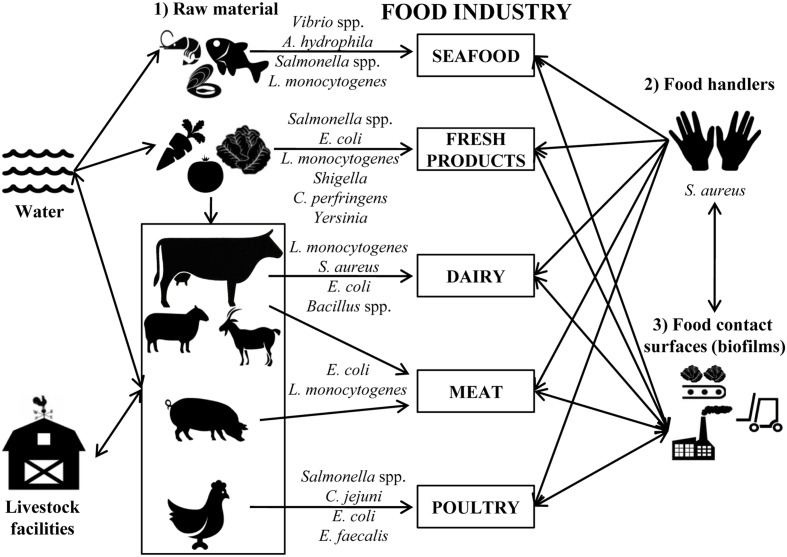
Biofilms mainly cause problems in the dairy (Latorre et al., 2010), meat (Giaouris et al., 2014), poultry (Silagyi et al., 2009), seafood (Thimothe et al., 2004), and vegetable processing industries (Liu et al., 2013). Depending on the food-processing industry, the type of bacteria and the route of access to foodstuffs differs (Figure 1).
FIGURE 1.
Schematic representation of the main sources of contamination in food industries. Most common bacteria detected in each agri-food industry are indicated.
In seafood industries, the most common bacterial pathogens that form biofilms are Vibrio spp., Aeromonas hydrophila, Salmonella spp., and L. monocytogenes (Mizan et al., 2015). Vibrio parahaemolyticus can form biofilms on different surfaces including the chitin of oysters, and this process is recognized as vital to the physiology of these microorganisms (Thompson et al., 2010). Vibrio cholerae can form biofilms attached to the surface of phytoplankton and zooplankton, from where they can contaminate seafood products after consumption (Mizan et al., 2015). A correlation between the persistence of Salmonella spp. in the fish-processing industry and the ability for biofilm formation was also reported (Vestby et al., 2009). It was also demonstrated that other bacteria such as L. monocytogenes isolated from seafood industries can form biofilms on stainless steel surfaces (Gudmundsdottir et al., 2006).
In the fresh produce industry, bacteria such as Salmonella, E. coli O157:H7, L. monocytogenes, Shigella, Bacillus cereus, Clostridium perfringens, and Yersinia go into the processing facilities adhered to the plant tissues where they can grow forming biofilms (Beuchat, 2002; Da Silva Felicio et al., 2015). The accessibility of sanitizers to these microorganisms is hindered, not only by the presence of biofilms, but also by the intrinsic structure of vegetables, making it necessary to optimize the decontamination methods to extend their shelf-life (López-Gálvez et al., 2010).
In the dairy industry, most contamination comes from inadequate cleaning of the equipment and the presence of pathogenic bacteria; e.g., L. monocytogenes in milking equipment was determined to be the cause of contamination of bulk tank milk (Latorre et al., 2010). In addition, biofilm formation by L. monocytogenes may be promoted by specific conditions in the dairy industry like those used in cheese manufacturing (low pH values during milk fermentation and increased salt concentration). Thus, some strains increased their adherence to polystyrene after salt adaptation, and the exposure to acid increased the survival of cells adhering to stainless steel (Adriao et al., 2008). Milk proteins are also able to increase the attachment of E. coli, L. monocytogenes, and S. aureus to stainless steel (Barnes et al., 1999). On the other hand, members of the Bacillus genus are very common in dairy plants, where biofilm formation is triggered during milk lipolysis (Pasvolsky et al., 2014).
E. coli O157:H7 is a pathogenic bacteria also related with contamination in the meat industry. The ability of this bacterium to attach to meat-contact surfaces is influenced by the type of meat residues and the temperature. In fact, this microorganism significantly increases its counts number during inactivity periods of facilities (15°C) and also during cold storage temperatures (4°C; Dourou et al., 2011). Recently, it has been reported that E. coli O157:H7 strains isolated from a “high event period” (period of time during which commercial meat plants undergo a higher rate of contamination with this pathogen than normal) have a significantly higher potential of mature biofilm formation after incubation for 4–6 days, and also exhibit significantly stronger resistance to sanitization (Wang et al., 2014). L. monocytogenes was also isolated from bovine carcasses and meat processing facilities (Peccio et al., 2003; von Laer et al., 2009). The ability of this bacterium to colonize materials used in food processing surfaces (Rodriguez et al., 2008; Hingston et al., 2013), and to survive in niches that are difficult to sanitize such as countertops, cutting blades, or joints is well known (Verran et al., 2008).
Salmonella spp. and Campylobacter spp. are the most common pathogens found in poultry industries. Salmonella adhesion is influenced by different physicochemical properties of surfaces; for instance, Salmonella is able to grow at 16°C on stainless steel, while adherence was hindered on glass (De Oliveira et al., 2014). Recent studies have found that chicken meat exudation increases Campylobacter jejuni biofilm formation on glass, polystyrene, and stainless steel surfaces by covering and conditioning the surface (Brown et al., 2014). In addition, aerobic or stressful conditions (Reuter et al., 2010) and the presence of other bacteria such as Enterococcus faecalis and Staphylococcus simulans, also found in poultry processing environments, increase the level of biofilm formation allowing the survival of C. jejuni under detrimental conditions (Teh et al., 2010).
Overall, the main concern about biofilms is their wide resistance to disinfectants commonly used in food industries, which include quaternary ammonium compounds such as benzalkonium chloride (BAC). The resistance to these compounds shown by several foodborne pathogens results in their reduced efficacy (Saa-Ibusquiza et al., 2011); e.g., L. monocytogenes is able to modify the physicochemical properties of the cell surface as a response to low concentrations of BAC resulting in a higher resistance to this compound (Bisbiroulas et al., 2011). Biofilm resistance to antimicrobials is attributed to several intrinsic biofilm properties such as reduced diffusion, physiological changes of cells, reduced growth rates, and the production of enzymes that degrade the antimicrobial compounds (Bridier et al., 2011). In this regard, it has been shown that the extracellular material constitutes a physical barrier for biocides and the chemical interaction with this material reduces the rate of diffusion to the biofilm inside. Besides the physical barrier found by the antimicrobial compounds to penetrate into the biofilm, there is a physiological resistance due to the altered growth rate of cells forming the biofilm, which grow more slowly than planktonic cells and consequently are less affected by the biocide (Evans et al., 1991). The presence of persister cells, which are tolerant to antimicrobials, could also explain the resistance of biofilm to biocides along with an adaptive tolerance (Simoes et al., 2011). Thus, it has been suggested that exposure to sublethal concentrations of biocides allows bacterial adaptation and survival at the level of biocide concentrations used in the food environment (Capita et al., 2014). In many bacteria, such as S. aureus, multidrug efflux pumps are responsible for this biocide resistance (Rouch et al., 1990). In fact, prolonged exposure to sublethal concentrations of biocides can lead to the overexpression of these efflux pumps and hence to the increased multidrug resistance in bacteria (Gilbert and McBain, 2003). In this regard, in vitro cross-resistance with antibiotics has been described for some biocide-resistant foodborne pathogens (Davin-Regli and Pages, 2012; Gnanadhas et al., 2013) supporting the need for monitoring and regulating the usage of biocides. The maturation stage of biofilms may also enhance resistance to disinfectants, since it has been reported that sodium hypochlorite, sodium hydroxide, and BAC failed to eradicate mature Salmonella biofilms (Corcoran et al., 2014).
Impact of Food-Processing Conditions on Biofilm Development
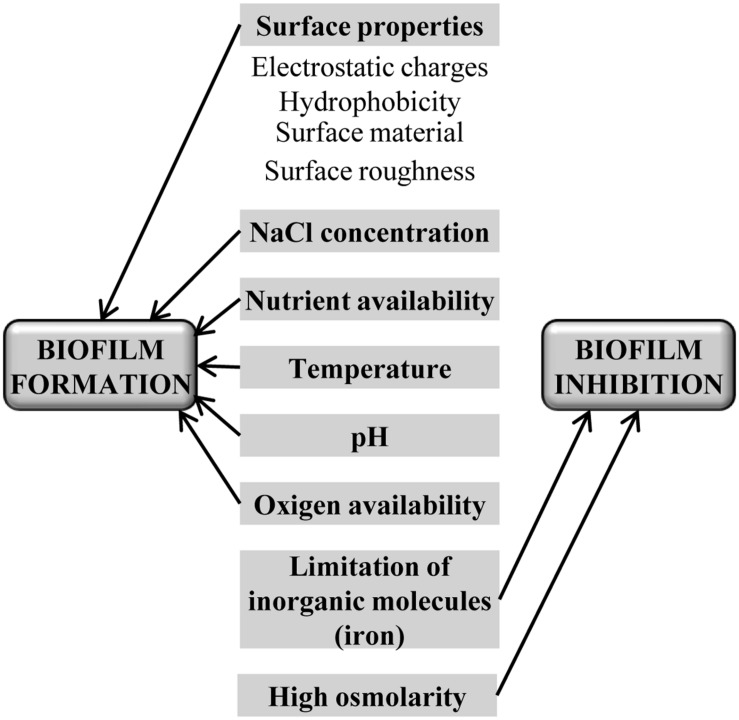
In food processing environments, there are a number of variable conditions such as temperature, pH, oxygen and nutrients availability, and surface type, which can modulate biofilm development (Figure 2). Surface properties such as electrostatic charges, hydrophobicity, and roughness influence biofilm development in some species. For instance, hydrophilic surfaces are more quickly colonized by L. monocytogenes (Chavant et al., 2002), whereas S. aureus have not shown any differences between hydrophobic and hydrophilic surfaces (da Silva Meira et al., 2012), and Salmonella has a higher ability to adhere to some materials used in food-contact surfaces like Teflon, followed by stainless steel, glass, Buna-N rubber, and polyurethane (Chia et al., 2009). In some cases, biofilm retention is more affected by the surface roughness than by the chemical composition (Tang et al., 2011). Other components of food environments such as NaCl also contribute to increase the adhesion of L. monocytogenes to surfaces (Jensen et al., 2007), although it is influenced by temperature and nutrients as well (Moltz and Martin, 2005), and even by the presence of other bacteria in the food-processing environment (Carpentier and Chassaing, 2004).
FIGURE 2.
Main food industry’s parameters that can influence biofilm development.
Food-related environmental factors have a variable impact on biofilm development. Bacteria sense these factors through sophisticated intracellular and extracellular signaling networks resulting in a negative or positive response (Karatan and Watnick, 2009). For instance, nutrient limitation induces Salmonella enterica serovar Typhimurium to biofilm formation (Gerstel and Romling, 2001), whereas V. cholerae needs a nutrient-rich environment to develop a biofilm structure (Yildiz et al., 2004). Similarly, in S. aureus an increase in biofilm formation was observed in a nutrient-rich growth media (Herrera et al., 2007) and at high incubation temperatures (Vázquez-Sánchez et al., 2013). Secondary metabolites such as antibiotics may also induce biofilm formation (Hoffman et al., 2005). Another example was recently reported by Nesse et al. (2014), where potentially human-pathogenic E. coli from the ovine reservoir can form biofilms under conditions used in the food production chain [on different surfaces such as stainless steel, glass, and polystyrene and at temperatures relevant for food production and handling (12, 20, and 37°C)]. Of note, for most bacteria, limitation of inorganic molecules such as iron and inorganic phosphate has an inhibitory effect on biofilm formation (Mey et al., 2005; Monds et al., 2007), and high osmolarity inhibits in general, biofilm formation, although this effect is clearly dependent on the osmolyte (Jubelin et al., 2005).
Bacteriophages Properties as Antimicrobials
Bacteriophages are viruses of prokaryotes widespread in all habitats where their hosts are located. Classifications of bacteriophages are based on their shape, size, and kind of nucleic acid. The most abundant belong to the Caudovirales order (tailed-bacteriophages), which is divided into three families (Myoviridae, Podoviridae, and Siphoviridae) according to the microscopic features of the tail morphology. Bacteriophages belonging to the Siphoviridae family are the most abundant (57.3%; Ackermann and Prangishvili, 2012).
Bacteriophages can infect bacteria by following two different life cycles, lytic and lysogenic (Kutter et al., 2004). In most phages, the lytic cycle ends with the lysis of host bacteria and the progeny release. Thus, antimicrobial properties of bacteriophages are linked to the lytic cycle (lytic phages) since the infected host is intended to die. On the contrary, the lysogenic cycle followed by temperate bacteriophages implies the survival and establishment of the phage genome into the bacterial chromosome (prophage) until environmental signals trigger a lytic cycle, thereby killing only a part of the infected population. In addition, lysogenic bacteria, carrying a prophage, are resistant to infection for a related phage (superinfection immunity; Kutter et al., 2004).
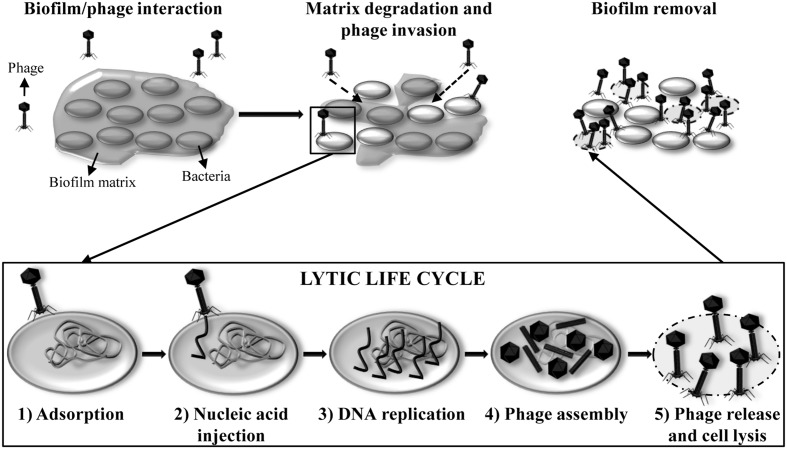
In nature, most bacteria are living in biofilms (Hall-Stoodley et al., 2004). The interaction between the host bacteria and the lytic phages occurs in six different steps (Figure 3). The adsorption of the bacteriophage and release of the new phage progeny play a key role in the bacteriophage infection process. When host bacteria are included in a biofilm, the biofilm matrix can constitute a first physical barrier to the phage. To solve this problem, some phages possess polysaccharide depolymerases which are specific hydrolytic enzymes that can use polysaccharides or polysaccharides derivatives as substrate (Pires et al., 2016; Figure 4A). Numerous studies have shown that polysaccharide depolymerase activity is related to tail-spike proteins which are components of the tail of many bacteriophages (Barbirz et al., 2009). The presence of polysaccharide depolymerases confers the phage an important advantage since it enhances the process of invasion and dispersion through the biofilm to start the infection process of new bacteria. Moreover, some phages are provided with lytic enzymes which are named VAPGHs, with a role in the first step of the infection cycle (Figure 4B). Their activity produces a small hole in the cell wall through which phage genetic material reaches the cytoplasm, being responsible for the “lysis from without” caused by the adsorption of a high number of phages to the cell at the initial infection step (Moak and Molineux, 2004). Recently, these proteins have also been proposed as new antimicrobials due to their lytic activity (Rodríguez-Rubio et al., 2013).
FIGURE 3.
Lytic life cycle of phages inside a biofilm. (1) Adsorption of the phage particle onto the host bacterial cell surface. Tail fibers bind to specific receptors on the cell surface. (2) Injection of the nucleic acid into the cytoplasm of the host bacterium. (3) Replication of the phage genome in multiple copies. Phage early genes are expressed to regulate the host metabolic machinery to be subjected to phage propagation. (4) Formation of new phage particles by expression of the phage late genes and assembly of the phage heads and tails, packaging of the nucleic acid inside the heads and maturation of the virions. (5) Lysis of the host bacterium and release of the new phage progeny ready to infect other cells in the biofilm and start a new cycle.
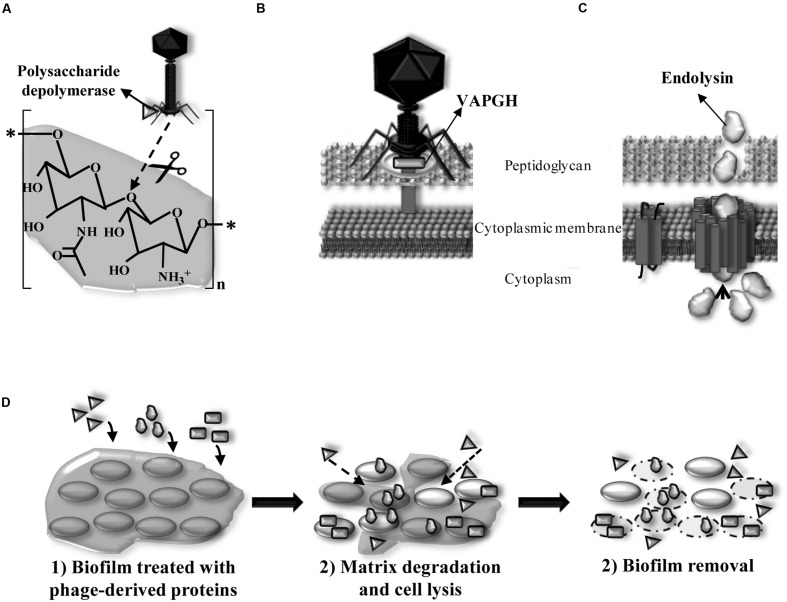
FIGURE 4.
(A) Location of exopolysaccharide depolymerase degrading β-(1,6) bonds of the biofilm extracellular matrix (PIA/PNAG) of staphylococcal species in the phage particle and mode of action. (B) Location of virion-associated peptidoglycan hydrolase (VAPGH) at the phage particle and its role in the infection process. (C) Structure of Gram-positive bacteria cell wall and role of the endolysin during the bacterial lysis. (D) Activity of phage derived proteins when added exogenously and their application as anti-biofilm agents degrading polysaccharidic matrices (polysaccharide depolymerases) and lysing bacteria (VAPGHs and endolysins).
Double-stranded phages encode lytic proteins, named endolysins, which act along with holins to disrupt the cell wall and lyse the host bacteria at the last step of the lytic infection cycle (Figure 4C). Endolysins, which are also peptidoglycan hydrolases, access the periplasmic space through holes formed by holins in the cytoplasmic membrane. Hydrolysis of covalent bonds in the peptidoglycan molecule produces the destabilization of the cell wall structure and lysis by the increase of the osmotic pressure inside the cytoplasm. In Gram-positive bacteria, endolysins are able to degrade the peptidoglycan when they are added from outside the cell, which gives them an antimicrobial activity (enzybiotics; Fischetti, 2008). In Gram-negative bacteria, peptidoglycan is protected by the outer membrane, these bacteria being insensitive to endolysins. Nowadays, research efforts made into endolysin applications against Gram-negative pathogens are changing this rule. This is the case of Artilysins that combine a polycationic peptide able to penetrate the outer membrane with an endolysin, which renders a protein with high bactericidal activity against Gram-negative pathogens (Briers et al., 2014a,b).
The use of phage-encoded proteins as antimicrobials has some advantages over the use of the viral particles; e.g., no resistant bacteria to phage lytic proteins has been described to date (Nelson et al., 2012; Rodríguez-Rubio et al., 2013). Additionally, the spectrum of activity of endolysins is usually broader than the host range of bacteriophages and, there are no risks of transferring virulence genes (Fischetti, 2008).
Due to the above described working mechanisms, bacteriophages and phage-derived proteins could be used in the production of foodstuffs against unwanted bacteria to ensure quality, safety, and good hygienic conditions, covering the entire food chain production (“from farm to fork”). This includes strategies to improve animal health (phage therapy), decontamination of fresh-food and ready-to-eat products, disinfection of food-contact surfaces, as well as their use as biopreservatives to inhibit the development of pathogenic or spoilage bacteria during storage, and also as tools for detecting undesirable bacteria through the different manufacturing steps (García et al., 2010).
Control of Biofilms Using Bacteriophages and Phage-Derived Proteins
The recent interest in phage therapy as an alternative to conventional antibiotics has fostered the use of phages in multiple applications, among which their use as anti-biofilm agents should be noticed. Biofilms are the lifestyle of bacteria in nature and therefore, phages have evolved to infect cells at this stage (Hall-Stoodley et al., 2004). There are two putative limitations for phage infection of cells inside the biofilm. First, the accessibility of phages to cells due to the structure of biofilm and the presence of extracellular material. Briandet et al. (2008) demonstrated that phage c2 was able to penetrate inside the Lactococcus lactis biofilm structure through water channels and cell clusters; in addition, the infection of E. coli surface-attached cells was confirmed by using T4 fluorescently labeled phages (Doolittle et al., 1996). Some phages, provided with exopolysaccharide depolymerases, can degrade the extracellular polymeric material, thus facilitating the entrance of phages into the deeper layers of the biofilms with subsequent lysis of the target bacteria (Parasion et al., 2014). The second limitation in the treatment of biofilms with bacteriophages is the metabolic status of a proportion of the population, persister cells and stationary phase bacteria, which have a slow metabolism. Bacteriophages infect preferably exponentially growing bacteria but recently, it has been demonstrated that persister bacteria can be infected by phages when bacteria switch to normal growth rate (Pearl et al., 2008). Moreover, persister cells can be removed by phage lytic proteins (Gutiérrez et al., 2014) due to these proteins are able to easily penetrate into the biofilms (Shen et al., 2013). Furthermore, bacteriophages can be engineered to express proteins intended to enhance their anti-biofilm properties. For instance, phage T7 was genetically engineered to incorporate the gene dspB encoding a polysaccharide depolymerase from Actinobacillus actinomycetemcomitans, which was more effective at reducing the bacterial count in E. coli biofilms (Lu and Collins, 2007). In addition, it has been demonstrated that engineered bacteriophages overexpressing proteins able to suppress bacterial SOS response network in E. coli are more effective against persister cells (Lu and Collins, 2009).
Several studies using biofilms preformed in laboratory conditions confirm the potential of phages in biofilm removal (Table 1). For biofilms formed by pathogenic bacteria with relevance in the food industry, there is evidence of effective removal in different conditions and using materials similar to those found in food-contact surfaces. Regarding this, three phages LiMN4L, LiMN4p, and LiMN17 infecting L. monocytogenes were assayed, individually and as a cocktail, against 7-day biofilms formed by a mixture of three strains on stainless steel coupons (104 cfu/cm2), previously covered with a fish broth layer that simulated seafood processing facilities. Treatments with the single phages (109 pfu/ml) reduced adhered bacterial cells to up 3 log units, whereas treatment with the phage cocktail reduced cell counts to undetectable levels after 75 min (Ganegama-Arachchi et al., 2013). Similarly, a treatment with phage P100 (109 pfu/ml) reduced biofilms formed by L. monocytogenes strains in 3.5–5.4 log/cm2, irrespective of the serotype, growth conditions and biofilm level (Soni and Nannapaneni, 2010). Despite the efficacy of bacteriophages to reduce L. monocytogenes biofilms, there is evidence that complete removal is not always achieved. By using epifluorescence microscopy, L. monocytogenes was monitored after treatment with phage P100 (108 pfu/ml) and, although disaggregation of biofilms could be observed after 8 h, viable cells were still present up to 48 h later, indicating that other sanitization methodologies should be used in combination with phages (Montañez-Izquierdo et al., 2012).
Table 1.
Application of bacteriophages and phage proteins for biofilm removal.
| Phage or phage protein | Scope of application | Bacteria | Efficacy of the treatment | Reference |
|---|---|---|---|---|
| Phages LiMN4L, LiMN4p, and LiMN17 | Stainless steel | L. monocytogenes | Phage cocktail reduced biofilm cell counts to undetectable levels after 75 min | Ganegama-Arachchi et al., 2013 |
| Phage P100 | Stainless steel | L. monocytogenes | Reduction in the cell counts from 3.5 to 5.4 log units/cm2 | Soni and Nannapaneni, 2010 |
| Phage P100 | Stainless steel | L. monocytogenes | Reduction of the biofilm cell counts to undetectable levels after 48 h | Montañez-Izquierdo et al., 2012 |
| Phage K and phage derivatives | Polystyrene | S. aureus | Complete elimination of the biomass after 72 h of incubation. Complete inhibition of biofilm formation was achieved when co-culturing phage mixture and bacteria | Kelly et al., 2012 |
| Phage K and DRA88 | Polystyrene | S. aureus | Complete elimination of the biomass after 48 h of treatment | Alves et al., 2014 |
| Phages ISP, Romulus, and Remus | Polystyrene | S. aureus | Biofilm reduction of 37.8, 34.4, and 60.4% after 24 h treatment when using phages ISP, Romulus, and Remus, respectively | Vandersteegen et al., 2013 |
| Phages phiIPLA-RODI and phiIPLA-C1C | Polystyrene | S. aureus | Reduction by 2 log units/well was achieved after 8 h of treatment | Gutiérrez et al., 2015b |
| Phage SAP-26 | Polystyrene | S. aureus | Reduction of bacteria about 28% after phage treatment, while a synergistic effect with rifampicin allows a reduction of about 65% | Rahman et al., 2011 |
| Phage CP8 and CP30 | Glass | C. jejuni | Reduction in the biofilm cell counts of 1–3 log units/cm2 | Siringan et al., 2011 |
| Phage KH1 | Stainless steel | E. coli O157:H7 | Reduction of 1.2 log units per coupon after 4 days treatment at 4°C | Sharma et al., 2005 |
| BEC8 (phage mixture) | Stainless steel, ceramic tile, and high density polyethylene | E. coli O157:H7 | Reduction of the biofilm cell counts to undetectable levels after 1 h of treatment at 37, 23, and 12°C | Viazis et al., 2011 |
| Phage mixture | Spinach harvester blade | E. coli O157:H7 | Reduction of biofilm cell counts by 4.5 log units per blade after 2 h of treatment | Patel et al., 2011 |
| Phage T4 | Polystyrene | E. coli O157:H7 | Complete elimination of the biomass after phage treatment combined with cefotaxime | Ryan et al., 2012 |
| Endolysin from phage phi11 | Polystyrene | S. aureus | Complete elimination of the biomass after 2 h of treatment at 37°C | Sass and Bierbaum, 2007 |
| Endolysin SAL-2 | Polystyrene | S. aureus | Reduction of the biomass after 2 h of treatment at 37°C | Son et al., 2010 |
| Endolysin LysH5 | Polystyrene | S. aureus | Reduction of biofilm cell counts by 1–3 log units after 3 h of treatment | Gutiérrez et al., 2014 |
| Domain CHAPK derived from endolysin LysK | Polystyrene | S. aureus | Complete elimination of the biomass after 4 h of incubation Complete inhibition of biofilm formation was achieved | Fenton et al., 2013 |
| Chimeric lysin ClyH | Polystyrene | S. aureus | Reduction of the biomass in more than 60% after 30 min of treatment | Yang et al., 2014 |
| Endolysin Lys68 | Polystyrene | S. Typhimurium | Reduction of biofilm cell counts by 1 log unit after 2 h of treatment in the presence of outer membrane permeabilizers | Oliveira et al., 2014 |
| Exopolysaccharide depolymerase Dpo7 | Polystyrene | S. aureus | Degradation of 30% of the polysaccharidic matrix of the biofilm | Gutiérrez et al., 2015a |
Staphylococcus aureus is another important foodborne pathogen with the ability to form biofilms on different surface’s materials. Staphylococcal phage K and a mixture of derivative phages with broader host ranges were used to effectively prevent S. aureus biofilm formation over incubation periods of 48 h. It was also shown that the removal of bacteria by the phage cocktail (109 pfu/ml) was time-dependent, with the highest reduction occurring after 72 h at 37°C (Kelly et al., 2012). A similar result was obtained using phage K combined with another staphylococcal phage, DRA88 (MOI 10), to treat established biofilms produced by three S. aureus isolates, which were significantly reduced after 4 h and completely removed after 48 h at 37°C (Alves et al., 2014). Other staphylococcal phages such as ISP, Romulus, and Remus applied individually at 109 phages per polystyrene peg were able to degrade by 37.8, 34.4, and 60.4%, respectively, an S. aureus PS47 biofilm after 24 h (Vandersteegen et al., 2013). Similar results were obtained after the application of phages phiIPLA-RODI, phiIPLA-C1C, and a mixture of both phages, against biofilms formed by S. aureus where a reduction by about 2 log units was achieved after 8 h of treatment at 37°C (Gutiérrez et al., 2015b). In some cases, however, it was also necessary to combine phages with other antimicrobials to increase their effectiveness. Thus, treatment of 1-day-old biofilms formed by S. aureus D43 strain with phage SAP-26 reduced live bacteria by about 28% while a synergistic effect with rifampicin allowed a reduction of about 65% (Rahman et al., 2011).
Bacteriophages were also assayed against C. jejuni biofilms. Two virulent phages, CP8 and CP30 led to 1–3 log cfu/cm2 reduction in viable counts after 24 h of treatment. However, a high percentage of bacteriophage-resistant bacteria in biofilms were observed for some C. jejuni strains (Siringan et al., 2011).
Sharma et al. (2005) assayed the lytic bacteriophage KH1 (7.7 log pfu/ml) against stainless steel coupons containing E. coli O157:H7 biofilms (2.6 log cfu/coupon). These were treated for 4 days at 4°C and a reduction of 1.2 log units per coupon was observed. Better results were obtained when treating E. coli O157:H7 biofilms preformed on other materials typically used in food processing surfaces (stainless steel, ceramic tile, and high density polyethylene), since a reduction to undetectable levels was observed after 1 h of treatment at 23°C with a phage mixture named BEC8 (MOI 100; Viazis et al., 2011). The use of a phage mixture to remove biofilms formed on blades used to harvest spinach was also demonstrated, a reduction of 4.5 log units of the viable cells of E. coli O157:H7 being achieved after 2 h of phage treatment (Patel et al., 2011). As it was previously reported, a combination of T4 bacteriophage and cefotaxime significantly enhanced the eradication of E. coli biofilms when compared to treatment with phage alone (Ryan et al., 2012).
Phage lytic proteins are also an alternative for removing bacterial biofilms in food-related environments (Figure 4D; Table 1). Endolysin from phage phi11 (10 μg/well) was able to remove biofilms formed by S. aureus strains on polystyrene surfaces after 2 h at 37°C (Sass and Bierbaum, 2007). Similarly, endolysin SAL-2 from bacteriophage SAP-2 eliminated S. aureus biofilms using 15 μg/well (Son et al., 2010). Recently, Gutiérrez et al. (2014) showed that endolysin LysH5 (0.15 μM) is able to remove staphylococcal biofilms after treatment of 12 h at 37°C and even to lyse persister cells. Engineered endolysins, by deletion or shuffling domains, have also been successfully used as anti-biofilm agents. For instance, peptidase CHAPk (31.25 μg/ml), derived from the staphylococcal endolysin LysK, was able to completely prevent biofilm formation. This protein also removed staphylococcal biofilms after treatment of 4 h at 37°C (Fenton et al., 2013). In addition, the minimum concentration (6.2–50 mg/l) of ClyH, a staphylococcal chimeric lysin, required for S. aureus biofilm eradication was lower than that needed when antibiotics were used (Yang et al., 2014). This protein contains the catalytic domain of endolysin Ply187 and the cell wall binding domain of phiNM3 lysin. Regarding biofilms formed by Gram-negative bacteria, removal of these structures by using endolysins needs an additional component to disestablish the outer membrane. Biofilms formed by S. enterica serovar Typhimurium were treated with endolysin Lys68 (2 μM), which reduced by 1 log unit the viable cells in preformed biofilms after 2 h of incubation in the presence of outer membrane permeabilizers (Oliveira et al., 2014).
Regarding phage-encoded exopolysaccharide depolymerases, there is scarce data about the biofilm dispersion mediated by these proteins but they seem to be very promising. Cornelissen et al. (2011) identified an exopolysaccharide-degrading activity associated to a tail spike protein from Pseudomonas putida phage Φ15, which is involved in the hydrolysis of extracellular material. However, the addition of the purified tail spike protein did not result in biofilm removal. Since the addition of 106 phages yielded a significant biofilm degradation of 37% in 24 h, this seems to require phage amplification (Cornelissen et al., 2012). Recently, an exopolysaccharide depolymerase named Dpo7 was identified in the S. epidermidis phage phiIPLA7. Purified protein was used to treat S. aureus biofilms, showing its ability to degrade up to 30% of the polysaccharidic matrix formed by S. aureus 15981 (Gutiérrez et al., 2015a).
Overall, these results showed a noticeable potential of phages and phage-derived proteins, but undoubtedly additional studies are necessary to transfer this knowledge to the food industry. For instance, application of these anti-biofilm compounds would be feasible as long as their application can be implemented as part of the standard processes of cleaning in the industrial facilities. Therefore, the study of synergy/antagonism with disinfectants and the effectiveness at temperatures commonly used in the industry could be relevant. It should be also noticed the scarce data available about the use of phages and phage lytic proteins against mixed biofilms formed by different strains from several species in food industrial surfaces. This gap should be filled in to go further into the control of bacterial biofilms.
Future Perspectives for Phage-Based Disinfectants
The most important issues to address before the implementation of phages and phage-derived proteins as disinfectants are the following.
Safety
Beyond efficiency, safety of phage-based products must be a priority to take into account. Only phages fully characterized at molecular level and with the complete genome sequenced should be taken into consideration as potential components of disinfectants to avoid the presence of virulence and antibiotic resistance genes. These phages must be lytic, since temperate bacteriophages have the ability to integrate their genomes into their host bacterium’s chromosome, and non-transducers, i.e., without the ability to transfer genetic material from host bacteria. One of the most important characteristics of bacteriophages, their high specificity for the host bacteria, could be a potential limitation in their use as disinfectants. A cocktail of different phages with overlapping host ranges or the use of polyvalent phages with a wide host range would solve this problem. Finally, in the selection of phages to be included in the cocktail, the presence of those encoding polysaccharide depolymerase enzymes should be preferred.
Regarding the safety of engineered phages, the main hurdle for their use is the generalized opposition of consumers to genetic manipulation, despite of engineered phages can overcome the limitations of phages as antimicrobials and even specific modifications can eliminate some of their risks such as virulence genes or gene transfer (Nobrega et al., 2015).
Before the extensive use of bacteriophages as disinfectants, the absence of an ecological impact on the environment must be also guaranteed. In this regard, bacteriophages should be inactivated before their release outside the industry settings. Some commercial sanitizers and disinfectants commonly used in the food industry can be effective to inactivate phages, oxidizing agents and quaternary ammonium compounds being the most efficient ones (Campagna et al., 2014). Other treatments such as CO2, high pressure and UV light could be evaluated for each phage (Guglielmotti et al., 2011; Cheng et al., 2013). On the other hand, development of phage insensitive bacteria could be a cause of concern, since they may hamper the effectiveness of the phage disinfection process. Generally, the rate at which bacteria develop resistance is very low, especially when a cocktail of different phages is used. Moreover, phage-insensitive bacteria are associated with a reduced fitness (Gutiérrez et al., 2015b); therefore, this question is expected to have minor relevance.
In this context, endolysins have some important advantages compared to phages due to their proteinaceous nature, which is easily degraded in the environment. Regarding safety, the most important is their inability to transfer virulence genes.
Large-Scale Production
Implementation of phages as disinfectants in the food industry implies obtaining large volumes of phage suspensions with high titer using an inexpensive protocol. Therefore, some work is still necessary to optimize propagation and purification processes for each phage (Bourdin et al., 2014). In this regard, phages should be propagated in a non-pathogenic bacterium and then purified in order to remove cell debris or other contaminating substances. For preparations of bacteriophages infecting Gram-negative bacteria some procedures to remove endotoxin have been reported (Boratynski et al., 2004), and several companies also sell kits for endotoxin detection. However, the importance of these contaminating components in medical applications is more crucial than for disinfection. In the latter, undesirable effects of phages could be related with allergy by skin contact or by inhalation of aerosols. At present, nevertheless, there are no reported side effects of the use of bacteriophages in animal models of phage therapy applications (Golkar et al., 2014), which is not surprising as phages are abundant in human microbiota (De Paepe et al., 2014) and in the environment (Díaz-Munoz and Koskella, 2014). Overall, the purification methods used at laboratory-scale are well defined and consist of the precipitation of phages by polyethylene glycol followed by purification of phages in a cesium chloride gradient. However, these procedures are neither easy to scale up, nor cheap for the large-scale production required for the application of phages as disinfectants. New purification alternatives are being studied, e.g., suitable methods designed for purification of bionanoparticles, based on anion-exchange chromatography, with a 60% recovery of viable phages (Adriaenssens et al., 2012). Alternatives to centrifugation such as tangential flow filtration and specific membrane materials could also be explored (Hambsch et al., 2012).
The main drawback in the extensive use of endolysins might be the difficulty of their effective expression in E. coli (Rosano and Ceccarelli, 2014). Other bacteria like L. lactis have been proposed as suitable cell factories (D’Souza et al., 2012), but even the expression might need to be optimized (Rodríguez-Rubio et al., 2012). Moreover, large-scale production and purification of proteins is a costly process in itself. A similar scenario might be drawn for exopolysaccharide depolymerases due to the requirement of having large amounts of pure protein. In addition, more research is necessary to find out how specific the activity is for its substrate.
Regarding phage-derived proteins, thermostability seems to be a challenge, and a big concern, when applying enzymes for disinfection. However, heat stability seems to be a recurring property of phage structural lysins or VAPGHs (Rodríguez-Rubio et al., 2013). On the other hand, although the thermolabile nature of endolysins is well known (Varea et al., 2004; Obeso et al., 2008; Filatova et al., 2010; Heselpoth and Nelson, 2012), there are exceptions to the rule. In fact, two novel thermostable endolysins have recently been described, Lys68 from Salmonella phage phi68 (Oliveira et al., 2014) and Ph2119 from bacteriophage Ph2119 infecting Thermus scotoductus strain MAT2119 (Plotka et al., 2014). This thermostability supports the potential use of these phage-derived enzymes as disinfectants.
In addition to propagation and purification of phages and phage-derived proteins, other parameters such as a proper formulation, stability under non-refrigerated conditions, and lytic activity under usual conditions for the food industry should also be studied. No data about phage formulations and storage other than lyophilization (Merabishvili et al., 2013) and spray drying (Vandenheuvel et al., 2013) is available. Survival of phages in both processes is strictly dependent on an appropriate protector, in most cases sucrose being the most effective agent to protect phages (Merabishvili et al., 2013). However, this sugar is not suitable as excipient for disinfection processes. The controlled delivery of phages and their stability in encapsulated microspheres are worth studying. In fact, strategies under development for medical applications include phage encapsulation using different materials suitable for oral delivery or inhalation (Puapermpoonsiri et al., 2009; Dini et al., 2012; Balcao et al., 2014). Furthermore, bacteriophages could be useful to develop specific antimicrobial packaging materials for use in the food industry (Han et al., 2014).
Market and Regulatory
The potential of phages in the food industry is so extensive that several companies have developed phage-based products against important foodborne pathogens that could be used as disinfectants on surfaces and as food-processing aids. OmniLytics Inc. (Sandy, UT, USA) has developed two washing products, BacWashTM against Salmonella, and FinalyseTM against E. coli O157:H7, marketed by Elanco (Greenfield, IN, USA). Intralytix Inc. (Baltimore, MD, USA) developed three phage products, ListShieldTM, EcoShieldTM, and SalmoFreshTM, to be used in the food industry against L. monocytogenes, E. coli, and Salmonella, respectively. In Europe, Micreos BV (Wageningen, Netherlands) has commercialized ListexTM (P100) against L. monocytogenes, and SalmonelexTM against Salmonella. All these products are setting a precedent for future approval of phages as disinfectants. In fact, one of the most important drawbacks in the use of phage-based products might be the specific regulatory framework of each country. The US Department of Agriculture and FDA have already approved the use of several phage-based products, mentioned above, in food production environments, including their application as both food biopreservatives and disinfectants of food-contact surfaces. In Europe, however, the EFSA has argued that it is not clear whether bacteriophages can protect food against a re-contamination in spite of having been reported that bacteriophages are effective in the elimination of pathogens (EFSA, 2012). Finally, it is worth noticing that bacteriophages have also been approved as processing-aids in food processing and handling in several countries, but nothing has been reported about the use of phages as antimicrobial agents for the cleaning of industrial surfaces.
Concluding Remarks
The development of new disinfection products, non-toxic to humans and friendly to the environment, has good prospects for the future. Bacteriophage-based disinfectants fulfill all the requirements regarding effectiveness and safety. However, two main challenges have to be overcome before the implementation of phages in the food industry: (i) more research is necessary to solve the technical problems in manufacturing, such as the scaling up of the processes of propagation or expression, and purification of phages and proteins, and (ii) a regulatory framework for phage applications should be established, which would boost investment in these new products.
Author Contributions
PG, AR, and BM conceived the revision work. DG and LR-R designed the figures. DG, LR-R, BM, AR, and PG wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research study was supported by grants AGL2012-40194-C02-01 (Ministry of Science and Innovation, Spain), GRUPIN14-139 (Program of Science, Technology and Innovation 2013–2017 and FEDER EU funds, Principado de Asturias, Spain) and bacteriophage network FAGOMA. PG, BM, and AR are members of the FWO Vlaanderen funded “Phagebiotics” research community (WO.016.14). DG is a fellow of the Ministry of Science and Innovation, Spain. LR-R is a FWO Pegasus Marie Curie Fellow.
References
- Ackermann H. W., Prangishvili D. (2012). Prokaryote viruses studied by electron microscopy. Arch. Virol. 157 1843–1849. 10.1007/s00705-012-1383-y [DOI] [PubMed] [Google Scholar]
- Adriaenssens E. M., Lehman S. M., Vandersteegen K., Vandenheuvel D., Philippe D. L., Cornelissen A., et al. (2012). CIM(®) monolithic anion-exchange chromatography as a useful alternative to CsCl gradient purification of bacteriophage particles. Virology 434 265–270. 10.1016/j.virol.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriao A., Vieira M., Fernandes I., Barbosa M., Sol M., Tenreiro R. P., et al. (2008). Marked intra-strain variation in response of Listeria monocytogenes dairy isolates to acid or salt stress and the effect of acid or salt adaptation on adherence to abiotic surfaces. Int. J. Food Microbiol. 123 142–150. 10.1016/j.ijfoodmicro.2007.12.016 [DOI] [PubMed] [Google Scholar]
- Alkawash M. A., Soothill J. S., Schiller N. L. (2006). Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 114 131–138. 10.1111/j.1600-0463.2006.apm_356.x [DOI] [PubMed] [Google Scholar]
- Alves D. R., Gaudion A., Bean J. E., Perez Esteban P., Arnot T. C., Harper D. R., et al. (2014). Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl. Environ. Microbiol. 80 6694–6703. 10.1128/AEM.01789-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S., Singh D. (2013). Resistance of the constitutive microflora of biofilms formed on whey reverse-osmosis membranes to individual cleaning steps of a typical clean-in-place protocol. J. Dairy Sci. 96 6213–6222. 10.3168/jds.2013-7012 [DOI] [PubMed] [Google Scholar]
- Balcao V. M., Glasser C. A., Chaud M. V., del Fiol F. S., Tubino M., Vila M. M. (2014). Biomimetic aqueous-core lipid nanoballoons integrating a multiple emulsion formulation: a suitable housing system for viable lytic bacteriophages. Colloids Surf. B Biointerfaces 123 478–485. 10.1016/j.colsurfb.2014.09.045 [DOI] [PubMed] [Google Scholar]
- Barbirz S., Becker M., Freiberg A., Seckler R. (2009). Phage tailspike proteins with beta-solenoid fold as thermostable carbohydrate binding materials. Macromol Biosci. 9 169–173. 10.1002/mabi.200800278 [DOI] [PubMed] [Google Scholar]
- Barnes L. M., Lo M. F., Adams M. R., Chamberlain A. H. (1999). Effect of milk proteins on adhesion of bacteria to stainless steel surfaces. Appl. Environ. Microbiol. 65 4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchat L. R. (2002). Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes Infect. 4 413–423. 10.1016/S1286-4579(02)01555-1 [DOI] [PubMed] [Google Scholar]
- Bisbiroulas P., Psylou M., Iliopoulou I., Diakogiannis I., Berberi A., Mastronicolis S. K. (2011). Adaptational changes in cellular phospholipids and fatty acid composition of the food pathogen Listeria monocytogenes as a stress response to disinfectant sanitizer benzalkonium chloride. Lett. Appl. Microbiol. 52 275–280. 10.1111/j.1472-765X.2010.02995.x [DOI] [PubMed] [Google Scholar]
- Boles B. R., Horswill A. R. (2011). Staphylococcal biofilm disassembly. Trends Microbiol. 19 449–455. 10.1016/j.tim.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratynski J., Syper D., Weber-Dabrowska B., Lusiak-Szelachowska M., Pozniak G., Gorski A. (2004). Preparation of endotoxin-free bacteriophages. Cell Mol. Biol. Lett. 9 253–259. [PubMed] [Google Scholar]
- Bourdin G., Schmitt B., Marvin Guy L., Germond J. E., Zuber S., Michot L., et al. (2014). Amplification and purification of T4-like Escherichia coli phages for phage therapy: from laboratory to pilot scale. Appl. Environ. Microbiol. 80 1469–1476. 10.1128/AEM.03357-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briandet R., Lacroix-Gueu P., Renault M., Lecart S., Meylheuc T., Bidnenko E., et al. (2008). Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl. Environ. Microbiol. 74 2135–2143. 10.1128/AEM.02304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridier A., Briandet R., Thomas V., Dubois-Brissonnet F. (2011). Comparative biocidal activity of peracetic acid, benzalkonium chloride and ortho-phthalaldehyde on 77 bacterial strains. J. Hosp. Infect. 78 208–213. 10.1016/j.jhin.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Briers Y., Walmagh M., Grymonprez B., Biebl M., Pirnay J. P., Defraine V., et al. (2014a). Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58 3774–3784. 10.1128/AAC.02668-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briers Y., Walmagh M., Van Puyenbroeck V., Cornelissen A., Cenens W., Aertsen A., et al. (2014b). Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio 5:e1379-14. 10.1128/mBio.01379-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. L., Reuter M., Salt L. J., Cross K. L., Betts R. P., van Vliet A. H. (2014). Chicken juice enhances surface attachment and biofilm formation of Campylobacter jejuni. Appl. Environ. Microbiol. 80 7053–7060. 10.1128/AEM.02614-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna C., Villion M., Labrie S. J., Duchaine C., Moineau S. (2014). Inactivation of dairy bacteriophages by commercial sanitizers and disinfectants. Int. J. Food Microbiol. 171 41–47. 10.1016/j.ijfoodmicro.2013.11.012 [DOI] [PubMed] [Google Scholar]
- Capita R., Riesco-Pelaez F., Alonso-Hernando A., Alonso-Calleja C. (2014). Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 80 1268–1280. 10.1128/AEM.02283-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier B., Chassaing D. (2004). Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 97 111–122. 10.1016/j.ijfoodmicro.2004.03.031 [DOI] [PubMed] [Google Scholar]
- Cegelski L., Pinkner J. S., Hammer N. D., Cusumano C. K., Hung C. S., Chorell E., et al. (2009). Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5 913–919. 10.1038/nchembio.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerca N., Gomes F., Bento J. C., Franca A., Rolo J., Miragaia M., et al. (2013). Farnesol induces cell detachment from established S. epidermidis biofilms. J. Antibiot. (Tokyo). 66 255–258. 10.1038/ja.2013.11 [DOI] [PubMed] [Google Scholar]
- Chavant P., Martinie B., Meylheuc T., Bellon-Fontaine M. N., Hebraud M. (2002). Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 68 728–737. 10.1128/AEM.68.2.728-737.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Imai T., Teeka J., Hirose M., Higuchi T., Sekine M. (2013). Inactivation of bacteriophages by high levels of dissolved CO2. Environ. Technol. 34 539–544. 10.1080/09593330.2012.704403 [DOI] [PubMed] [Google Scholar]
- Chia T. W., Goulter R. M., McMeekin T., Dykes G. A., Fegan N. (2009). Attachment of different Salmonella serovars to materials commonly used in a poultry processing plant. Food Microbiol. 26 853–859. 10.1016/j.fm.2009.05.012 [DOI] [PubMed] [Google Scholar]
- Corcoran M., Morris D., De Lappe N., O’Connor J., Lalor P., Dockery P., et al. (2014). Commonly used disinfectants fail to eradicate Salmonella enterica biofilms from food contact surface materials. Appl. Environ. Microbiol. 80 1507–1514. 10.1128/AEM.03109-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen A., Ceyssens P. J., Krylov V. N., Noben J. P., Volckaert G., Lavigne R. (2012). Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 434 251–256. 10.1016/j.virol.2012.09.030 [DOI] [PubMed] [Google Scholar]
- Cornelissen A., Ceyssens P. J., T’Syen J., Van Praet H., Noben J. P., Shaburova O. V., et al. (2011). The T7-related Pseudomonas putida phage phi15 displays virion-associated biofilm degradation properties. PLoS ONE 6:e18597 10.1371/journal.pone.0018597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva E. P., De Martinis E. C. (2013). Current knowledge and perspectives on biofilm formation: the case of Listeria monocytogenes. Appl. Microbiol. Biotechnol. 97 957–968. 10.1007/s00253-012-4611-1 [DOI] [PubMed] [Google Scholar]
- Da Silva Felicio M. T., Hald T., Liebana E., Allende A., Hugas M., Nguyen-The C., et al. (2015). Risk ranking of pathogens in ready-to-eat unprocessed foods of non-animal origin (FoNAO) in the EU: initial evaluation using outbreak data (2007-2011). Int. J. Food Microbiol. 195 9–19. 10.1016/j.ijfoodmicro.2014.11.005 [DOI] [PubMed] [Google Scholar]
- da Silva Meira Q. G., de Medeiros Barbosa I., Alves Aguiar Athayde A. J., de Siqueira-Júnior J. P., de Souza E. L. (2012). Influence of temperature and surface kind on biofilm formation by Staphylococcus aureus from food-contact surfaces and sensitivity to sanitizers. Food Control. 25 469–475. 10.1016/j.foodcont.2011.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin-Regli A., Pages J. M. (2012). Cross-resistance between biocides and antimicrobials: an emerging question. Rev. Sci. Technol. 31 89–104. [PubMed] [Google Scholar]
- De Oliveira D. C., Fernandes Junior A., Kaneno R., Silva M. G., Araujo Junior J. P., Silva N. C., et al. (2014). Ability of Salmonella spp. to produce biofilm is dependent on temperature and surface material. Foodborne Pathog. Dis. 11 478–483. 10.1089/fpd.2013.1710 [DOI] [PubMed] [Google Scholar]
- De Paepe M., Leclerc M., Tinsley C. R., Petit M. A. (2014). Bacteriophages: an underestimated role in human and animal health? Front. Cell Infect. Microbiol. 4:39 10.3389/fcimb.2014.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVita M. D., Whadhera R. K., Theis M. L., Ingham S. C. (2007). Assessing the potential of Streptococcus pyogenes and Staphylococcus aureus transfer to foods and customers via a survey of hands, hand-contact surfaces and food-contact surfaces at foodservice facilities. J. Foodserv. 18 76–79. 10.1111/j.1745-4506.2007.00049.x [DOI] [Google Scholar]
- Díaz-Munoz S. L., Koskella B. (2014). Bacteria-phage interactions in natural environments. Adv. Appl. Microbiol. 89 135–183. 10.1016/B978-0-12-800259-9.00004-4 [DOI] [PubMed] [Google Scholar]
- Dini C., Islan G. A., de Urraza P. J., Castro G. R. (2012). Novel biopolymer matrices for microencapsulation of phages: enhanced protection against acidity and protease activity. Macromol. Biosci. 12 1200–1208. 10.1002/mabi.201200109 [DOI] [PubMed] [Google Scholar]
- Donlan R. M., Costerton J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15 167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle M. M., Cooney J. J., Caldwell D. E. (1996). Tracing the interaction of bacteriophage with bacterial biofilms using fluorescent and chromogenic probes. J. Ind. Microbiol. 16 331–341. 10.1007/BF01570111 [DOI] [PubMed] [Google Scholar]
- Dourou D., Beauchamp C. S., Yoon Y., Geornaras I., Belk K. E., Smith G. C., et al. (2011). Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures, on various food-contact surfaces encountered in beef processing. Int. J. Food Microbiol. 149 262–268. 10.1016/j.ijfoodmicro.2011.07.004 [DOI] [PubMed] [Google Scholar]
- D’Souza R., Pandeya D. R., Rahman M., Seo Lee H., Jung J. K., Hong S. T. (2012). Genetic engineering of Lactococcus lactis to produce an amylase inhibitor for development of an anti-diabetes biodrug. New Microbiol. 35 35–42. [PubMed] [Google Scholar]
- EFSA (2012). Scientific opinion on the evaluation of the safety and efficacy of ListexTM P100 for the removal of Listeria monocytogenes surface contamination of raw fish. EFSA J. 10:43. [Google Scholar]
- EFSA and ECDC (2016). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13:4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J., Allison D. G., Brown M. R., Gilbert P. (1991). Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27 177–184. 10.1093/jac/27.2.177 [DOI] [PubMed] [Google Scholar]
- Fenton M., Keary R., McAuliffe O., Ross R. P., O’Mahony J., Coffey A. (2013). Bacteriophage-derived peptidase CHAP(K) eliminates and prevents staphylococcal biofilms. Int. J. Microbiol. 2013 625341 10.1155/2013/625341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatova L. Y., Becker S. C., Donovan D. M., Gladilin A. K., Klyachko N. L. (2010). LysK, the enzyme lysing Staphylococcus aureus cells: specific kinetic features and approaches towards stabilization. Biochimie 92 507–513. 10.1016/j.biochi.2010.01.026 [DOI] [PubMed] [Google Scholar]
- Fischetti V. A. (2008). Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11 393–400. 10.1016/j.mib.2008.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- Ganegama-Arachchi G. J., Cridge A. G., Dias-Wanigasekera B. M., Cruz C. D., McIntyre L., Liu R., et al. (2013). Effectiveness of phages in the decontamination of Listeria monocytogenes adhered to clean stainless steel, stainless steel coated with fish protein, and as a biofilm. J. Ind. Microbiol. Biotechnol. 40 1105–1116. 10.1007/s10295-013-1313-3 [DOI] [PubMed] [Google Scholar]
- Gao G., Lange D., Hilpert K., Kindrachuk J., Zou Y., Cheng J. T., et al. (2011). The biocompatibility and biofilm resistance of implant coatings based on hydrophilic polymer brushes conjugated with antimicrobial peptides. Biomaterials 32 3899–3909. 10.1016/j.biomaterials.2011.02.013 [DOI] [PubMed] [Google Scholar]
- García P., Rodríguez L., Rodríguez A., Martínez B. (2010). Food biopreservation: promising strategies using bacteriocins, bacteriophages and endolysins. Trends Food Sci. Technol. 21 373–382. 10.1016/j.tifs.2010.04.010 [DOI] [Google Scholar]
- Gerstel U., Romling U. (2001). Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3 638–648. 10.1046/j.1462-2920.2001.00235.x [DOI] [PubMed] [Google Scholar]
- Giaouris E., Heir E., Hebraud M., Chorianopoulos N., Langsrud S., Moretro T., et al. (2014). Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 97 298–309. 10.1016/j.meatsci.2013.05.023 [DOI] [PubMed] [Google Scholar]
- Gilbert P., McBain A. J. (2003). Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin. Microbiol. Rev. 16 189–208. 10.1128/CMR.16.2.189-208.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadhas D. P., Marathe S. A., Chakravortty D. (2013). Biocides–resistance, cross-resistance mechanisms and assessment. Exp. Opin. Investig. Drugs 22 191–206. 10.1517/13543784.2013.748035 [DOI] [PubMed] [Google Scholar]
- Golkar Z., Bagasra O., Pace D. G. (2014). Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 8 129–136. 10.3855/jidc.3573 [DOI] [PubMed] [Google Scholar]
- Gounadaki A. S., Skandamis P. N., Drosinos E. H., Nychas G. J. (2008). Microbial ecology of food contact surfaces and products of small-scale facilities producing traditional sausages. Food Microbiol. 25 313–323. 10.1016/j.fm.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir S., Gudbjornsdottir B., Einarsson H., Kristinsson K. G., Kristjansson M. (2006). Contamination of cooked peeled shrimp (Pandalus borealis) by Listeria monocytogenes during processing at two processing plants. J. Food Prot. 69 1304–1311. [DOI] [PubMed] [Google Scholar]
- Guglielmotti D. M., Mercanti D. J., Reinheimer J. A., Quiberoni Adel L. (2011). Review: efficiency of physical and chemical treatments on the inactivation of dairy bacteriophages. Front. Microbiol. 2:282 10.3389/fmicb.2011.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez D., Briers Y., Rodríguez-Rubio L., Martínez B., Rodríguez A., Lavigne R., et al. (2015a). Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in staphylococcal Species. Front. Microbiol. 6:1315 10.3389/fmicb.2015.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez D., Delgado S., Vázquez-Sánchez D., Martínez B., Cabo M. L., Rodríguez A., et al. (2012a). Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl. Environ. Microbiol. 78 8547–8554. 10.1128/AEM.02045-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez D., Martínez B., Rodríguez A., García P. (2012b). Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential. BMC Genomics 13:228 10.1186/1471-2164-13-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez D., Ruas-Madiedo P., Martínez B., Rodríguez A., García P. (2014). Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 9:e107307 10.1371/journal.pone.0107307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez D., Vandenheuvel D., Martínez B., Rodríguez A., Lavigne R., García P. (2015b). Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species Staphylococcal biofilms. Appl. Environ. Microbiol. 81 3336–3348. 10.1128/AEM.03560-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hambsch B., Bosl M., Eberhagen I., Muller U. (2012). Removal of bacteriophages with different surface charges by diverse ceramic membrane materials in pilot spiking tests. Water Sci Technol. 66 151–157. 10.2166/wst.2012.141 [DOI] [PubMed] [Google Scholar]
- Han J. H., Wang M. S., Das J., Sudheendra L., Vonasek E., Nitin N., et al. (2014). Capture and detection of T7 bacteriophages on a nanostructured interface. ACS Appl. Mater Interf. 6 4758–4765. 10.1021/am500655r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M., Eberl L., Nielsen J., Givskov M. (2003). Quorum sensing : a novel target for the treatment of biofilm infections. BioDrugs 17 241–250. 10.2165/00063030-200317040-00003 [DOI] [PubMed] [Google Scholar]
- Herrera J. J., Cabo M. L., Gonzalez A., Pazos I., Pastoriza L. (2007). Adhesion and detachment kinetics of several strains of Staphylococcus aureus subsp. aureus under three different experimental conditions. Food Microbiol. 24 585–591. 10.1016/j.fm.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Heselpoth R. D., Nelson D. C. (2012). A new screening method for the directed evolution of thermostable bacteriolytic enzymes. J. Vis. Exp. 4216 10.3791/4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingston P. A., Stea E. C., Knochel S., Hansen T. (2013). Role of initial contamination levels, biofilm maturity and presence of salt and fat on desiccation survival of Listeria monocytogenes on stainless steel surfaces. Food Microbiol. 36 46–56. 10.1016/j.fm.2013.04.011 [DOI] [PubMed] [Google Scholar]
- Hoffman L. R., D’Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., Miller S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436 1171–1175. 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- Jensen A., Larsen M. H., Ingmer H., Vogel B. F., Gram L. (2007). Sodium chloride enhances adherence and aggregation and strain variation influences invasiveness of Listeria monocytogenes strains. J. Food Prot. 70 592–599. [DOI] [PubMed] [Google Scholar]
- Jubelin G., Vianney A., Beloin C., Ghigo J. M., Lazzaroni J. C., Lejeune P., et al. (2005). CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187 2038–2049. 10.1128/JB.187.6.2038-2049.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E., Watnick P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73 310–347. 10.1128/MMBR.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D., McAuliffe O., Ross R. P., Coffey A. (2012). Prevention of Staphylococcus aureus biofilm formation and reduction in established biofilm density using a combination of phage K and modified derivatives. Lett. Appl. Microbiol. 54 286–291. 10.1111/j.1472-765X.2012.03205.x [DOI] [PubMed] [Google Scholar]
- Kesel S., Mader A., Seeberger P. H., Lieleg O., Opitz M. (2014). Carbohydrate coating reduces adhesion of biofilm-forming Bacillus subtilis to gold surfaces. Appl. Environ. Microbiol. 80 5911–5917. 10.1128/AEM.01600-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E., Raya R., Carlson K. (2004). “Molecular mechanisms of phage infection,” in Bacteriophages: Biology and Applications, eds Kutter E., Sulakvelidze A. (Boca Raton, FL: CRC press; ), 165–222. [Google Scholar]
- Latorre A. A., Van Kessel J. S., Karns J. S., Zurakowski M. J., Pradhan A. K., Boor K. J., et al. (2010). Biofilm in milking equipment on a dairy farm as a potential source of bulk tank milk contamination with Listeria monocytogenes. J. Dairy Sci. 93 2792–2802. 10.3168/jds.2009-2717 [DOI] [PubMed] [Google Scholar]
- Liu N. T., Lefcourt A. M., Nou X., Shelton D. R., Zhang G., Lo Y. M. (2013). Native microflora in fresh-cut produce processing plants and their potentials for biofilm formation. J. Food Prot. 76 827–832. 10.4315/0362-028X.JFP-12-433 [DOI] [PubMed] [Google Scholar]
- López-Gálvez F., Gil M. I., Truchado P., Selma M. V., Allende A. (2010). Cross-contamination of fresh-cut lettuce after a short-term exposure during pre-washing cannot be controlled after subsequent washing with chlorine dioxide or sodium hypochlorite. Food Microbiol. 27 199–204. 10.1016/j.fm.2009.09.009 [DOI] [PubMed] [Google Scholar]
- Lu T. K., Collins J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 104 11197–11202. 10.1073/pnas.0704624104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T. K., Collins J. J. (2009). Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. U.S.A. 106 4629–4634. 10.1073/pnas.0800442106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabishvili M., Vervaet C., Pirnay J. P., De Vos D., Verbeken G., Mast J., et al. (2013). Stability of Staphylococcus aureus phage ISP after freeze-drying (lyophilization). PLoS ONE 8:e68797 10.1371/journal.pone.0068797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey A. R., Craig S. A., Payne S. M. (2005). Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73 5706–5719. 10.1128/IAI.73.9.5706-5719.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizan M. F., Jahid I. K., Ha S. D. (2015). Microbial biofilms in seafood: a food-hygiene challenge. Food Microbiol. 49 41–55. 10.1016/j.fm.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Moak M., Molineux I. J. (2004). Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51 1169–1183. 10.1046/j.1365-2958.2003.03894.x [DOI] [PubMed] [Google Scholar]
- Moltz A. G., Martin S. E. (2005). Formation of biofilms by Listeria monocytogenes under various growth conditions. J. Food Prot. 68 92–97. [DOI] [PubMed] [Google Scholar]
- Monds R. D., Newell P. D., Gross R. H., O’Toole G. A. (2007). Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63 656–679. 10.1111/j.1365-2958.2006.05539.x [DOI] [PubMed] [Google Scholar]
- Montañez-Izquierdo V. Y., Salas-Vázquez D. I., Rodrígez-Jerez J. J. (2012). Use of epifluorescence microscopy to assess the effectiveness of phage P100 in controlling Listeria monocytogenes biofilms on stainless steel surfaces. Food Control. 23 470–477. 10.1016/j.foodcont.2011.08.016 [DOI] [Google Scholar]
- Myszka K., Czaczyk K. (2011). Bacterial biofilms on food contact surfaces - a review. Pol. J. Food Nutr. Sci. 61 73–180. 10.2478/v10222-011-0018-4 [DOI] [Google Scholar]
- Nelson D. C., Schmelcher M., Rodriguez-Rubio L., Klumpp J., Pritchard D. G., Dong S., et al. (2012). Endolysins as antimicrobials. Adv. Virus Res. 83 299–365. 10.1016/B978-0-12-394438-2.00007-4 [DOI] [PubMed] [Google Scholar]
- Nesse L. L., Sekse C., Berg K., Johannesen K. C., Solheim H., Vestby L. K., et al. (2014). Potentially pathogenic Escherichia coli can form a biofilm under conditions relevant to the food production chain. Appl. Environ. Microbiol. 80 2042–2049. 10.1128/AEM.03331-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega F. L., Costa A. R., Kluskens L. D., Azeredo J. (2015). Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23 185–191. 10.1016/j.tim.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Obeso J. M., Martínez B., Rodríguez A., García P. (2008). Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 128 212–218. 10.1016/j.ijfoodmicro.2008.08.010 [DOI] [PubMed] [Google Scholar]
- O’Flaherty S., Ross R. P., Coffey A. (2009). Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33 801–819. 10.1111/j.1574-6976.2009.00176.x [DOI] [PubMed] [Google Scholar]
- Oliveira H., Thiagarajan V., Walmagh M., Sillankorva S., Lavigne R., Neves-Petersen M. T., et al. (2014). A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram-negative pathogens in presence of weak acids. PLoS ONE 9:e108376 10.1371/journal.pone.0108376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasion S., Kwiatek M., Gryko R., Mizak L., Malm A. (2014). Bacteriophages as an alternative strategy for fighting biofilm development. Pol. J. Microbiol. 63 137–145. [PubMed] [Google Scholar]
- Pasvolsky R., Zakin V., Ostrova I., Shemesh M. (2014). Butyric acid released during milk lipolysis triggers biofilm formation of Bacillus species. Int. J. Food Microbiol. 181 19–27. 10.1016/j.ijfoodmicro.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Patel J., Sharma M., Millner P., Calaway T., Singh M. (2011). Inactivation of Escherichia coli O157:H7 attached to spinach harvester blade using bacteriophage. Foodborne Pathog. Dis. 8 541–546. 10.1089/fpd.2010.0734 [DOI] [PubMed] [Google Scholar]
- Pearl S., Gabay C., Kishony R., Oppenheim A., Balaban N. Q. (2008). Nongenetic individuality in the host-phage interaction. PLoS Biol. 6:e120 10.1371/journal.pbio.0060120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccio A., Autio T., Korkeala H., Rosmini R., Trevisani M. (2003). Listeria monocytogenes occurrence and characterization in meat-producing plants. Lett. Appl. Microbiol. 37 234–238. 10.1046/j.1472-765X.2003.01384.x [DOI] [PubMed] [Google Scholar]
- Periasamy S., Joo H. S., Duong A. C., Bach T. H., Tan V. Y., Chatterjee S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109 1281–1286. 10.1073/pnas.1115006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel-Filho N. J., Martins M. C., Nogueira G. B., Mantovani H. C., Vanetti M. C. (2014). Bovicin HC5 and nisin reduce Staphylococcus aureus adhesion to polystyrene and change the hydrophobicity profile and Gibbs free energy of adhesion. Int. J. Food Microbiol. 190 1–8. 10.1016/j.ijfoodmicro.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Pires D. P., Oliveira H., Melo L. D., Sillankorva S., Azeredo J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100 2141–2151. 10.1007/s00253-015-7247-0 [DOI] [PubMed] [Google Scholar]
- Plotka M., Kaczorowska A. K., Stefanska A., Morzywolek A., Fridjonsson O. H., Dunin-Horkawicz S., et al. (2014). Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl. Environ. Microbiol. 80 886–895. 10.1128/AEM.03074-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puapermpoonsiri U., Spencer J., van der Walle C. F. (2009). A freeze-dried formulation of bacteriophage encapsulated in biodegradable microspheres. Eur. J. Pharm. Biopharm. 72 26–33. 10.1016/j.ejpb.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Rahman M., Kim S., Kim S. M., Seol S. Y., Kim J. (2011). Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 27 1087–1093. 10.1080/08927014.2011.631169 [DOI] [PubMed] [Google Scholar]
- Ramasubbu N., Thomas L. M., Ragunath C., Kaplan J. B. (2005). Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 349 475–486. 10.1016/j.jmb.2005.03.082 [DOI] [PubMed] [Google Scholar]
- Reuter M., Mallett A., Pearson B. M., van Vliet A. H. (2010). Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76 2122–2128. 10.1128/AEM.01878-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Autio W. R., McLandsborough L. A. (2008). Effect of surface roughness and stainless steel finish on Listeria monocytogenes attachment and biofilm formation. J. Food Prot. 71 170–175. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rubio L., Gutiérrez D., Martínez B., Rodríguez A., García P. (2012). Lytic activity of LysH5 endolysin secreted by Lactococcus lactis using the secretion signal sequence of bacteriocin Lcn972. Appl. Environ. Microbiol. 78 3469–3472. 10.1128/AEM.00018-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rubio L., Martínez B., Donovan D. M., Rodríguez A., García P. (2013). Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. Crit. Rev. Microbiol. 39 427–434. 10.3109/1040841X.2012.723675 [DOI] [PubMed] [Google Scholar]
- Romling U., Balsalobre C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272 541–561. 10.1111/joim.12004 [DOI] [PubMed] [Google Scholar]
- Rosano G. L., Ceccarelli E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5:172 10.3389/fmicb.2014.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch D. A., Cram D. S., DiBerardino D., Littlejohn T. G., Skurray R. A. (1990). Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4 2051–2062. 10.1111/j.1365-2958.1990.tb00565.x [DOI] [PubMed] [Google Scholar]
- Ryan E. M., Alkawareek M. Y., Donnelly R. F., Gilmore B. F. (2012). Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 65 395–398. 10.1111/j.1574-695X.2012.00977.x [DOI] [PubMed] [Google Scholar]
- Saa-Ibusquiza P., Herrera J. J., Cabo M. L. (2011). Resistance to benzalkonium chloride, peracetic acid and nisin during formation of mature biofilms by Listeria monocytogenes. Food Microbiol. 28 418–425. 10.1016/j.fm.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Salwiczek M., Qu Y., Gardiner J., Strugnell R. A., Lithgow T., McLean K. M., et al. (2014). Emerging rules for effective antimicrobial coatings. Trends Biotechnol. 32 82–90. 10.1016/j.tibtech.2013.09.008 [DOI] [PubMed] [Google Scholar]
- Sass P., Bierbaum G. (2007). Lytic activity of recombinant bacteriophage φ11 and φ12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl. Environ. Microbiol. 73 347–352. 10.1128/AEM.01616-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher M., Donovan D. M., Loessner M. J. (2012). Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 7 1147–1171. 10.2217/fmb.12.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Ryu J. H., Beuchat L. R. (2005). Inactivation of Escherichia coli O157:H7 in biofilm on stainless steel by treatment with an alkaline cleaner and a bacteriophage. J. Appl. Microbiol. 99 449–459. 10.1111/j.1365-2672.2005.02659.x [DOI] [PubMed] [Google Scholar]
- Sharma S. K., Dai T., Kharkwal G. B., Huang Y. Y., Huang L., De Arce V. J., et al. (2011). Drug discovery of antimicrobial photosensitizers using animal models. Curr. Pharm. Des. 17 1303–1319. 10.2174/138161211795703735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Koller T., Kreikemeyer B., Nelson D. C. (2013). Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. J. Antimicrob. Chemother. 68 1818–1824. 10.1093/jac/dkt104 [DOI] [PubMed] [Google Scholar]
- Silagyi K., Kim S. H., Lo Y. M., Wei C. I. (2009). Production of biofilm and quorum sensing by Escherichia coli O157:H7 and its transfer from contact surfaces to meat, poultry, ready-to-eat deli, and produce products. Food Microbiol. 26 514–519. 10.1016/j.fm.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Simoes L. C., Lemos M., Pereira A. M., Abreu A. C., Saavedra M. J., Simoes M. (2011). Persister cells in a biofilm treated with a biocide. Biofouling 27 403–411. 10.1080/08927014.2011.579599 [DOI] [PubMed] [Google Scholar]
- Siringan P., Connerton P. L., Payne R. J., Connerton I. F. (2011). Bacteriophage-Mediated Dispersal of Campylobacter jejuni Biofilms. Appl. Environ. Microbiol. 77 3320–3326. 10.1128/AEM.02704-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J. S., Lee S. J., Jun S. Y., Yoon S. J., Kang S. H., Paik H. R., et al. (2010). Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl. Microbiol. Biotechnol. 86 1439–1449. 10.1007/s00253-009-2386-9 [DOI] [PubMed] [Google Scholar]
- Soni K. A., Nannapaneni R. (2010). Removal of Listeria monocytogenes biofilms with bacteriophage P100. J. Food Prot. 73 1519–1524. [DOI] [PubMed] [Google Scholar]
- Tang L., Pillai S., Revsbech N. P., Schramm A., Bischoff C., Meyer R. L. (2011). Biofilm retention on surfaces with variable roughness and hydrophobicity. Biofouling. 27 111–121. 10.1080/08927014.2010.544848 [DOI] [PubMed] [Google Scholar]
- Teh K. H., Flint S., French N. (2010). Biofilm formation by Campylobacter jejuni in controlled mixed-microbial populations. Int. J. Food Microbiol. 143 118–124. 10.1016/j.ijfoodmicro.2010.07.037 [DOI] [PubMed] [Google Scholar]
- Thimothe J., Nightingale K. K., Gall K., Scott V. N., Wiedmann M. (2004). Tracking of Listeria monocytogenes in smoked fish processing plants. J. Food Prot. 67 328–341. [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Thompson C. C., Vicente A. C., Klose K. E. (2010). Vibrio2009: the third international conference on the biology of Vibrios. Mol. Microbiol. 77 1065–1071. 10.1111/j.1365-2958.2010.07286.x [DOI] [PubMed] [Google Scholar]
- van Dyk B. N., de Bruin W., du Plessis E. M., Korsten L. (2016). Microbiological food safety status of commercially produced tomatoes from production to marketing. J. Food Prot. 79 392–406. 10.4315/0362-028X.JFP-15-300 [DOI] [PubMed] [Google Scholar]
- Van Houdt R., Michiels C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109 1117–1131. 10.1111/j.1365-2672.2010.04756.x [DOI] [PubMed] [Google Scholar]
- Vandenheuvel D., Singh A., Vandersteegen K., Klumpp J., Lavigne R., Van den Mooter G. (2013). Feasibility of spray drying bacteriophages into respirable powders to combat pulmonary bacterial infections. Eur. J. Pharm. Biopharm. 84 578–582. 10.1016/j.ejpb.2012.12.022 [DOI] [PubMed] [Google Scholar]
- Vandersteegen K., Kropinski A. M., Nash J. H., Noben J. P., Hermans K., Lavigne R. (2013). Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J. Virol. 87 3237–3247. 10.1128/JVI.02763-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varea J., Monterroso B., Saiz J. L., Lopez-Zumel C., Garcia J. L., Laynez J., et al. (2004). Structural and thermodynamic characterization of Pal, a phage natural chimeric lysin active against pneumococci. J. Biol. Chem. 279 43697–43707. 10.1074/jbc.M407067200 [DOI] [PubMed] [Google Scholar]
- Vázquez-Sánchez D., Habimana O., Holck A. (2013). Impact of food-related environmental factors on the adherence and biofilm formation of natural Staphylococcus aureus isolates. Curr. Microbiol. 66 110–121. 10.1007/s00284-012-0247-8 [DOI] [PubMed] [Google Scholar]
- Verran J., Airey P., Packer A., Whitehead K. A. (2008). Microbial retention on open food contact surfaces and implications for food contamination. Adv. Appl. Microbiol. 64 223–246. 10.1016/S0065-2164(08)00408-5 [DOI] [PubMed] [Google Scholar]
- Vestby L. K., Moretro T., Langsrud S., Heir E., Nesse L. L. (2009). Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet Res. 5:20 10.1186/1746-6148-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viazis S., Akhtar M., Feirtag J., Diez-Gonzalez F. (2011). Reduction of Escherichia coli O157:H7 viability on hard surfaces by treatment with a bacteriophage mixture. Int. J. Food Microbiol. 145 37–42. 10.1016/j.ijfoodmicro.2010.11.021 [DOI] [PubMed] [Google Scholar]
- von Laer A. E., de Lima A. S., Trindade Pdos S., Andriguetto C., Destro M. T., da Silva W. P. (2009). Characterization of Listeria monocytogenes isolated from a fresh mixed sausage processing line in Pelotas-RS by PFGE. Braz. J. Microbiol. 40 574–582. 10.1590/S1517-838220090003000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Kalchayanand N., King D. A., Luedtke B. E., Bosilevac J. M., Arthur T. M. (2014). Biofilm formation and sanitizer resistance of Escherichia coli O157:H7 strains isolated from “high event period” meat contamination. J. Food Prot. 77 1982–1987. 10.4315/0362-028X.JFP-14-253 [DOI] [PubMed] [Google Scholar]
- Yang H., Zhang Y., Huang Y., Yu J., Wei H. (2014). Degradation of methicillin-resistant Staphylococcus aureus biofilms using a chimeric lysin. Biofouling 30 667–674. 10.1080/08927014.2014.905927 [DOI] [PubMed] [Google Scholar]
- Yildiz F. H., Liu X. S., Heydorn A., Schoolnik G. K. (2004). Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53 497–515. 10.1111/j.1365-2958.2004.04154.x [DOI] [PubMed] [Google Scholar]