Abstract
Purpose
Microbial communities in or on the body (i.e., the microbiome) are highly physiologically active and influence human health. Although environmental scientists are increasingly aware of the gut microbiome, the respiratory microbiome's role in the human response to inhaled pollutants is largely unknown.
Methods
We reviewed the literature and present mechanisms by which the microbiome might mediate or modify human responses to inhaled pollutants.
Results
The respiratory microbiome has been shown to influence chronic lung disease exacerbations, and increasing evidence indicates a role in disease development. Research also suggests that the respiratory microbiome could plausibly metabolize inhaled pollutants or modulate host inflammatory responses to exposure. Because these responses depend on the microbes present, defining the composition of the resident microbiome and how microbial communities shift with exposure may help to explain variations in susceptibility to inhaled pollutants. More research is needed but significant measurement challenges remain for large epidemiological studies of the respiratory microbiome.
Conclusions
The respiratory microbiome is likely an underexplored intermediate and potential cause of individual susceptibility to inhaled irritants/toxicants. Characterizing the microbiome's role in the human response to inhaled exposures could improve our understanding of the casual agents of exposure and suggest novel public health interventions.
Inhaled irritants and toxicants represent an important environmental exposure that are linked to death and disease. Health effects associated with these exposures include increased risks of lung cancer, heart and respiratory diseases, as well as metabolic disorders.1 Given continuous exposures across the lifetimes of all people, it is estimated that 3.1 million deaths and 3.1% of disability-adjusted life years are lost globally per year due to exposures to outdoor particulate pollution. Exposures to air pollution from household combustion of solid fuels account for an additional 3.5 million deaths and 4.5% of disability-adjusted life years lost. Active and passive exposures to tobacco smoke further contribute 6.3 million deaths and 6.3% of disability adjusted life years lost. Collectively, these impacts place the inhalation of air pollutants within the top 10 risk factors for the Global Burden of Disease.2
Governmental regulations have successfully reduced outdoor air pollution concentrations and limited tobacco smoke exposures in the United States, with corresponding improvements in health.3-6 Excess risk remains, however, even at levels of pollution below existing standards. In addition, not all individuals bear the same burden of disease from inhaled pollution exposures. Enhanced susceptibility has been reported among children, seniors, and persons with obesity, diabetes, coronary artery disease, and asthma.7,8 Although these factors may be associated with a several-fold larger risk in any individual investigation, the characteristics conferring risk are not always consistent across studies. This observation suggests that traditional risk factors alone may be insufficient to identify those at enhanced risk.
In this article we hypothesize that microbial communities, especially those within the respiratory tract, may have an important, yet under-recognized, role in the human response to inhaled irritants/toxicants. Microbes, including bacteria, fungi, and viruses, reside on all human tissues exposed to the external environment and outnumber human cells by approximately ten to one. Collectively referred to as the microbiome, microbial communities in or on the body are highly physiologically active and known to influence the well-being of their host.9 While most research is focused on relationships between the microbiome and health, environmental scientists have begun to pay increasing attention to the gut microbiome since microbes of the gut have been shown to metabolize environmental toxicants,10-12 stimulate host inflammatory response, and affect risk of host infection.9 In spite of the clear physiological parallels, however, very little thought has been given to the role of the respiratory microbiome in the human response to inhaled irritants/toxicants. Here, we describe what is known about the respiratory microbiome, discuss how it and the gut microbiome might influence the human response to inhalation exposures, and encourage researchers to consider the respiratory microbiome as a mechanistic intermediate and potential cause of individual susceptibility to inhaled irritants/toxicants.
What Do We Know About the Respiratory Microbiome?
For over 100 years, traditional wisdom was that in those without lung diseases, microbial communities resided only in the upper (i.e., mouth and nose) but not the lower (i.e., lungs) airways. More recently, however, the use of culture-independent, sequence-based techniques has clearly shown that the lungs are not sterile.13,14 The current state of the science can be found summarized by several excellent review papers15-18 with brief highlights below.
The origins of the microbial communities in the lungs include inspired air, which contains around 100 bacteria/m3,19 as well as those microaspirated and/or dispersed from the oropharynx.18,20 With no physical barrier blocking bidirectional movement, the lungs also actively eliminate microbes via mucociliary clearance, cough, and innate and adaptive host immune responses. In health, alveolar macrophages, antibacterial surfactant, and other environmental conditions (e.g., temperature, pH, and nutrients) inhibit extensive bacterial growth, resulting in low colonization of the lungs in comparison to other compartments. For example, it is estimated that there are approximately 1,000 times fewer microbes in the lungs than the mouth and 1 million to 1,000 million times fewer microbes than in the gut.21,22 In spite of their low abundances, there are diverse and dynamic communities present. Bacterial species common to healthy lungs include Streptococcus, Prevotella, and Veillonella.19,23
In diseased lungs, conditions often become more favorable for bacterial reproduction. Evidence of this growth is provided by a small, but growing, literature documenting different bacterial communities between healthy individuals and those with chronic respiratory diseases such as cystic fibrosis, chronic obstructive pulmonary disease (COPD), and asthma.14,24-28 For example, individuals with asthma or COPD have been reported to have higher abundances of Proteobacteria than healthy individuals.13,28 This finding is important because this phylum includes known respiratory pathogens.
Interestingly, it appears as though the respiratory microbiome community structure may not just reflect the presence of a disease but may also correlate with disease severity. For example, differences in bacterial communities in the lungs have been associated with asthma severity29-32 and with the frequency of exacerbations in patients with bronchiectasis.33 There is also evidence that bacterial communities in the lungs are related to responsiveness to therapeutic interventions27 and administration of probiotics has been shown to reduce the frequency of cystic fibrosis exacerbations.33,34 Collectively, these findings suggest an important role for resident bacteria not only in disease development, as proposed by the hygiene hypothesis,35 but also for the initiation of exacerbations and potentially for treatments.
How Might the Respiratory Microbiome Influence the Human Response to Inhaled Irritants/Toxicants?
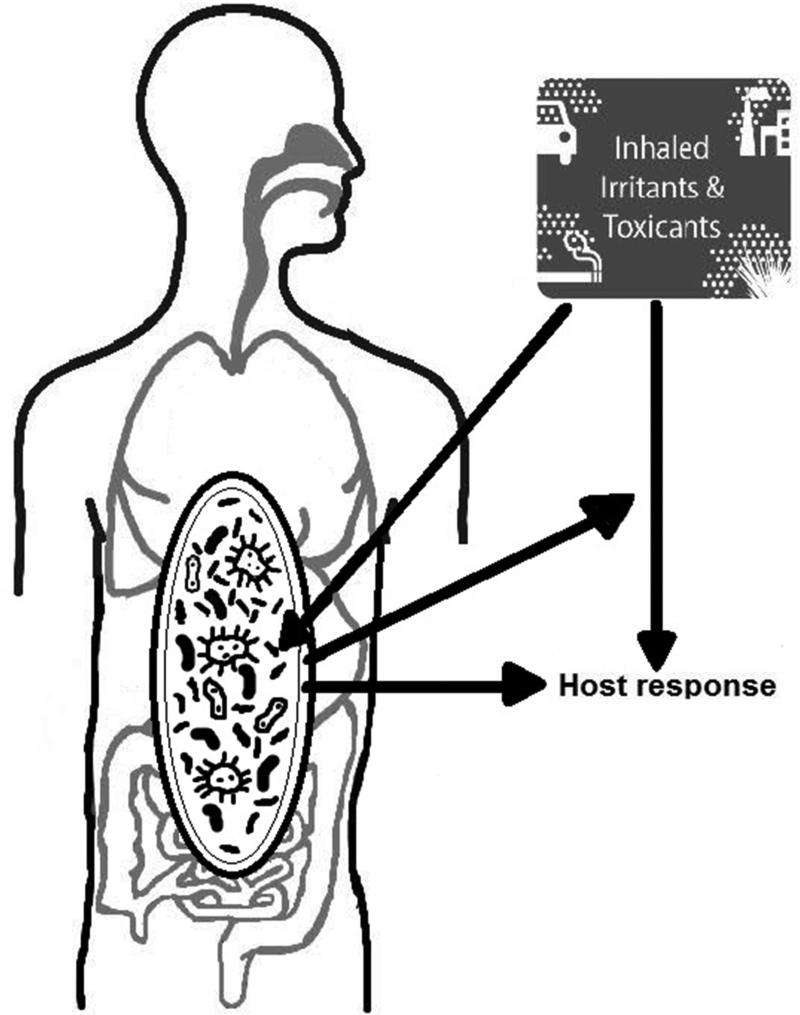
There is a growing understanding that our microbiota play a critical role in the development and mediation of many human processes. As some of the first cells in the body encountering inhaled environmental toxicants, it is likely that the respiratory microbiome is both affected by these exposures and affects these exposures (Figure 1). In the ideal world, the microbiome would serve as a protective shield for human host. However, it is also likely that the human host gets caught in the cross-hairs of the microbial response to inhaled pollutants and experience collateral damage from those interactions.
Figure 1.
Hypothesized interplay of inhaled irritants, the respiratory microbiome, and health
One possible mechanism underlying the hypothesis that the microbiome blunts the impacts of human exposure, or that it amplifies it, is that members of the respiratory microbiome can be selectively injured or killed by inhaled toxicants. Inhaled irritants/toxicants can deposit throughout the respiratory tract, with larger particles depositing more prominently in the upper airways (i.e., nose) and the smallest particles and gases reaching deep into the lower airways.36 If these inhaled exposures induce direct oxidative stress or changes to growing conditions such as local alterations in pH, a likely result could be a shift in which microbes are present (i.e., the microbiome community structure). Disrupting the community structure of the microbiome could then result in changes in the functions it performs, with downstream consequences for human health. Specifically, we hypothesize that changes to microbial function will include alterations to the balance of anti-oxidant and pro-inflammatory conditions. We propose such a mechanism based on the known modulation of the host immune response by the microbiome16,24. Additionally, there is strong evidence from controlled and observational studies implicating oxidative stress and inflammation as key mechanisms in the pathogenesis of inhaled pollutants.37 This conjecture is consistent with findings that chronic inflammatory conditions such as asthma and diabetes8,38-43, which themselves have been linked to the microbiome, are associated with differential susceptibility to inhaled pollution.
Although there is little known about the impacts of inhaled environmental exposures on the respiratory microbiome, research from the gut has demonstrated selective shifts in microbial community structure and function following the ingestion of environmental pollutants. For example, arsenic-treated mice experienced reductions in Firmicutes but not Bacteroidetes in the gut.44 These same exposed mice also exhibited bidirectional changes in key metabolites, including those related to bile acids, lipids, amino acids, and isoflavones. Given that many of the observed changes are related to altered absorption of nutrients from the gut, it is plausible that alterations to the microbiome may underlie observed associations between metals and metabolic diseases such as obesity and insulin resistance.45-47 New evidence also points to changes in the gut microbial community structure following chemical exposures, including polychlorinated biphenyls, that reduced bacterial abundance following ingestion by mice.48
With or without shifts in the microbial communities following exposure, a second plausible mechanism by which the microbiome might influence the human response inhaled exposures is based on its well-known ability to transform chemicals into forms that are more or less bioaccessible to humans. Bacteria have been known for decades to contribute to the biotransformation of environmental metals such as arsenic.49 Only recently, however, have scientists begun to characterize such biotransformations by the internal microbiome. One seminal study demonstrated that human intestinal bacteria were able in vitro to metabolize inorganic arsenic in contaminated soil into methylated arsenic compounds and thioarsenicals.50,51 More recently, evidence of these transformations has been extended to other pollutants such as polycyclic aromatic hydrocarbons. These typically non-estrogenic by-products of combustion can be converted to compounds with estrogenic-like activity by bacteria from the human colon.52 Assuming that similar reactions occur within the microbial communities of the respiratory tract, is likely that the toxicity of inhaled pollutants, which include metals and polycyclic aromatic hydrocarbons, is influenced by microbiome-mediated chemical conversions. In fact, a very recent study demonstrated that administration of antibiotics to mice altered their airway hyper-responsiveness following inhalation of the reactive gas, ozone.53
Ultimately, because transformations of inhaled pollutants will depend on the specific microbes present and microbial communities differ between individuals,9 it is likely that the microbiome contributes to variations in disease susceptibility. Interestingly, both early lifetime and recent ecological and social settings influence an individual's microbiome, 54 suggesting a novel mechanism for adaptation or exaggeration of human responsiveness to long-term exposures.
What Evidence is There of a Respiratory Microbiome Response to Environmental Pollutants?
An extremely small literature on smoking supports the hypothesis that inhaled air irritants/toxicants may impact the respiratory microbiome. Most such research, however, is short-term and from the subgingival environment. In this environment, lower abundances of health-promoting microbes and higher abundances of pathogens have been reported with smoking exposures.55 Interestingly, smoking appears to have a rapid impact on the oral microbiome with changes in bacterial colonization reported within 24-hours. 56 Smoking cessation has similarly been shown to shift the community composition of both dental and intestinal microbes within weeks.57,58
Chronic differences in the microbiome of smokers has also been suggested in the oropharynx and, to a lesser extent, the nasopharynx.59,60 By contrast, active smoking has not been shown to alter the community structure of the lower respiratory tract microbiome, at least in individuals with normal lung function defined spirometrically.14,59 We consider it highly unlikely that the same is true in those with established COPD or asthma of even mild severity but this is a crucial topic for further investigation. Finally, epidemiological studies have shown both cigarette smoke and indoor biomass burning exposures are associated with higher respiratory infection rates from pneumococcal pneumonia, Legionnaire's disease, influenza, and tuberculosis.61-63 Smoking also clearly depresses the ability to fend off respiratory viral infections.64-68 Hence, smoking and potentially other air pollutants may foster outgrowth of opportunistic respiratory bacterial species that can exploit the niche created by transient epithelial damage induced by respiratory viral infection and the immunosuppression that follows viral clearance.69-71
What Challenges Do We Face in Studying the Role of the Respiratory Microbiome?
One of the key challenges to clinical and epidemiological research on environmental exposures and the respiratory microbiome relates to existing measurement techniques. Approaches to study microbial communities of the lungs have traditionally used bronchoalveolar lavage and induced sputum.31 Although previous concerns about the potential for contamination during the bronchoscopic process have been allayed based on results of protected specimen brushing,20 both of these methods are high-burden techniques for healthy individuals. This burden limits the ability of researchers to pursue the large scale studies likely required to understand the complex interactions that may be at play. For this reason, we and others are actively working to identify a less invasive media that can reliably quantify the respiratory microbiome.
Another important issue for respiratory microbiome research is that of low microbial biomass. Given that microbial communities are present in low abundances in the lungs, extra care must be taken to prevent confusion between the true signal and that from other sources (i.e., “background”). Distortion of the microbial community structure can occur due to the technical aspects of high-throughput sequencing including the presence of nucleic acids from human cells or bacteria in the sampling reagents, differences in extraction protocols, as well as PCR efficiency.72-75 Although careful accounting of these sources of noise will produce meaningful results, this is not always the default procedure. Thus researchers must be diligent in adopting adequate quality assurance protocols for their own research and acknowledging the limitations of findings from laboratories that have not used such procedures.
Finally, we have focused this review on the respiratory microbiome but would be remiss not to acknowledge the potential role of the gut microbiome in the human response to inhaled exposures. First, the theory of a “common mucosal response” implies that lymphoid cells can travel between the gut and lungs to create a widespread inflammatory response.76 In fact, there is a rich evidence to support the importance of the gut microbiome to the airways with reports of modulation of the immune response to respiratory infection by the gut and associations between the gut microbiome and airway inflammation following allergen or viral challenges.77-80 There are also direct exposures of the gut to airborne irritants/toxicants as the body clears particles deposited in the lungs to the intestine by way of mucociliary transport.36 Moreover, air pollutants can deposit on our food and water supplies81 with recent evidence that the ingestion of food contaminated by air pollution shifts of the gut microbiome towards increased Firmicutes and Verrucomicrobia and decreased Bacteroidetes.82,83 Thus, the respiratory microbiome may be only part of the story regarding the role of the microbiome on inhaled pollutants.
Summary and Implications
There is a growing understanding that the human responsiveness to external assaults can be shaped by our microbiome. In this article, we presented evidence that the respiratory microbiome is an active player in human health and proposed mechanisms by which the microbiome might plausibly mediate or modify the human health response to inhaled irritants/toxicants. Such a role would have several important implications for our understanding of how inhaled irritants/toxicants impact health and who is most susceptible. First, it would imply new avenues for exploring toxicity as well as shine a new light on old ones. Characterizing which microbial communities and functions are influenced by or influence exposures may lead to new insights as to which pollutants pose the greatest risk to humans but may cause new considerations in the applicability of in vitro and in vivo animal testing in the absence of realistic human microbial communities. Second, this line of evidence could provide new insight as to who may be at greatest risk from exposures and suggest novel public health interventions to protect these individuals. For example, the importance of the microbiome to differentiate who is at elevated risk from exposures could open the door for the use of probiotics (live bacteria), prebiotics (to target the growth of certain beneficial bacteria), or targeted antibiotics to shift individuals at heightened susceptibility towards a more protective microbiome. For all of these reasons, we encourage investigators to consider the microbiome in future studies of the human response to inhaled irritants/toxicants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
References
- 1.Brunekreef B, Holgate ST. Air pollution and health. The Lancet. 2002;360(9341):1233–1242. doi: 10.1016/S0140-6736(02)11274-8. [DOI] [PubMed] [Google Scholar]
- 2.Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380(9859):2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laden F, Schwartz J, Speizer FE, Dockery DW. Reduction in fine particulate air pollution and mortality - Extended follow-up of the Harvard six cities study. American Journal of Respiratory and Critical Care Medicine. 2006;173(6):667–672. doi: 10.1164/rccm.200503-443OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones M, Barnoya J, Stranges S, Losonczy L, Navas-Acien A. Cardiovascular events following smoke-free legislations: An updated systematic review and meta-analysis. Current Environmental Health Reports. 2014;1(3):239–249. doi: 10.1007/s40572-014-0020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pope CA, Ezzati M, Dockery DW. Fine particulate air pollution and life expectancy in the United States. New England Journal of Medicine. 2009;360(4):376–386. doi: 10.1056/NEJMsa0805646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correia AW, Pope CAI, Dockery DW, Wang Y, Ezzati M, Dominici F. Effect of air pollution control on life expectancy in the United States: An analysis of 545 U.S. Counties for the period from 2000 to 2007. Epidemiology. 2013;24(1):23–31. doi: 10.1097/EDE.0b013e3182770237. 10.1097/EDE.1090b1013e3182770237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell ML, Zanobetti A, Dominici F. Evidence on vulnerability and susceptibility to health risks associated with short-term exposure to particulate matter: a systematic review and meta-analysis. American journal of epidemiology. 2013:kwt090. doi: 10.1093/aje/kwt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacks JD, Stanek LW, Luben TJ, et al. Particulate matter-induced health effects: Who Is susceptible? Environmental Health Perspectives. 2011;119(4):446–454. doi: 10.1289/ehp.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho I, Blaser MJ. The Human Microbiome: at the interface of health and disease. Nature reviews. Genetics. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van de Wiele T, Vanhaecke L, Boeckaert C, et al. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environmental health perspectives. 2005:6–10. doi: 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhaecke L, Van Hoof N, Van Brabandt W, et al. Metabolism of the food-associated carcinogen 2-amino-1-methyl-6-phenylimidazo [4, 5-b] pyridine by human intestinal microbiota. Journal of agricultural and food chemistry. 2006;54(9):3454–3461. doi: 10.1021/jf053170+. [DOI] [PubMed] [Google Scholar]
- 12.Tuohy KM, Hinton DJ, Davies SJ, Crabbe MJC, Gibson GR, Ames JM. Metabolism of Maillard reaction products by the human gut microbiota–implications for health. Molecular nutrition & food research. 2006;50(9):847–857. doi: 10.1002/mnfr.200500126. [DOI] [PubMed] [Google Scholar]
- 13.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Translational Research. 2012;160(4):258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384(9944):691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. American journal of respiratory and critical care medicine. 2013;187(12):1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickson RP, Huffnagle GB. The lung microbiome: New principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11(7):e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lighthart B. Mini-review of the concentration variations found inthe alfresco atmospheric bacterial populations. Aerobiologia. 2000;16(1):7–16. [Google Scholar]
- 20.Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Annals of the American Thoracic Society. 2015;12(6):821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson RP, Erb-Downward JR, Freeman CM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS One. 2014;9(5):e97214. doi: 10.1371/journal.pone.0097214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6(2) doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology. 2011;127(2):372–381. e371–373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. Journal of Allergy and Clinical Immunology. 2013;131(2):346–352. e343. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goleva E, Jackson LP, Harris JK, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188(10):1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang YJ, Kim E, Cox MJ, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. Omics : a journal of integrative biology. 2010;14(1):9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1(1):19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. Journal of Allergy and Clinical Immunology. 2011;127(2):372–381. e373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med. 2013;187(12):1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One. 2014;9(6):e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers GB, Zain NM, Bruce KD, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc. 2014;11(4):496–503. doi: 10.1513/AnnalsATS.201310-335OC. [DOI] [PubMed] [Google Scholar]
- 34.Weiss B, Bujanover Y, Yahav Y, Vilozni D, Fireman E, Efrati O. Probiotic supplementation affects pulmonary exacerbations in patients with cystic fibrosis: a pilot study. Pediatric pulmonology. 2010;45(6):536–540. doi: 10.1002/ppul.21138. [DOI] [PubMed] [Google Scholar]
- 35.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Reports. 2012;13(5):440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.USEPA . Integrated Science Assessment for Particulate Matter. Research Triangle Park, NC; NCEARTP Office; Dec, 2009. EPA/600/R-08/139F. [Google Scholar]
- 37.Brook RD, Rajagopalan S, Pope CA, et al. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation. 2010;121:2331–2378. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- 38.Adar SD, Adamkiewicz G, Gold DR, Schwartz J, Coull BA, Suh H. Ambient and microenvironmental particles and exhaled nitric oxide before and after a group bus trip. Environmental Health Perspectives. 2007;115(4):507–512. doi: 10.1289/ehp.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubowsky SD, Suh H, Schwartz J, Coull BA, Gold DR. Diabetes, obesity, and hypertension may enhance associations between air pollution and markers of systemic inflammation. Environmental Health Perspectives. 2006;114(7):992–998. doi: 10.1289/ehp.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirabelli MC, Golan R, Greenwald R, et al. Modification of traffic-related respiratory response by asthma control in a population of car commuters. Epidemiology. 2015;20:00–00. doi: 10.1097/EDE.0000000000000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanobetti A, Schwartz J. Are diabetics more susceptible to the health effects of airborne particles? American Journal of Respiratory and Critical Care Medicine. 2001;164(5):831–833. doi: 10.1164/ajrccm.164.5.2012039. [DOI] [PubMed] [Google Scholar]
- 42.Zanobetti A, Schwartz J. Cardiovascular damage by airborne particles: are diabetics more susceptible? Epidemiology. 2002;13(5):588–592. doi: 10.1097/00001648-200209000-00016. [DOI] [PubMed] [Google Scholar]
- 43.Zanobetti A, Schwartz J. Are diabetics more susceptible to CVD health effects of airborne particles? Results from four cities. Epidemiology. 2002;13(4):S168–S168. [Google Scholar]
- 44.Lu K, Abo RP, Schlieper KA, et al. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect. 2014;122(3):284–291. doi: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: uman gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 46.Bervoets L, Van Hoorenbeeck K, Kortleven I, et al. Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathogens. 2013;5:10–10. doi: 10.1186/1757-4749-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Alimentary Pharmacology & Therapeutics. 2012;36(10):909–921. doi: 10.1111/apt.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M. Exercise Attenuates PCB-Induced Changes in the Mouse Gut Microbiome. Environmental Health Perspectives. 2013;121(6):725–730. doi: 10.1289/ehp.1306534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silver S, Phung LT. Genes and Enzymes Involved in Bacterial Oxidation and Reduction of Inorganic Arsenic. Applied and Environmental Microbiology. 2005;71(2):599–608. doi: 10.1128/AEM.71.2.599-608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van de Wiele T, Gallawa CM, Kubachka KM, et al. Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environ Health Perspect. 2010;118(7):1004–1009. doi: 10.1289/ehp.0901794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.SS DCR, Alava P, Zekker I, Du Laing G, Van de Wiele T. Arsenic thiolation and the role of sulfate-reducing bacteria from the human intestinal tract. Environ Health Perspect. 2014;122(8):817–822. doi: 10.1289/ehp.1307759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van de Wiele T, Vanhaecke L, Boeckaert C, et al. Human Colon Microbiota Transform Polycyclic Aromatic Hydrocarbons to Estrogenic Metabolites. Environmental Health Perspectives. 2005;113(1):6–10. doi: 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Youngji C, David IK, Joel AM, Jeffrey DB, Stephanie AS. D102. American Thoracic Society; 2014. The Effects Of A Depleted Gut Microbiome on ozone induced airway hyperresponsiveness and inflammation in mice. pp. A6390–A6390. [Google Scholar]
- 54.Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: Health and disease. Frontiers in Immunology. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adachi Y, Nakajima Y, Satomoto M, Morita K, Doi M, Sato S. The heart rate variability in mice: telemetric evaluation of endotoxin shock. Masui. The Japanese journal of anesthesiology. 2006;55(4):436–440. [PubMed] [Google Scholar]
- 56.Kumar PS, Matthews CR, Joshi V, de Jager M, Aspiras M. Tobacco Smoking affects bacterial acquisition and colonization in oral biofilms. Infection and Immunity. 2011;79(11):4730–4738. doi: 10.1128/IAI.05371-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delima SL, McBride RK, Preshaw PM, Heasman PA, Kumar PS. Response of subgingival bacteria to smoking cessation. Journal of Clinical Microbiology. 2010;48(7):2344–2349. doi: 10.1128/JCM.01821-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. American Journal of Respiratory and Critical Care Medicine. 2013;187(10):1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Straus WL, Plouffe JF, File TM, et al. Risk factors for domestic acquisition of Legionnaires disease. Arch. Intern. Med. 1996;156(15):1685–1692. [PubMed] [Google Scholar]
- 62.Arcavi L, Benowitz NL. Cigarette smoking and infection. Arch. Intern. Med. 2004;164(20):2206–2216. doi: 10.1001/archinte.164.20.2206. [DOI] [PubMed] [Google Scholar]
- 63.Smith KR, Samet JM, Romieu I, Bruce N. Indoor air pollution in developing countries and acute lower respiratory infections in children. Thorax. 2000;55(6):518–532. doi: 10.1136/thorax.55.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arcavi L, Benowitz NL. Cigarette smoking and infection. Arch Intern Med. 2004;164(20):2206–2216. doi: 10.1001/archinte.164.20.2206. [DOI] [PubMed] [Google Scholar]
- 65.Aronson MD, Weiss ST, Ben RL, Komaroff AL. Association between cigarette smoking and acute respiratory tract illness in young adults. JAMA. 1982;248(2):181–183. [PubMed] [Google Scholar]
- 66.Bensenor IM, Cook NR, Lee IM, et al. Active and passive smoking and risk of colds in women. Ann Epidemiol. 2001;11(4):225–231. doi: 10.1016/s1047-2797(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 67.Stampfli MR, Anderson GP. How cigarette smoke skews immune responses to promote infection, lung disease and cancer. Nat Rev Immunol. 2009;9(5):377–384. doi: 10.1038/nri2530. [DOI] [PubMed] [Google Scholar]
- 68.Marcy TW, Merrill WW. Cigarette smoking and respiratory tract infection. Clin Chest Med. 1987;8(3):381–391. [PubMed] [Google Scholar]
- 69.Erickson JJ, Gilchuk P, Hastings AK, et al. Viral acute lower respiratory infections impair CD8+ T cells through PD-1. J Clin Invest. 2012;122(8):2967–2982. doi: 10.1172/JCI62860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molyneaux PL, Mallia P, Cox MJ, et al. Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188(10):1224–1231. doi: 10.1164/rccm.201302-0341OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKendry RT, Spalluto CM, Burke H, et al. Dysregulation of anti-viral function of CD8+T cells in the COPD lung: Role of the PD1/PDL1 Axis. Am J Respir Crit Care Med. 2015 doi: 10.1164/rccm.201504-0782OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jervis-Bardy J, Leong LEX, Marri S, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biesbroek G, Sanders EAM, Roeselers G, et al. Deep sequencing analyses of low density microbial communities: Working at the boundary of accurate microbiota detection. PLoS One. 2012;7(3):e32942. doi: 10.1371/journal.pone.0032942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willner D, Daly J, Whiley D, Grimwood K, Wainwright CE, Hugenholtz P. Comparison of DNA extraction methods for microbial community profiling with an application to pediatric bronchoalveolar lavage samples. PLoS One. 2012;7(4):e34605. doi: 10.1371/journal.pone.0034605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mestecky J. The common mucosal immune system and current strategies for induction of immune responses in external secretions. J Clin Immunol. 1987;7(4):265–276. doi: 10.1007/BF00915547. [DOI] [PubMed] [Google Scholar]
- 77.Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yasui H, Kiyoshima J, Hori T. Reduction of influenza virus titer and protection againstinfluenza virus infection in infant mice fed Lactobacillus Casei Shirota. Clinical and Diagnostic Laboratory Immunology. 2004;11(4):675–679. doi: 10.1128/CDLI.11.4.675-679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang YJ, Boushey HA. The microbiome in asthma. Journal of Allergy and Clinical Immunology. 2015;135(1):25–30. doi: 10.1016/j.jaci.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 81.De Brouwere K, Buekers J, Cornelis C, Schlekat CE, Oller AR. Assessment of indirect human exposure to environmental sources of nickel: oral exposure and risk characterization for systemic effects. Sci Total Environ. 2012;419:25–36. doi: 10.1016/j.scitotenv.2011.12.049. [DOI] [PubMed] [Google Scholar]
- 82.Salim SY, Kaplan GG, Madsen KL. Air pollution effects on the gut microbiota: A link between exposure and inflammatory disease. Gut Microbes. 2014;5(2):215–219. doi: 10.4161/gmic.27251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kish L, Hotte N, Kaplan GG, et al. Environmental particulate matter induces murine intestinal inflammatory responses and alters the gut microbiome. PLoS One. 2013;8(4):e62220. doi: 10.1371/journal.pone.0062220. [DOI] [PMC free article] [PubMed] [Google Scholar]