Abstract
Objective:
To determine the phenotypic spectrum caused by mutations in GRIN1 encoding the NMDA receptor subunit GluN1 and to investigate their underlying functional pathophysiology.
Methods:
We collected molecular and clinical data from several diagnostic and research cohorts. Functional consequences of GRIN1 mutations were investigated in Xenopus laevis oocytes.
Results:
We identified heterozygous de novo GRIN1 mutations in 14 individuals and reviewed the phenotypes of all 9 previously reported patients. These 23 individuals presented with a distinct phenotype of profound developmental delay, severe intellectual disability with absent speech, muscular hypotonia, hyperkinetic movement disorder, oculogyric crises, cortical blindness, generalized cerebral atrophy, and epilepsy. Mutations cluster within transmembrane segments and result in loss of channel function of varying severity with a dominant-negative effect. In addition, we describe 2 homozygous GRIN1 mutations (1 missense, 1 truncation), each segregating with severe neurodevelopmental phenotypes in consanguineous families.
Conclusions:
De novo GRIN1 mutations are associated with severe intellectual disability with cortical visual impairment as well as oculomotor and movement disorders being discriminating phenotypic features. Loss of NMDA receptor function appears to be the underlying disease mechanism. The identification of both heterozygous and homozygous mutations blurs the borders of dominant and recessive inheritance of GRIN1-associated disorders.
NMDA receptors (NMDARs) are tetrameric ligand-gated ion channels permeable to Na+, K+, and Ca2+, composed of 2 glycine binding GluN1 subunits and 2 glutamate binding GluN2/3 subunits (GluN2A, GluN2B, GluN2C, GluN2D, GluN3A, GluN3B).1–3
In contrast to GluN2/3 subunits, which each have specific spatial and temporal expression patterns throughout the CNS, GluN1 encoded by GRIN1 is a ubiquitous component of the receptor.4,5
Mutations of the NMDAR subunits are associated with a variety of different neurodevelopmental phenotypes,3 including intellectual disability (ID),6–8 epilepsy,9–13 and autism spectrum disorders (ASD), and different psychiatric diseases.14,15
While several studies have focused on the phenotypic spectrum of GRIN2A- and GRIN2B-associated disorders, little is known about the clinical presentations of individuals with GRIN1 mutations. We reviewed clinical data on individuals with GRIN1 mutations from different research cohorts as well as from diagnostic laboratories.
In addition, we reviewed available clinical and genetic information from previously reported patients with GRIN1 mutations. Nine heterozygous de novo mutations had previously been reported with rudimentary phenotypic information: 4 individuals were described with nonsyndromic and nonspecific ID (mental retardation, autosomal dominant 8, OMIM 614254),8,16,17 1 individual had epileptic encephalopathy of Lennox-Gastaut type,9 and 4 individuals were described with severe developmental delay and associated movement disorders.18
Our study aimed at a comprehensive delineation of the associated phenotype and at an investigation of the functional consequences of the identified GRIN1 mutations.
METHODS
Patients.
We reviewed clinical and genetic information on patients in whom GRIN1 mutations were detected through different next-generation sequencing approaches in diagnostic and research settings including targeted panel sequencing19 or whole-exome sequencing (table e-1, a and b, on the Neurology® Web site at Neurology.org). Depending on the availability of parental samples, the de novo status of each mutation was confirmed through conventional Sanger sequencing.
Standard protocol approvals, registration, and patient consents.
Written informed consent for genetic testing was obtained for every individual. Genetic testing within research settings was approved by the Commission of Medical Ethics of the University of Antwerp, Belgium; the Ethics Committee of Western Zealand, Denmark; the Kantonale Ethikkommission Zürich, Switzerland; and additional local ethics committees of the contributing centers.
In silico prediction.
Predictions of the functional impact of identified coding variants were assessed by different in silico analysis programs (PolyPhen2, http://genetics.bwh.harvard.edu/pph2/ and MutationTaster, http://www.mutationtaster.org) using the GRIN1 transcript number ENST00000371561 (table e-2). Variants were compared to 60,706 controls of the ExAC browser (http://exac.broadinstitute.org/). Conservation of mutated positions was evaluated using sequence alignment of different species (figure e-1). Molecular modeling of the NMDAR subunits was performed according to Endele et al.7
DNA constructs, oocyte expression, and electrophysiology.
For Xenopus laevis oocyte experiments, GluN1 and GluN2B constructs and capped cRNAs were generated as described previously.11,12 Several mutations were introduced into these constructs using the QuikChange site-directed mutagenesis kit (Stratagene; Agilent Technologies, Santa Clara, CA) and confirmed by DNA sequencing. Oocytes were isolated and maintained as described previously.20 Glutamate, glycine, and Zn2+ dose–response curves of wild-type GluN1/GluN2B and mutant NMDARs were analyzed by 2-electrode voltage clamp recording as described.21 Concentration–response curves and current traces shown in the figures were drawn using KaleidaGraph (Synergy Software, Reading, PA). Data represent means ± SE. Statistical significance was evaluated using a Student 2-tailed unpaired t test if not otherwise stated. All chemicals used were obtained from Sigma.
RESULTS
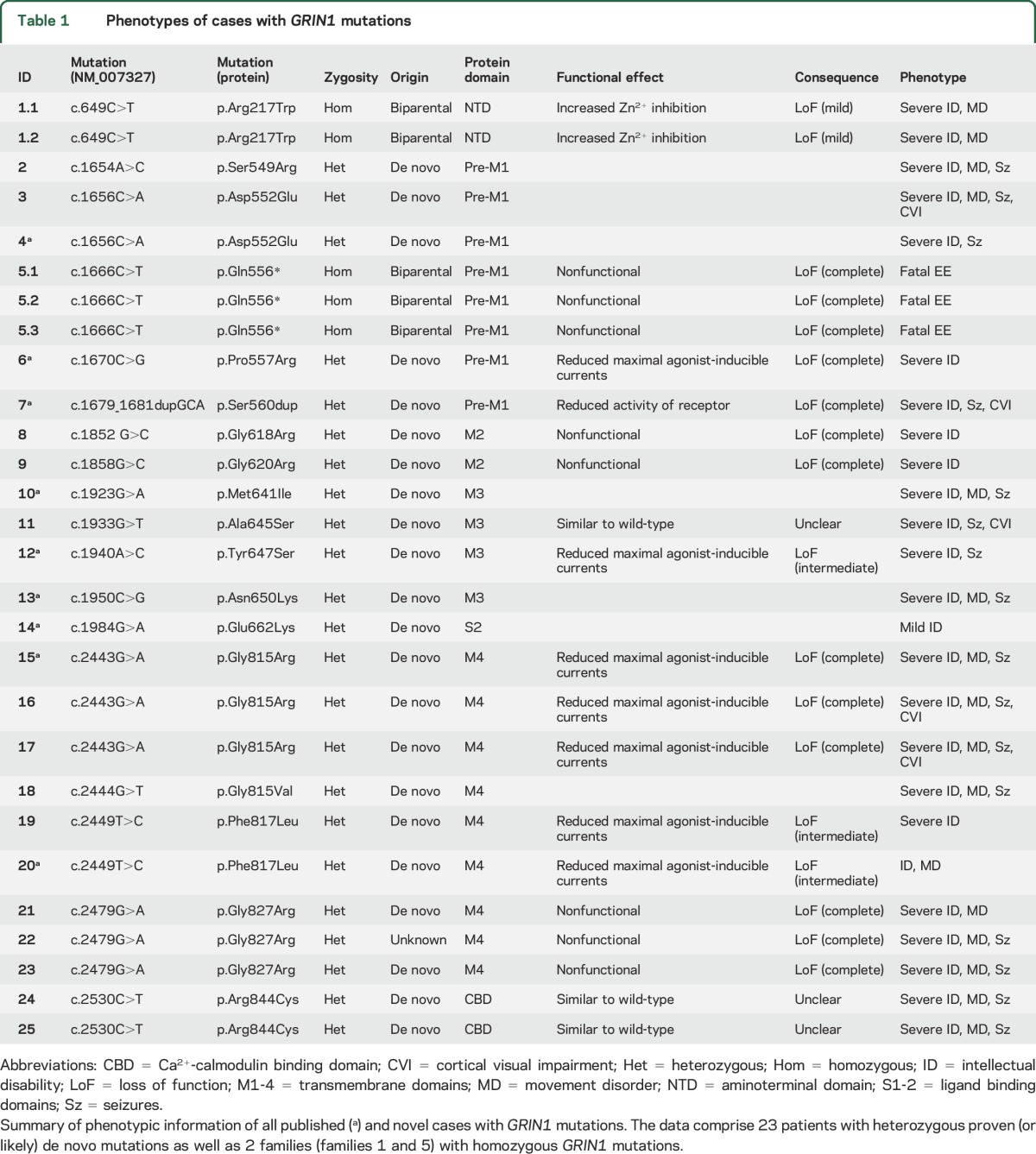
We identified 14 previously unreported patients with heterozygous GRIN1 mutations. In 13 individuals, the mutation was confirmed to be de novo, whereas in patient 22 parental samples were not available for segregation analysis. However, given that the particular mutation in patient 22 was recurrent in 2 other patients (patients 21 and 23) and absent in large control datasets, we concluded that the mutation is pathogenic and included this patient in our analysis. In addition to these 14 novel cases, we reviewed phenotypic data of all 9 previously reported cases8,9,16–18 and present additional clinical information on 4 of these patients.8,9,16 Thus, we were able to collectively review the clinical data of 23 patients (table 1). In addition to patients with de novo GRIN1 mutations, we identified 2 homozygous GRIN1 mutations segregating in 2 unrelated families with severe neurodevelopmental disorders (1.1–2 and 5.1–3). In both cases, family members carrying heterozygous GRIN1 variants were unaffected.
Table 1.
Phenotypes of cases with GRIN1 mutations
Phenotypes associated with heterozygous de novo GRIN1 mutations.
Almost all patients carrying a de novo GRIN1 mutation presented with profound global developmental delay, usually already apparent in the neonatal period and resulting in severe intellectual disability (21/22; 95%). (table e-1, a and b) Patients usually never acquired the ability to walk and had absent or extremely limited verbal communication skills.
Of the 21 patients in whom information regarding muscular tone could be retrieved, 15 (71%) had severe truncal and initial appendicular hypotonia. Many patients developed corticospinal signs, such as hyperreflexia (6/21; 29%) or spasticity (6/21; 29%), consistent with a diagnosis of spastic quadriparesis. The majority of patients showed choreatic, dystonic, or dyskinetic movement disorders (14/23; 61%), including oculomotor abnormalities such as oculogyric crises (5/23; 22%). Nonspecific stereotypic movements were noted in 7/21 patients (33%).
A significant proportion of patients (16/23; 70%) had epilepsy. The epilepsy phenotype of GRIN1 mutation carriers was variable with respect to age at onset (day of life 1–11 years), seizure semiology (infantile spasms, tonic and atonic seizures, hypermotor seizures, focal dyscognitive seizures, febrile seizures, generalized seizures, status epilepticus), and the associated EEG pattern (hypsarrhythmia, focal, multifocal and generalized spikes and waves). In addition to the variability of the epilepsy phenotype, the outcome relating to the control of seizures was variable: while at least 5 patients (5/16; 31%) had therapy-resistant epilepsy, 2 patients became seizure-free or had long periods of seizure freedom on valproate (VPA), whereas 2 additional patients responded well on a combination of topiramate, levetiracetam, and clobazam or the introduction of vigabatrin and clonazepam in addition to VPA. In most other epilepsy patients, clinical data were not available or seizures were not reported as a prominent phenotype (table e-3, a and b). Given this variability, the epilepsy phenotype of GRIN1 mutation carriers usually did not resemble specific forms of epileptic encephalopathies, which is in contrast to GRIN2A- and some GRIN2B-associated disorders.9–13
At least 8 GRIN1 patients were diagnosed with ASD or ASD-like features (8/23; 35%), acknowledging that the severe level of ID in most patients made a separate diagnosis of ASD challenging. This is in line with previous reports in GRIN2B-associated disorders14: 4 individuals presented with aggression and self-injurious behavior or disturbed pain perception (4/23; 17%). Eight patients (8/23; 35%) had nonspecified sleep disorders. Several patients had feeding difficulties (9/23; 39%), likely due to their underlying hypotonia and spasticity, requiring tube feeding in some patients. Several patients presented with cortical visual impairment or delayed visual maturation (5/23; 22%).
MRI findings were available for review in 19 patients, and nonspecific volume loss or atrophy was seen in 11/19 (58%) patients. These MRI findings were considered insufficient to explain the presence of quadriplegia or movement disorder seen in patients. Cortical atrophy was apparent already at young ages (e.g., cases 18 at 20 months and 25 at 3 years of age) without evidence for a significant age-dependent progression.
Two patients (2/21; 10%) had a marfanoid habitus, whereas several had normal stature or growth retardation or low body weight. Microcephaly was observed in 6/23 (26%) patients. Dysmorphic features, if present, were subtle. Deep-set eyes, which were previously reported as a common feature,18 could not be consistently seen in our patients.
While 21/22 patients had severe ID, one previously reported patient carrying a GRIN1 p.Glu662Lys mutation presented with a less severe phenotype.8 Patient 14 was able to walk independently at age 18 months and had speech delay with adequate first words at 9 months and 2-word phrases only at 5 years of age. Additionally, the patient did not present with any other comorbid conditions including epilepsy, movement disorder, and abnormalities of muscle tone or vision.
GRIN1 mutations with recessive inheritance.
In a consanguineous family with ID and ASD, a homozygous GRIN1 p.Arg217Trp missense variant was found in 2 affected brothers. Both parents were healthy heterozygous carriers. The variant was predicted to be pathogenic (table e-2) and was absent in ExAC controls.
In a second consanguineous family, we identified a homozygous p.Gln556* truncation mutation in 3 siblings with severe neonatal epileptic encephalopathy. All 3 siblings died between 5 days of life and 5 months secondary to intractable seizures. Both parents were unaffected heterozygous carriers of the p.Gln556* mutation.
Among the 110 ExAC-annotated missense and 8 truncating GRIN1 variants, none was reported to be homozygous in 60,706 control samples.
Spectrum and clustering of GRIN1 mutations.
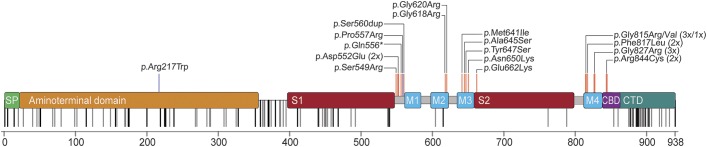
All 16 different de novo mutations identified in the 23 novel and published cases are missense alterations and cluster within or in close proximity to the transmembrane domains forming the intrinsic ion channel pore of the receptor (figure 1). Interestingly, this region is widely spared from genetic variation in controls and shows a high level of conservation in different species (figure e-1). In contrast, there is extensive enrichment of reported single nucleotide variants (SNV) in controls in the N-terminal domain as well as the C-terminus of GRIN1. De novo mutations have not been observed in either domain. The reported mutations do not allow us to assess the genetic variability of S1 and S2 ligand binding sites. However, the S1 domain contains numerous and frequent SNV but no disease-causing mutations so far, whereas S2 showed less variability with only 2 nonrecurrent SNV but one mutation (figure 1). Moreover, this single mutation detected within S2 was associated with the mildest phenotype (patient 14) when compared to all other GRIN1 de novo mutations closely related to the transmembrane domains. Five de novo mutations have been detected recurrently in independent patients within our cohort (p.Asp552Glu, p.Gly815Arg, p.Phe817Leu, p.Gly827Arg, p.Arg844Cys).
Figure 1. Domains of GRIN1 and distribution of variants.
Signal peptide (SP), the extracellular N-terminal domain, and ligand binding sites (S1, S2), the transmembrane domains (M1-4), as well as the intracellular C-terminal domain (CTD) with the proximal Ca2+ calmodulin binding domain (CBD). De novo mutations (red) cluster within or in very close proximity to M1-4. In addition, this region is particularly spared from nonsynonymous genetic variation according to the ExAC browser (rare/single variants, gray; repeated/frequent variants, black). The 2 homozygous GRIN1 variants are marked in blue.
The only mutation being far outside the transmembrane cluster is the p.Arg217Trp missense variant that was identified in a recessive family with unaffected heterozygous carriers.
Functional investigations.
Ectopic expression of the GluN1 mutants with wild-type GluN2B subunits revealed that 4 of the mutants gave no response to up to 10 mM glutamate and glycine and were hence rated as nonfunctional (p.Gln556*, p.Gly618Arg, p.Gly620Arg, p.Gly827Arg; n = 10, each). For all other mutants, changes in maximal inducible currents and agonist affinities were examined. Four mutations (p.Pro557Arg, p.Tyr647Ser, p.Gly815Arg, p.Phe817Leu) resulted in a highly significant reduction of agonist Imax values (p < 0.001, n = 5–9; figure e-2a). Our model of the GRIN1/GRIN2B NMDAR11 predicts strong intersubunit and intrasubunit interactions between adjacent helical structures M3 and M4 for mutated positions p.Pro557, p.Tyr647, p.Gly815, p.Phe817, and p.Gly827, thereby destabilizing the quaternary structure of the receptor transmembrane domains and disturbing assembly of NMDARs. Consistent with an impaired function of GRIN1 de novo mutations, analysis of the agonist dose-response curves for mutations p.Gly815Arg and p.Phe817Leu additionally revealed a highly significant reduction in the affinities for both glutamate and glycine (figure e-2b). For positions p.Gly618 and p.Gly620, our model predicts a location in the membrane re-entrant loop (the P-loop) of the GluN1 subunit, which determines the narrow constriction and ion selectivity of the channel. Thus, conversion of the pore residues p.Gly618 and p.Gly620 to a charged arginine side chain likely has strong impact on NMDA channel properties. Coexpression of wild-type GluN1 with mutants GluN1 p.Tyr647Ser or p.Phe817Leu resulted in intermediate effects of agonist Imax values (p < 0.01, n = 5), supporting the idea of a negative impact of the substitutions on the function of hetero-oligomeric NMDARs (figure e-2c). Mutations p.Arg217Trp, p.Ala645Ser, and p.Arg844Cys generated functional receptors with agonist Imax values (p > 0.05, n = 5) similar to wild-type (data not shown). Functional analysis of the missense alteration GluN1 p.Arg217Trp in the N-terminal Zn2+-binding domain with wild-type GluN2A subunits showed a significant increase in Zn2+ inhibition (figure e-2d), suggesting impaired activation in vivo due to increased tonic inhibition of GRIN1Arg217Trp–GRIN2A receptors at physiologic concentrations of Zn2+. However, functional analysis of mutations p.Ala645Ser and p.Arg844Cys revealed no significant effects on agonist Imax values or affinity, suggesting that these 2 substitutions may alter NMDAR function through other mechanisms. For example, since the recurrent substitution p.Arg844Cys is located in the intracellular Ca2+-calmodulin-binding domain, disturbed interactions with intracellular proteins may impair receptor function. Our analyses of de novo mutations are consistent with a dominant-negative effect resulting in a significant loss of receptor function.
DISCUSSION
We reviewed 14 novel and 9 previously published individuals carrying heterozygous GRIN1 de novo mutations associated with neurodevelopmental phenotypes.
Profound developmental delay associated with severe ID and lack of speech development are predominant features, and muscular hypotonia leading to spastic quadriplegic cerebral palsy, hyperkinetic movement disorders including dystonia and chorea, and oculomotor abnormalities as well as cortical visual impairment appeared to be recurrent findings in individuals with GRIN1 encephalopathy. While some of these features can be seen in other genetic epileptic encephalopathies such as SCN2A and SCN8A encephalopathies (OMIM 613721, 600702), these findings are novel in the context of NDMAR encephalopathies. Seizures occurred in about two-thirds of cases. However, there was no obvious epilepsy pattern with respect to age at onset, seizure semiology, EEG features, or outcome. Furthermore, the frequency of epilepsy in GRIN1 patients may be an overestimate due to inclusion of a high proportion of epilepsy patients in the screening cohorts.
GRIN1 encodes GluN1 and autoantibodies primarily targeting an epitope within the N-terminal domain of GluN1 result in the increasingly recognized clinical entity of NMDAR encephalitis.22,23 NMDAR encephalitis is an acute paraneoplastic or parainfectious neurologic disorder where decay of NMDARs is considered to be the underlying pathomechanism, partially paralleling the pathophysiology of GluN1 loss of function seen in our patients with GRIN1 encephalopathy. While the acuity of the clinical presentation is vastly different, with NMDAR encephalitis presenting as an acute acquired condition while GRIN1 encephalopathy is a chronic neurodevelopmental disease, we would like to emphasize a shared group of symptoms including choreatic and dystonic movements, seizures, and sleep cycle dysregulation, which we observe in our patient cohort. Further research will be able to address the question whether the GRIN1 encephalopathy phenotype is specific enough to hypothesize a spectrum of NMDAR impairment in human disease.
GRIN1 de novo mutations cluster within or in direct proximity to the transmembrane domains of GRIN1 (figure 1). These regions are largely spared from genetic variation, underlining the crucial importance of these domains. The sole mutation outside this cluster is p.Arg217Trp, which appears to be only pathogenic when present homozygously and thus might mediate its effect through different effects compared to the heterozygous variants near the transmembrane domains.
All reported de novo mutations are missense variants. In contrast to GRIN2A and GRIN2B, heterozygous truncation of GRIN1 apparently does not result in a neurologic phenotype. Furthermore, both deletions encompassing GRIN1 as well as truncating or splice-site variants are seen in control databases, suggesting that haploinsufficiency in GRIN1, albeit rare, is tolerated in the human population.
In addition to 16 different heterozygous de novo mutations, we describe 2 recessive GRIN1 mutations. The homozygous loss-of-function p.Arg217Trp mutation segregated with severe ID, ASD, and movement disorders in 2 affected siblings in a consanguineous family (family 1), and a homozygous p.Gln556* truncation mutation was found in 3 individuals with fatal epileptic encephalopathy in family 5 (table 1).
As the NMDAR obligatorily contains 2 GluN1 subunits, truncation or lack of both GRIN1 alleles would result in a knock-out and complete deprivation of the NMDAR. The almost continuous seizure activity with suppression-burst EEG and early death of all 3 affected siblings in family 5 underlines the vital role of GluN1 in NMDAR functioning.
This disease mechanism as well as functional data point towards a dominant-negative effect. In contrast to the gain of function frequently detected in other GRIN-associated neurodevelopmental disorders,11,12 GRIN1 de novo mutations p.Pro557Arg, p.Ser560dup, p.Gly618Arg, p.Gly620Arg, p.Tyr647Ser, p.Gly815Arg, p.Phe817Leu, and p.Gly827Arg result in a loss of function.
The extent and nature of the loss of NMDAR function due to de novo GRIN1 mutations corresponded only marginally to the uniformly severe phenotypes. One possible explanation for this lack of genotype–phenotype correlation may be effects on the subunit composition or trafficking of the NMDAR that potentially lead to a shared secondary mechanism compensating GluN1 defects independent of the individual severity of the respective GRIN1 mutation eventually escaping visualization using standard artificial in vitro models.
Given the multiple observations of heterozygous de novo GRIN1 loss-of-function mutations, GRIN1-associated disorders are a recurrent cause of severe and complex neurodevelopmental disorders. Additionally, homozygous GRIN1 mutations segregating with severe phenotypes display phenotypic overlap with individuals carrying heterozygous GRIN1 de novo mutations. This observation blurs the borders of autosomal-dominant and autosomal-recessive inheritance. The constellation of severe ID and associated findings including prominent hypotonia, movement disorders, oculogyric crises, cortical visual impairment or blindness, absent speech, behavioral issues, sleep disorder, and seizures may allow for a phenotypic differentiation from other frequent forms of severe ID. Together with the catastrophic phenotype seen in homozygous truncation mutation carriers, our findings underscore the essential role of the NMDAR subunits in neurodevelopment.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the patients and family members for their participation in this study.
GLOSSARY
- ASD
autism spectrum disorder
- ID
intellectual disability
- NMDAR
NMDA receptor
- SNV
single nucleotide variant
- VPA
valproate
Footnotes
Supplemental data at Neurology.org
AUTHOR AFFILIATIONS
From the Institute of Human Genetics (J.R.L., H.O.H., J.H.) and Integrated Research and Treatment Center (IFB) Adiposity Diseases (H.O.H.), University of Leipzig Hospitals and Clinics; Department of Neurophysiology and Neurosensory Systems (K.G., H.S., B.L.), Technical University Darmstadt, Germany; Division of Clinical Genomics (K.L.H.), Ambry Genetics, Aliso Viejo, CA; Division of Human Genetics (C.C.), University Children's Hospital, Inselspital, Bern, Switzerland; Folkhälsan Institute of Genetics (C.C.), Helsinki, Finland; INSERM (C.D., C.N., S.W.), U 1127, Sorbonne Universités, UPMC Université Paris 06, CNRS, UMR 7225, Institut du Cerveau et de la Moelle Épinière (ICM); Département de Génétique (C.D., C.N., D.H.) and Centre de Reference Épilepsies Rares, Epilepsy Unit (S.W.), AP-HP, Hôpital de la Pitié-Salpêtrière, Paris, France; Danish Epilepsy Centre (R.S.M., H.H.), Dianalund; Institute for Regional Health Services (R.S.M., H.H.), University of Southern Denmark, Odense; Cologne Center for Genomics (CCG) (D.L., P.N., H.T.) and Center for Molecular Medicine Cologne (CMMC) (P.N.), University of Cologne, Germany; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston; Program in Medical and Population Genetics (D.L.) and Stanley Center for Psychiatric Research (D.L.), Broad Institute of MIT and Harvard, Cambridge, MA; Department of Neuropediatrics (B.A.N.), University Medical Center Giessen and Marburg, Giessen; Department of Neuropediatrics (G.K.), University Medical Center Münster, Germany; Division of Medical Genetics, Department of Pediatrics, University of Pittsburgh School of Medicine (G.L.A.), and Division of Child Neurology (A.G., D.S.R.), Children's Hospital of Pittsburgh, PA; Department of Medical Genetics (V.B.), Children's Hospitals and Clinics of Minnesota, Minneapolis; Institute of Clinical Genetics (D.B.), Klinikum Stuttgart, Germany; Department of Medical Genetics (C.R.J.P., D.M., E.F.) and Department of Clinical Neurosciences for Children, Women and Children's Division (P.S.), Oslo University Hospital and University of Oslo, Norway; Division of Neurology (D.L.D., I.H.), The Children's Hospital of Philadelphia, PA; Section of Clinical Genetics (E.S.D.), Carilion Clinic Children's Hospital, Roanoke, VA; Department of Clinical Genetics (E.K.B., C.A.R., M.J.V.H.), Leiden University Medical Center, the Netherlands; The Center for Rare Childhood Disorders (V.N., K.R., N.B., I.S., R.R.), Translational Genomics Research Institute (TGen), Phoenix, AZ; Department of Medical Genetics (B.P.C.K., C.G.F.d.K.), UMCU, Utrecht, the Netherlands; Centro Hospitalar do Algarve (J.S., C.M.), Faro, Portugal; Neurogenetics Group, VIB-Department of Molecular Genetics (S.W., K.H., P.D.J.), and Laboratory of Neurogenetics, Institute Born-Bunge (S.W., K.H., P.D.J.), University of Antwerp; Division of Neurology (P.D.J. and S.W.), Antwerp University Hospital, Belgium; Department of Paediatric Neurology (L.D.M.), University Hospital, Vrije Universiteit Brussel, Bruxelles, Belgium; Aix Marseille Université (M.M.), INSERM, GMGF UMR_S 910; APHM, Departement de Genetique Medicale (C.B.), and Département de Génétique Médicale (T.B.), Hopital d'Enfants de la Timone, Marseille; Génétique Clinique (M.L.), Chromosomique et Moléculaire, CHU Hôpital Nord, St Pirest en Jarez; Service de Génétique Médicale (C.F.), Centre Hospitalier Universitaire de Clermont-Ferrand; Département de Médicine Translationnelle et Neurogénétique (A.P.), IGBMC, CNRS UMR 7104/INSERM U964/Université de Strasbourg, Illkirch; Laboratoire de Diagnostic Génétique (A.P.), Hôpitaux Universitaires de Strasbourg, France; CeGaT GmbH (E.R., S.B.), Tübingen, Germany; Swiss Epilepsy Center (E.R., H.V., T.D.), Zurich, Switzerland; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Kiel Campus, Germany; Centre de Recherche du Centre Hospitalier Universitaire Sainte-Justine and Departments of Pediatrics and Neurosciences (J.L.M.), Université de Montréal, Canada; and the Department of General Paediatrics (S.S.), Division of Child Neurology and Inherited Metabolic Diseases, Centre for Paediatrics and Adolescent Medicine, University Hospital Heidelberg, Germany.
AUTHOR CONTRIBUTIONS
Study concept and design: Johannes R. Lemke, Bodo Laube, Steffen Syrbe. Acquisition of data: Johannes R. Lemke, Kirsten Geider, Katherine L. Helbig, Hannah Schütz, Carolina Courage, Christel Depienne, Caroline Nava, Delphine Heron, Rikke S. Møller, Helle Hjalgrim, Dennis Lal, Bernd A. Neubauer, Peter Nürnberg, Holger Thiele, Gerhard Kurlemann, Georgianne L. Arnold, Vikas Bhambhani, Deborah Bartholdi, Christeen Ramane J. Pedurupillay, Doriana Misceo, Eirik Frengen, Petter Strømme, Dennis J. Dlugos, Emily S. Doherty, Emilia K. Bijlsma, Claudia A. Ruivenkamp, Mariette J.V. Hoffer, Amy Goldstein, Deepa S. Rajan, Vinodh Narayanan, Keri Ramsey, Newell Belnap, Isabelle Schrauwen, Ryan Richholt, Bobby P.C. Koeleman, Joaquim Sá, Carla Mendonça, Carolien G. de Kovel, Sarah Weckhuysen, Katia Hardies, Peter De Jonghe, Linda De Meirleir, Mathieu Milh, Catherine Badens, Marine Lebrun, Tiffany Busa, Christine Francannet, Amélie Piton, Erik Riesch, Saskia Biskup, Heinrich Vogt, Thomas Dorn, Jacques L. Michaud. Analysis and interpretation: Johannes R. Lemke, Kirsten Geider, Katherine L. Helbig, Henrike O. Heyne, Hannah Schütz, Julia Hentschel, Erik Riesch, Saskia Biskup, Ingo Helbig, Bodo Laube, Steffen Syrbe. Writing of manuscript: Johannes R. Lemke, Katherine L. Helbig, Ingo Helbig, Bodo Laube, Steffen Syrbe. Critical revision of manuscript for intellectual input: all authors. Study supervision: Johannes R. Lemke, Bodo Laube, Steffen Syrbe.
STUDY FUNDING
Johannes R. Lemke (32EP30_136042/1) and Peter De Jonghe (G.A.136.11.N and FWO/ESF-ECRP) received financial support within the EuroEPINOMICS-RES network (www.euroepinomics.org) within the Eurocores framework of the European Science Foundation (ESF). Saskia Biskup and Henrike Heyne received financial support from the German Federal Ministry for Education and Research (BMBF IonNeurONet: 01 GM1105A and FKZ: 01EO1501). Katia Hardies is a PhD fellow of the Institute for Science and Technology (IWT) Flanders. Ingo Helbig was supported by intramural funds of the University of Kiel, by a grant from the German Research Foundation (HE5415/3-1) within the EuroEPINOMICS framework of the European Science Foundation, and additional grants of the German Research Foundation (DFG, HE5415/5-1, HE 5415/6-1), German Ministry for Education and Research (01DH12033, MAR 10/012), and grant by the German chapter of the International League against Epilepsy (DGfE). The project also received infrastructural support through the Institute of Clinical Molecular Biology in Kiel, supported in part by DFG Cluster of Excellence “Inflammation at Interfaces” and “Future Ocean.” The project was also supported by the popgen 2.0 network (P2N) through a grant from the German Ministry for Education and Research (01EY1103) and by the International Coordination Action (ICA) grant G0E8614N. Christel Depienne, Caroline Nava, and Delphine Heron received financial support for exome analyses by the Centre National de Génotypage (CNG, Evry, France).
DISCLOSURE
J. Lemke and K. Geider report no disclosures relevant to the manuscript. K. Helbig is employed by and receives a salary from Ambry Genetics. H. Heyne, H. Schütz, J. Hentschel, C. Courage, C. Depienne, C. Nava, D. Heron, R. Møller, H. Hjalgrim, D. Lal, B. Neubauer, P. Nürnberg, H. Thiele, G. Kurlemann, G. Arnold, V. Bhambhani, D. Bartholdi, C. Pedurupillay, D. Misceo, E. Frengen, P. Strømme, D. Dlugos, E. Doherty, E. Bijlsma, C. Ruivenkamp, M. Hoffer, A. Goldstein, D. Rajan, V. Narayanan, K. Ramsey, N. Belnap, I. Schrauwen, R. Richholt, B. Koeleman, J. Sá, C. Mendonça, C. de Kovel, and S. Weckhuysen report no disclosures relevant to the manuscript. K. Hardies, since mid-October 2015, is under employment of UCB Pharma (Braine-l'Alleud, Belgium). The company had no part in this study. P. De Jonghe, L. De Meirleir, M. Milh, C. Badens, M. Lebrun, T. Busa, C. Francannet, and A. Piton report no disclosures relevant to the manuscript. E. Riesch is employed by and receives a salary from CeGaT GmbH. Exome sequencing is among commercially available tests of both institutions S. Biskup is employed by and receives a salary from CeGaT GmbH. Exome sequencing is among commercially available tests of both institutions. H. Vogt, T. Dorn, I. Helbig, J. Michaud, B. Laube, and S. Syrbe report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Laube B, Kuhse J, Betz H. Evidence for a tetrameric structure of recombinant NMDA receptors. J Neurosci 1998;18:2954–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 2013;14:383–400. [DOI] [PubMed] [Google Scholar]
- 3.Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF. Ionotropic GABA and glutamate receptor mutations and human neurologic diseases. Mol Pharmacol 2015;88:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci 2010;11:682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paoletti P. Molecular basis of NMDA receptor functional diversity. Eur J Neurosci 2011;33:1351–1365. [DOI] [PubMed] [Google Scholar]
- 6.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012;367:1921–1929. [DOI] [PubMed] [Google Scholar]
- 7.Endele S, Rosenberger G, Geider K, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet 2010;42:1021–1026. [DOI] [PubMed] [Google Scholar]
- 8.Hamdan FF, Gauthier J, Araki Y, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 2011;88:306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nature 2013;501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvill GL, Regan BM, Yendle SC, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet 2013;45:1073–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemke JR, Hendrickx R, Geider K, et al. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol 2014;75:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemke JR, Lal D, Reinthaler EM, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet 2013;45:1067–1072. [DOI] [PubMed] [Google Scholar]
- 13.Lesca G, Rudolf G, Bruneau N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet 2013;45:1061–1066. [DOI] [PubMed] [Google Scholar]
- 14.O'Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 2011;43:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarabeux J, Kebir O, Gauthier J, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry 2011;1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redin C, Gerard B, Lauer J, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 2014;51:724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med 2015;17:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohba C, Shiina M, Tohyama J, et al. GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia 2015;56:841–848. [DOI] [PubMed] [Google Scholar]
- 19.Lemke JR, Riesch E, Scheurenbrand T, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia 2012;53:1387–1398. [DOI] [PubMed] [Google Scholar]
- 20.Laube B, Hirai H, Sturgess M, Betz H, Kuhse J. Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron 1997;18:493–503. [DOI] [PubMed] [Google Scholar]
- 21.Laube B, Kuhse J, Rundstrom N, Kirsch J, Schmieden V, Betz H. Modulation by zinc ions of native rat and recombinant human inhibitory glycine receptors. J Physiol 1995;483:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalmau J, Tuzun E, Wu HY, et al. Paraneoplastic anti-N-methyl-D-aspartate receptor encephalitis associated with ovarian teratoma. Ann Neurol 2007;61:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gleichman AJ, Spruce LA, Dalmau J, Seeholzer SH, Lynch DR. Anti-NMDA receptor encephalitis antibody binding is dependent on amino acid identity of a small region within the GluN1 amino terminal domain. J Neurosci 2012;32:11082–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.