Abstract
Background
The low-phosphate-tolerant maize mutant Qi319-96 was obtained from Qi319 through cellular engineering. To elucidate the molecular mechanisms underlying the low-phosphate tolerance of this mutant, we performed comparative proteome analyses of the leaves of Qi319-96 and Qi319 under inorganic phosphate (Pi)-sufficient and Pi-deficient conditions.
Results
Low-phosphorus levels limit plant growth and metabolism. Although the overall phosphorus contents of shoots were not significantly different between Qi319 and Qi319-96, the Pi level of Qi319-96 was 52.94 % higher than that of Qi319. Under low phosphorus conditions, Qi319-96 had increased chlorophyll levels and enhanced photosynthesis. The changes in starch and sucrose contents under these conditions also differed between genotypes. The proteomic changes included 29 (Pi-sufficient) and 71 (Pi-deficient) differentially expressed proteins involved in numerous metabolic processes. Proteome and physiological analyses revealed that Qi319-96 could better remodel the lipid composition of membranes and had higher V-ATPase activity levels than Qi319 under low-phosphate starvation, which enhanced the recycling of intracellular Pi, as reflected by its increased Pi levels. Chlorophyll biosynthesis was improved and the levels, and activities, of several Calvin cycle and “CO2 pump” enzymes were greater in Qi319-96 than in Qi319, which led to a higher rate of photosynthesis under low-phosphate stress in this line compared with in Qi319.
Conclusions
Our results suggest that the increased tolerance of the maize mutant Qi319-96 to low-phosphate levels is owing to its ability to increase Pi availability. Additionally, inbred lines of maize with low-P-tolerant traits could be obtained effectively through cellular engineering.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0825-1) contains supplementary material, which is available to authorized users.
Keywords: Maize, Inorganic phosphorus, Low-phosphate-tolerant, Proteome, Leaf
Background
Phosphorus is a vital plant macronutrient, functioning as a component in essential biomolecules such as phospholipids and ATP. Inorganic phosphate (Pi) plays central roles in virtually all of the major metabolic processes in plants, particularly photosynthesis [1, 2]. To further increase crop yields will require improving photosynthesis [3]. Thus, the efficient use of phosphorus during photosynthesis is a potentially important determinant of crop growth and yield.
Plants have evolved a series of strategies to cope with inadequate phosphate conditions while maintaining a proper balance of internal phosphorus levels [4]. These adjustments include (1) reducing phosphorus consumption by the plant [5], (2) increasing plant internal phosphorus recycling [6], and (3) improving phosphorus use in metabolic pathways [7].
Physiological and molecular adaptations that improve the phosphorus use efficiency include accelerated leaf senescence combined with the redirection of resources to growing tissues, as well as changes to metabolic pathways, such as primary carbon metabolism and phospholipid metabolism [8]. The release of phosphorus from membrane phospholipids by lipid remodeling is an important mechanism used by plants to adapt to low-phosphate conditions [9–11]. Sulfoquinovosyl diglyceride (SQDG) is a non-phosphorus lipid associated with several protein complexes in photosynthetic membranes, such as chloroplasts CF0-CF1 of ATPase, light harvesting complex II-apoproteins, and D1/D2 heterodimers [12]. The glycerophosphodiester-mediated lipid metabolic pathway may be involved in phosphorus release from phospholipids under low-phosphate stress. Sulfolipids and galactolipids, rather than phospholipids, are the major lipids of the thylakoid membrane in plants subjected to phosphate-deficiency stress. Under these conditions, plants can replace the phospholipids in photosynthetic membranes with specific non-phosphorus lipids [13]. These changes prolong and enhance the productive use of phosphorus during photosynthesis. Starch accumulation in the shoots is another common reaction of all plants to long-term phosphate deficiency [2]. One of the effects associated with starch accumulation is the release of phosphorus from chloroplasts to the cytoplasm for phosphorus recycling [14]. The accumulation of starch in phosphate-deficient leaves may help maintain the phosphorus balance between the cytoplasm and chloroplasts [15].
Increasing phosphorus recycling and phosphorus release from the vacuole may increase the phosphorus use efficiency. The vacuole is an important organelle involved in maintaining cytoplasmic phosphorus homeostasis [14, 16]. Excess cellular phosphorus in the cytoplasm is stored in the vacuole and is used to buffer the phosphorus demands of the cytoplasm [7]. The influx of phosphorus into the vacuole moderates phosphorus fluctuations by controlling the external intake of phosphorus and influencing cell metabolism. Under phosphorus deficiency, the V-ATPase gene may improve the proton transport to maintain an electrochemical gradient across the tonoplast by increasing its expression level, thereby providing the required energy to facilitate phosphorus transport [17].
Previously, we obtained the low-phosphate-tolerant mutant Qi319-96 from Qi319 using cellular engineering, therefore, they have a common genetic background. A comparison of the low-phosphate responses in these two maize genotypes indicated that low-phosphate tolerance is greater in the Qi319-96 genotype than in Qi319. The light energy conversion efficiency and Pi contents are higher in Qi319-96 than in Qi319 under low-phosphate conditions [18]. We previously performed a systematic proteome analysis of Qi319 maize leaves in response to phosphate starvation, finding that the phosphate starvation response is a complicated process involving several metabolic reactions, such as photosynthesis, carbohydrate metabolism, energy metabolism, secondary metabolism, signal transduction, and protein synthesis. After being subjected to a long period of phosphorus stress, the internal phosphorus use efficiency in Qi319 maize may increase through altered photorespiration and lipid composition, along with increased starch synthesis [19]. To elucidate the molecular mechanisms of the different tolerance levels to low-P conditions between Qi319-96 and Qi319, a comparative proteome analysis should be performed.
In this study, we performed comparative proteome analyses of leaves from mutant Qi319-96 and wild-type Qi319 plants treated with 1000 μM (+P, Pi-sufficient) and 5 μM (–P, Pi-deficient) KH2PO4 over a long time period. The objectives of this study were (i) to determine the reasons behind the differences in leaf Pi levels between the two genotypes; (ii) to investigate the mechanism behind the high photosynthetic efficiency levels in Qi319-96; and (iii) to provide information for further research into the functions of genes involved in phosphate-stress responses.
Results
Differential growth and physiological responses to phosphate deprivation between Qi319 and Qi319-96
After treatment with 5 μmol KH2PO4 for 25 days, Qi319 and Qi319-96 maize plants grew to the six- to seven-leaf stage but exhibited apparent phosphorus deficiency symptoms, such as reduced overall phosphorus contents, marked changes in biomass (Table 1), heliotrope-colored stems, and restricted growth (Fig. 1).
Table 1.
Influence of different phosphate treatments on biomass, inorganic phosphorus concentration and phosphorus contents of Qi319 and Qi319-96
| Inbred lines | Treatment | Inorganic phosphorus content (μg g-1 FW) | Biomass (g/three plant) | P contents (mg P g-1DW) | ||||
|---|---|---|---|---|---|---|---|---|
| Root | Shoot | total | Root | Shoot | total | |||
| Qi319-96 | +P | 24.29 ± 1.01a | 4.36 ± 0.08a | 8.70 ± 0.09a | 13.06 ± 0.17a | 1.71 ± 0.05a | 2.56 ± 0.03a | 2.27 ± 0.04a |
| -P | 17.39 ± 0.99c | 4.01 ± 0.06b | 5.65 ± 0.15b | 9.66 ± 0.21b | 0.58 ± 0.01b | 0.87 ± 0.01b | 0.75 ± 0.02b | |
| Qi319 | +P | 20.22 ± 1.02b | 4.47 ± 0.03a | 8.84 ± 0.06a | 13.31 ± 0.15a | 1.61 ± 0.07a | 2.48 ± 0.06a | 2.19 ± 0.07a |
| -P | 11.37 ± 0.86d | 3.35 ± 0.03c | 5.37 ± 0.09b | 8.72 ± 0.07c | 0.44 ± 0.01c | 0.86 ± 0.02b | 0.68 ± 0.02c | |
Three seedlings per bottle were cultured in phosphorus saturation solution (1000 μM KH2PO4) to the three-leaf stage, followed by low phosphate stress solution (5 μM KH2PO4) for an additional 25 days to the six–seven-leaf stage. Values represent the means of nine seedlings ± SD. Values with different letters within a row are significantly different (P < 0.05) by multiple comparison analysis
Fig. 1.
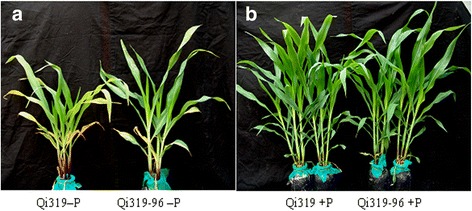
Qi319 and Qi319-96 maize plants grown in two growth conditions + P and -P. a: Qi319 and Qi319-96 grew under -P conditions (5 μmol KH2PO4). b: Qi319 and Qi319-96 grew under + P conditions (1000 μmol KH2PO4)
The phosphorus contents, and root and shoot biomasses, were not significantly different between Qi319-96 and Qi319 under the sufficient phosphate (+P) treatment. However, the root biomasses were significantly higher in Qi319-96 than in Qi319 after phosphorus stress treatments. The phosphorus content in roots was significantly higher in Qi319-96 than in Qi319, while the overall phosphorus content of the shoots did not significantly differ between genotypes (Table 1). However, the Pi levels of Qi319-96 and Qi319 decreased by 28.41 % and 43.77 %, respectively, after the phosphorus deficiency treatment, but the Pi content in shoots was still 52.95 % higher in Qi319-96 than in Qi319 (Table 1).
Low-phosphate stress limits plant photosynthesis (Table 2). Under phosphate-deficiency conditions, the net photosynthesis rate (Pn) decreased by 49.88 % in Qi319 and 41.33 % in Qi319-96. Under low-phosphate stress, the Pn of Qi319-96 was 26.81 % higher than that of Qi319. Phosphate deficiency also reduced the stomatal limitation value (Ls). The Ls values of the two genotypes, when subjected to the sufficient phosphorus treatment, were not significantly different, but the intercellular CO2 concentration (Ci) increased in both genotypes after low-phosphate stress (Table 2). The chlorophyll content in leaves was 50.00 % higher in Qi319-96 than in Qi319 (Table 3), indicating that photo-absorption by the chloroplasts was better in Qi319-96 under low-phosphate conditions.
Table 2.
Influence of different phosphate treatments on photosynthesis in Qi319 and Qi319-96
| Inbred lines | P treatment | Pn (μmol CO2 m-2 s-1) | Ci (μmol mol-1) | Ls |
|---|---|---|---|---|
| Qi319-96 | +P | 22.09 ± 0.35a | 76.49 ± 1.56c | 0.79 ± 0.03a |
| -P | 12.96 ± 0.40c | 108.51 ± 3.58b | 0.69 ± 0.02b | |
| Qi319 | +P | 20.39 ± 0.14b | 71.21 ± 5.57c | 0.80 ± 0.03a |
| -P | 10.22 ± 0.39d | 143.06 ± 2.29a | 0.60 ± 0.01c |
Three seedlings per bottle were cultured in phosphorus saturation solution (1000 μM KH2PO4) to the three-leaf stage, followed by low phosphate stress solution (5 μM KH2PO4) for an additional 25 days to the six–seven-leaf stage. Values represent the means of nine seedlings ± SD. Values with different letters within a row are significantly different (P < 0.05) by multiple comparison analysis
Table 3.
Influence of different phosphate treatments on sucrose, starch and chlorophyll contents
| Inbred lines | P treatment | Sucrose content (mg g-1 DW) | Starch content (mg g-1 DW) | Chorophyll content (mg g-1 FW) |
|---|---|---|---|---|
| Qi319-96 | +P | 46.61 ± 1.22a | 200.22 ± 1.45b | 2.54 ± 0.18a |
| -P | 31.51 ± 1.32c | 210.34 ± 1.56a | 1.47 ± 0.16c | |
| Qi319 | +P | 40.84 ± 1.33b | 189.25 ± 1.48c | 2.13 ± 0.15b |
| -P | 25.63 ± 1.24d | 209.39 ± 1.55a | 0.98 ± 0.09d |
Three seedlings per bottle were cultured in phosphorus saturation solution (1000 μM KH2PO4) to the three-leaf stage, followed by low phosphate stress solution (5 μM KH2PO4) for an additional 25 days to the six–seven-leaf stage. Values represent the means of nine seedlings ± SD. Values with different letters within a row are significantly different (P < 0.05) by multiple comparison analysis
The sucrose contents in the leaves significantly declined under low-phosphate stress, but the starch contents were still higher than the contents detected under phosphate-sufficient conditions (Table 3). The sucrose contents in the leaves were higher in Qi319-96 than in Qi319. More of the photosynthetic products were used for sucrose biosynthesis in Qi319-96 than in Qi319. These data indicated that the distribution of the photosynthetic carbon metabolism between sucrose and starch was altered in both genotypes under low-phosphate conditions. Thus, the low-phosphate-tolerant mutant Qi319-96 had a higher photosynthetic CO2 fixation rate and plant biomass compared with wild-type Qi319. Although the phosphorus level in shoots did not differ between genotypes, Qi319-96 had significantly higher levels of Pi than Qi319.
Differential analysis of leaf protein profiles
We performed comparative proteomic studies of Qi319 and Qi319-96 maize leaves subjected to two different phosphate levels using immobilized pH gradient (IPG) strips (pH 5–8), with three biological replicates. The number of protein spots detected in the gels and the proteins that differentially accumulated in the two genotypes are summarized in Table 4. Approximately 680 spots were detected under the phosphate saturation treatment. Of these, 29 (4.26 %) spots differentially accumulated between Qi319 and Qi319-96. Of the 29 spots, nine spots (including proteins not visible in the Qi319-96 gels) accumulated to a greater extent in Qi319 than in Qi319-96, whereas 20 spots (including proteins not visible in the Qi319 gels) accumulated to a greater extent in Qi319-96 (Fig. 2a, b). After the phosphate-deficiency treatment, 592 spots were detected, 71 (11.99 %) of which differentially accumulated between Qi319 and Qi319-96. Of these, 55 spots (including proteins not visible in the Qi319 gels) accumulated to a greater extent in Qi319-96 than in Qi319, whereas 16 spots (including proteins not visible in the Qi319-96 gels) accumulated to a greater extent in Qi319 (Fig. 2c, d).
Table 4.
Number of differentially accumulated proteins between the two genotypes and in response to phosphate stress on 2-DE gels (Fig. 2)
| Qi319 + P versus Qi319-96 + P | Qi319 – P versus Qi319-96 – P | |
|---|---|---|
| No. of proteins over-accumulated in Qi319-96 | 9 | 55 |
| No. of proteins over-accumulated in Qi319 | 20 | 16 |
| Total no. of differentially accumulated proteins | 29 | 71 |
Fig. 2.
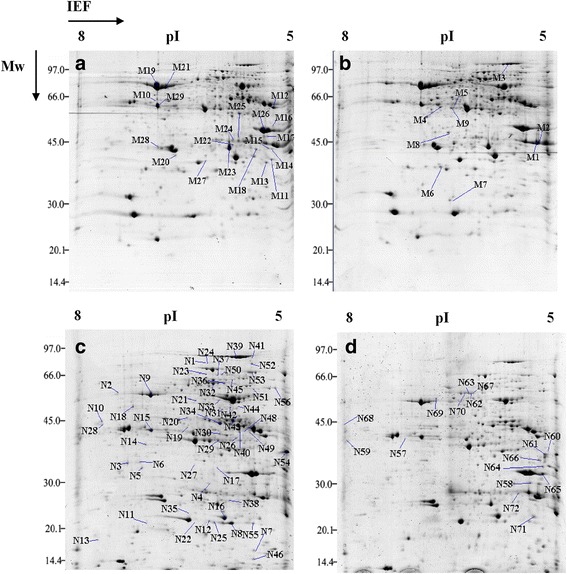
Comparison 2-DE protein gel maps taken from Qi319 and Qi319-96 maize leaves that had been subjected to two different phosphorus concentrations. The proteins were extracted by TCA/acetone from the middle of the fourth leaf and the 1.2 mg protein samples were separated in IEF using 17 cm pH 5–8 IPG strips. Then they were put on a 12 % polyacrylamide gel for the second dimensional separation and stained with CBB. The gel image analysis was carried out using PDQuest software (version 7.2.0; Bio-Rad). The spots marked with numbers (M1–M29) indicated proteins that were differentially accumulated in leaves of Qi319-96 and Qi-319 under + P conditions identified by MALDI-TOF MS; The spots marked with numbers (N1-N72) indicate proteins that were differentially accumulated in leaves of Qi319-96 and Qi-319 under -P conditions identified by MALDI-TOF MS. a: the image of the + P treatment Qi319-96 protein; b: the image of the + P treatment Qi319 protein; c: the image of the –P treatment Qi319-96 protein; d: the image of the –P treatment Qi319 protein
Identification and classification of phosphate-stress-responsive proteins
We identified the proteins that were differentially expressed after the two phosphate treatments using matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI-TOF MS) to gain a better understanding of the mechanisms involved in phosphorus stress and the differences in phosphorus tolerance between Qi319-96 and Qi319 maize plants. In total, 100 proteins were identified using the NCBI databases (Table 5 and Additional file 1). The detailed peptide sequences are shown in Additional file 2. We classified these proteins based on the TAIR (http://www.arabidopsis.org/) and KEGG (http://www.genome.jp/kegg) databases. The proteins were classified into protein fate, protein synthesis, cell rescue/defense/virulence, metabolism, energy and transcription/signal transduction mechanisms (Table 5). To confirm the results produced by peptide mass finger printing (PMF), eight randomly selected spots from among these proteins were subjected to MALDI-TOF-TOF MS analysis. The results for all eight were consistent with the PMF results (Additional file 3 and Additional file 4), which confirmed their reliability.
Table 5.
Differentially accumulated proteins with similar functionsin Qi319-96 and Qi319 leaves under both + P and − P conditions
| Matched protein namea | gi Numberb | Spot no.c | Patternd | Qi319-96– P versus Qi319-96 + Pe | Qi319– P versus Qi319 + Pf |
|---|---|---|---|---|---|
| Qi319-96 + P/Qi319 + P | |||||
| Metabolism | |||||
| Chlorophyll A-B binding protein | gi|242088861 | M1 | Increase | 1:1.04 | 1.07:1 |
| Chlorophyll A-B binding protein | gi|194700378 | M2 | Increase | 1:1.70 | 1: 5.05 |
| Chain A, Pyruvate phosphate dikinase | gi|62738111 | M3 | Increase | 1.62:1 | 1.78:1 |
| malate dehydrogenase | gi|226498728 | M4 | Increase | 1:1.74 | 1:2.03 |
| Coproporphyrinogen III oxidase | gi|308081534 | M5 | Increase | 6.93:1 | 1:60.35 |
| Triosephosphate isomerase | gi|194702698 | M9 | Increase | 1.75:1 | 1:2.96 |
| hydroxypyruvate reductase | gi|194697898 | M10 | Decrease | 1.30:1 | 1.07:1 |
| thylakoid lumenal 19 KDa protein | gi|226491484 | M11 | Decrease | 4.89:1 | 2.72:1 |
| sedoheptulose bisphosphatase1 | gi|226506366 | M12 | Decrease | 1:1.12 | 1:1.72 |
| Cyanobacterial and plastid NDH-1 subunit M | gi|194701566 | M13 | Decrease | 1:7.25 | 1:2.25 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|144583482 | M19 | Decrease | 1:19.87 | 5.91:1 |
| Ribulose bisphosphate carboxylase large chain | gi|132061 | M20 | Decrease | 1:6.24 | 1:3.99 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|11467200 | M21 | Decrease | 1:1.11 | 1:3.09 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|260677427 | M22 | Decrease | 1.30:1 | 1.08:1 |
| chlorophyll a-b binding protein 8 | gi|195613254 | M23 | Decrease | 1:3.41 | 1:2.12 |
| oxygen-evolving enhancer protein 1 | gi|195619530 | M26 | Decrease | 1:2.45 | 1:4.89 |
| NADH dehydrogenase subunit I | gi|11467259 | M28 | Decrease | 1:8.84 | 1:1.71 |
| Energy | |||||
| GlyoxalaseI | gi|212274373 | M16 | Decrease | 1:1.27 | 8.09:1 |
| glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic | gi|162461856 | M17 | Decrease | 1:1.49 | 1:1.39 |
| GADPH (383 AA) | gi|22240 | M29 | Decrease | 7.99:1 | 2.09:1 |
| Protein fate | |||||
| peptidyl-prolyl cis-trans isomerase | gi|226491656 | M6 | Increase | 1:18.53 | 1:35.85 |
| chaperonin | gi|195623400 | M14 | Decrease | 1:1.19 | 2.20:1 |
| CPN10 | gi|194688414 | M15 | Decrease | 1:1.50 | 3.19:1 |
| Pepsin-like aspartate proteases | gi|168041407 | M24 | Decrease | 1:1.39 | 1:16.85 |
| peptide methionine sulfoxide reductase | gi|226532399 | M27 | Decrease | 4.14:1 | 3.16:1 |
| Protein synthesis | |||||
| ribonucleoprotein | gi|22942613 | M18 | Decrease | 2.47:1 | 1:17.3 |
| Transcription/cellular communication/signal transduction | |||||
| glycine-rich RNA binding protein 2 | gi|195609654 | M7 | Increase | 1:10.72 | 1:15.2 |
| Unknown | |||||
| hypothetical protein SORBIDRAFT_09g001130 | gi|242086601 | M8 | Increase | 4.21:1 | 1.17:1 |
| LOC542632 | gi|162462462 | M25 | Decrease | 10.09:1 | 1.24:1 |
| Qi319-96 – P/Qi319 – P | |||||
| Metabolism | |||||
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|144583490 | N3 | Increase | 1:7.29 | 1:3.06 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|27448357 | N4 | Increase | 1:1.35 | 1:2.99 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|290086785 | N5 | Increase | 1:1.89 | 2.41:1 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|135991663 | N6 | Increase | 1:6.23 | 1:1.87 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|3560858 | N7 | Increase | 1.86:1 | 1:12.11 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|224382434 | N8 | Increase | 4.21:1 | 6.33:1 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|11467200 | N9 | Increase | 1:16.96 | 1:19.00 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|144583486 | N11 | Increase | 1:4.14 | 1:3.37 |
| Ribulose bisphosphate carboxylase large chain | gi|261279200 | N12 | Increase | 1:4.78 | 1:6.44 |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | gi|164565217 | N14 | Increase | 1:6.53 | 1:3.59 |
| chlorophyll a-b binding protein 8 | gi|226496924 | N15 | Increase | 1:7.07 | 9.89:1 |
| Chlorophyll a-b binding protein6A | gi|223973225 | N16 | Increase | 1:1.33 | 1.33:1 |
| chlorophyll a-b binding protein 8 | gi|195613254 | N17 | Increase | 3.79:1 | 1:10.14 |
| UDP-sulfoquinovose synthase | gi|238014584 | N18 | Increase | 4.69:1 | 3.63:1 |
| NDH-dependent cyclic electron flow 1 NAD-dependent epimerase | gi|194704742 | N19 | Increase | 1:1.33 | 1:2.93 |
| sucrose-phosphatase 1 | gi|194699500 | N21 | Increase | 1:1.79 | 1:6.51 |
| NADP-dependent malic enzyme,chloroplastic | gi|162459265 | N32 | Increase | 1.27:1 | 1:5.15 |
| NADP-dependent malic enzyme, chloroplastic precursor | gi|162459265 | N35 | Qi319-96 only | 1:1.59 | 0:1 |
| NADP-dependent malic enzyme, chloroplastic | gi|162459265 | N36 | Increase | 1.05:1 | 1:8.35 |
| pyruvate orthophosphate dikinase | gi|285013667 | N38 | Increase | 2.24:1 | 4.35:1 |
| pyruvateorthophosphate dikinase | gi|168586 | N39 | Increase | 1:1.66 | 1:7.35 |
| pyruvate orthophosphate dikinase | gi|219819651 | N41 | Qi319-96 only | 1:2.26 | 0:1 |
| delta-aminolevulinic acid dehydratase | gi|226496321 | N43 | Increase | 1:2.07 | 1:2.96 |
| transferase, transferring glycosyl groups, putative | gi|255562878 | N44 | Increase | 1:4.52 | 1:6.58 |
| glucose-6-phosphate isomerase | gi|226530882 | N51 | Increase | 1:1.31 | 1:1.91 |
| Low PSII Accumulation 3 | gi|242060200 | N54 | Increase | 2.07:1 | 2.59:1 |
| subunit NDH-M of NAD(P)H:plastoquinone dehydrogenase | gi|226497418 | N55 | Increase | 1:1.85 | 1.08:1 |
| carbonate dehydratase | gi|293332983 | N69 | Decrease | 1:2.58 | 1:3.07 |
| thylakoid lumenal 19 kDa protein | gi|226491484 | N71 | Decrease | 1:1.83 | 3.33:1 |
| Energy | |||||
| NADP+-dependent non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase B | gi|194688752 | N1 | Increase | 1:1.72 | 1:1.98 |
| NADP+-dependent non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase B | gi|194700892 | N2 | Increase | 2.04:1 | 1:1.68 |
| 6-phosphogluconate dehydrogenase family protein | gi|162463282 | N10 | Increase | 1.99:1 | 1:1.50 |
| alcohol dehydrogenase 2 | gi|195613268 | N20 | Increase | 1:1.02 | 1:1.68 |
| phosphoglucomutase, cytoplasmic 1 | gi|162463106 | N23 | Increase | 2.21:1 | 1:3.65 |
| fructose-bisphosphate aldolase | gi|195622374 | N26 | Increase | 8.56:1 | 1:4.94 |
| fructose-bisphosphate aldolase | gi|223975775 | N27 | Increase | 1:4.39 | 1:3.22 |
| fructose-bisphosphate aldolase | gi|194690156 | N28 | Increase | 2.90:1 | 2.18:1 |
| fructose-bisphosphate aldolase | gi|195634659 | N29 | Increase | 1.41:1 | 1.38:1 |
| aconitase | gi|238014964 | N33 | Increase | 2.05:1 | 1:4.53 |
| Glyceraldehyde-3-phosphate dehydrogenase B, chloroplast | gi|108705994 | N34 | Increase | 1:5.96 | 1:2.29 |
| phosphoglucomutase, cytoplasmic 1 | gi|162463106 | N37 | Increase | 1.26:1 | 1:9.30 |
| phosphoglycerate mutase | gi|293336560 | N45 | Increase | 1:1.40 | 1:3.19 |
| pyruvate dehydrogenase E1 component subunit beta | gi|195621752 | N48 | Increase | 4.55:1 | 1:2.99 |
| pyruvate dehydrogenase E1 beta subunit isoform 3 | gi|162458813 | N49 | Increase | 1.20:1 | 2.92:1 |
| vacuolar ATP synthase catalytic subunit A(V-ATPase A subunit) | gi|195658441 | N50 | Increase | 2.04:1 | 11.1:1 |
| 6-phosphogluconolctonase | gi|226493090 | N58 | Decrease | 6.78:1 | 1:1.14 |
| ATP synthase CF1 alpha subunit | gi|260677417 | N62 | Decrease | 1:1.97 | 1:3.01 |
| ATPase alpha subunit from chromosome 10 chloroplast | gi|19920165 | N63 | Decrease | 2.96:1 | 1:1.28 |
| ATP synthase CF1 alpha subunit | gi|50812525 | N67 | Decrease | 1.27:1 | 1:4.11 |
| Protein fate | |||||
| peptidylprolyl cis-trans isomerase | gi|226531796 | N40 | Qi319-96 Only | 1:3.50 | 0:1 |
| hsp70 | gi|308081377 | N53 | Increase | 4.02:1 | 3.06:1 |
| cpn60 chaperonin family protein | gi|242090109 | N56 | Increase | 1.69:1 | 4.71:1 |
| chaperonin | gi|195623400 | N72 | Decrease | 1.22:1 | 1.59:1 |
| Protein synthesis | |||||
| elongation factor G | gi|242076604 | N52 | Increase | 1.21:1 | 1:11.66 |
| Transcription/cellularcomm-unication/signal transduction | |||||
| GTP binding protein | gi|242061356 | N24 | Increase | 4.54:1 | 1:5.16 |
| translation initiation factor | gi|162462542 | N42 | Increase | 1:0 | 1.96:1 |
| RNA binding protein | gi|212722236 | N59 | Decrease | 1.97:1 | 2.81:1 |
| Cell rescue, defense and virulence | |||||
| CBS domain protein | gi|226490863 | N13 | Increase | 1:2.68 | 2.58:1 |
| peptide methionine sulfoxide reductase | gi|293332177 | N22 | Increase | 2.75:1 | 81.14:1 |
| peptide methionine sulfoxide reductase | gi|226532399 | N25 | Increase | 1.09:1 | 10.74:1 |
| hydroxyproline-rich glycoprotein family protein | gi|226507242 | N57 | Decrease | 1.19:1 | 1.89:1 |
| Unknown | |||||
| predicted protein | gi|224157625 | N60 | Decrease | 62.39:1 | 18.20:1 |
| unknown protein | gi|18419782 | N61 | Qi319 only | 2.00:1 | 1:0 |
| hypothetical protein | gi|218185826 | N65 | Decrease | 1:1.78 | 2.97:1 |
| hypothetical protein VOLCADRAFT_104026 | gi|302834273 | N66 | Decrease | 1:3.02 | 31.68:1 |
| clamp (CC)-tetratricopeptide repeat (TPR) proteins | gi|115446205 | N68 | Decrease | 3.84:1 | 1:7.29 |
| Secondary metabolism | |||||
| Caffeic acid O-methyltransferase | gi|33641714 | N30 | Increase | 1:3.35 | 1:1.86 |
| 3-N-debenzoyl-2-deoxytaxol N-benzoyltransferase | gi|226500072 | N31 | Increase | 1.81:1 | 1:2.71 |
| dehydroquinate dehydratase | gi|255070969 | N46 | Increase | 1.52:1 | 1:81.95 |
| ornithine carbamoyltransferase | gi|255538702 | N64 | Decrease | 84.38:1 | 30.71:1 |
| ketol-acid reductoisomerase | gi|212722020 | N70 | Decrease | 4.35:1 | 1:9.89 |
a: Name of protein identified by MALDI-TOF MS
b: Database accession numbers from NCBInr
c: Assigned spot number, as indicated in Fig. 2
d: “Increase” indicates significance at P < 0.05 and an increase in amount of at least 1.5-fold on the Qi319-96 gel under + P or –P conditions; “decrease”indicates significance at P < 0.05 and a decrease in amount of at least 1.5-fold on the Qi319-96 gel under + P or –P conditions compared withQi319
e: Specificity indicates the ratio of accumulation of a particular protein in leavesbetween Qi319-96 + P and Qi319-96 –P
f: Specificity indicates the ratio of accumulation of a particular protein in leavesbetween Qi319 + P and Qi319 – P
Differentially accumulated proteins and their effects on photosynthesis in the low-phosphate-tolerant mutant and wild-type maize
The levels of several proteins that participate in photosynthesis were significantly different between the two genotypes under the two phosphate treatments. Under low phosphate conditions, the levels of RuBisCO (N3, N4, N5, N6, N7, N8, N9, N11, N12 and N14), NADP-malic enzyme (NADP-ME; N32 (Fig. 3), N35 and N36), pyruvate orthophosphate dikinase (PPDK; N38 and N39 (Fig. 3)), delta-aminolevulinic acid dehydratase (N43, Fig. 3), sucrose-phosphatase (N21), cytoplasm- phosphoglucomutase (PGM; N23 and N37), fructose-bisphosphate (FBP) aldolase (N26, N27, N28 and N29 (Fig. 3)), NADP-glyceraldehyde-3-phosphate dehydrogenase (NADP-GP3DH; N34), NADPH dihydroethidium (N19), plastoquinone-dehydrogenase (NADPH dehydrogenase; N55), and chlorophyll a/b binding protein (N15, N16 and N17) increased significantly compared with Qi319. To verify these differences, we performed several physiological and biochemical experiments. The RuBisCO, PGM, FBP aldolase, NADP-ME and PPDK activities were also higher in Qi319-96 than in Qi319 under low phosphate stress (Table 6), which was consistent with the two-dimensional gel electrophoresis (2-DE) results. FBP aldolase catalyzes the reaction between fructose-1,6-diphosphate and sedoheptulose-1,7-diphosphate during ribulose-1,5-bisphosphate (RuBP) regeneration. NADP-malic enzyme and PPDK play important roles in CO2 fixation in bundle sheath cells. The chlorophyll content in leaves from Qi319-96 was higher than from Qi319 by 50 % (Table 3), which may be related to the significant increase in delta-aminolevulinic acid dehydratase expression. Furthermore, the photosynthetic rate was 26.81 % higher in Qi319-96 than in Qi319 (Table 2). The proteome and physiological data showed that Qi319-96 has a higher photosynthetic rate due to its higher chlorophyll content, and the higher expression levels and activities of Calvin cycle and “CO2 pump” enzymes during phosphate stress.
Fig. 3.
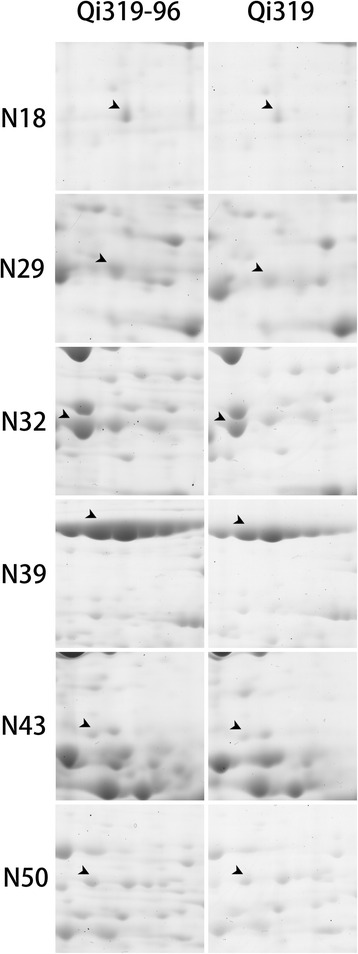
Comparison of 6 important protein between Qi319 and Qi319-96 under Pi-deficient condition. N18, uridine-5’-diphospho-sulfoquinovose (UDP-SQ) synthase; N29, fructose-bisphosphate (FBP) aldolase; N32, NADP-malic enzyme; N39, pyruvate orthophosphate dikinase; N43, delta-aminolevulinic acid dehydratase; N50, V-ATPase
Table 6.
Influence of different phosphorus concentrations on the activities of several enzymes involved in photosynthesis
| Inbred line | Pi treatment | RuBisCO (μmol CO2 mg pr-1 min-1) | PGM (μmol NADPH*mg-1 pr*min-1) | FBPaldose (μmol NADH/mg pr*min) | NADP-malic enzyme (μmol NADPH*mg-1 pr*min-1) | PPDK (μM AMP/mg Pr. min) |
|---|---|---|---|---|---|---|
| Qi319-96 | +P | 0.24 ± 0.03a | 2.19 ± 0.12a | 0.55 ± 0.02a | 7.76 ± 0.24b | 85.44 ± 2.71a |
| –P | 0.18 ± 0.08b | 0.84 ± 0.04c | 0.29 ± 0.08b | 8.47 ± 0.22a | 55.48 ± 2.02b | |
| Qi319 | +P | 0.23 ± 0.05a | 2.33 ± 0.12a | 0.54 ± 0.04a | 7.11 ± 0.05c | 83.27 ± 1.28a |
| –P | 0.13 ± 0.06c | 0.47 ± 0.01b | 0.16 ± 0.01c | 6.75 ± 0.11d | 45.35 ± 1.59c |
Three seedlings per bottle were cultured in phosphorus saturation solution (1000 μM KH2PO4) to the three-leaf stage, followed by low phosphate stress solution (5 μM KH2PO4) for an additional 25 days to the six–seven-leaf stage. Values represent the means of nine seedlings ± SD. Values with different letters within a row are significantly different (P < 0.05) by multiple comparison analysis
Differentially accumulated proteins are involved in energy metabolism between the low-phosphate-tolerant mutant and wild-type maize
The levels of several proteins that participate in energy metabolism were significantly different between the two genotypes under the two phosphorus treatments. These proteins are involved in the tricarboxylic acid (TCA) cycle, the pentose phosphate pathway and glycolysis (EMP). Under the phosphorus deficiency treatment, NADP-non-phosphorylated glyceraldehyde-3-phosphate dehydrogenase (NADP-non-GAPDH; N1 and N2) accumulated, which may have allowed EMP to proceed smoothly under very low phosphate conditions [20]. Increases in the levels of aconitase (N33) and the pyruvate dehydrogenase complex (N48 and N49) in Qi319-96 may accelerate ATP synthesis through the TCA cycle in this line compared with in Qi319. The physiological data showed that the amount of ATP in maize leaves was 78.28 % higher in Qi319-96 than in Qi319 under low phosphate stress (Table 7).
Table 7.
Influence of different phosphorus concentrations on ATP, SQDG, PG content and V-ATPase activity
| Inbred line | Pi treatment | ATP (nmol*g-1 Fw) | SQDG (mol/%) | PG (mol/%) | V-ATPase (μM Pi mg-1 Pro.h) |
|---|---|---|---|---|---|
| Qi319-96 | +P | 395.22 ± 22.23a | 9.25 ± 0.43a | 8.22 ± 0.47a | 8.36 ± 0.21a |
| –P | 134.53 ± 8.52b | 13.45 ± 0.76b | 6.12 ± 0.41b | 12. 74 ± 0.32b | |
| Qi319 | +P | 374.68 ± 17.45a | 9.02 ± 0.24a | 8.15 ± 0.55a | 8.17 ± 0.11a |
| –P | 75.46 ± 4.47c | 11.25 ± 0.88c | 5.68 ± 0.33c | 10.33 ± 0.23c |
Three seedlings per bottle were cultured in phosphorus saturation solution (1000 μM KH2PO4) to the three-leaf stage, followed by low phosphate stress solution (5 μM KH2PO4) for an additional 25 days to the six–seven-leaf stage. Values represent the means of nine seedlings ± SD. Values with different letters within a row are significantly different (P < 0.05) by multiple comparison analysis
Differentially accumulated proteins associated with increased phosphorus utilization between the low-phosphate-tolerant mutant and wild-type maize
Under low phosphate stress, the level of uridine-5’-diphospho-sulfoquinovose (UDP-SQ) synthase (N18, Fig. 3) increased significantly in Qi319-96 leaves compared with in Qi319 leaves. UDP-SQ synthase may increase the production of UDP-SQ, leading to an increase in available SQ, which is then used to produce SQDG. The increase in UDP-SQ synthase in Qi319-96 suggests that Qi319-96 may produce more SQDG than Qi319 under low phosphate stress. The accumulation of SQDG in the photo-membrane may displace phosphatidylglycerols (PG), which would accelerate the transformation of organic phosphorus and the utilization of internal phosphorus. The SQDG contents were 19.55 % higher in Qi319-96 than in Qi319, which is consistent with the UDP-SQ expression patterns for Qi319-96 and Qi319 (Table 7).
V-ATPase levels were not significantly different between Qi319-96 and Qi319 under the high phosphate treatment. However, under the low phosphate treatment, the increase in V-ATPase (N50, Fig. 3) levels was greater in Qi319-96 than in Qi319, suggesting that Qi319-96 might release phosphorus from the vacuole to increase the metabolic reaction rate in the cell, which would mitigate the symptoms caused by low phosphorus stress. The V-ATPase activity in the leaves was 23.33 % higher in Qi319-96 than in Qi319, which is consistent with the 2-DE results (Table 7).
Other differentially accumulated proteins between the low-phosphate-tolerant mutant and wild-type maize
The abundance of some proteins, including molecular chaperones (M14, M15, M27, N22, N25 and N56), resistance-related proteins (M6, N13, N40 and N57), and proteins involved in protein synthesis (M18, N42 and N52), signal transduction (M7, N24 and N59), and secondary metabolism (N30, N46 and N64), were significantly different between Qi319-96 and Qi319 under different phosphate levels. Several proteins involved in protein folding and assembly, protein synthesis and mediating signal transduction accumulated differently between the two genotypes under different phosphate levels.
Discussion
In this study, we applied comparative proteomics to gain insights into the phosphate-stress tolerance of Qi319-96 compared with Qi319. Compared with Qi319, the majority of the phosphate-stress-responsive proteins in Qi319-96 are involved in photosynthesis and internal phosphorus mobilization.
Altered membrane lipid compositions, and increased V-ATPase activities and abundances in Qi319-96, increased the phosphorus use efficiency under low phosphate stress
To counteract the detrimental effects of phosphate stress, higher plants have evolved a series of mechanisms for maintaining internal phosphorus levels. An important adaptive strategy used by plants subjected to low phosphate conditions is to increase the internal phosphorus utilization efficiency by remodeling the lipid membrane [9]. One-third of the total organophosphate contents in plants is present as phospholipids. When plants are subjected to low phosphate stress, membrane phospholipids are replaced by non-phosphorus glycerolipids, which promotes the mobilization of organophosphates [10]. SQDG and PG are thought to be involved in maintaining phosphorus levels in the thylakoid membrane, and their contents are known to be regulated by the phosphorus level [21]. Under the low phosphate treatment, the expression of UDP-SQ synthase increased in Qi319-96 (compared with Qi319) to supply SQ polar groups for SQDG biosynthesis. Indeed, the physiological data showed that the SQDG content was higher in Qi319-96 than in Qi319 under phosphate deprivation, indicating that SQDG accumulates in the photo-membrane, which could then be used to supplement PG levels. These changes may accelerate organophosphate conversion and increase available phosphorus recycling during periods of low phosphate stress. There is a great interest in the ability of Qi319 to increase internal phosphorus utilization through lipid remodeling [19]. Compared with wild-type Qi319, Qi319-96 exhibited an increase in UDP-SQ expression in response to low phosphate stress, suggesting that UDP-SQ synthase plays a crucial role in maintaining an internal Pi balance in this maize mutant.
Phosphorus in the vacuole acts as a cushion under fluctuating external and internal phosphorus levels. Phosphorus may move from the vacuole to the cytoplasm and chloroplasts, thereby participating in metabolic reactions during phosphate stress [7]. Therefore, the vacuole plays a key role in maintaining cytoplasmic phosphorus homeostasis [22]. Phosphorus movement across the membrane depends on an electrochemical H+-gradient across the membrane, which is maintained by V-ATPase [2]. V-ATPase is more highly accumulated in Qi319-96 than in Qi319 during phosphate stress. The mutant may utilize the additional V-ATPase to intensify vacuolar-membrane proton transport, forming an electrochemical gradient across the membrane, and thus, increasing the energy supply for phosphorus transport across the membrane. This would significantly improve the tolerance of Qi319-96 to low phosphate stress.
Qi319-96 plants exhibited much better lipid composition remodeling and higher V-ATPase expression and activity levels than Qi319 plants, which could facilitate phosphorus utilization during periods of phosphate stress. Furthermore, these changes could be responsible for the increased Pi levels in Qi319-96 leaves compared with in Qi319 leaves.
Increased expression of enzymes involved in the Calvin cycle and CO2 fixation may enhance photosynthesis in Qi319-96 compared with in Qi319
Phosphate deprivation has an adverse effect on plant photosynthesis [23]. The decline in photosynthetic efficiency in maize leaves under low phosphate stress is related to the reduced levels of proteins involved in CO2 enrichment, the Calvin cycle and the electron transport system. The reduced abundance of these proteins correlates well with the rates of photochemical reactions, CO2 assimilation, the Calvin cycle and RuBP regeneration [19]. Several enzymes that play key roles in photosynthesis, such as RuBisCO and 3-phosphoglyceric phosphokinase, are involved in plant responses to low phosphate conditions [24]. RuBisCO plays an important role in carbon fixation during the Calvin cycle [25]. Reduced RuBisCO activity is one of the non-stomatal factors that leads to a substantial decline in photosynthesis [26]. In the current study, RuBisCO was more highly expressed in Qi319-96 than in Qi319, and RuBisCO carboxylase activity was significantly higher in Qi319-96 than in Qi319, which may be related to the increased rate of photosynthesis in Qi319-96.
A reduction in RuBP regeneration has a considerable impact on photosynthesis. The rate of photosynthesis may decrease if RuBP regeneration declines [26]. Reductions in RuBP regeneration may be due to reduced ATP levels when plants are subjected to low phosphate conditions. Reduced ATP levels inhibit the reduction of 3-phosphoglycerate to triose phosphate through 3-phosphoglyceric phosphokinase, and they also inhibit the phosphorylation of ribulose-5-phosphate to RuBP through phosphoribulokinase [27, 28]. The increased accumulation of ATP in the leaves of Qi319-96 compared with Qi319 under low phosphate stress may be beneficial for RuBP regeneration. In addition, the reduced RuBP regeneration may be due to the reduced initial activities of several enzymes, such as NADP-GAPDH and FBP aldolase. RuBP regeneration is also affected by the increased amount of photosynthetic carbon that is diverted to starch formation and away from RuBP regeneration (RuBP is only found in chloroplasts) [29]. The higher level and activity of FBP aldolase in the chloroplasts of Qi319-96 versus Qi319 under phosphorus deprivation conditions may have led to a greater regeneration of RuBP in Qi319-96 compared with in Qi319.
Maize is a C4 plant with “CO2 pumps” that have a “floral hoop structure”. The “CO2 pump” mechanism in C4 plants increases the level of CO2 around RuBisCO, which may increase the carboxylation reaction rate and reduce the alternative fixation of O2. C4 photosynthesis is more efficient than C3 photosynthesis under low atmospheric CO2 conditions, which suggests that the “CO2 pump” is an important mechanism that leads to an increase in photosynthesis [30]. The accumulation and activity levels of NADP-ME and PPDK were higher in Qi319-96 than in Qi319 under phosphate deprivation conditions, which favored a greater capture rate and level of CO2 in vascular bundle sheath cells. Higher CO2 levels not only increase RuBisCO carboxylase activity, but they also suppress the oxidation of RuBisCO and reduce photorespiration. The higher CO2 level in Qi319-96 helped the plants increase carbon assimilation and counteract phosphate deprivation.
In conclusion, Qi319-96 had a higher rate of photosynthesis than Qi319 under low phosphate conditions, which could be attributed to its higher chlorophyll content and increased levels/activities of photosynthesis-related enzymes, such as RuBisCO, PPDK and NADP-ME.
Conclusions
Qi319-96 had a significantly higher level of Pi than Qi319 under low-phosphate stress, which may be related to Qi319-96’s remodeled membrane lipids and the increase in V-ATPase activity, which releases phosphorus from plant organophosphorus to the cytoplasm. A physiological analysis showed that the carbon assimilation rate of Qi319-96 was significantly higher than that of Qi319 under low-phosphate stress, which may be owing to the increased accumulation of several photosynthesis-related in the mutant Qi319-96 compared with wild-type Qi319. Our results clearly indicated that the differences in increasing the internal P-use efficiency are the main reasons for the higher tolerance to low-P conditions in the mutant compared with the wild-type. At the same time, this study suggests that the inbred lines of maize with low-P tolerant traits could be obtained effectively through cellular engineering.
Methods
Plant growth and treatments
Maize mutant Qi319-96 with a low-P tolerance was obtained using cellular engineering technology. The immature embryo of maize inbred line Qi-319 were transferred to inducing medium to produce calli. Embryogenic calli were subcultured on subculture medium for 6 months, and then screened continuously on selecting medium without phosphate. After selection for 5 generations, we transferred the survival embryogenic calli to differentiation medium for the regeneration of plantlets (R0). The survival plants (R0) were transplanted to the field, and self-pollinated to harvest seeds (R1). Low-P tolerant R1 plants and their inbred progeny were selected by low phosphorus stress. After six generations, low-P tolerant inbred lines, including Qi319-96, were obtained [31, 32]. Seeds of the maize inbred line Qi319 and mutant Qi319-96 were sterilized with ethanol and HgCl2, and germinated in the dark at 28 °C. Three-day-old seedlings were grown in nutrient solution (+P, Pi-sufficient, 1,000 μM KH2PO4) for 15 days (to the three-leaf stage). Half of the seedlings were then cultured in low-phosphate nutrient solution (–P, Pi-deficient, 5 μM MKH2PO4). For the low-phosphate treatment, the 1,000 μM KH2PO4 in the + P nutrient solution was substituted with 1,000 μM KCl. The control nutrient solutions were the same as those used by Li et al. [31]. Seedlings were positioned in a completely randomized design in a greenhouse, and three batches of seedlings were cultured separately to provide biological replicates.
Measuring the biomass, total phosphorus content, and Pi level
The maize seedlings were transferred to a phosphate-deficiency solution (5 μM KH2PO4), cultured for an additional 25 days (to the six- to seven-leaf stage), and washed with distilled water. The shoots and roots were weighed after drying at 80 °C to a constant weight. The phosphorus contents of the roots and shoots were determined according to Murphy and Riley [33]. The Pi level in the shoots was determined by measuring the molybdenum complex content, as described by Taussky and Shorr [34].
Measuring ATP levels
Fresh samples (1 g) from the middle of the fourth leaves were boiled in 5 mL of MgSO4 for 15 min and centrifuged at 5,000 × g for 15 min at 4 °C. The supernatant was stored on ice until analyzed. The maize leaf ATP level was determined using the method described by Fan et al. [35].
Measuring chlorophyll, sucrose, and starch contents
Fresh samples (0.1 g) were extracted in 80 % acetone, and chlorophyll levels were analyzed according to Arnon [36]. The sucrose and starch levels were assayed with resorcinol as previously described [19].
Determining photosynthetic performance
To characterize photosynthetic performance in the maize plants, a portable photosynthesis system (LI-6400; LI-COR, Inc., Lincoln, NE, USA) was used to detect Pn, ambient carbon dioxide (Co), and Ci levels in the fourth expanded leaf of each sample. The Ls value was then calculated using the following formula: Ls = 1 – Ci/Co. The photon flux density was kept at 800 μmol m–2 s–1 using an internal LED source, the temperature in the leaf chamber was maintained at 25 °C, and the relative humidity was 55–60 %. The CO2 level was approximately 360 μmol CO2 mol–1. All of the measurements were carried out between 09:30 am and 11:30 am.
Enzyme activity assays
Fresh samples (1 g) were rapidly collected from the third leaf (from the bottom) of each plant and rapidly ground in 4 mL of buffer (0.1 mM Hepes-NaOH, pH 7.5, 50 mM MgCl2, 2 mM EDTA, 2 % polyvinylpyrrolidone, and 1 % β-mercaptoethanol) pre-cooled on ice. The homogenates were centrifuged at 15,000 × g for 20 min at 4 °C, and the supernatant was used for the enzyme assays [37]. PPDK was determined by assaying for NADH oxidation in a mixture containing 0.15 M Tris-HCl, 18 μM MgCl2, 30 μM dithiothreitol (DTT), 0.45 μM NADH, 3 μM phosphoenolpyruvate, 3 μM AMP, 3 μM sodium pyrophosphate, 6 units of lactic dehydrogenase, and an aliquot of leaf extract. The assays were initiated by adding 3 μM sodium pyrophosphate [37]. For the NADP-ME assay, an aliquot of leaf extract was added to a mixture containing 50 mM Hepes-KOH (pH 8.0), 5 mM DTT, and 0.5 mM NADP. The reaction was initiated by adding MgCl2 [37]. The mixture for the FBP aldolase assay contained 30 mM Hepes-KOH (pH 7.6), 10 mM FBP, 0.25 mM NADH, and 2–4 units mL-1 of alpha-glycerol-3-phosphate dehydrogenase and triose phosphate isomerase. The reaction was initiated by adding FBP [29]. PGM activity was determined after its reaction with NADP by measuring the change in absorbance at 340 nm. The reaction mixture contained 30 mM Hepes-KOH, 4 mM MgCl2, 0.5 mM NADP, and 2–4 units of glucose-6-phosphate dehydrogenase. The reaction was initiated by adding 1.2 mM glucose-1-phosphate [38]. The RuBisCO assay reaction mixture contained 50 mM Hepes-KOH (pH 8.0), 1 mM EDTA–2Na, 20 mM MgCl2, 25 mM DTT, 10 mM NaHCO3, 5 mM ATP, 0.15 mM NADH, 5 mM creatine phosphate, 0.6 mM RuBP, 10 units of phosphocreatine kinase, 10 units of glyceraldehyde-3-phosphate dehydrogenase, and 10 units of phosphoglycerate kinase. RuBisCO activity was determined by monitoring the absorbance change at 340 nm owing to the oxidation of NADH according to the method of Sawada et al. [39]. For the V-ATPase assay, vesicle membranes were isolated by sucrose density gradient ultracentrifugation according to Wang et al. [40].
Lipid extraction, purification, and analysis
Fresh samples (0.5 g) were ground to a powder in liquid nitrogen and suspended in chloroform and methanol. The lipid was extracted and purified according to Blihg and Dyer [41]. The mixture was separated into individual lipids by two-dimensional thin-layer silica gel chromatography (G model, 10 cm × 10 cm). The first dimension was composed of acetone/methylbenzene/H2O2 (91:30:8 v/v/v), and the second dimension was composed of chloroform/methanol/isopropamide/ammonia (65:35:0.5:5 v/v/v/v). The thin-layer chromatography plates were sprayed with 0.01 % Primulin in acetone/water (3:2 v/v) and analyzed under a ultraviolet light (366 nm) to identify the locations of individual lipids. Spots corresponding to the lipid classes were removed and methylated. The lipid contents were determined using gas chromatography with heptadecanoic acid as an internal standard. The relative contents of individual lipids are presented as molar percentages (mol %) [42].
All physiological experimental data represent the means of three biological replicates ± SD. A significance analysis was performed using Duncan’s multiple range tests. All graphs were constructed using Sigma Plot 13.0.
Protein sample preparation and 2-DE mapping
The fourth leaves from maize seedlings exhibiting phosphorus-stress symptoms were collected for protein extraction. Fresh samples (2 g) were ground to a powder in liquid nitrogen and combined with 20 mL of acetone containing 10 % TCA, 10 mM DTT, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The mixture was precipitated at −20 °C overnight and then centrifuged at 19,000 × g for 20 min at 4 °C. The pellet was carefully washed twice in acetone containing 10 mM DTT and 1 mM PMSF to remove any pigment [43], and vacuum dried with a vacuum pump. The pellet was then dissolved in 2.5 mL of protein solubilization buffer [7 M urea, 2 M thiourea, 4 % CHAPS, 0.5 % v/v carrier ampholyte (pH 3–10), 10 mM DTT and 1 mM PMSF] for 2.5 h. The insoluble material was removed by centrifugation at 40,000 × g for 25 min. The protein level in the supernatant was measured using the Bradford assay and sub-sampled for 2-DE analysis [44].
The 2-DE was performed using pH 5–8 IPG strips (Bio-Rad, California, USA). Liquid rehydration buffer containing 1.2 mg of protein (7 M urea, 2 M thiourea, 4 % CHAPS, 1.5 % v/v carrier ampholyte, and 65 mM DTT) was used to hydrate the strips for 13 h using a GE Healthcare III (GE Healthcare, Buckinghamshire, United Kingdom). The voltage procedure was as follows: (1) grade voltage increased to 100 V for 30 min; (2) grade voltage increased to 250 V for 1 h; (3) step voltage increased to 1,000 V for 1 h; (4) step voltage increased to 5,000 V for 3 h; (5) grade voltage increased to 10,000 V for 6 h; and finally (6) step voltage increased to 10,000 V with the focus increased to 100 kVh. After isoelectric focusing, the IPG strips were equilibrated before sodium dodecyl sulfate polyacrylamide gel electrophoresis(SDS-PAGE) according to Yan et al. [45]. The strips were loaded onto 12 % denaturing acrylamide gels and sealed with 0.5 % agarose solution. The electrophoresis was carried out using a PROTEANII Ready Gel System (20 cm × 20 cm; Bio-Rad) at 10 mA gel–1 for 1 h and 25 mA gel–1 for 6 h. The gels were stained with Coomassie brilliant blue according to Katam et al. [46] and scanned using a GS-800 calibrated densitometer (Bio-Rad). The 2-DE experiment was carried out with three replications using independent samples. The images were analyzed using PDQuest software (version 7.2.0; Bio-Rad). After background subtraction and spot detection, the spots were matched and normalized using the total density in the gel image method.
The statistical significances of quantitative data were determined using the Student’s t-test (n = 3, P < 0.05) at a 95 % confidence level, and proteins with a 1.5-fold or more change at this confidence level were considered differentially accumulated.
In-gel digestion and MALDI-TOF/MALDI-TOF-TOF MS analysis
Several protein spots were excised from gels and washed twice with distilled water to remove the redundant sodium dodecyl sulfate. The spots were destained in 25 mM NH4HCO3 [dissolved in 50 % acetonitrile (ACN)] and dehydrated in 100 % ACN. The protein spots were reduced, alkylated, and washed thoroughly, as described by Yan et al. [45]. The spots were then digested with 5–8 μL of trypsin (proteome grade trypsin; Sigma). The samples were dissolved in 40 mM ammonium bicarbonate and 9 % ACN at 20 ng mL–1 (pH 8.0) for 30 min at 4 °C. The redundant trypsin solutions were removed, and the gel pieces were dipped in 15 μL of 25 mM NH4HCO3 solution (pH 8.0) and incubated at 37 °C overnight. The supernatant was then transferred to new centrifuge tubes and the combined with 25 μL of solution containing 67 % ACN and 3.3 % trifluoroacetic acid (TFA). The two supernatant liquids were combined, dried under a vacuum, dissolved in 4–5 μL of 0.1 % TFA, and stored in 0.5-μL aliquots at −80 °C. Before analysis, the samples were mixed with 0.6 μL of 10 mg ml–1 w/v cyano-4-hydroxycinnamic acid in 0.1 % TFA/50 % ACN and dried on a metal plate. After air drying, the samples were subjected to MALDI-TOF MS and MALDI-TOF/TOF MS on a Bruker Ultraflex TOF/TOF controlled by the Flexcontrol 2.4 package using default parameters (Bruker Daltonics, Karlsruhe, Germany).
Protein identification and database searching
After calibration and a monoisotopic peak analysis using GPS Explorer (Applied Biosystems 2006), the monoisotopic peak lists obtained were compared against the National Center for Biotechnology Information database using the MASCOT program (http://www.matrixscience.com), allowing one trypsin cleavage error. Carbamidomethylation of cysteine and oxidation of methionine were recognized as the fixed modifications, and pyro-glutamic acid formation of N-terminal glutamine was the variable modification. To obtain highly accurate identification results, the proteins had to fulfill the following criteria: (1) molecular weight search score > 73 (P < 0.05); (2) more than six peptides matched the theoretical result; (3) sequence coverage was greater than 15 %; and (4) the proteins had a peptide mass tolerance of at least 100 ppm. The search criteria for the MALDI-TOF-TOF/MS results were similar to the PMF criteria: (1) individual ions scores were > 43 (P < 0.05); (2) the peptide mass tolerance was 100 ppm; and (3) the fragment mass tolerance was at least 0.3 Da. The proteins identified using MALDI-TOF MS were categorized using the TAIR (http://www.arabidopsis.org/) and KEGG (http://www.genome.jp/kegg/) databases.
Abbreviations
2-DE, two-dimensional gel electrophoresis; ACN, acetonitrile; EMP, glycolysis; IPG, immobilized pH gradient; MALDI-TOF MS, matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry; PG, phosphatidylglycerol; Pi, inorganic phosphate; PMF, peptide mass finger printing; PMSF, phenylmethylsulfonyl fluoride; PPDK, pyruvate orthophosphate dikinase; RuBP, ribulose-1,5-bisphosphate; SQDG, sulfoquinovosyl diglyceride; TFA, trifluoroacetic acid; UDP-SQ, uridine-5’-diphospho-sulfoquinovose.
Acknowledgments
We thank Bioedit (Specialized English Editing, Writing and Publisher Services in the Life Sciences) and Edanz Language Editing for assistance in language editing. We also thank the BGI Company for MADLI-TOF-TOF MS.
Funding
This research was supported by the National Natural Science Foundation of China (31172028), Shandong Province Agricultural Breeding Project (Research on Utilization of Agricultural Biological Resources Innovation), and Programs Involved in Research on Transgenic Plants in China (2014ZX0800504B).
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files.
Authors’ contributions
KZ participated in conceiving the project and designing the experiments. In addition, KZ provided assistance with physiological methodology, participated in data analyses, and revised the manuscript. HL carried out the proteome and physiological experiments and wrote the manuscript. JS provided help with analysis methodology and revised the manuscript. WW provided help with physiological experiments. KL performed proteome experiments and helped with analysis methodology. JZ participated in proofreading the manuscript. All authors reviewed and contributed to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Differentially accumulated proteins with similar functions present in the leaves of Qi319-96 and Qi319 under both + P and − P conditions. (DOC 2104 kb)
Identification of differentially accumulated proteins and sequences of the identified peptides. (XLS 203 kb)
MS/MS patterns of sequenced peptides. (DOC 1467 kb)
List of differentially expressed proteins in leaves of Qi319 and Qi319-96 seedlings under − P and + P conditions by MALDI-TOF-TOF/MS. (XLS 26 kb)
References
- 1.Malboobi MA, Zamani K, Lohrasebi T, Sarikhani MR, Samaian A, Sabet MS. Phosphate: the silent challenge. Prog Bio Sci. 2014;4(1):1–32. [Google Scholar]
- 2.Plaxton WC, Tran HT. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011;156(3):1006–1015. doi: 10.1104/pp.111.175281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 4.Elanchezhian R, Krishnapriya V, Pandey R, Rao AS, Abrol YP. Physiological and molecular approaches for improving phosphorus uptake efficiency of crops. Curr Sci. 2015;108(7):1271–1279. [Google Scholar]
- 5.Cheng L, Bucciarelli B, Liu J, Zinn K, Miller S, Patton-Vogt J, Allan D, Shen J, Vance CP. White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol. 2011;156(3):1131–1148. doi: 10.1104/pp.111.173724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol. 2014;65:95–123. doi: 10.1146/annurev-arplant-050213-035949. [DOI] [PubMed] [Google Scholar]
- 7.Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelck CE, Plaxton WC, Price CA, Scheible WR, Shane MW, White PJ, Raven JA. Opportunities for improving phosphorus‐use efficiency in crop plants. New Phytol. 2012;195(2):306–320. doi: 10.1111/j.1469-8137.2012.04190.x. [DOI] [PubMed] [Google Scholar]
- 8.Richardson AE, Lynch JP, Ryan PR, Delhaize E, Smith FA, Smith SE, Harvey PR, Ryan MH, Veneklaas EJ, Lambers H. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil. 2011;349(1-2):121–156. doi: 10.1007/s11104-011-0950-4. [DOI] [Google Scholar]
- 9.Nakamura Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res. 2013;52(1):43–50. doi: 10.1016/j.plipres.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, Kusano M, Aoki T, Hirai MY, Saito K. A new class of plant lipid is essential for protection against phosphorus depletion. Nature Communi. 2013;4:1510–1520. doi: 10.1038/ncomms2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siebers M, Dormann P, Holzl G. Membrane Remodelling in Phosphorus-deficient Plants. Annu Plant Rev. 2015;48:237–263. [Google Scholar]
- 12.Kern J, Zouni A, Guskov A, Krauβ N. Lipids in the structure of photosystem I, photosystem II and the cytochrome b6f complex. In: Wada H, Murata N, editors. Lipids in photosynthesis: essential and regulatory functions. The Netherlands: Springer; 2009. p. 203-242.
- 13.Cheng Y, Zhou W, Peters C, Li M, Wang X, Huang J. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid‐localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 2011;66(5):781–795. doi: 10.1111/j.1365-313X.2011.04538.x. [DOI] [PubMed] [Google Scholar]
- 14.Marchive C, Yehudai-Resheff S, Germain A, Fei Z, Jiang X, Judkins J, Wu H, Fernie AR, Fait A, Stern DB. Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol. 2009;151(2):905–924. doi: 10.1104/pp.109.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasaki J, Shinano T, Onishi K, Yonetani R, Yazaki J, Fujii F, Shimbo K, Ishikawa M, Shimatani Z, Nagata Y. Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J Exp Bot. 2006;57(9):2049–2059. doi: 10.1093/jxb/erj158. [DOI] [PubMed] [Google Scholar]
- 16.Aziz T, Finnegan PM, Lambers H, Jost R. Organ‐specific phosphorus‐allocation patterns and transcript profiles linked to phosphorus efficiency in two contrasting wheat genotypes. Plant, Cell Environ. 2014;37(4):943–960. doi: 10.1111/pce.12210. [DOI] [PubMed] [Google Scholar]
- 17.Xia M, Wang X, Li H, Wu P. Identification of the rice vacuolar ATPase B subunit gene and its expression pattern analysis under phosphorus deficiency. Acta Bot Sin. 2001;44(5):573–578. [Google Scholar]
- 18.Liu H, Zhong W, Zhang K. Effect of low phosphorous stress on photosynthesis and chlorophyll fluorescence characteristics of maize inbred lines Qi319 and low-phosphorus-tolerant mutant Qi319-96. Chinese Agricultural Science Bulletin. 2014;30(27):21–28. [Google Scholar]
- 19.Zhang K, Liu H, Tao P, Chen H. Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. Plos One. 2014;9(5):e98215. doi: 10.1371/journal.pone.0098215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duff SM, Moorhead GB, Lefebvre DD, Plaxton WC. Phosphate starvation inducible ‘bypasses’ of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989;90(4):1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu B, Benning C. Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J. 2003;36(6):762–770. doi: 10.1046/j.1365-313X.2003.01918.x. [DOI] [PubMed] [Google Scholar]
- 22.Pratt J, Boisson A-M, Gout E, Bligny R, Douce R, Aubert S. Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol. 2009;151(3):1646–1657. doi: 10.1104/pp.109.144626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond JP, White PJ. Sucrose transport in the phloem: integrating root responses to phosphorus starvation. J Exp Bot. 2008;59(1):93–109. doi: 10.1093/jxb/erm221. [DOI] [PubMed] [Google Scholar]
- 24.Usuda H, Shimogawara K. Phosphate deficiency in maize. I. Leaf phosphate status, growth, photosynthesis and carbon partitioning. Plant Cell Physiol. 1991;32(4):497–504. [Google Scholar]
- 25.Andersson I, Backlund A. Structure and function of Rubisco. Plant Physiol Biochem. 2008;46(3):275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Arulanantham AR, Rao IM, Terry N. Limiting factors in photosynthesis VI. Regeneration of ribulose 1, 5-bisphosphate limits photosynthesis at low photochemical capacity. Plant Physiol. 1990;93(4):1466–1475. doi: 10.1104/pp.93.4.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giersch C, Robinson SP. Regulation of photosynthetic carbon metabolism during phosphate limitation of photosynthesis in isolated spinach chloroplasts. Photo Res. 1987;14(3):211–227. doi: 10.1007/BF00032706. [DOI] [PubMed] [Google Scholar]
- 28.Usuda H, Shimogawara K. Phosphate deficiency in maize III. Changes in enzyme activities during the course of phosphate deprivation. Plant Physiol. 1992;99(4):1680–1685. doi: 10.1104/pp.99.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao IM, Terry N. Leaf phosphate status, photosynthesis, and carbon partitioning in sugar beet I. Changes in growth, gas exchange, and Calvin Cycle enzymes. Plant Physiol. 1989;90(3):814–819. doi: 10.1104/pp.90.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Sun N, Yang S, Zhao Y, Wang X, Hao X, Qiao Z. Evolutionary transition from C3 to C4 photosynthesis and the route to C4 rice. Biologia. 2013;68(4):577–586. doi: 10.2478/s11756-013-0191-5. [DOI] [Google Scholar]
- 31.Li K, Xu Z, Zhang K, Yang A, Zhang J. Efficient production and characterization for maize inbred lines with low-phosphorus tolerance. Plant Sci. 2007;172(2):255–264. doi: 10.1016/j.plantsci.2006.09.004. [DOI] [Google Scholar]
- 32.Zhang K, Yin X, Shang M, Quan R, Zhang J. Selection of maize cells with low-phosphorus tolerance and study on the progeny of regenerated plants. Sci Agric Sin. 2000;33:124–131. [Google Scholar]
- 33.Murphy J, Riley JP. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta. 1962;27:31–36. doi: 10.1016/S0003-2670(00)88444-5. [DOI] [Google Scholar]
- 34.Taussky HH, Shorr E. A microcolorimetric method for the determination of inorganic phosphorus. J Biological Chem. 1953;202(2):675–685. [PubMed] [Google Scholar]
- 35.Fan G, Gu S, Ning T, Li X. Determination of ATP in maize kernel by bio-luminescence. Modern Instruments. 2005;3:34–35. [Google Scholar]
- 36.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usuda H, Ku MS, Edwards GE. Activation of NADP-malate dehydrogenase, pyruvate, Pi dikinase, and fructose 1, 6-bisphosphatase in relation to photosynthetic rate in maize. Plant Physiol. 1984;76(1):238–243. doi: 10.1104/pp.76.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai C, Salamini F, Nelson O. Enzymes of carbohydrate metabolism in the developing endosperm of maize. Plant Physiol. 1970;46(2):299–306. doi: 10.1104/pp.46.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawada S, Sato M, Kasai A, Yaochi D, Kameya Y, Matsumoto I, Kasai M. Analysis of the feed-forward effects of sink activity on the photosynthetic source-sink balance in single-rooted sweet potato leaves. I. Activation of RuBPcase through the development of sinks. Plant Cell Physiol. 2003;44(2):190–197. doi: 10.1093/pcp/pcg024. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Lüttge U, Ratajczak R. Effects of salt treatment and osmotic stress on V‐ATPase and V‐PPase in leaves of the halophyte Suaeda salsa. J Exp Bot. 2001;52(365):2355–2365. doi: 10.1093/jexbot/52.365.2355. [DOI] [PubMed] [Google Scholar]
- 41.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Xu Y. Phosphatidylglycerol degradation is one crucial reason for the decrease of its concentration in wheat leaves under phosphate deprivation stress. Plant Growth Regul. 2006;49(1):105–112. [Google Scholar]
- 43.Kim ST, Cho KS, Jang YS, Kang KY. Two‐dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis. 2001;22(10):2103–2109. doi: 10.1002/1522-2683(200106)22:10<2103::AID-ELPS2103>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 44.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics. 2006;5(3):484–496. doi: 10.1074/mcp.M500251-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Katam R, Basha SM, Suravajhala P, Pechan T. Analysis of peanut leaf proteome. J Proteome Res. 2010;9(5):2236–2254. doi: 10.1021/pr901009n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional files.


