Abstract
Background
Germline mutations in the succinate dehydrogenase complex genes SDHB, SDHC, and SDHD predispose to pheochromocytomas and paragangliomas. Here, we examine the SDHB, SDHC, and SDHD mutation spectrum in the Danish population by screening of 143 Danish pheochromocytoma and paraganglioma patients.
Methods
Mutational screening was performed by Sanger sequencing or next-generation sequencing. The frequencies of variants of unknown clinical significance, e.g. intronic, missense, and synonymous variants, were determined using the Exome Aggregation Consortium database, while the significance of missense mutations was predicted by in silico and loss of heterozygosity analysis when possible.
Results
We report 18 germline variants; nine in SDHB, six in SDHC, and three in SDHD. Of these 18 variants, eight are novel. We classify 12 variants as likely pathogenic/pathogenic, one as likely benign, and five as variants of unknown clinical significance.
Conclusions
Identifying and classifying SDHB, SDHC, and SDHD variants present in the Danish population will augment the growing knowledge on variants in these genes and may support future clinical risk assessments.
Keywords: Classification, Germline, Mutation, Paraganglioma, Pheochromocytoma, SDHB, SDHC, SDHD
Background
Pheochromocytomas (PCC; OMIM 171300) and paragangliomas (PGL; OMIM 115310) are neuroendocrine neoplasms localized in the medulla of the adrenal gland and the extra-adrenal paraganglia, respectively. Both neoplasms stem from the neural crest and are characterized by excess catecholamine production. Approximately 60 % of PCC and PGL are sporadic; however, up to 40 % are due to genetic predisposition [1, 2]. The familial PCC/PGL are associated with germline mutations in at least 11 susceptibility genes, including multiple endocrine neoplasia 2A and 2B (RET), von Hippel-Lindau (VHL), neurofibromatosis type 1 (NF1), transmembrane protein 127 (TMEM127), MYC-associated factor X (MAX), fumarate hydratase (FH), and succinate dehydrogenase complex subunit A, B, C, D, and AF2 (SDHA, SDHB, SDHC, SDHD, SDHAF2) [3–13]. These genes form the basis for genetic testing and risk assessment in patients suffering from PCC or PGL [1, 2]. Among the first genes to be identified in the tumorigenesis of familial PCC and PGL were SDHB, SDHC, and SDHD, all of which belong to the succinate dehydrogenase gene family [14]. These genes encode a protein complex that is involved in the oxidation of succinate in the respiratory chain and are classified as tumor suppressors. The succinate dehydrogenase complex (complex II) itself is composed of four nuclear-encoded subunits, SDHA, SDHB, SDHC, and SDHD, and functions in the inner mitochondrial membrane where it oxidizes succinate to fumarate. The SDHB gene encodes the iron-sulfur catalytic subunit of complex II. It is located on chromosome 1 and comprises eight exons encoding a 280-amino acid protein. The SDHC and SDHD genes encode the two membrane integral components that anchor the complex to the mitochondrial membrane. The SDHC gene is located on chromosome 1 and consists of six exons encoding a protein of 169 amino acids, while the SDHD gene is located on chromosome 11 and comprises four exons encoding a protein consisting of 159 amino acids [15–17]. Here, we report the SDHB, SDHC, and SDHD variants identified in Danish PCC and PGL families.
Methods
Patients
In agreement with national guidelines in endocrinology (Danish Endocrine Society: http://www.endocrinology.dk/), patients were referred for genetic screening from the Departments of Endocrinology or the Departments of Clinical Genetics throughout the regions of Denmark. After obtaining verbal and written consent from each patient, blood samples were collected for germline variant screening. In total, 143 individuals were screened between 2006 and 2015 for SDHB, SDHC, and SDHD germline variants. This investigation was approved by the local ethics committee in the capital region of Denmark (H-4-2010-050).
SDH screening
Genomic DNA was isolated from whole blood or formalin-fixed paraffin-embedded (FFPE) tumor tissue using a QIAamp DNA mini kit (Qiagen) or QIAamp DNA FFPE tissue kit according to the instructions provided by the manufacturer. From 2006 to 2014, the coding exons and adjacent intronic sequences of SDHB, SDHC, and SDHD were amplified by PCR followed by Sanger sequencing using an ABI3730 DNA analyzer (Applied Biosystems). Moreover, genomic DNA was examined for large genomic rearrangements by multiplex ligation-dependent probe amplification (MLPA) analysis using a SALSA MLPA P226 SDH kit (MRC-Holland). Since 2014, the analysis has been performed using targeted next-generation sequencing and a library designed to capture all exons from the three genes. Library construction was carried out from 50–500 ng of genomic DNA and adaptor ligation of Illumina’s adaptors included in the TruSeq DNA sample preparation kit (Illumina) was performed using the SPRIworks System I (Beckman Coulter). Sequence capture was conducted using a double capture protocol (Roche) whereby 8–10 samples were pooled prior to hybridization. Sequencing was performed on a MiSeq (llumina) to an average depth of at least 100×. Sequencing data were analyzed using SequencePilot (JSI medical systems), where variants were called if the allele frequency was above 25 %. Moreover, the samples were analyzed for copy number variations. SDHB, SDHC, and SDHD variants are numbered according to GenBank accession numbers NM_003000.2, NM_003001.3, and NM_003002.3, in which the A in the AUG start codon has number 1, using the guidelines from the Human Genome Variation Society (http://varnomen.hgvs.org/). Sequence variants, except well-known polymorphisms, were verified by Sanger sequencing in a new blood sample.
In silico data analysis
The integrated Alamut Visual software (v.2.6.1) (http://www.interactive-biosoftware.com) including Align GVGD (A-GVGD), PolyPhen-2, and SIFT was used to predict the pathogenicity of specific variants in SDHB, SDHC, and SDHD. The in silico effect of variants on splicing was examined as previously described [18]. Default settings were used in all predictions. The frequency of the variants was obtained from the Exome Aggregation Consortium (ExAC) or the Exome Sequencing Project (ESP) databases. Moreover the frequency of novel missense variants was examined in data from 2000 Danish exomes [19]. A combined assessment on the pathogenicity of each variant was used according to the five-tiered scheme, where Class 5 is pathogenic, Class 4 is likely pathogenic, Class 3 is uncertain due to insufficient evidence, Class 2 is likely benign, and Class 1 is benign [20].
Results
During the last 9 years, we have performed genetic screening of the entire coding region and the exon-intron boundaries of the SDHB, SDHC, and SDHD genes on genomic DNA from Danish PCC/PGL patients. Up until May 2015, we have screened 143 individuals and so far 18 germline variants have been identified, of which eight are novel (Table 1). Moreover, 14 well-known polymorphisms were identified, including six in SDHB, five in SDHC, and three in SDHD (Table 2).
Table 1.
SDHB, SDHC, and SDHD variants identified in Danish patients with neuroendocrine cancer. PCC pheochromocytoma, PGL paraganglioma, LGR large genomic rearrangement; a identified in homozygote state; N/A not accessible. Results from In silico data analyses from A-GVGD, SIFT, and PolyPhen-2 are listed in the corresponding columns. Each variant is classed according to the five-tiered scheme
| Family ID | Disease | Age (yr) | Gene | Exon | Nucleotide | Protein | Mutation type | A-GVGD | SIFT | PolyPhen-2 | Frequency (ExAC) | Class | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PGL | 51 | SDHB | 1 | c.-1-?_72+?del | - | LGR | - | - | - | - | 5 | [21, 22] |
| 2 | PGL | 36 | SDHB | 1–7 | c.-1-?_765+?del | - | LGR | - | - | - | - | 5 | Novel |
| 3 | PGL | 29 | SDHB | 1 | c.3G > A | p.Met1? | Start loss | - | - | - | - | 4 | [24, 32, 43] |
| 4 | PGL | 34 | SDHB | 3 | c.203G > A | p.Cys68Tyr | Missense | C65 | Deleterious | Possibly damaging | - | 4 | [30, 43] |
| 5 | PGL | 16 | SDHB | 4–5 | c.287-?_540+?del | - | LGR | - | - | - | - | 5 | Novel |
| 6 | PCC | 51 | SDHB | 4 | c.300T > C | p.Ser100Ser | Synonymous | - | - | - | 0.15 % | 3 | [30] |
| 7 | PCC | 29 | SDHB | 4 | c.416T > C | p.Leu139Pro | Missense | C65 | Deleterious | Possibly damaging | - | 4 | Novel |
| 8 | PGL | 12 | SDHB | 6 | c.566G > A | p.Cys189Tyr | Missense | C65 | Deleterious | Possibly damaging | - | 3 | Novel |
| 9 | PGL | 39 | SDHB | 7 | c.653G > A | p.Trp218Ter | Nonsense | - | - | - | - | 5 | [30] |
| 10 | PGL | 41 | SDHC | 2 | c.43C > T | p.Arg15Ter | Nonsense | - | - | - | 0.00082 % | 5 | [31–33] |
| 11 | PGL | 75 | SDHC | - | c.78-19C > T | - | - | - | - | - | 0.021 % | 3 | Novel |
| 12 | PGL | N/A | SDHC | 3 | c.148C > T | p.Arg50Cys | Missense | C25 | Deleterious | Probably damaging | - | 4 | [32, 43] |
| 13 | PGL | 64 | SDHC | 4 | c.191_207del17 | p.Pro64LeufsTer28 | Frameshift | - | - | - | - | 5 | Novel |
| 14 | PCC | 31 | SDHC | 5 | c.242G > Ta | p.Gly81Val | Missense | C0 | Deleterious | Probably damaging | 0.00082 % | 3 | Novel |
| 15 | PCC | 74 | SDHC | 6 | c.496C > G | p.Leu166Val | Missense | C0 | Tolerated | Benign | - | 3 | Novel |
| 16 | PGL | 41 | SDHD | 2 | c.149A > G | p.His50Arg | Missense | C0 | Tolerated | Probably damaging | 0.65 % | 2 | [22, 30, 43–48] |
| 17 | PCC | 37 | SDHD | 3 | c.274G > T | p.Asp92Tyr | Missense | C65 | Deleterious | Probably damaging | - | 5 | [4, 36, 38–43] |
| 18 | PGL | 41 | SDHD | 4 | c.341A > G | p.Tyr114Cys | Missense | C65 | Deleterious | Probably damaging | - | 5 | [24, 37, 42, 49–53] |
Table 2.
SDHB, SDHC, and SDHD polymorphisms identified in Danish patients with neuroendocrine cancer
| Gene | Nucleotide | Protein | Frequency (ESP)a | Frequency (ExAC)b |
|---|---|---|---|---|
| SDHB | c.18C > A | p.Ala6Ala | 2.70 % | 2.99 % |
| SDHB | c.24C > Tc | p.Ser8Ser | 0.76 % | 0.65 % |
| SDHB | c.200 + 33G > A | - | 7.79 % | 8.42 % |
| SDHB | c.200 + 35G > A | - | 3.74 % | 3.93 % |
| SDHB | c.201-36G > T | - | 97.38 % | 96.99 % |
| SDHB | c.487T > C | p.Ser163Pro | 1.30 % | 1.52 % |
| SDHC | c.20 + 11_20 + 12dup | - | 6.25 % | 6.37 % |
| SDHC | c.77 + 36T > A | - | 2.08 % | 2.44 % |
| SDHC | c.77 + 43del | - | - | 3.15 % |
| SDHC | c.77 + 48C > T | - | 5.31 % | 4.69 % |
| SDHC | c.406-14del | - | - | 41.08 % |
| SDHD | c.170-29A > G | - | 1.28 % | 1.16 % |
| SDHD | c.204C > T | p.Ser68Ser | 1.28 % | 1.16 % |
| SDHD | c.315-32T > C | - | 1.25 % | 1.17 % |
The frequency of the polymorphism was obtained from the Exome Sequencing Project (ESP) among aEuropean American and the Exome Aggregation Consortium (ExAC) databases among bEuropean (Non-Finnish); cObserved in 1.35 % of the Finnish population obtained from the ExAC database
Variants in the SDHB gene were the most frequent, with a total of nine germline variants of which four are novel. The variants included a p.Trp218Ter nonsense mutation, as well as three large genomic rearrangements (LGR) deleting exon 1 (c.-1-?_72+?del), exons 1–7 (c.-1-?_765+?del), and exons 4–5 (c.287-?_540+?del), of which the two latter are novel. Moreover, we identified a start-loss mutation (p.Met1?) and three missense variants (p.Cys68Tyr, p.Leu139Pro, and p.Cys189Tyr) in SDHB, which were all highly conserved and predicted to be pathogenic by A-GVGD, SIFT, or PolyPhen-2 (Table 1). None of the missense variants have been described before in the ExAC database or in the Danish population. Sequencing analysis of DNA from tumor tissue from the p.Leu139Pro carrier revealed loss of heterozygosity (LOH) (Fig. 1). Finally, we identified a synonymous variant in SDHB located in exon 4 (p.Ser100Ser). This variant is reported with a frequency of 0.15 % in the ExAC database.
Fig. 1.
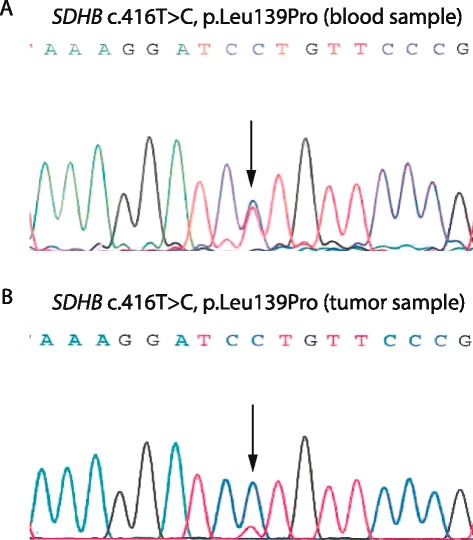
LOH analysis of DNA from the p.Leu139Pro variant carrier. DNA was purified from blood and tumor samples from the patient and SDHB exon 4 was amplified using intronic primer pairs flanking exon 4 and sequenced. DNA sequences from blood (a) and from tumor tissue (b) are shown. The SDHB c.416T > C, p.Leu139Pro position is indicated by an arrow
Moreover, we identified six variants in the SDHC gene, including one nonsense mutation, one frameshift mutation, three missense variants, and one intronic variant. Four of the six variants are novel (Table 1). Both the nonsense mutation (p.Arg15Ter) and the frameshift mutation (c.191_207del17) result in premature stop codons. The p.Arg15Ter mutation is reported in one individual out of more than 60,000 (0.00082 %) in the ExAC database. Of the three SDHC missense variants (p.Arg50Cys, p.Gly81Val, and p.Leu166Val), only the p.Gly81Val variant is reported in one South Asian individual (0.00082 %) in the ExAC database, while none of the missense variants have been identified in the Danish population. In silico analysis predicted that the p.Arg50Cys and p.Gly81Val variants to be pathogenic, while p.Leu166Val was suggested to be benign. Moreover, we identified an intronic variant (c.78-19C > T) in SDHC, which is reported with a frequency of 0.021 % in the ExAC database.
Finally, we identified three SDHD missense variants (p.His50Arg, p.Asp92Tyr, and p.Tyr114Cys) all of which have previously been described. Two of the missense variants were identified in a single patient (p.His50Arg and p.Tyr114Cys). Only the p.His50Arg variant is reported with a frequency of 0.65 % in the ExAC database. In silico analysis predicted the p.Asp92Tyr and p.Tyr114Cys variants to be pathogenic, whereas the p.His50Arg variant was predicted to be benign.
Discussion
Screening for SDHB, SDHC, and SDHD germline variants in Danish PCC and PGL patients revealed the identification of 18 different germline variants of which eight are novel.
Four of the nine variants identified in SDHB are novel, including two LGR. The deletion of exons 1–7 (c.-1-?_765+?del) was identified in a patient who suffered from a PGL at the age of 36, while the deletion of exons 4–5 (c.287-?_540+?del) was identified in a 16-year-old patient diagnosed with PGL. In addition, we found an exon 1 deletion in a patient with PGL and severe hypertension recognized at the age of 51. This deletion has previously been reported in patients with PGL [21, 22]. All LGRs in SDHB introduce frameshifts and premature stop codons and were classified as pathogenic (Class 5). The SDHB p.Trp218Ter nonsense mutation was identified in a patient with PGL at the age of 39 and was classified as pathogenic (Class 5) in accordance with a previous report [23]. Moreover, we identified a start-loss mutation (p.Met1?) in SDHB in a patient diagnosed with a PGL at the age of 29. This mutation destroys the methionine start codon and is thought to prevent SDHB from being translated from this position. Although an alternative downstream in-frame methionine start codon is present at codon 58, the mutation has previously been associated with the development of hereditary PGL [24]. Furthermore, immunohistochemical (IHC) staining shows loss of SDHB in association with this variant [25] and thus we classify this variant as likely pathogenic (Class 4). All three SDHB missense variants are predicted as pathogenic by in silico analysis (Table 1). The p.Cys68Tyr variant has previously been described in patients with PCC or PGL, where IHC analysis of tumor tissue showed no SDHB staining or LOH of the wild-type SDHB allele [26, 27]. Based on these findings, the variant is classified as likely pathogenic (Class 4). The p.Leu139Pro variant has not previously been reported. Sequencing analysis of DNA from FFPE tumor tissue showed LOH, indicating that the mutation is likely pathogenic (Class 4). The final missense variant (p.Cys189Tyr) has not previously been reported, although two different amino acid changes at this position (p.Cys189Arg and p.Cys189Phe) have been associated with hereditary PGL [23, 28]. We currently classify this variant as a variant of uncertain significance (VUS) (Class 3) since no co-segregation or functional studies have been performed. Finally, the synonymous variant p.Ser100Ser has previously been reported in patients with PGL [29, 30] but since no functional studies have been performed, we classify this variant as a VUS (Class 3).
The p.Arg15Ter nonsense mutation and the c.191_207del17 frameshift mutation in SDHC both result in premature stop codons and are thus classified as pathogenic (Class 5). The p.Arg15Ter variant has previously been reported [31–33], whereas the frameshift mutation is novel. Moreover, we identified three missense variants in SDHC, of which p.Gly81Val and p.Leu166Val are novel. In silico analysis indicates that the former variant is pathogenic while the latter is predicted to be benign. The nucleotide substitution leading to p.Gly81Val is present on the first base in exon 5 and in silico splicing analysis indicates that this change could affect the splicing of exon 5. Recently, another change at position 81 (p.Gly81Arg) has been reported to affect splicing via the splice donor site [23]. The p.Gly81Arg variant was identified in the homozygous state in a patient who developed PCC at the age of 31. Homozygous pathogenic germline mutations usually result in a much more severe phenotype or syndrome, and since no functional studies have been performed at either the RNA or protein level, the p.Gly81Val variant is classified as a VUS (Class 3). The p.Leu166Val variant is located close to the stop codon in SDHC. Another variant at position 166 (p.Leu166Arg) has been reported in the LOVD database but is not described in the literature. Since no functional studies have been performed, we classify this variant as a VUS (Class 3). The p.Arg50Cys variant has previously been described, and in silico and functional analyses indicate that this variant has some effect on the SDH activity [32, 34]. Moreover, since the variant was identified in an offspring of the proband, who suffered from PGL at a young age (<25 years), the variant is classified as likely pathogenic (Class 4). Finally, we identified a novel intronic variant (c.78-19C > T) in SDHC in an elderly patient with PGL at age 75. In silico analysis does not predict any effect of this variant on splicing; however, since no functional studies have been performed, it is classified as a VUS (Class 3).
The missense variants identified in SDHD have all previously been described. The p.His50Arg variant is predicted to be benign by in silico analysis. The variant has been described several times in the literature and functional studies in yeast show that this variant has no effect on the phenotype [35]. In agreement with these data and taking the high frequency in the control population into consideration, we classify the variant as likely benign (Class 2). The p.His50Arg variant was identified alongside a pathogenic mutation p.Tyr114Cys, which is highly conserved and has been shown to co-segregate with disease [4, 34, 36–38] and therefore are classified as pathogenic (Class 5). Finally, the p.Asp92Tyr is a well-known Dutch founder mutation and was indeed identified in a Dutch individual, who had inherited the mutation from his father. Functional analysis has shown that the mutation impairs the function of the SDHD protein [4, 36, 38–42] and in agreement with these findings we classify this variant as pathogenic (Class 5).
In conclusion, we have examined the mutation spectrum associated with familial PCC and PGL in the Danish population. Our investigation uncovered 18 germline variants in SDHB, SDHC, and SDHD, of which eight are novel. Based on the literature, in silico data, and LOH analysis, we classify 12 variants as likely pathogenic/pathogenic, one variant as likely benign, while five variants are classified as variants of unknown clinical significance. Further co-segregation or functional studies are needed before these variants can be classified.
Abbreviations
IHC, immunohistochemical; LOH, loss of heterozygosity; PCC, pheochromocytomas; PGL, paragangliomas; VUS, variant of uncertain significance
Acknowledgements
We thank Aseeba Ayub, Bettina M. Andersen, Lene W. Pedersen, and Søren Petersen for excellent technical assistance.
Authors’ contributions
Clinical data was provided by ÅKR, AMG, ABS, and UBJ, while TvOH, FCN, and MR supervised the molecular analyses and performed the interpretation of variants. The manuscript was drafted by MB, MR and TVOH. All authors approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval
The study was approved by the local Ethics Committee, Capital Region of Denmark, Denmark.
References
- 1.Dahia PL. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat. Rev. Cancer. 2014;14:108–119. doi: 10.1038/nrc3648. [DOI] [PubMed] [Google Scholar]
- 2.Favier J, Amar L, Gimenez-Roqueplo AP. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat. Rev. Endocrinol. 2015;11:101–111. doi: 10.1038/nrendo.2014.188. [DOI] [PubMed] [Google Scholar]
- 3.Astuti D, Latif F, Dallol A, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baysal BE, Ferrell RE, Willett-Brozick JE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 5.Burnichon N, Briere JJ, Libe R, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro-Vega LJ, Buffet A, de Cubas AA, et al. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet. 2014;23:2440–2446. doi: 10.1093/hmg/ddt639. [DOI] [PubMed] [Google Scholar]
- 7.Comino-Mendez I, Gracia-Aznarez FJ, Schiavi F, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat. Genet. 2011;43:663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 8.Kunst HP, Rutten MH, de Monnink J, et al. SDHAF2 (PGL2-SDH5) and hereditary head and neck paraganglioma. Clin Cancer Res. 2011;17:247–254. doi: 10.1158/1078-0432.CCR-10-0420. [DOI] [PubMed] [Google Scholar]
- 9.Neumann HP, Berger DP, Sigmund G, et al. Pheochromocytomas, multiple endocrine neoplasia type 2, and von Hippel-Lindau disease. N Engl J Med. 1993;329:1531–1538. doi: 10.1056/NEJM199311183292103. [DOI] [PubMed] [Google Scholar]
- 10.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat. Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 11.Qin Y, Yao L, King EE, et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat. Genet. 2010;42:229–233. doi: 10.1038/ng.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith DP, Eng C, Ponder BA. Mutations of the RET proto-oncogene in the multiple endocrine neoplasia type 2 syndromes and Hirschsprung disease. J. Cell Sci. Suppl. 1994;18:43–49. doi: 10.1242/jcs.1994.Supplement_18.6. [DOI] [PubMed] [Google Scholar]
- 13.Viskochil D, Buchberg AM, Xu G, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- 14.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim. Biophys Acta. 2011;1807:1432–1443. doi: 10.1016/j.bbabio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Jain-Ghai S, Cameron JM, Al Maawali A, et al. Complex II deficiency--a case report and review of the literature. Am J Med Genet A. 2013;161A:285–294. doi: 10.1002/ajmg.a.35714. [DOI] [PubMed] [Google Scholar]
- 16.Mailloux RJ, Willmore WG. S-glutathionylation reactions in mitochondrial function and disease. Front Cell Dev.Biol. 2014;2:68. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Yu L, Yu CA. Generation of superoxide anion by succinate-cytochrome c reductase from bovine heart mitochondria. J Biol Chem. 1998;273:33972–33976. doi: 10.1074/jbc.273.51.33972. [DOI] [PubMed] [Google Scholar]
- 18.Ahlborn LB, Dandanell M, Steffensen AY, et al. Splicing analysis of 14 BRCA1 missense variants classifies nine variants as pathogenic. Breast Cancer Res. Treat. 2015;150:289–298. doi: 10.1007/s10549-015-3313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohmueller KE, Sparso T, Li Q, et al. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet. 2013;93:1072–1086. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWhinney SR, Pilarski RT, Forrester SR, et al. Large germline deletions of mitochondrial complex II subunits SDHB and SDHD in hereditary paraganglioma. J. Clin. Endocrinol. Metab. 2004;89:5694–5699. doi: 10.1210/jc.2004-0769. [DOI] [PubMed] [Google Scholar]
- 22.Cascon A, Montero-Conde C, Ruiz-Llorente S, et al. Gross SDHB deletions in patients with paraganglioma detected by multiplex PCR: a possible hot spot? Genes Chromosomes. Cancer. 2006;45:213–219. doi: 10.1002/gcc.20283. [DOI] [PubMed] [Google Scholar]
- 23.Burnichon N, Rohmer V, Amar L, et al. The succinate dehydrogenase genetic testing in a large prospective series of patients with paragangliomas. J. Clin. Endocrinol. Metab. 2009;94:2817–2827. doi: 10.1210/jc.2008-2504. [DOI] [PubMed] [Google Scholar]
- 24.Piccini V, Rapizzi E, Bacca A, et al. Head and neck paragangliomas: genetic spectrum and clinical variability in 79 consecutive patients. Endocr. Relat Cancer. 2012;19:149–155. doi: 10.1530/ERC-11-0369. [DOI] [PubMed] [Google Scholar]
- 25.Sjursen W, Halvorsen H, Hofsli E, et al. Mutation screening in a Norwegian cohort with pheochromocytoma. Fam. Cancer. 2013;12:529–535. doi: 10.1007/s10689-013-9608-0. [DOI] [PubMed] [Google Scholar]
- 26.Burnichon N, Vescovo L, Amar L, et al. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Hum Mol Genet. 2011;20:3974–3985. doi: 10.1093/hmg/ddr324. [DOI] [PubMed] [Google Scholar]
- 27.van Nederveen FH, Gaal J, Favier J, et al. An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. Lancet Oncol. 2009;10:764–771. doi: 10.1016/S1470-2045(09)70164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fokkema IF, Taschner PE, Schaafsma GC, et al. LOVD v. 2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 29.Brouwers FM, Eisenhofer G, Tao JJ, et al. High frequency of SDHB germline mutations in patients with malignant catecholamine-producing paragangliomas: implications for genetic testing. J. Clin. Endocrinol. Metab. 2006;91:4505–4509. doi: 10.1210/jc.2006-0423. [DOI] [PubMed] [Google Scholar]
- 30.Lima J, Feijao T, Ferreira da Silva A, et al. High frequency of germline succinate dehydrogenase mutations in sporadic cervical paragangliomas in northern Spain: mitochondrial succinate dehydrogenase structure-function relationships and clinical-pathological correlations. J Clin Endocrinol Metab. 2007;92:4853–4864. doi: 10.1210/jc.2007-0640. [DOI] [PubMed] [Google Scholar]
- 31.Pasini B, McWhinney SR, Bei T, et al. Clinical and molecular genetics of patients with the Carney-Stratakis syndrome and germline mutations of the genes coding for the succinate dehydrogenase subunits SDHB, SDHC, and SDHD. Eur. J. Hum. Genet. 2008;16:79–88. doi: 10.1038/sj.ejhg.5201904. [DOI] [PubMed] [Google Scholar]
- 32.Neumann HP, Erlic Z, Boedeker CC, et al. Clinical predictors for germline mutations in head and neck paraganglioma patients: cost reduction strategy in genetic diagnostic process as fall-out. Cancer Res. 2009;69:3650–3656. doi: 10.1158/0008-5472.CAN-08-4057. [DOI] [PubMed] [Google Scholar]
- 33.Vandy FC, Sisk G, Berguer R. Synchronous carotid body and thoracic paraganglioma associated with a germline SDHC mutation. J. Vasc. Surg. 2011;53:805–807. doi: 10.1016/j.jvs.2010.09.064. [DOI] [PubMed] [Google Scholar]
- 34.Panizza E, Ercolino T, Mori L, et al. Yeast model for evaluating the pathogenic significance of SDHB, SDHC and SDHD mutations in PHEO-PGL syndrome. Hum Mol Genet. 2013;22:804–815. doi: 10.1093/hmg/dds487. [DOI] [PubMed] [Google Scholar]
- 35.Yu W, He X, Ni Y, et al. Cowden syndrome-associated germline SDHD variants alter PTEN nuclear translocation through SRC-induced PTEN oxidation. Hum Mol Genet. 2015;24:142–153. doi: 10.1093/hmg/ddu425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dannenberg H, Dinjens WN, Abbou M, et al. Frequent germ-line succinate dehydrogenase subunit D gene mutations in patients with apparently sporadic parasympathetic paraganglioma. Clin. Cancer Res. 2002;8:2061–2066. [PubMed] [Google Scholar]
- 37.Schiavi F, Dematte S, Cecchini ME, et al. The endemic paraganglioma syndrome type 1: origin, spread, and clinical expression. J. Clin. Endocrinol. Metab. 2012;97:E637–E641. doi: 10.1210/jc.2011-2597. [DOI] [PubMed] [Google Scholar]
- 38.Taschner PE, Jansen JC, Baysal BE, et al. Nearly all hereditary paragangliomas in the Netherlands are caused by two founder mutations in the SDHD gene. Genes Chromosomes. Cancer. 2001;31:274–281. doi: 10.1002/gcc.1144. [DOI] [PubMed] [Google Scholar]
- 39.Badenhop RF, Jansen JC, Fagan PA, et al. The prevalence of SDHB, SDHC, and SDHD mutations in patients with head and neck paraganglioma and association of mutations with clinical features. J. Med. Genet. 2004;41:e99. doi: 10.1136/jmg.2003.011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douwes Dekker PB, Corver WE, Hogendoorn PC, et al. Multiparameter DNA flow-sorting demonstrates diploidy and SDHD wild-type gene retention in the sustentacular cell compartment of head and neck paragangliomas: chief cells are the only neoplastic component. J. Pathol. 2004;202:456–462. doi: 10.1002/path.1535. [DOI] [PubMed] [Google Scholar]
- 41.Neumann HP, Bausch B, McWhinney SR, et al. Germ-line mutations in nonsyndromic pheochromocytoma. N Engl J Med. 2002;346:1459–1466. doi: 10.1056/NEJMoa020152. [DOI] [PubMed] [Google Scholar]
- 42.Neumann HP, Pawlu C, Peczkowska M, et al. Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations. JAMA. 2004;292:943–951. doi: 10.1001/jama.292.8.943. [DOI] [PubMed] [Google Scholar]
- 43.Evenepoel L, Papathomas TG, Krol N, et al. Toward an improved definition of the genetic and tumor spectrum associated with SDH germ-line mutations. Genet. Med. 2015;17:610–620. doi: 10.1038/gim.2014.162. [DOI] [PubMed] [Google Scholar]
- 44.Gimenez-Roqueplo AP, Favier J, Rustin P, et al. Mutations in the SDHB gene are associated with extra-adrenal and/or malignant phaeochromocytomas. Cancer Res. 2003;63:5615–5621. [PubMed] [Google Scholar]
- 45.Perren A, Barghorn A, Schmid S, et al. Absence of somatic SDHD mutations in sporadic neuroendocrine tumors and detection of two germline variants in paraganglioma patients. Oncogene. 2002;21:7605–7608. doi: 10.1038/sj.onc.1205812. [DOI] [PubMed] [Google Scholar]
- 46.Riemann K, Sotlar K, Kupka S, et al. Chromosome 11 monosomy in conjunction with a mutated SDHD initiation codon in nonfamilial paraganglioma cases. Cancer Genet Cytogenet. 2004;150:128–135. doi: 10.1016/j.cancergencyto.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 47.Kytola S, Nord B, Elder EE, et al. Alterations of the SDHD gene locus in midgut carcinoids, Merkel cell carcinomas, pheochromocytomas, and abdominal paragangliomas. Genes Chromosomes. Cancer. 2002;34:325–332. doi: 10.1002/gcc.10081. [DOI] [PubMed] [Google Scholar]
- 48.Castellano M, Mori L, Giacche M, et al. Genetic mutation screening in an italian cohort of nonsyndromic pheochromocytoma/paraganglioma patients. Ann N.Y Acad Sci. 2006;1073:156–165. doi: 10.1196/annals.1353.016. [DOI] [PubMed] [Google Scholar]
- 49.Antonello M, Piazza M, Menegolo M, et al. Role of the genetic study in the management of carotid body tumor in paraganglioma syndrome. Eur. J. Vasc. Endovasc. Surg. 2008;36:517–519. doi: 10.1016/j.ejvs.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 50.Benn DE, Gimenez-Roqueplo AP, Reilly JR, et al. Clinical presentation and penetrance of pheochromocytoma/paraganglioma syndromes. J. Clin. Endocrinol. Metab. 2006;91:827–836. doi: 10.1210/jc.2005-1862. [DOI] [PubMed] [Google Scholar]
- 51.Braun S, Riemann K, Kupka S, et al. Active succinate dehydrogenase (SDH) and lack of SDHD mutations in sporadic paragangliomas. Anticancer Res. 2005;25:2809–2814. [PubMed] [Google Scholar]
- 52.Fish JH, Klein-Weigel P, Biebl M, et al. Systematic screening and treatment evaluation of hereditary neck paragangliomas. Head Neck. 2007;29:864–873. doi: 10.1002/hed.20638. [DOI] [PubMed] [Google Scholar]
- 53.Milunsky JM, Maher TA, Michels VV, et al. Novel mutations and the emergence of a common mutation in the SDHD gene causing familial paraganglioma. Am J Med Genet. 2001;100:311–314. doi: 10.1002/ajmg.1270. [DOI] [PubMed] [Google Scholar]


