Abstract
Bacterial persisters are phenotypic variants that survive extraordinary concentrations of antibiotics, and are thought to underlie the propensity of biofilm infections to relapse. Unfortunately many aspects of persister physiology remain ill-defined, which prevents progress toward eradicating the phenotype. Recently, we identified respiration within non-growing Escherichia coli populations as a potential target for the elimination type I persisters, which are those that arise from passage through stationary phase. Here we discovered that nitric oxide (NO) treatment at the onset of stationary phase significantly reduced type I persister formation through its ability to inhibit respiration. NO decreased protein and RNA degradation in stationary phase cells, and produced populations that were more fit for protein synthesis and growth resumption upon introduction into fresh media than untreated controls. Overall, this data shows that NO, which is a therapeutically-relevant compound, has the potential to decrease the incidence of recurrent infections from persisters.
Introduction
Bacterial persisters are rare phenotypic variants with impressive abilities to survive antibiotic treatments. It has been hypothesized that persisters underlie the proclivity of biofilm infections to relapse (Lewis, 2007). The majority of hospital-treated infections are associated with biofilms, which suggests that persisters constitute a large burden on healthcare systems (Lewis, 2007). In general, persister tolerances have been attributed to inactivity of antibiotic primary targets, which serves to limit antibiotic-induced damage (Balaban, et al., 2013; Balaban, et al., 2004; Lewis, 2007; Maisonneuve and Gerdes, 2014), though several exceptions to this model exist (Dörr, et al., 2010; Völzing and Brynildsen, 2015; Wakamoto, et al., 2013). In a seminal study of persistence, Balaban and colleagues discovered two types of Escherichia coli persister based on growth-rate and formation period, which they designated as type I and type II persisters (Balaban, et al., 2004). Notably, type I persisters form from passage through stationary phase, whereas type II persisters are continuously generated during growth.
In a recent study, we examined stationary phase E. coli cultures, and found that respiration was necessary for type I persister formation (Orman and Brynildsen, 2015). We found that treatment of cultures with KCN or transfer to an anaerobic chamber immediately prior to the onset of stationary phase reduced type I persister formation up to 1,000-fold. Although this study suggested that respiration of non-growing bacterial populations was a potential target for anti-persister therapies, a therapeutically-relevant way to inhibit respiration was not explored.
Here we investigated the capacity of nitric oxide (NO), a well-known inhibitor of bacterial respiration (Mason, et al., 2009; Robinson and Brynildsen, 2015; Yu, et al., 1997), to impair type I persister formation in E. coli. In addition to the role of NO as a mammalian signaling molecule (Nathan, 1992), it is a potent antimicrobial produced by immune cells (Bowman, et al., 2011; Fang, 2004; Robinson, et al., 2014). Further, administration of exogenous NO with the use of nanoparticles, probiotic patches, and other technologies, has shown promise for the treatment of different infections (Friedman, et al., 2011; Jones, et al., 2010; Sulemankhil, et al., 2012; Sun, et al., 2012). Inspired by the therapeutic relevance of NO, we tested whether NO could prevent type I persister formation in stationary phase cultures of E. coli. We found that treatment of E. coli cultures with NO at the onset of stationary phase significantly reduced type I persister formation, decreased protein and RNA degradation in stationary phase, and produced bacterial populations that were more fit to synthesize protein and resume growth upon exposure to fresh nutrients than untreated controls, which mirror the previous results obtained with KCN and transfer to anaerobic conditions. Collectively, these data demonstrate that NO can inhibit type I persister formation in E. coli, and suggest that direct delivery of NO to infection sites prior to antibiotic exposure could improve treatment outcomes.
Materials and Methods
Bacterial strains, chemicals, and growth conditions
E. coli MG1655 strain was used in this study. MO001 has a chromosomally-integrated mCherry expression cassette and it was generated from E. coli MG1655 in a previous study (Orman and Brynildsen, 2013). A variant of the pQE-80L plasmid (Qiagen, Valencia, CA) was used to express GFP under the control of a synthetic T5 promoter (Orman and Brynildsen, 2015). The same plasmid was used to overexpress SodA and SodB as described previously (Orman and Brynildsen, 2015). Δhmp, ΔnorV, ΔnsrR, and ΔdksA were transferred to E. coli MG1655 from their respective mutants in the Keio collection using the standard P1 phage method (Baba, et al., 2006). ΔhmpΔnorV was generated by transferring ΔnorV to Δhmp background using the same P1 phage method. ΔkatEΔkatG was generated in a previous study (Adolfsen and Brynildsen, 2015). When needed, the kanamycin resistance gene (KanR) was removed using FLP recombinase (Datsenko and Wanner, 2000). All mutations and plasmid insertions were checked with PCR and/or DNA sequencing (Genewiz, South Plainfield, NJ).
NO donor (Z)-1-[N-(3-aminopropyl)-N-(3-ammoniopropyl)amino]diazen-1-ium-1,2-diolate, otherwise known as DPTA NONOate (DPTA), and the Nitrate/Nitrite Colorimetric Assay Kit were purchased from Cayman Chemical Company. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology. All other chemicals and reagents were purchased from Sigma Aldrich or Fisher Scientific. LB medium (10 g/L Tryptone, 5 g/L Yeast Extract, and 10 g/L NaCl) and LB-agar plates (LB + 15 g/L agar) were prepared from components and autoclaved for 30 min at 121°C to achieve sterilization. For selection (deletion mutants and plasmids), 50 μg/mL kanamycin was used. For persister assays, 200 μg/mL ampicillin or 5 μg/mL ofloxacin were used (Orman and Brynildsen, 2015). To induce GFP expression, 1mM IPTG was used. All chemical solutions were filter-sterilized with 0.22μm filters. The stock DPTA solutions (200 mM) were freshly prepared by dissolving DPTA powder in 10 mM NaOH solution. Unless stated otherwise, overnight cultures were prepared from a 25% glycerol, −80 °C stock in 2 mL LB medium in a test tube (glass and/or 17×100 mm 14 mL polypropylene tubes) and cultured aerobically at 37 °C with shaking (250 rpm) for 24 h. Then overnight cultures were diluted 1000-fold in 2 mL LB medium in a test tube and cultured at 37 °C with shaking. After 4 h, these cultures were treated with DPTA at the indicated concentrations. For controls, the cell cultures were treated with equal volumes of solvent (NaOH solution). The final concentration of NaOH in cultures was 0.15 mM. When necessary, cells at early stationary phase (t=4 h) were transferred to an anaerobic environment (Coy Hypoxic Glovebox with Anoxic Upgrade, Grass Lake, MI) to inhibit aerobic respiration, and cultured anaerobically (98% N2 and 2% H2) at 37 °C with shaking (250 rpm). When necessary, 40 mM NaNO3 and/or 3 mM DPTA were added to cell cultures before transfer to the anaerobic chamber. At t=24 h, cells from treated or untreated cultures were washed with fresh LB by centrifugation at 15,000 rpm (21,130xg, Eppendorf 5424) for 3 minutes, and diluted 100-fold in 1 mL LB in a test tube for persister, cell-division, and transcription/translation activity assays. When necessary, cell growth (OD600) was measured with a Synergy H1 Hybrid Multi-Mode microplate reader (BioTek, Winooski, VT).
Persister assay
DPTA-treated or untreated stationary phase cells (t=24 h) were washed to remove the chemicals and diluted 100-fold in 1 mL LB with 200 μg/mL ampicillin or 5 μg/mL ofloxacin in test tubes and incubated aerobically in a shaker at 37 °C and 250 rpm for 7 h. Just before antibiotic treatments, 10 μL of the samples from the cell cultures were serially diluted in phosphate buffered- saline (PBS), and 10 μL of these diluted samples were plated on LB agar to enumerate the initial concentration of colony formation units (CFU). During the antibiotic treatments, 100 μL samples at indicated time points (1, 3, 5 and 7 h) were mixed with 900 μL of PBS, pelleted by centrifugation at 15,000 rpm for 3 minutes and 900 μL of the supernatant was removed. This washing procedure was repeated three times to decrease the antibiotic concentrations to below their respective minimum inhibitory concentrations (MIC) (Andrews, 2001). Following the washing procedure, 10 μL of the samples from 100 μL cell suspensions were serially diluted in PBS and spotted on LB agar to enumerate CFUs. To improve the limit of detection, the remaining 90 μL sample was also plated on LB agar. The plates were incubated aerobically at 37 °C for 16 h before counting CFUs.
Cell division assay
An E. coli strain with an inducible mCherry cassette (MO001) was cultured with inducer (1mM IPTG) in both the overnight culture and the following growth period. Cells from both treated (DPTA) cultures and untreated controls were then washed to remove chemicals and diluted 100-fold in 1 mL LB without the inducer in a test tube. These cultures were incubated aerobically for 2.5 h at 37 °C and 250 rpm. At t=0 h and 2.5 h, cells were washed and suspended in PBS and analyzed with a flow cytometer to quantify non-growing cell population. Wild-type cells without mCherry expression system were used for negative controls. The gates of the non-growing cell population were determined using mCherry positive and negative cells as described previously (Orman and Brynildsen, 2013; Orman and Brynildsen, 2015).
Transcription/translation assay
An E. coli strain carrying a pQE-80L plasmid variant with an inducible gfp expression cassette (Orman and Brynildsen, 2015) was cultured in both the overnight culture and the following growth period without inducer. Cells from both cultures treated with DPTA and untreated controls were washed to remove the chemicals and diluted 100-fold in 1 mL LB in a test tube with inducer (1 mM IPTG). Cells were cultured aerobically at 37 °C and 250 rpm for 30 min. At indicated time points (0, 10, 20, and 30 min); cells were pelleted to remove the supernatant and suspended in PBS for analysis by flow cytometry.
For a negative control, we used MG1655 carrying the same plasmid but without an inducible gfp expression cassette. These cells did not fluoresce when introduced to fresh media with inducer (1mM IPTG). For a positive control, we used a culture of MG1655 with the inducible gfp expression cassette that was induced throughout the overnight. These cells were highly fluorescent when introduced to fresh media with inducer (Supplementary Figure S1).
As additional controls, we assessed whether GFP expression required IPTG and translation. To do this, DPTA-treated and untreated cells carrying a pQE-80L plasmid variant with an inducible gfp expression cassette were diluted in fresh media without inducer or fresh media with inducer and 100 μg/mL chloramphenicol. The GFP levels were monitored with flow cytometry for 30 min (Supplementary Figure S2).
Flow cytometry
Both GFP and mCherry levels were determined with an LSRII flow cytometer (BD Biosciences, San Jose, CA). Cells were exposed to a laser emitting at 488 nm for GFP and 561 nm for mCherry. Fluorescence was collected using green (525/50 nm) and red (610/20 nm) fluorescence bandpass filters. Data were acquired with FACSDiVa software (BD Biosciences, San Jose, CA). FlowJo (Tree Star Software, Ashland, OR) was used to analyze the data.
Protein degradation assay
An E. coli strain carrying pQE-80L plasmid variants with an inducible ssrA-tagged gfp was cultured in both the overnight culture and the following growth period with 1 mM IPTG. At t= 4h cells were washed to remove the IPTG and resuspended in 2 mL filter-sterilized spent media (without inducer) from WT cultures grown under the same conditions. Then, cells were treated with 3 mM DPTA, and cultured aerobically at 37 °C and 250 rpm. At indicated time points, cells were diluted to OD600 0.1 in PBS to measure GFP levels with a plate reader (485/515 excitation/emission). Background fluorescence was determined by using PBS without cells.
RNA isolation
Total RNA from the indicated cultures were purified with RNeasy extraction kits using the manufacturer’s protocol (Qiagen). Briefly, ~108–109 cells were lysed with lysozyme and treated with Proteinase K to inactivate nucleases. Cells were further mechanically disrupted with acid-washed glass beads (Sigma) to lyse the remaining intact stationary phase cells as described in the manufacturer’s protocol. Isolated total RNA samples were analyzed with a bioanalyzer using an RNA 6000 Nano kit (Agilent Technologies, Inc, Santa Clara, CA) in order to calculate the RNA integrity number (RIN). RIN provides a quantitative measure of the integrity of total RNA samples, based on a numbering system from 1 to 10, with 1 being the most degraded and 10 being the most intact. The RIN algorithm, which was introduced by Agilent Technologies, is a trained artificial neural network model that uses the most informative features of electrophoretic traces including the total RNA ratio, the heights of 18S and 28S peaks, the fast area ratio, and the height of the lower marker (Imbeaud, et al., 2005; Mueller, et al., 2004).
Dissolved oxygen measurements
The FireStingO2 fiber-optic O2 meter with the OXYROB 10-CL2 robust oxygen miniprobe (PyroScience, GmbH) was used to measure dissolved oxygen concentrations in cell cultures. Dissolved oxygen levels were reported as the percentage of dissolved oxygen with respect to cell-free media in equilibrium with the atmosphere. A detailed description of the method was provided in a previous study (Orman and Brynildsen, 2015). Briefly, samples were removed from the shaking incubator and the oxygen probe was inserted into the media at indicated time points. The oxygen levels in all cultures were reported exactly 2 min after insertion of the probe into cultures. As described previously (Orman and Brynildsen, 2015), these measurements reflect relative respiratory activity, rather than the oxygen concentrations in the shaking test tubes (measurements with this probe had to be conducted with non-shaking systems). According to the manufacturer, the maximum measuring range of the oxygen sensor is 0–45mg/L dissolved oxygen.
Nitrate and nitric oxide measurements
At early stationary phase (t=4 h), 40 mM NaNO3 were added into cultures, and then the cultures were treated with 3 mM DPTA or solvent and transferred to an anaerobic chamber. At t=24 h, the cell cultures were filter-sterilized, and the cell-free media were diluted 1000-fold in Nitrate/Nitrite Assay Buffer (Cayman Chemical, MI). Nitrate concentrations in diluted cell-free media were measured using a Nitrate/Nitrite Colorimetric Assay Kit (Cayman Chemical) according to manufacturer’s protocols.
To measure NO concentrations, cell cultures at early stationary phase (t=4 h) in test tubes were removed from the shaker and immersed in a stirred water bath that was maintained at 37°C. A sterilized magnetic stirring bar was added to the culture for mixing and cultures were treated with 3 mM DPTA. Control cultures (untreated) were treated with the solvent only. NO concentrations were monitored continuously using an ISO-NOP NO sensor (World Precision Instruments, Inc.). The measurement range of the sensor is 1 nM - 40 μM NO. The electrode was calibrated daily according to the manufacturer’s protocols.
Statistical Analysis
At least three biological replicates were performed for each experimental condition. Each data point depicts the mean value ± standard error. A two-tailed t-test with unequal variance was used for pairwise comparisons. The final time points of the persister data (kill curves) were utilized for statistical analysis. P-values ≤ 0.05 were considered significant.
Results
Nitric oxide treatment impairs formation of type I persisters
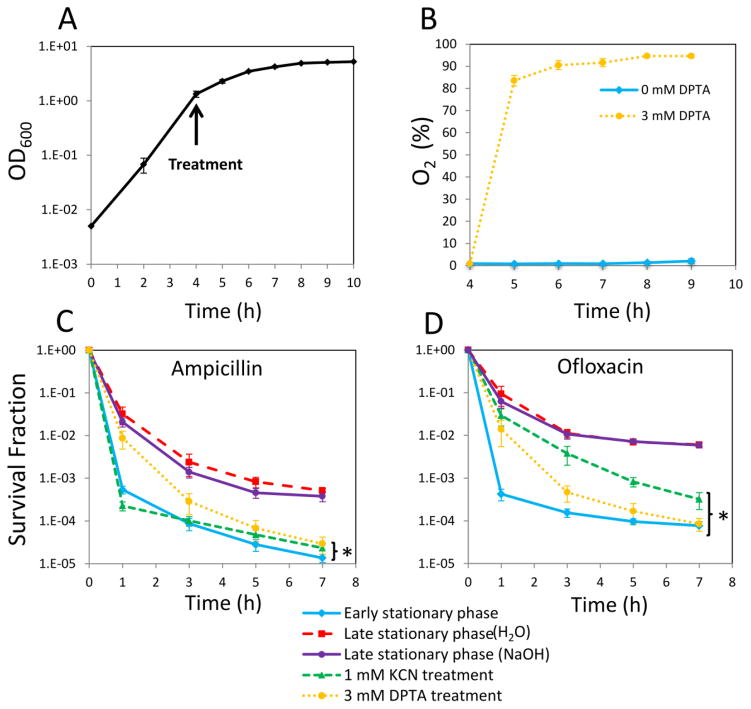
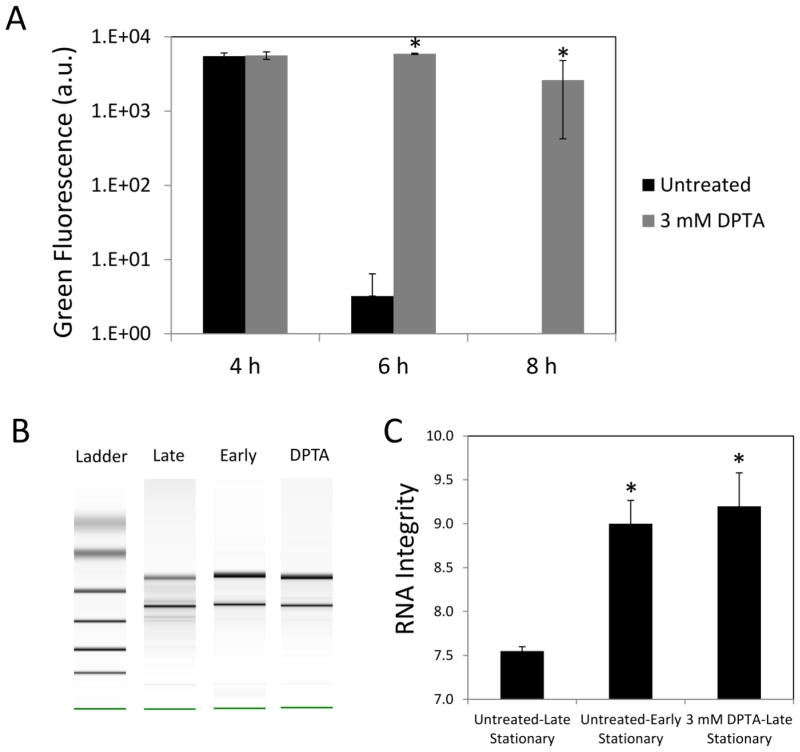
Stationary phase respiration is critical to the formation of type I persisters in E. coli (Orman and Brynildsen, 2015). When respiration was inhibited with KCN or transfer to an anaerobic environment upon entry into stationary phase, persister formation was significantly reduced (Orman and Brynildsen, 2015). Since one of the cytotoxic effects of NO is respiratory inhibition (Mason, et al., 2009; Robinson and Brynildsen, 2013; Robinson, et al., 2014; Yu, et al., 1997), we sought to test if NO treatment could inhibit type I persister formation. To do this, we used a similar experimental approach to that previously described (Orman and Brynildsen, 2015) with slight modifications. Herein, overnight cultures were diluted 1000-fold in fresh media to provide the same initial cell population before any treatment and grown until they reached early stationary phase (t=4 h) (Figure 1A), at which point cultures were treated with KCN, DPTA, or left untreated. DPTA is a NO donor that can effectively release NO under the conditions used here (Supplementary Figure S3). Similar to KCN (Orman and Brynildsen, 2015), NO can also inhibit the respiration (Figure 1B). Bacterial respiration in untreated cultures consumes oxygen at a rate that yields low oxygen concentrations in the media. However, as illustrated in Figure 1B, when respiration is inhibited (e.g., with NO), the oxygen concentration increases significantly and approaches to saturation. At t=24 h, inhibitors were removed and cells were diluted in fresh media with antibiotics to quantify the levels of type I persisters. Consistent with previous reports (Balaban, et al., 2004; Luidalepp, et al., 2011; Orman and Brynildsen, 2015), passage through stationary phase resulted in significant persister formation (comparison of early and late stationary phase persister levels, P-value < 0.05, t-test) (Figure 1CD). In addition, KCN treatment at 1 mM significantly reduced persister formation (Figure 1CD), which confirmed that the phenomenon described previously was also observed under the modified conditions used here (Methods). When cultures were treated with 3 mM DPTA at the onset of stationary phase both ampicillin and ofloxacin persister levels were significantly reduced compared to untreated controls (P-value < 0.05, t-test). Further, persister levels from NO-treated cultures were quantitatively comparable to those of KCN-treated and early stationary phase cultures (Figure 1CD). These data suggested that NO had the ability to impair persister formation during the stationary phase of E. coli cultures. To assess whether NO treatment at later times could reduce persister levels, we treated cultures with 3 mM DPTA at 4 h, 5 h, 6 h and 8 h after inoculation. NO treatment at 4 and 5 h significantly reduced both ampicillin and ofloxacin persister levels; however, treatments performed at t= 6 and 8 h did not (Supplementary Figure S4). This demonstrates that the timing of NO exposure is an important treatment parameter.
Figure 1. Impact of NO on respiration and type I persister formation.
A. Overnight cultures were diluted in fresh media, and cultured as described in Methods. Cell growth was monitored by measuring optical density at 600 nm (OD600). B. Percentages of dissolved oxygen concentrations with respect to saturated media in cultures with and without 3 mM DPTA treatment at t=4 h were quantified with an oxygen probe. C–D. Cell cultures at t=4 h were treated with 1 mM KCN or 3 mM DPTA. For controls, cell cultures were treated with equal volumes of solvent (0.15mM NaOH for DPTA, H2O for KCN). At t=24 h, cultures were washed to remove the chemical inhibitors, diluted (100-fold) in fresh LB, and treated with ampicillin or ofloxacin. Cell cultures at t=4 h (untreated early stationary phase) were similarly diluted and treated with ampicillin or ofloxacin in LB. Cell survival fractions were monitored for 7 h during the treatments. * indicates a statistical difference between control groups and DPTA-treated, KCN-treated, or early stationary phase cultures (P-value<0.05, t-test). The 7 hour time points of the survival data (final time points) were used for statistical analysis. At least three biological replicates were performed for each experimental condition. Each data point represents the mean value ± standard error.
Catalase inhibition, DksA modulation, and superoxide do not impact the ability of NO to impair type I persister formation
Although NO is a potent inhibitor of bacterial respiration (Figure 1B), its broad reactivity produces a range of biological effects (Bowman, et al., 2011; Robinson, et al., 2014). Several of those effects that could affect persister formation include inhibition of catalase activity (Brown, 1995), alteration of DksA activity (Henard, et al., 2014), and formation of peroxynitrite through reaction of NO with superoxide (Brunelli, et al., 1995). Since oxidative stress can increase persister levels (Vega, et al., 2012; Wu, et al., 2012) and NO impairs catalase activity, we treated an E. coli strain devoid of catalases (ΔkatEΔkatG) with DPTA at 4 h of growth, and measured type I persistence at 24 h. We observed that NO reduces type I persister levels of ΔkatEΔkatG to a similar extent as wild type (Supplementary Figure S5A), which indicated that inhibition of catalase activity by NO does not explain the phenomenon described here. Since removal of DksA lowers type I persister levels (Amato, et al., 2013; Orman and Brynildsen, 2015) and NO interferes with DksA function, we assessed whether NO could reduce persister formation in ΔdksA. We found that persister levels of ΔdksA were significantly lower than those of wild type (Supplementary Figure S5B) and that treatment with DPTA lowered ΔdksA even further; though, we note that the levels were found to be at or near the limit of detection, which impaired our ability to quantify the complete reduction in persistence (Supplementary Figure S5B). These data show that NO does not require DksA to reduce type I persister levels.
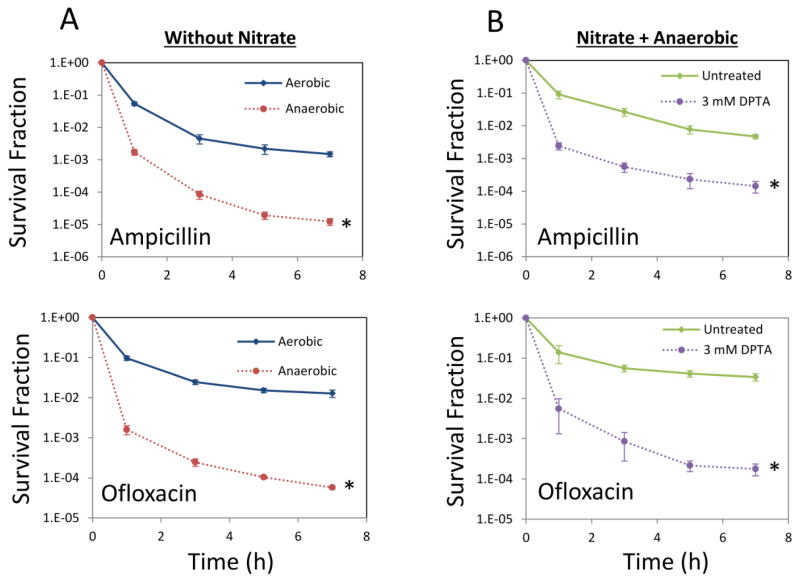
To assess whether superoxide or its reaction product peroxynitrite participate in the phenomenon reported here, we performed experiments under anaerobic conditions. Cultures were transferred to an anaerobic chamber at early stationary phase (t=4 h) and at t=24 h they were assayed for type I persistence. Consistent with our previous study (Orman and Brynildsen, 2015), in the absence of stationary phase respiration, type I persister levels were low, and when nitrate was provided to support anaerobic respiration, formation of type I persisters was restored (Figure 2AB). Further, when nitrate reductases were inhibited with NO under anaerobic conditions (Supplementary Figure S6), type I persister formation was significantly reduced (Figure 2B). Collectively, these results provide evidence that the ability of NO to inhibit type I persister formation does not depend on superoxide or peroxynitrite. To provide additional evidence for this conclusion, superoxide dismutases (sodA and sodB) were over-expressed starting at t= 0 h, cultures were treated with DPTA at t= 4 h, and persister levels were assessed at 24 h. As expected, overexpression of these antioxidant enzymes failed to alter persister levels (Supplementary Figure S7). We note that in our previous study (Orman and Brynildsen, 2015), the catalytic competencies of the over-expressed SODs were established by observing that they could eliminate the growth defect of an SOD-deficient mutant in an O2-dependent manner (Carlioz and Touati, 1986). Overall, these results discount a role for catalase inhibition, DksA modulation, and superoxide in the ability of NO to impair type I persister formation.
Figure 2. Persister levels in anaerobic cultures.
A. WT cell cultures at t=4 h were transferred to an anaerobic chamber without adding an electron acceptor and cultured anaerobically until t=24 h. For comparison, cells were cultured aerobically throughout the time course of the experiment. At t=24 h, cultures were diluted in fresh LB and treated with ampicillin or ofloxacin aerobically for 7 h. * indicates a statistical difference between aerobically and anaerobically grown cultures (P-value<0.05, t-test). B. NaNO3 (electron acceptor) at 40 mM was added to cultures at t=4 h. Then the cultures were treated with either 3 mM DPTA or the solvent (untreated), and transferred to an anaerobic chamber. At t=24 h, cultures were washed to remove the chemicals and diluted (100-fold) in fresh LB and treated with ampicillin or ofloxacin aerobically. Survival fractions were monitored for 7 h during the treatments. * indicates a statistical difference between control groups and DPTA treated cultures (P-value<0.05, t-test). The 7 hour time points of the survival data (final time points) were used for statistical analysis. At least three biological replicates were performed for each experimental condition. Each data point was denoted by mean value ± standard error.
Decreased persister levels correlate with decreased levels of non-growing cells
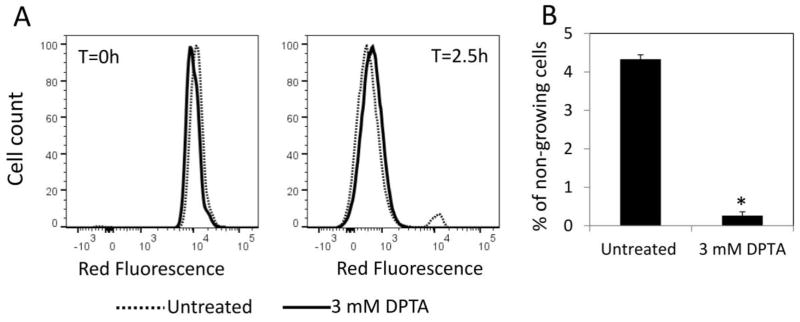
Previously we found that inhibiting stationary phase respiration impairs type I persister formation by producing bacterial populations that are more fit to rapidly resume cell division upon introduction into fresh media (Orman and Brynildsen, 2015). In comparison to untreated controls, respiratory-inhibited cultures exhibited less degradation of RNA and protein, synthesized protein more quickly when exposed to fresh nutrients, and contained far fewer cells that had difficulty resuming growth in new media (Orman and Brynildsen, 2015). We note that in earlier work, the majority of persisters in growing E. coli populations were found to arise from non-growing cells, which were those that failed to rapidly resume growth in fresh media (Orman and Brynildsen, 2013). In that study (Orman and Brynildsen, 2013), late stationary phase cells (24 h) were diluted in fresh media, grown for 2.5 h, and flow cytometry was applied to determine growing and non-growing cell subpopulations using a fluorescent protein dilution method (Roostalu, et al., 2008). Then those subpopulations were segregated with FACS, and persistence to ampicillin and ofloxacin was quantified, which demonstrated that non-growing cells were far more likely to give rise to persisters than their growing counterparts, and that the non-growing subpopulation was the main source of persisters in those cultures (Orman and Brynildsen, 2013). When stationary phase respiration was inhibited, we observed that the abundances of both non-growing cells and persisters were significantly reduced (Orman and Brynildsen, 2015). To test if NO impairs persister formation in a manner that was similar to other respiratory inhibitors (Orman and Brynildsen, 2015), we first tracked cell division using an E. coli strain with an inducible mCherry expression cassette integrated into its chromosome as described previously (Orman and Brynildsen, 2013). To monitor cell division, mCherry-positive stationary phase cells were diluted in fresh media, and the proportions of cells that retained high red fluorescence were quantified with flow cytometry. As depicted in Figure 3A, mCherry abundance in both DPTA-treated and untreated cells were very high upon introduction into fresh media (t=0 h). As cells divided, the red fluorescence of the majority of cells declined, except for small subpopulations that remained non-growing and thus maintained their abundance of mCherry (t=2.5 h) (Figure 3A). When the non-growing cell levels in DPTA-treated cultures were compared to untreated cultures, a significant decrease (~16 fold) was observed (P-value <0.05, t-test) (Figure 3B). We note that overexpressing a fluorescent protein (mCherry) under these conditions did not alter the effect of NO on persister formation (Supplementary Figure S8). This observation, which was consistent with the physiology previously described for KCN-treated and anaerobically-transferred cultures (Orman and Brynildsen, 2015), indicated a clear correlation between reduced persister levels in NO-treated cultures and lower abundances of non-growing cells.
Figure 3. Quantification of non-growing cell population after NO treatment.
Cell cultures grown in LB with 1mM IPTG to induce mCherry protein were treated with 3 mM DPTA at t=4 h. For control groups (untreated), solvent was added to cultures. At t=24 h, cells were washed and diluted in fresh LB without inducer, and cultured for 2.5 h. A. mCherry at single cell level was determined by flow cytometry. At t=2.5 h mCherry levels were reduced in growing cells due to cell division, whereas it remained constant in non-growing cell populations. B. Percentages of non-growing cell populations in untreated or DPTA-treated cultures were determined at t=2.5 h. * indicates a statistical difference between untreated and DPTA treated groups (P-value<0.05, t-test). At least three biological replicates were performed for each experimental condition. Each data point represents the mean value ± standard error.
NO treatment improves protein synthesis upon introduction into fresh media
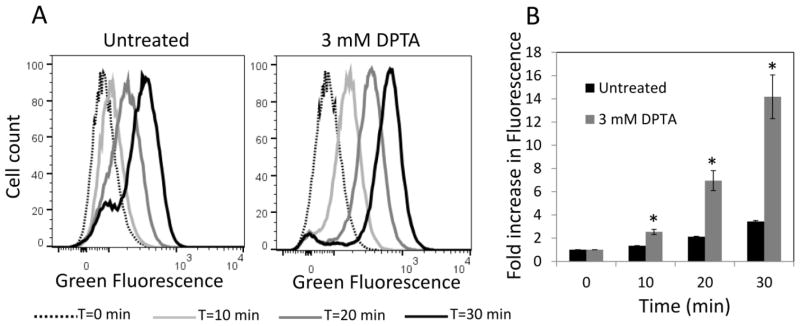
Next, we tested whether NO treatment improves the ability of cells to rapidly synthesize protein when exposed to fresh nutrients. We measured transcriptional and translational capabilities of DPTA-treated cultures shortly after inoculation into fresh media using E. coli that contained a pQE-80L plasmid variant expressing gfp from an inducible promoter (Orman and Brynildsen, 2015). Cultures were treated with DPTA upon entry into stationary phase, and then washed and diluted in fresh media with inducer (1 mM IPTG). As shown in Figure 4AB, NO-treated cells synthesized GFP much more rapidly than untreated controls shortly after inoculation (P-value <0.05, t-test). As control experiments, DPTA-treated and untreated cultures were diluted in fresh media without inducer or with inducer and 100 μg/mL of chloramphenicol. The GFP levels were monitored with flow cytometry, and we observed that cells were not capable of expressing GFP in the absence of inducer or in the presence of chloramphenicol, which inhibits translation (Supplementary Figure S2). We note that use of this plasmid-bearing strain did not change the impact of NO on persister formation (Supplementary Figure S9). Overall, these results indicate that NO treatment at the beginning of stationary phase produced cell populations with more robust transcription and translation capabilities when introduced into fresh media, which mirrors the physiology of populations treated with other respiratory inhibitors (Orman and Brynildsen, 2015).
Figure 4. Protein production upon inoculation in fresh media.
Cultures treated with 3 mM DPTA at t=4 h and untreated controls were grown until 24 h and then washed and diluted in fresh LB with inducer for GFP expression. A. GFP expression was monitored at indicated time points with flow cytometry. B. Fold increase in mean fluorescence values were plotted with respect to time. Fold increase has been quantified as the ratio of fluorescence at any time point to the fluorescence at t=0. * indicates a statistical difference between untreated and DPTA treated groups (P-value<0.05, t-test). At least three biological replicates were performed for each experimental condition. Each data point represents the mean value ± standard error.
NO treatment inhibits protein and RNA degradation during stationary phase
Previously, we demonstrated a correlation between respiration, self-digestion, and the formation of type I persisters (Orman and Brynildsen, 2015). Namely, when stationary phase respiration was inhibited, protein and RNA degradation were impaired and persister formation was reduced (Orman and Brynildsen, 2015). To determine if NO treatment inhibited protein degradation; we used an IPTG-inducible ssrA-tagged gfp (Orman and Brynildsen, 2015). To enhance the degradation of super-folder GFP, which is very stable, we added an ssrA-degradation tag to its C-terminus, which facilitates degradation by one of the major proteases in E. coli (ClpP) (Gottesman, et al., 1998). GFP was expressed until cells reached early stationary phase (t= 4 h), and then the inducer was removed and cultures were treated with DPTA. As shown in Figure 5A, treatment with 3 mM DPTA significantly reduced the degradation of GFP when compared to the untreated control. To demonstrate the functionality of this reporter, we further showed that GFP is stable in stationary phase cells without the degradation tag, and GFP-ssrA is also stable in a ΔclpP strain (Supplementary Figure S10). These experiments confirm that loss of GFP fluorescence in the experiments performed was caused by degradation by ClpP protease; though, we note that this assay does not report on global protease activity.
Figure 5. Impact of NO on protein and RNA degradation in stationary phase.
A. Cells carrying an IPTG-inducible, ssrA-tagged GFP were grown in the presence of inducer in both overnight and following cultures. At t=4 h, the inducer was removed, and the cultures were treated with 3 mM DPTA or left untreated. GFP levels were measured with a plate reader (Methods). Note that GFP in untreated cultures at t=8 h was not detectable. B–C. Total RNA were isolated from early stationary phase, DPTA-treated, and untreated late stationary phase cultures, and analyzed with a bioanalyzer. RNA integrity values range from 10 (intact) to 1 (totally degraded). For control groups (untreated), solvent was added to cultures. * indicates a statistical difference between untreated and DPTA treated groups (P-value<0.05, t-test). At least three biological replicates were performed for each experimental condition. Each data point represents the mean value ± standard error.
To test the effects of NO on RNA degradation, we analyzed the electrophoretic traces and RNA integrity (RIN) values obtained from early stationary phase, DPTA-treated late stationary phase, and untreated late stationary phase cultures (Figure 5BC). We observed that untreated, late stationary phase cells contained more significantly degraded RNA compared to early stationary phase or DPTA-treated late stationary phase cultures. These results confirmed that NO had a similar effect on protein and RNA degradation in stationary phase as other respiratory inhibitors (Orman and Brynildsen, 2015).
NO detoxification enzymes temper its effects on persister formation
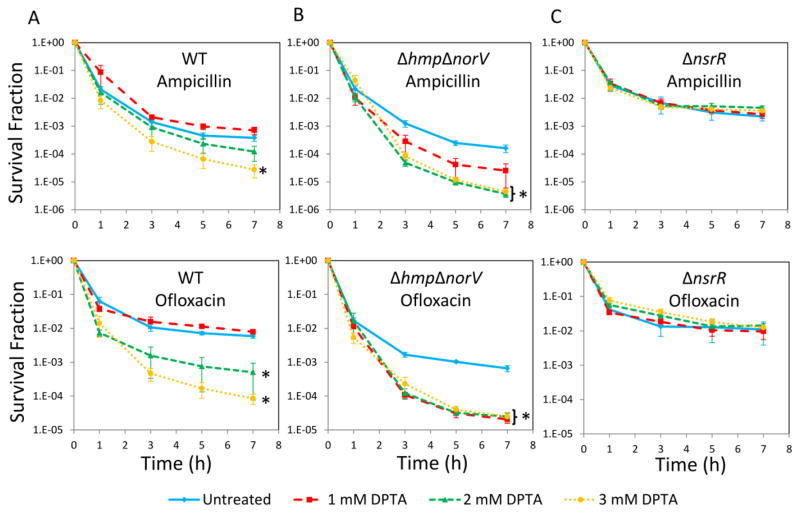
Since E. coli can metabolically detoxify NO with Hmp (NO dioxygenase) and NorV (NO reductase) (Gardner, et al., 1998; Gomes, et al., 2002; Robinson, et al., 2014), we hypothesized that the abundance of those enzymes might dictate the amount of NO required to impair type I persister formation. To test this hypothesis, we investigated the sensitivity of two E. coli mutants, one that was devoid of both Hmp and NorV, ΔhmpΔnorV (Gardner and Gardner, 2002; Gardner, et al., 2002; Robinson and Brynildsen, 2013), and another that constitutively expressed them, ΔnsrR (Bodenmiller and Spiro, 2006; Filenko, et al., 2007). When 1, 2, and 3mM DPTA were tested on WT, the effect of NO on persistence was found to be concentration dependent with the most significant reductions occurring at 3 mM DPTA (P-value <0.05, t-test), and 1mM DPTA resembled the untreated control (Figure 6A). When ΔhmpΔnorV was assayed, persister formation was significantly inhibited at concentrations as low as 1mM DPTA (Figure 6B), which demonstrated that the NO detoxification systems were attenuating the impact of NO on persister formation. When ΔnsrR was tested, all DPTA concentrations tested here failed to perturb persister formation (Figure 6C).
Figure 6. Persister levels of WT, ΔhmpΔnorV, and ΔnsrR.
Cell cultures at t=4 h were treated with DPTA at indicated concentrations. For control groups (untreated), solvent was added to cultures. At t=24 h, persister assays were performed. A. Survival fractions of WT were monitored for 7 h during ampicillin and ofloxacin treatments. B. Survival fractions of ΔhmpΔnorV were monitored for 7 h during ampicillin and ofloxacin treatments. C. Survival fractions of ΔnsrR were monitored for 7 h during ampicillin and ofloxacin treatments. * indicates a statistical difference between untreated and DPTA treated groups (P-value<0.05, t-test). The 7 hour time points of the survival data (final time points) were used for statistical analysis. At least three biological replicates were performed for each experimental condition. Each data point represents the mean value ± standard error.
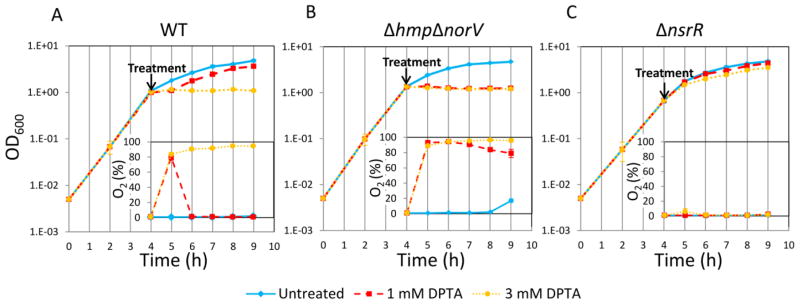
To complement the survival data, we tested the extent to which 1mM and 3mM DPTA impaired growth and respiration in early stationary phase cultures of WT, ΔhmpΔnorV, and ΔnsrR. As illustrated in Figure 7A, 3 mM DPTA treatment successfully inhibited oxygen utilization and cell growth in WT cultures, whereas growth and oxygen utilization after 1 mM DPTA treatment were only transiently inhibited. With ΔhmpΔnorV, both 1 and 3 mM DPTA significantly reduced the oxygen utilization and growth, whereas respiration and growth of ΔnsrR was not perturbed by either DPTA concentration (Figure 7BC). Overall, these results show that NO detoxification systems exert considerable influence on the ability NO to impair type I persister formation, which suggest that inhibitors of Hmp and NorV could synergize with direct NO delivery methods to reduce persister formation in nutrient-deprived bacterial populations.
Figure 7. Growth and oxygen consumption of WT, ΔhmpΔnorV, and ΔnsrR following NO treatment.
Cell cultures at t=4 h were treated with DPTA at indicated concentrations. A. Cell growth and oxygen utilization of WT were monitored at indicated time points. B. Cell growth and oxygen utilization of ΔhmpΔnorV were monitored at indicated time points. C. Cell growth and oxygen utilization of ΔnsrR were monitored at indicated time points. At least three biological replicates were performed for each experimental condition. Each data point represents the mean value ± standard error.
Discussion
Type I persisters were first described by Balaban and colleagues after analysis of the growth and ampicillin-induced death of the hipA7 high persistent mutant (Balaban, et al., 2004). Type I persisters are a pre-existing subpopulation that are generated by passage through stationary phase, and when introduced into fresh media they revert back to normal physiology and resume replication with a characteristic time scale that exceeds that of normal cells (Balaban, et al., 2004; Fridman, et al., 2014; Levin-Reisman, et al., 2010). In addition to type I persisters, which are found in growth-arrested states, Balaban and colleagues described type II persisters based on the physiology of the hipQ mutant, which are cells that grow very slowly compared to normal cells (~order of magnitude less) (Balaban, et al., 2004). Further, Balaban and colleagues demonstrated that wild-type populations contained both persister types, and their results depicted that type I persisters dominate the persister pool at the point when cultures are exposed to fresh media (Balaban, et al., 2004).
Type I persisters are enumerated by introduction of bacterial populations into fresh media containing antibiotics, and evidence has suggested that it is the timescale of awakening that is critical for their survival. For example, Joers and colleagues inoculated the same stationary-phase culture into different fresh media and found that type I persister levels were related to the rates at which populations resumed cell division (Joers, et al., 2010). In a different study by Fridman and colleagues, where evolution experiments were performed, stationary phase cultures were introduced into fresh media with ampicillin for defined periods of time, followed by removal of the antibiotic, growth to stationary phase, and repetition of the assay (Fridman, et al., 2014). They observed that the characteristic time of cells to resume cell division was tuned in evolved populations to the time period of antibiotic exposure (Fridman, et al., 2014). Collectively, these and other studies suggest that an effective means to eliminate type I persisters is to increase the ability bacteria to resume replication upon exposure to fresh nutrients (Fridman, et al., 2014; Joers, et al., 2010; Orman and Brynildsen, 2013; Orman and Brynildsen, 2015; Roostalu, et al., 2008).
In previous work, we discovered that type I persisters mainly originated from stationary phase cells with high levels of redox activity (Orman and Brynildsen, 2015). In addition, we demonstrated that inhibiting cellular respiration with either chemical (KCN), environmental (anaerobic), or genetic means (Δmdh, ΔsdhC, ΔsucB, ΔubiF) largely prevented type I persister formation (Orman and Brynildsen, 2015). The evidence we obtained from our previous work and current study suggested that respiration is critical to persister formation during stationary phase. Respiration is powered by reducing equivalents, and during stationary phase reducing power is primarily derived from accumulated acetate and the digestion of endogenous cellular components, such as ribosomes and proteins (Gonidakis, et al., 2010; Nyström, 2004; Orman and Brynildsen, 2015). We have shown that cultures respiring in stationary phase are less fit to rapidly resume translation when presented with fresh nutrients and that they contain a significant subpopulation of cells that fail to resume cell division (primary source of type I persisters). Using KCN, transfer to anaerobic conditions, and NO, we observed a striking correlation between respiratory inhibition and prevention of RNA and protein degradation that occurs in stationary phase. We postulate that continued respiration provides a metabolic demand for reducing equivalents, which is met in part by degradation of RNA and proteins. This self-digestion produces cells that are less fit to resume growth when presented with fresh media, and the associated lag protects them from antibiotics that are present in the media. When respiration is inhibited, that metabolic demand is eliminated, cells reduce their degradation of RNA and proteins, and thereby, when fresh media is provided, they rapidly resume growth and die if antibiotics are present.
Unlike KCN treatment or transfer to anaerobic conditions, NO treatment is more therapeutically-relevant. NO is a critical antimicrobial used by the human immune system (Hibbs, et al., 1988), and exogenous administration of NO to infection sites using a number of different delivery mechanisms has produced promising results (Friedman, et al., 2011; Jones, et al., 2010; Robinson, et al., 2014; Sulemankhil, et al., 2012; Sun, et al., 2012) This molecule with diverse functions easily diffuses through the bacterial membranes and reacts with iron-sulfur clusters, superoxide, and oxygen, whereas its oxidized forms damage thiols, DNA, and residues on proteins (Robinson, et al., 2014). This reactivity provides NO with an ability to inhibit or disrupt a diverse set of enzymes and regulatory proteins, which include catalases, respiratory cytochromes, and bacterial reductases (Brown, 1995; Brunelli, et al., 1995; Henard, et al., 2014; Mason, et al., 2009). Such pleotropic effects lead to a complex relationship between NO and antibiotic tolerance. For instance, Gusarov and co-workers demonstrated that NO generated endogenously by bacterial NOSs increase the tolerance of certain pathogens, such as Bacillus anthracis and Staphylococcus aureus, to a broad spectrum of antibiotics (Gusarov, et al., 2009). McCollister and co-workers showed that repression of aerobic respiration by NO reduced aminoglycoside uptake and led to tolerance in several bacteria including Salmonella enterica, Pseudomonas aeruginosa, and Staphylococcus aureus (McCollister, et al., 2011). Whereas, here we demonstrated that treatment of E. coli cultures with NO upon the onset of nutrient depirvation leads to an impressive reduction in type I persistence. Further, we showed that the timing of NO exposure was an important treatment parameter for the reduction of persister levels. Given such complexity and the differences that exist between in vitro and in vivo settings, additional studies, such as those involving animal models, are needed to assess the potential of NO to reduce antibiotic persistence within clinical settings.
Supplementary Material
Highlights.
NO• treatment at the onset of stationary phase reduces persister levels.
NO• inhibits respiration and self-digestion during stationary phase.
NO• yields a population with robust protein synthesis and cell division abilities.
Acknowledgments
We are grateful to Christina J. DeCoste, John J. Grady, Jessica B. Wiggins, and Jonathan L. Robinson for their technical assistance, and thank the National BioResource Project (NIG, Japan) for their distribution of the Keio collection. This research was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R21AI115075), the Department of the Army (W81XWH-12-2-0138), and Princeton University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, Department of the Army, or Princeton University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that no competing interests exist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsen KJ, Brynildsen MP. A Kinetic Platform to Determine the Fate of Hydrogen Peroxide in Escherichia coli. Plos Computational Biology. 2015;11 doi: 10.1371/journal.pcbi.1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato SM, Orman MA, Brynildsen MP. Metabolic Control of Persister Formation in Escherichia coli. Molecular Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. Journal of Antimicrobial Chemotherapy. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2006;2:11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nature Reviews Microbiology. 2013;11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. Journal of Bacteriology. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman LAH, McLean S, Poole RK, Fukuto JM. The Diversity of Microbial Responses to Nitric Oxide and Agents of Nitrosative Stress: Close Cousins but Not Identical Twins. In: Poole RK, editor. In Advances in Microbial Physiology. Vol. 59. London: Academic Press Ltd-Elsevier Science Ltd; 2011. pp. 135–219. [DOI] [PubMed] [Google Scholar]
- Brown GC. Reversible binding and inhibition of catalase by nitric-oxide. European Journal of Biochemistry. 1995;232:188–191. doi: 10.1111/j.1432-1033.1995.tb20798.x. [DOI] [PubMed] [Google Scholar]
- Brunelli L, Crow JP, Beckman JS. The comparative toxicity of nitric-oxide and peroxynitrite to escherichia-coli. Archives of Biochemistry and Biophysics. 1995;316:327–334. doi: 10.1006/abbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? The EMBO Journal. 1986;5:623. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS biology. 2010;8:427. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nature Reviews Microbiology. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- Filenko N, Spiro S, Browning DF, Squire D, Overton TW, Cole J, Constantinidou C. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. Journal of Bacteriology. 2007;189:4410–4417. doi: 10.1128/JB.00080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- Friedman AJ, Blecher K, Schairer D, Tuckman-Vernon C, Nacharaju P, Sanchez D, Gialanella P, Martinez LR, Friedman JM, Nosanchuk JD. Improved antimicrobial efficacy with nitric oxide releasing nanoparticle generated S-nitrosoglutathione. Nitric Oxide-Biology and Chemistry. 2011;25:381–386. doi: 10.1016/j.niox.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Gardner AM, Gardner PR. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli - Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. Journal of Biological Chemistry. 2002;277:8166–8171. doi: 10.1074/jbc.M110470200. [DOI] [PubMed] [Google Scholar]
- Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification Escherichia coli. Journal of Biological Chemistry. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: An enzymic function for flavohemoglobin. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CM, Giuffre A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. A novel type of nitric-oxide reductase - Escherichia coli flavorubredoxin. Journal of Biological Chemistry. 2002;277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- Gonidakis S, Finkel SE, Longo VD. Genome-wide screen identifies Escherichia coli TCA-cycle-related mutants with extended chronological lifespan dependent on acetate metabolism and the hypoxia-inducible transcription factor ArcA. Aging Cell. 2010;9:868–881. doi: 10.1111/j.1474-9726.2010.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Development. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous Nitric Oxide Protects Bacteria Against a Wide Spectrum of Antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henard CA, Tapscott T, Crawford MA, Husain M, Doulias PT, Porwollik S, Liu L, McClelland M, Ischiropoulos H, Vazquez-Torres A. The 4-cysteine zinc-finger motif of the RNA polymerase regulator DksA serves as a thiol switch for sensing oxidative and nitrosative stress. Molecular Microbiology. 2014;91:790–804. doi: 10.1111/mmi.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs JB, Taintor RR, Vavrin Z, Rachlin EM. Nitric-oxide - a cyto-toxic activated macrophage effector molecule. Biochemical and Biophysical Research Communications. 1988;157:87–94. doi: 10.1016/s0006-291x(88)80015-9. [DOI] [PubMed] [Google Scholar]
- Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic acids research. 2005;33:e56–e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joers A, Kaldalu N, Tenson T. The Frequency of Persisters in Escherichia coli Reflects the Kinetics of Awakening from Dormancy. Journal of Bacteriology. 2010;192:3379–3384. doi: 10.1128/JB.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Ganopolsky JG, Labbé A, Prakash S. A novel nitric oxide producing probiotic patch and its antimicrobial efficacy: preparation and in vitro analysis. Applied microbiology and biotechnology. 2010;87:509–516. doi: 10.1007/s00253-010-2490-x. [DOI] [PubMed] [Google Scholar]
- Levin-Reisman I, Gefen O, Fridman O, Ronin I, Shwa D, Sheftel H, Balaban NQ. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nature Methods. 2010;7:737–U100. doi: 10.1038/nmeth.1485. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nature Reviews Microbiology. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Luidalepp H, Joers A, Kaldalu N, Tenson T. Age of Inoculum Strongly Influences Persister Frequency and Can Mask Effects of Mutations Implicated in Altered Persistence. Journal of Bacteriology. 2011;193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nature Chemical Biology. 2009;5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- McCollister BD, Hoffman M, Husain M, Vazquez-Torres A. Nitric Oxide Protects Bacteria from Aminoglycosides by Blocking the Energy-Dependent Phases of Drug Uptake. Antimicrobial Agents and Chemotherapy. 2011;55:2189–2196. doi: 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller O, Lightfoot S, Schroeder A. RNA integrity number (RIN)–standardization of RNA quality control. Agilent Application Note, Publication. 2004:1–8. [Google Scholar]
- Nathan C. Nitric-oxide as a secretory product of mammalian-cells. Faseb Journal. 1992;6:3051–3064. [PubMed] [Google Scholar]
- Nyström T. Stationary-phase physiology. Annual Review of Microbiology. 2004;58:161–181. doi: 10.1146/annurev.micro.58.030603.123818. [DOI] [PubMed] [Google Scholar]
- Orman MA, Brynildsen MP. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrobial agents and chemotherapy. 2013;57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman MA, Brynildsen MP. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nature communications. 2015;6 doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Adolfsen KJ, Brynildsen MP. Deciphering nitric oxide stress in bacteria with quantitative modeling. Current Opinion in Microbiology. 2014;19:16–24. doi: 10.1016/j.mib.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Brynildsen MP. A Kinetic Platform to Determine the Fate of Nitric Oxide in Escherichia coli. Plos Computational Biology. 2013;9:19. doi: 10.1371/journal.pcbi.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Brynildsen MP. An ensemble-guided approach identifies ClpP as a major regulator of transcript levels in nitric oxide-stressed Escherichia coli. Metabolic engineering. 2015;31:22–34. doi: 10.1016/j.ymben.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Miller RV, Brynildsen MP. Model-driven identification of dosing regimens that maximize the antimicrobial activity of nitric oxide. Metabolic Engineering Communications. 2014;1:12–18. doi: 10.1016/j.meteno.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Joers A, Luidalepp H, Kaldalu N, Tenson T. Cell division in Escherichia coli cultures monitored at single cell resolution. Bmc Microbiology. 2008;8:14. doi: 10.1186/1471-2180-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulemankhil I, Ganopolsky JG, Dieni CA, Dan AF, Jones ML, Prakash S. Prevention and Treatment of Virulent Bacterial Biofilms with an Enzymatic Nitric Oxide-Releasing Dressing. Antimicrobial Agents and Chemotherapy. 2012;56:6095–6103. doi: 10.1128/AAC.01173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Slomberg DL, Chudasama SL, Lu Y, Schoenfisch MH. Nitric Oxide-Releasing Dendrimers as Antibacterial Agents. Biomacromolecules. 2012;13:3343–3354. doi: 10.1021/bm301109c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nature Chemical Biology. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völzing KG, Brynildsen MP. Stationary-Phase Persisters to Ofloxacin Sustain DNA Damage and Require Repair Systems Only during Recovery. mBio. 2015;6:e00731–00715. doi: 10.1128/mBio.00731-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- Wu YX, Vulic M, Keren I, Lewis K. Role of Oxidative Stress in Persister Tolerance. Antimicrobial Agents and Chemotherapy. 2012;56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Sato EF, Nagata K, Nishikawa M, Kashiba M, Arakawa T, Kobayashi K, Tamura T, Inoue M. Oxygen-dependent regulation of the respiration and growth of Escherichia coli by nitric oxide. Febs Letters. 1997;409:161–165. doi: 10.1016/s0014-5793(97)00494-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.