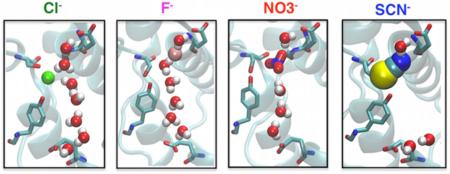
Abstract
Cl−/H+ transporters of the CLC superfamily form a ubiquitous class of membrane proteins that catalyze stoichiometrically coupled exchange of Cl− and H+ across biological membranes. CLC transporters exchange H+ for halides and certain polyatomic anions, but exclude cations, F−, and larger physiological anions, such as and . Despite comparable transport rates of different anions, the H+ coupling in CLC transporters varies significantly depending on the chemical nature of the transported anion. Although the molecular mechanism of exchange remains unknown, studies on bacterial ClC-ec1 transporter revealed that Cl− binding to the central anion-binding site (Scen) is crucial for the anion-coupled H+ transport. Here, we show that Cl−, F−, , and SCN− display distinct binding coordinations at the Scen site and are hydrated in different manners. Consistent with the observation of differential bindings, ClC-ec1 exhibits markedly variable ability to support the formation of the transient water wires, which are necessary to support the connection of the two H+ transfer sites (Gluin and Gluex), in the presence of different anions. While continuous water wires are frequently observed in the presence of physiologically transported Cl−, binding of F− or leads to the formation of pseudo-water-wires that are substantially different from the wires formed with Cl−. Binding of SCN−, however, eliminates the water wires altogether. These findings provide structural details of anion binding in ClC-ec1 and reveal a putative atomic-level mechanism for the decoupling of H+ transport to the transport of anions other than Cl−.
INTRODUCTION
The chloride channel (CLC) proteins1,2 constitute a superfamily of membrane transport proteins ubiquitous across species ranging from bacteria to mammals, where they play central roles in a broad range of fundamental biological processes.1,3 The CLC superfamily was originally thought to include only Cl− channels that catalyze passive diffusion of ions, but it is now well established to include transporters that catalyze the transmembrane exchange of Cl− for H+ with a 2:1 stoichiometry.4–6 One intriguing transport property of CLCs—both channels and transporters—is their ability to conduct various anionic species,7–12 namely, Cl−, Br−, I−, , and SCN−, with the permeability ratios varying within a factor of 10 for different anions and different CLC proteins.7–15
Characterizing the interaction of different permeant anions is of special significance in mechanistic studies of CLC transporters, due to widely different levels of H+ coupling, or complete lack thereof, among different anions. While Cl−, Br−, I−, , and SCN− all can pass through the anion selectivity filter with similar efficiencies, they exhibit strikingly different couplings to H+. In ClC-ec1, a prokaryotic homologue representing the best structurally and biophysically characterized CLC transporter, Cl− is transported with strict 2Cl−/1H+ stoichiometry, whereas shows a weaker coupling of 7−10 for each H+, and SCN− is transported without any measurable H+ transport, i.e., complete lack of coupling.12 It is notable that the 2Cl−/1H+ stoichiometry has been shown not only for bacterial ClC-ec1, but also for cyanobacterial syCLC,16 algal cmCLC,17 mammalian endosomal ClC-4, and ClC-5,4,6 and mammalian lysosomal ClC-7.18 Moreover, anion-dependent uncoupling has also been observed in ClC-4 and ClC-5, which show less H+ countertransport with , and almost complete loss of H+ transport with SCN−.14,15,19
Characterizing the interaction of the impermeant anion F− is also of interest. For many years, it was thought that the F− ion's lack of detectable permeation through CLC transporters20 and channels7,21 was due to the ion's strong hydration. However, a recent study demonstrated that the dehydrated F− ion binds in the anion-permeation pathway with similar affinity to Cl−.22 While crystallography has pinpointed binding interactions,22 an understanding of the dynamics of these interactions will add critical detail to our understanding of the mechanism by which F− inhibits the transport cycle. Worth noting, CLCs from a recently discovered prokaryotic clade, named CLCF, function as F−/H+ exchangers.23–25 These homologues are phylogenetically distant from the canonical CLCs: they share only ~20% sequence identity even to other prokaryotic CLCs, and they lack key signature-sequence residues. Structures have not yet been determined for these unique CLCs. The focus of the work presented here is on the interaction of F− with the canonical CLC transporters.
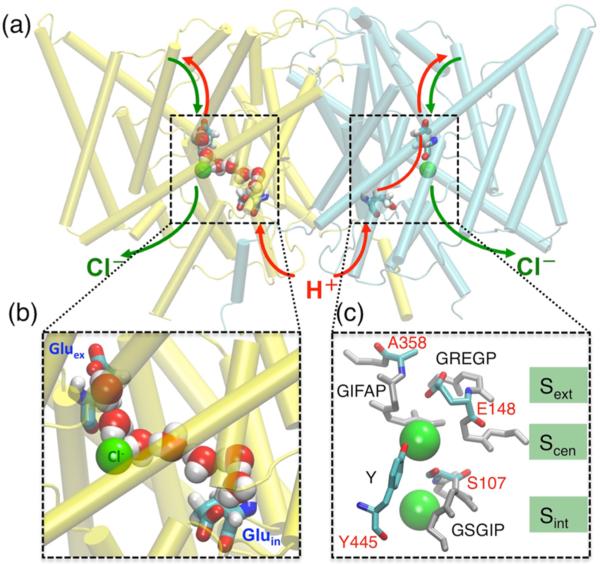
ClC-ec1, like all canonical CLCs, is a homodimeric protein in which each subunit operates independently (Figure 1(a)). The Cl− pathway is characterized by an hourglass-shaped selectivity filter with two aqueous vestibules extending from the extracellular and intracellular sides of the membrane and three anion binding sites.26 These anion binding sites are formed by four conserved motifs spread out in sequence space, namely, (ClC-ec1 numbering) GSGIP (106–110), GREGP (146–150), GIFAP (355–359), and Y445 (Figure 1(c)). The external anion-binding site (named Sext) can be occupied either by anion or by the deprotonated carboxylate of E148, a conserved residue in the GREGP motif.26 This residue, which is known as “Gluex”, obstructs the extracellular end of the anion-permeation pathway when positioned at Sext, as seen in the wild-type crystal structure (Figure 1(c)). When Gluex is neutralized by protonation or mutation, the side chain swings upward and Sext becomes occupied by Cl−, which comes in contact exclusively with backbone nitrogen atoms of motif-residues F357, A358, R147, E148, and G149.27 At the central anion-binding site (Scen), the ion is coordinated by backbone nitrogen atoms of I356 and F357, and by the side-chain oxygen atoms of S107 and Y445 (Figure 1(c)). Lastly, the anion at the internal site (Sint) is coordinated by backbone nitrogen atoms of S107 and G108 on one side, and exposed to the intracellular solution on the other side (Figure 1(c)).
Figure 1.
Key structural and functional aspects of CLC antiporters. (a) Cartoon representation of the ClC-ec1 homodimer (extracellular side on top), with the two identical subunits shown in yellow and cyan, respectively. Approximate Cl− and H+ permeation pathways are indicated by green and red arrows, respectively. The H+ pathway deviates from the Cl− pathway based on the identification of two key residues, Gluex and Gluin (see panel (b)), required for H+ transport. The Cl− ions bound at Scen are represented as green spheres. (b) Representative structure of the most common water wire connecting Gluex and Gluin observed in MD simulations of Cl−-bound ClC-ec1.29 (c) The selectivity filter and the three Cl− binding sites (Sext, Scen, and Sint) formed by the four highly conserved motifs (GSGIP, GREGP, GIFAP, and Y). In this structure (WT, PDB 1OTS), Sext is occupied by residue E148 (Gluex), and Scen and Sint are occupied by Cl− ions (green spheres). Residues forming the main constriction points (E148/A358 and S107/Y445) that occlude Cl− at Scen from the extracellular and intracellular solutions are drawn in color and individually labeled.
Structural and functional studies on ClC-ec1 indicate that Scen has unique characteristics worthy of special attention.12,28,29 Specifically, experimental and modeling studies have demonstrated a strong connection between Cl− occupancy of this site and Cl−/H+ coupling,12,28,29 although the exact molecular mechanism of coupled anion/H+ exchange remains unresolved. Scen is isolated from the extracellular and intracellular aqueous solutions by the structures containing E148 and S107/Y445, respectively (Figure 1(c)). Electrophysiological studies showed that substitution of small residues at Y445 leads to a loss of H+ coupling to Cl− transport,28,30 which strikingly correlates to the decrease of apparent halide occupancy at Scen measured crystallographically.28 The parallel loss of electrophysiologically measured H+ coupling and crystallographically determined Scen occupancy is also observed in wild-type (Y445-containing) ClC-ec1 when Cl− is replaced by small nonhalide anions.12 For example, X-ray crystallographic analysis of ClC-ec1 demonstrated that SeCN− (the crystallographic analog of the uncoupling anion SCN−) does not detectably occupy the Scen binding site.12 Presumably, the partial uncoupler may partially occupy this site; however, the crystallographic resolution of ClC-ec1 structures are too low to unambiguously determine occupancy. The impermeant F− can be discerned crystallographically at the Scen binding site. Its strong H+-bond to the protonated Gluex (mimicked by a Gluex–Gln mutation) provides a nice rationale for how this ion can halt the transport cycle.22 However, there may be additional effects of the F− ion not detectable in static crystal structures. To fully understand the relationship between anion binding and H+ transport (and therefore the mechanism of coupling itself) we need to develop a deeper understanding of the detailed atomic interactions at the Scen binding site.
Elucidating the degree of H+ coupling to different anions in ClC-ec1 depends on the understanding of the H+ transport pathway across the membrane. In contrast to the well-defined Cl− transport pathway,26,28,31 the H+ pathway remains relatively under-investigated. Two glutamate residues, E148 (Gluex) and E203 (Gluin), have been identified as critical H+ transfer sites in ClC-ec15,27,30,32–34 (Figure 1(a)). These two H+ sites, however, are separated by a ~15-Å hydrophobic region within the central part of ClC-ec1 void of any polar/charged groups capable of supporting H+ transfer.
Previous studies examining the H+ transport mechanism suggested that water networks occupying the hydrophobic regions of proteins could be utilized as H+ translocation pathways,35–40 most prominently in bacteriorhodopsin41 and other bioenergetic membrane proteins,42–44 as well as calcium pumps.45 Wang and Voth proposed a transient water-mediated H+ transport in CLCs through a pathway connecting E148 and E203, and successfully showed the H+ propagation along such a pathway using multistate empirical valence bound (MS-EVB) simulations.40 Ko and Jo reported spontaneous formation of a continuous water network that links E203 with E148 using MD simulations.46 A similar water network was captured by Cheng and Coalson in their simulations of a eukaryotic CLC transporter homologue CmCLC when E148 was protonated and occupied Scen.17,47 Using extended molecular dynamics (MD) simulations we showed that water wires that form transiently can connect Gluex and Gluin across this void (Figure 1(b)).29 We provided evidence for the role of these water wires in the H+ transport mechanism by identifying a residue that is conformationally coupled to water-wire formation and experimentally demonstrating the impact of its mutations. We additionally demonstrated the importance of the Cl− ion bound at the Scen site for stabilizing the water wires, thereby proposing a putative mechanism underlying the coupling of the two ions.29
In the present study, we investigate the binding mode and dynamics of various anions transported by CLCs. We examine through multiple extended MD simulations (350–400 ns for each) the structure and dynamics of ClC-ec1 in the presence of different anionic species, namely, Cl−, F−, , and SCN− bound at Scen. The different anions adopt distinct binding coordinations and hydration patterns, and, more importantly, exhibit differential abilities to support the formation of water wires between the H+ transfer sites (Gluin and Gluex). In contrast to the continuous water wires observed in the presence of Cl−, the wires formed in the presence of F− or directly incorporate the anions (pseudo-water-wires). This intervention by the anion may reduce the ability of the wire to conduct H+. Binding of SCN−, however, eliminates the water wires altogether. These findings explain the variable H+ coupling measured experimentally for these four representative anions, including fully coupled (Cl−), intermediately coupled (), absolutely uncoupled (SCN−), as well as impermeant (F−). Our results highlight the importance of anion binding for coupled H+ transport in ClC-ec1, thus providing deeper mechanistic insight into the function of CLC proteins.
MATERIALS AND METHODS
Simulation Protocols and Systems
The ClC-ec1 crystal structure (PDB ID:1OTS, solved at 2.51 Å),27 including all crystal water molecules, was used for the MD simulations. The program DOWSER48 was used to guess additional water molecules within the protein. DOWSER placed 49 additional water molecules (referred to as “solvation” water molecules, hereafter) in energetically favorable positions. As suggested by multiple computational studies40,49,50 using various force fields, one additional molecule was placed between the carboxylate group of E148 and the anion at Scen in each subunit, in order to stabilize the two negative charges in close proximity. For the solvation water molecules (i.e., those added by DOWSER), those in the central hydrophobic lumen of the protein (referred to as the “hydrophobic region”, hereafter) were removed prior the simulations to ensure that the hydration of the hydrophobic region is not artificially biased by our initial setup. E148 and E203 were modeled in their deprotonated (charged) forms, while E113 was protonated according to the thorough pKa analysis on titratable residues of the protein using Poison-Boltzmann electrostatic calculations.51 The protein was embedded into a POPE lipid bilayer, fully solvated with TIP3P water52 and buffered in 0.15 M NaCl. The resulting system, contained in a 105 × 105 × 110 Å3 simulation box comprising of ~110 000 atoms.
All MD simulations were carried out with NAMD 2.953 using the CHARMM-CMAP54 and CHARMM3655 force fields to model the proteins and lipids, respectively. The particle mesh Ewald (PME) method56 was used to calculate long-range electrostatic interactions every 4 fs. A smoothing function was employed for van der Waals and short-range electrostatic interactions at a distance of 10 Å with a cutoff of 12 Å. The bonded interactions and the short-range nonbonded interactions were calculated every 2 fs. The pairs of atoms whose interactions needed to be evaluated (neighbor list) were updated every 20 fs. A cutoff (13.5 Å) slightly longer than the nonbonded cutoff was applied to search for the atom pairs. All simulation systems were subjected to Langevin dynamics and the Nosé–Hoover Langevin piston method57,58 for constant pressure (P = 1 atm) and temperature (T = 310 K) (NPT).
ClC-ec1 systems with either Cl−, F−, , or SCN− bound at Scen in both subunits were simulated. The simulation system of Cl−-bound ClC-ec1 was adopted from our previous study.29 The systems with the other anions were generated by replacing the Cl− at the Scen site with either F−, (N replacing Cl−), or SCN− (C replacing Cl−). All simulation systems included ClC-ec1 dimers, therefore providing two independent copies of the anion-bound systems to be examined (we note that the two subunits in the crystal structure of ClC-ec1 are not identical). For each of these four systems, 5000 steps of energy minimization were performed, followed by an initial equilibration of 1 ns, during which the protein atoms and oxygen atoms of the crystallographic water molecules were positionally restrained (k = 2 kcal/mol/Å2). Then 350–400 ns of unrestrained simulations were performed for each system.
To ensure that the absence of water wires in the case of SCN− was not caused by the limited simulation time, an additional set of 20 short simulations in the presence of SCN− were performed. Each simulation was initiated with a Cl−-bound ClC-ec1 structure containing the most representative water wire (obtained from the Cl−-bound simulation).29 The Cl− was then replaced by SCN− while keeping the preformed water wire. The system was then energy-minimized (5000 steps), and simulated for 0.5 ns with positional restraints on all protein atoms and oxygen atoms of the water wire (pre-equilibrium). Then 20 1 ns simulations with different initial velocities were carried out during which the stability and lifetime of the water wires were evaluated.
Pore radius profiles were calculated using HOLE.59 Radial distribution function (RDF) calculations were performed using g(r) embedded in VMD. The hydrated radius was defined as a distance cutoff which encompassed the first density peak in the RDF. The coordination number was determined as the average number of protein or water hydrogen atoms within the hydrated radius.
RESULTS
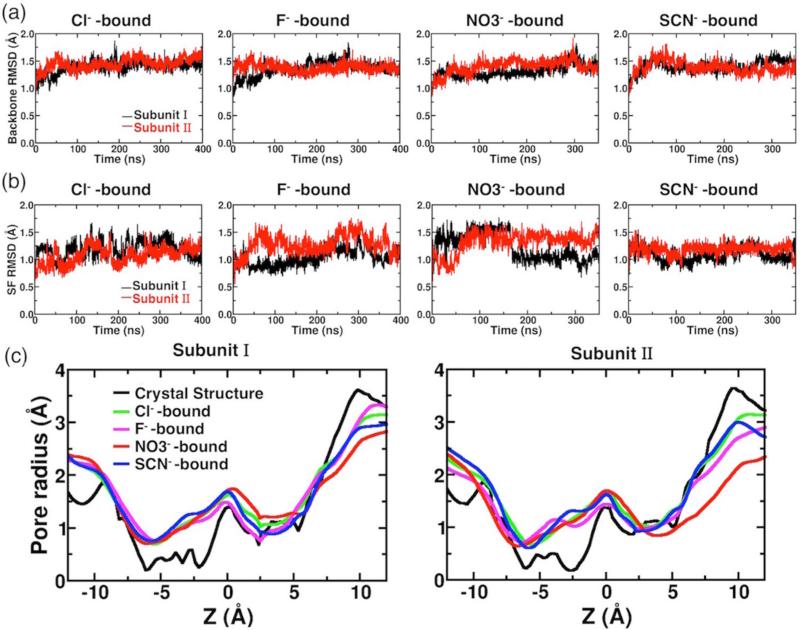
ClC-ec1 was simulated in the presence of four different anions (Cl−, F−, , or SCN−), in order to investigate and compare the mode of binding of these ions and their impacts on protein structure and dynamics, and on the hydration pattern of the protein lumen. During the simulations, the overall protein structure remains stable (average backbone RMSD < 1.5 Å for each subunit), regardless of the type of anion bound at the Scen site (Figure 2(a)). The configuration of the anion conduction pore is also intact in the presence of different anions (Figure 2, parts (b) and (c)). In all the cases, the average pore radius profile of the anion conduction pathway follows the trend observed for the crystal structure (Figure 2(c)). The two main bottlenecks, corresponding to the constriction regions that occlude the anion from the aqueous solutions, are clearly preserved, although the extracellular bottleneck shows a moderate (~0.5 Å) expansion relative to the crystal structure in all the simulated systems (Figure 2(c)). In general, the overall structure and dynamics of the protein do not appear to be largely affected by the chemical nature of the bound anion.
Figure 2.
Protein structural dynamics in the presence of different anions. (a) Backbone RMSDs of individual subunit shown as a function of simulation time for anion-bound simulations. (b) The RMSDs of all heavy (non-hydrogen) atoms of the selectivity filter (SF) in the presence of different anions. (c) Comparison of pore radius profiles of the crystal structure (black) and those of MD simulations (averaged over the whole trajectory) with different anions bound at Scen (colored traces). The center of the bound anion at t = 0, which corresponds to the Cl− position in the crystal structure, is set as the origin. The extracellular side is toward the left (z < 0), and the intracellular side is toward the right (z > 0).
Continuous Water Wires Arise in the Presence of Cl−
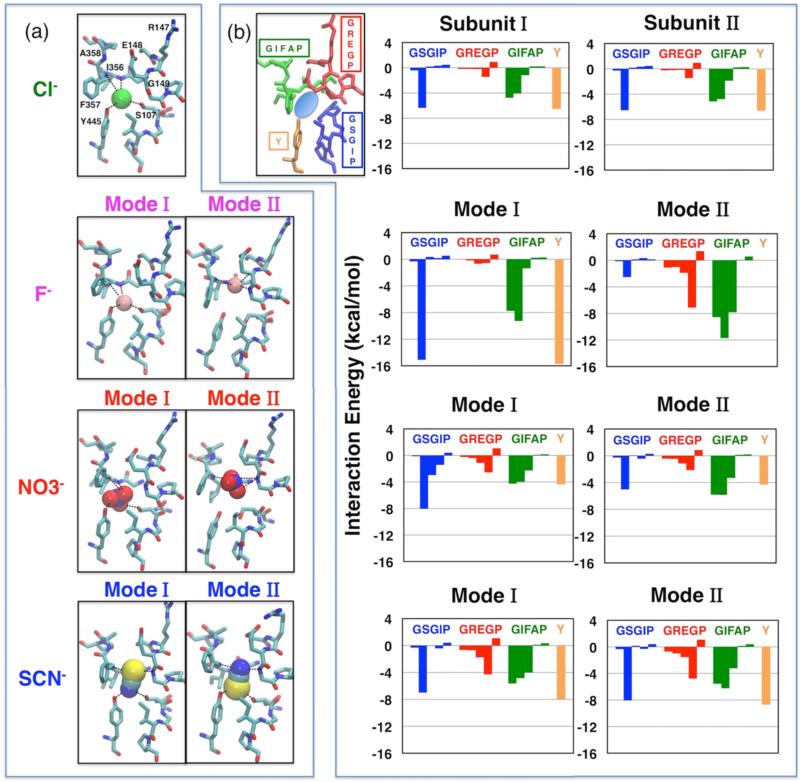
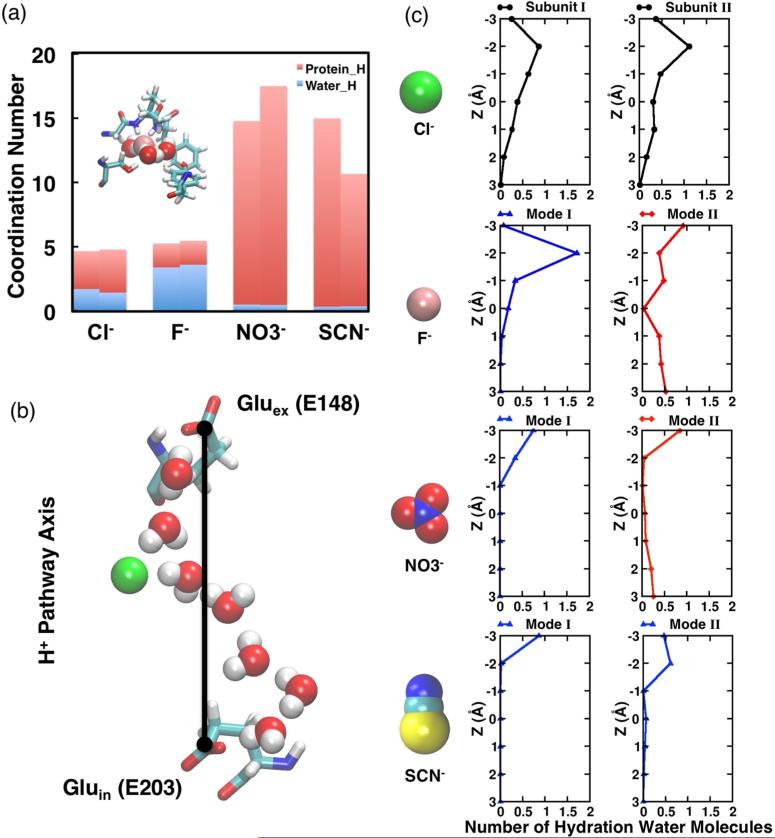
Cl− binds firmly at the Scen site in both subunits, where it is coordinated by the backbone amide nitrogen atoms of I356 and F357, and by the side-chain oxygen atoms of S107 and Y445 (Figure 3(a)). The average interaction energies between the Cl− at Scen and individual residues throughout the protein indicate that the Cl− binding is mainly supported by residues from three motifs: GIFAP, GSGIP, and Y445 (Figure 3(b) and Figure S1). The Cl− coordination shell, involving both surrounding protein residues and water molecules, is examined in detail by calculating the anion-hydrogen RDF for protein and water hydrogen atoms, respectively (Figure S2). The RDF analysis reveals that the bound Cl− is coordinated, on the average, by five hydrogen atoms, with ~1.6 of them originating from water molecules (Figure 4(a)). To monitor the fluctuations of the Cl− coordination shell, we quantified separately the involvement of protein residues and water molecules in the coordination shell during the course of the simulation (Figure S3). In both subunits, the Cl− ion is coordinated mainly by protein residues during the first 100 ns. Later, the ion becomes hydrated by 2–4 water molecules located nearby Scen, usually lining the central hydrophobic region between Gluex and Gluin.
Figure 3.
Anion binding modes. (a) Cl−, F−, , and SCN− (shown in van der Waals) bind distinctively in the selectivity filter. For F−, , and SCN−, two binding modes (Modes I and II) are observed in the two subunits. Cl− exhibits only one binding mode. Selected residues in the vicinity of the bound anions are shown. Coordinations of the bound anions by the protein residues are shown as dashed lines. (b) Interaction energy between the bound anion and the four highly conserved motifs. Motifs (colored and labeled in the left panel) forming the selectivity filter contribute the majority of the interaction energy between the anion and protein. Residues in the rest of the protein show negligible anion-interaction energies (absolute value <0.3 kcal/mol per residue, with the exception of K131 (−0.3 to −0.7 kcal/mol)).
Figure 4.
Anion solvation by protein and water. (a) Anion coordination numbers obtained from radial distribution function (RDF) analysis (see Figure S2) with protein or water hydrogen atoms. Inset shows an example of anion coordination, depicting F− surrounded by at least three water molecules on the average. (b) The putative H+ pathway axis is defined by the Gluex and Gluin carboxylate groups. (c) The distributions of anion hydration water molecules along the H+ pathway axis. Blue curves (▲) indicate the anion hydration water is only located above the anion (z > 0, extracellular side); red curves (◆) indicate the anion hydration water is observed both above (z > 0, extracellular side) and below (z < 0, intracellular side) the anion; and black curves (●) indicate the anion hydration water exists along the H+ pathway axis including the region right next to the anion (z = 0). The center of the bound anion is set as the origin.
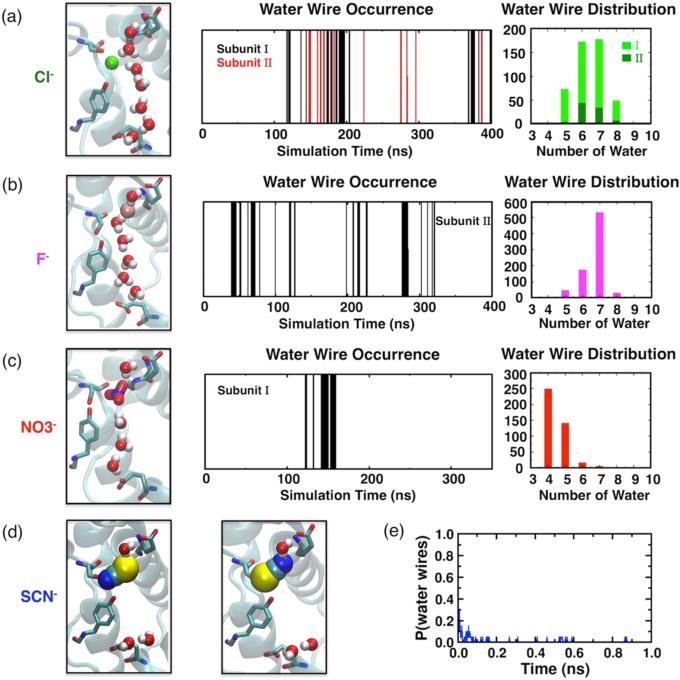
To understand in detail the influence of Cl− on the surrounding water molecules that fall between the two H+ sites, we characterize further the distribution of the Cl− hydration water molecules along the axis defined by the carboxylate groups of Gluex and Gluin that is, the putative H+ pathway axis, see Figure 4(b)). A water molecule was considered as a hydration water molecule if the shortest Cl−-water distance falls within 3 Å. For both subunits, hydration water molecules are observed uninterruptedly along the H+ pathway axis, including the region right next to the Cl− ion (z = 0 Å, Figure 4(c)). The presence of a continuous array of hydration water molecules implies a favorable environment for hydrogen-bonded water wire to arise nearby the Cl− ion. Consistent with this expectation, formation of continuous water wires is observed in both subunits of Cl−-bound ClC-ec1 during the simulation.29 Although the frequency of water wire formation in Subunit I is almost three times of that in Subunit II, in both subunits water wires containing 6–7 water molecules arise frequently (Figure 5(a)). The different behaviors observed in the simulations for the two subunits may be due to the minor structural asymmetry of the subunits in the crystal structure (Figure 2, parts (a) and (b)), which results in differences in the microenvironments of the binding sites and individual conduction pores. We note that the two subunits mediate Cl−/H+ exchange independently.60
Figure 5.
Effect of anion bound at Scen on formation of water wires. (a) Continuous water wires arise in both subunits in the presence of Cl−,29 following a normal distribution dominated by configurations including 7 water molecules. (b) Pseudo-water-wires are observed only in Subunit II when F− binds in binding Mode II. Those intervened wires follow a normal distribution dominated by 7 water molecules. (c) Pseudo-water-wires formed in Subunit I when binds in binding Mode II and follow an exponential distribution dominated by 4 water molecules due to the large size of . (d) Neither water wires nor pseudo-water-wires are observed in the presence of SCN−. (e) Average probability of water wires obtained from short (1 ns) simulations of ClC-ec1 with SCN− bound, starting from a preformed configuration of the most representative wire structure.
Pseudo-Water-Wires Form in the Presence of F−
F−, which has a radius ~0.5 Å smaller than Cl−, displays two binding modes around the Scen site (Figure 3(a)). In Subunit I, F− binds firmly at Scen (similar to what was observed for Cl−), while in Subunit II, it moves upward after ~40 ns and adopts a stable position somewhere between Scen and E148 for ~340 ns, with E148 side chain moving slightly toward the extracellular side (Figure 3(a)). The upward movement of F− results in hydrogen bond interactions between the F− ion and backbone nitrogen atoms of E148 and G149; meanwhile, F− loses its direct contacts with S107 and Y445 hydroxyl groups on the opposite side. Switching between these two binding modes arises only in Subunit II on the time scale of our simulation.
The average coordination number for F− is slightly over 5 in both binding modes, which is comparable to that of Cl−. However, a relatively larger portion of the coordination is contributed by water molecules (~3.5, Figure 4(a)). Moreover, the simulation trajectory in the presence of F− shows no apparent dehydration phase for F− as seen in the first 100 ns simulation with Cl−. The F− ion becomes hydrated quickly after the simulation starts and remains coordinated by ~3 water molecules throughout the simulation (Figure 4(a) inset and Figure S3). Due to its strong tendency for hydration, F− exhibits a different arrangement of hydration water molecules when compared to Cl−. In the Cl−-like binding mode (Mode I), F− is coordinated by water molecules from above (z > 0; extracellular side), while contacting the side chains of S107 and Y445 on the opposite side. In binding Mode II, F− is sandwiched by water molecules both from above (z > 0) and below (z < 0) (Figure 4(c)).
The distinct pattern of hydration of F− appears to be the main reason for not observing the formation of continuous water wires between Gluex and Gluin in either subunit during the simulation. Detailed inspection revealed that in binding Mode II, F− can be actually connected to Gluex and Gluin by separate, shorter water wires, involving about 7 water molecules. The separated water wires together with the F− ion form pseudo-water-wires connecting the two H+ sites that are however intervened by the anion (Figure 5(b)), indicating an essential difference between F− and Cl−.
Shorter Pseudo-Water-Wires Intervened by
is a polyatomic anion and exhibits a more complex chemical structure involving three oxygen atoms at corners of a triangle sharing the negative charge around a central nitrogen atom. Similar to the F−-bound system, the simulation of ClC-ec1 with reveals two possible binding modes for the anion. In Mode I, is fitted into Scen with one of its oxygen atoms coordinated by the backbone nitrogen atoms of I356 and F357, and the other two oxygen atoms coordinated by the side chain hydroxyl groups of S107 and Y445, respectively (Figure 3). Mode I is relatively more populated and is observed in both subunits. In Mode II, exhibits a similar behavior to F− in its binding Mode II, in that moves upward to a position between Scen and E148 and pushes away E148 side chain by ~2.30 Å. This binding mode involves hydrogen bond interactions between the oxygen atoms of and the backbone nitrogen atom of G149, instead of direct interaction with S107 and Y445 hydroxyl groups (Figure 3). Binding Mode II only occurs in Subunit I for ~150 ns (t = 10–160 ns), before moves back to the original, Scen binding site.
According to the RDF profiles, establishes a more extensive coordination to its environment compared to Cl− and F−, but much fewer of them (~0.5) arise from anion–water interactions (Figure 4(a) and Figure S2), suggesting that this large polyatomic anion relies less on interaction with water in order to be stabilized when bound at Scen. During the simulation, is only minimally hydrated (Figure S3), resulting in differential hydration configurations along the H+ pathway axis (Figure 4(c)).
In binding Mode II, hydration water molecules are observed along the H+ pathway axis except for the area right beside the anion (z = 0), while in binding Mode I, only the region above the bound is hydrated (z > 0, Figure 4(c)). As a result, pseudo-water-wires only arise when the anion is in binding Mode II. Compared to the pseudo-water-wires formed in the presence of F−, those involving favor shorter water wires, often containing only four water molecules (Figure 5(c)). The difference between the pseudo-water-wires involving F− and can be attributed to the different sizes of the anions. In particular, due to its geometry involving multiple negatively charged sites (oxygen atoms), appears larger effectively reducing the spatial gap between the Gluex and Gluin for formation of pseudo-water-wires.
SCN− Eliminates the Water Wires
The other polyatomic anion investigated, SCN−, has a linear chemical structure which extends to 4.77 Å in length.61 Due to its long shape, SCN−, which was initially centered at Scen in both subunits, rotates and shifts immediately after the simulation starts in order to optimally fit into the available space, resulting in two different binding orientations in the two independent subunits (Figure 3(a)). Both binding orientations of SCN− were stable over the simulation time.
Because of its larger size, SCN− occupies Scen while extending almost to Sext in both orientations, stabilized by its interaction (7–8 kcal/mol) with motif GREGP (Figure 3, parts (a) and (b)). In one orientation (Mode I), the sulfur atom of SCN− is coordinated by the backbone nitrogen atoms of G149, I356, and F357, while the nitrogen atom of the anion is coordinated by the side-chain hydroxyl groups of S107 and Y445. In the alternative orientation (Mode II), SCN− adopts an opposite configuration and binding coordination (Figure 3, parts (a) and (b)).
SCN− fills up a larger portion of the space within the binding region, resulting in very large coordination to its environment (Figure 4(a)). RDF calculations revealed that SCN− is only weakly hydrated in either binding mode, with only ~0.3 of the coordination arising from anion-water interactions (Figure 4(a) and Figure S2). Specifically, the hydration water molecules are only observed above the bound SCN− ion (z > 0) in either binding mode (Figure 4(c)). Hydration from the other side of SCN− (below it, z < 0) is not observed, since the central hydrophobic region remains dehydrated in the presence of this anion. Consequently, neither water wires nor pseudo-water-wires are observed to form in the presence of SCN− (Figure 5(d)), suggesting that water wires necessary for H+ are unlikely to be supported by this anion.
To preclude the possibility that the absence of water wires in the case of SCN− was merely due to the limited simulation time, we also examined the stability of preformed water wires in the presence of SCN− through a set of short simulations. These simulations all started from a configuration in which the water wires had been formed (adopted from Cl−-bound simulations), and were done in the presence of SCN−. The preformed water wires diminish quickly in all the simulations, and SCN− breaks the water wires immediately after the simulations start, by inserting either its sulfur or nitrogen atom into the water wires (Figure 4(e)). The lifetime of water wires averaged over 20 1 ns runs is 0.024 ± 0.026 ns with SCN−, which is significantly shorter than 0.6 ± 0.2 ns for Cl−-bound ClC-ec1.29
DISCUSSION
The molecular origin of coupled Cl−/H+ exchange is a central mechanistic aspect of CLC transporters. Understanding the atomic details underlying differential H+ coupling of various anions constitutes a major question that motivated this study. To address this question, we performed extended equilibrium MD simulations to investigate differential binding modes of different anions transported by ClC-ec1 to Scen and to determine whether and how they may affect the formation of water wires within the lumen, which in turn may be an important component of the H+ transport reaction.29,40
X-ray crystallographic studies of ClC-ec1 suggest that the bound anion does not induce major changes in the overall structure of the protein.12 Our RMSD results and pore profile calculations (Figure 2) confirm that the protein structure is not compromised by binding of the studied anions, which also include large, nonhalide species, and that the binding site/region can accommodate both the physiologically relevant Cl− ion as well as other anions, namely F−, , and SCN− (Figure 2). Although the overall protein structure does not seem to be affected by the type of anion bound at Scen, the details of the coordination and interaction between the bound anion and protein/water vary significantly among the anions examined (Figure 3 and 4). X-ray crystallography of anion-bound ClC-ec1 suggests that F− binds to Scen similarly to the biological substrate Cl−.22 and SCN− have not been unambiguously localized, and their binding modes not well characterized in ClC-ec1, due to low crystallographic resolution.12
The present study provides a detailed description of the binding modes of these anions in ClC-ec1. In contrast to Cl−, which binds stably at Scen, the same site observed in the Cl−-bound crystal structures,26,27 both F− and are found to populate two different binding positions (modes), one located at the original Scen site, and the other located between Scen and E148. SCN−, due to its extended shape, occupies Scen while extending almost to Sext with two possible (opposite) binding orientations. Despite the differential binding modes, all anions stay at or around the Scen site, during the simulations, where they are stabilized by several conserved motifs (Figure 3). However, the cumulative interaction energy between the anion and the protein varies in the order of F− > SCN− > > Cl− in both subunits, with the stabilization of F− almost twice as favorable as that of Cl− (Figure S1).
In a previous computational study, Ko and Jo showed that Cl− was coordinated by protein hydrogen atoms and no water molecules were observed along the Cl− conduction pore when E148 was protonated and Cl− was pulled from the intracellular side to the extracellular side along the pore axis.62 Several other simulation studies focusing on anion conduction, however, suggested that Cl− is partially hydrated.40,49,50,63 Our recent study revealed that water molecules can enter the anion-binding region via the H+ pathway and hydrate the initially dehydrated Cl−, a feature which we proposed to be critical to H+ transport in ClC-ec1.29 Therefore, the configuration of the anion coordination shell, including the number of water molecules in the first coordination shell, as well as the size and shape of the anion within this shell, can be critical to anion-coupled H+ transport by the protein.64,65 RDF and related calculations reveal that the anions at Scen exhibit two types of coordination shell structures, with distinguishable number of coordinations from protein and water molecules (Figure 4(a)). Halide anions (Cl− and F−) both exhibit an average coordination number of ~5, while the polyatomic anions ( and SCN−) establish a more extensive coordination (number of coordination >10) to their environment. Under equilibrium conditions, the physiologically transported anion, Cl−, is the only one that supports full hydration along the H+ pathway axis (Figure 4(c)), which we showed to be a prerequisite for water wires to arise and connect Gluex and Gluin.29
The observation that the anions at Scen exhibit distinct coordination structures and hydration shells suggests that the functionally important water wires, which need to arise nearby the anions, could experience different environments. Thus, we investigated further the effect of various anions at Scen on the formation of water wires. Different types of water wires were observed for various anions (Figure 5). They can be classified into two types based on the pattern of interaction with the bound anions as well as their configurations: (1) continuous water wires, supported by the anion located nearby, which connect the two H+ sites seamlessly, and (2) pseudo-water-wires, where the anion participates in the wire and prevents water molecules from forming a continuous hydrogen-bonded chain connecting the two H+ sites. The continuous water wires are only observed in the presence of Cl−, while the pseudo-water-wires were observed for both F− and in their alternate binding modes (Mode II).
Lim and colleagues recently showed that F− is coordinated much like Cl− at Scen.22 They also showed that F− binding to this site forms a strong hydrogen-bond to the protonated Gluex, as revealed by the short distance between the F− ion and the Gln side-chain in the E148Q mutant (where Gln mimicks the protonated Gluex). This strong hydrogen bond likely plays a key role in slowing the transport cycle. However, there may be additional factors contributing to the complete lack of F− transport. A hint that other factors may be involved is suggested by the fact that the inner-gate mutant Y445A, which destabilizes Cl− (and presumably F−) binding to Scen (see Introduction) retains wild-type selectivity against F−.22 Our observation of the binding Mode II for F− provides a microscopic view of how F− can bind in the permeation pathway away from the Scen site. The pseudo-water-wires arising in this binding mode are likely an additional key contributor to the transport selectivity against F−. This picture provides a compelling rationale for the wild-type selectivity against F− observed in the Y445A mutant: since the pseudo-water-wires only occur in binding Mode II, in which F− loses its direct contacts with S107 and Y445 hydroxyl groups, the Y445A mutation will not affect the behavior of the protein toward F−. In the future, it will be of interest to determine whether water wires play a role in the mechanism of the CLCF exchangers. Such an idea is speculative, given that the exchange stoichiometry of 1F−:1H+ implies a significantly different exchange mechanism from that occurring in the canonical 2:1 Cl−:H+ CLCs. Addressing this important question awaits a high-resolution structure of the CLCF homologues.
, like F−, induces the formation of pseudo-water-wires that are intervened by itself. Such water wires could allow occasional transport of H+, but with less fidelity than the pure water wires observed with Cl−. These pseudo-water-wires arise in binding Mode II, where the ion moves upward and has no direct interaction with the Scen-coordinating residues S107 and Y445, which in losing their interaction with the ion form a hydrogen bond one to another (Figure 3(a)). Such a binding mode is never observed with Cl−. Thus, the simulation predicts that these two residues are determinants of Cl−/ selectivity. Consistent with the prediction, certain plant CLC homologues that function as 2:1 exchangers66 have a proline at position 107, which is a serine in all 2:1 Cl−:H+ exchangers. In ClC-5, mutating the serine to a proline partially reproduces the plant phenotype, reducing the /H+ stoichiometry from ~10–25 to ~2–8 (over the +60–120 mV voltage range examined).67,68 In ClC-ec1 mutating this serine to a proline increases the binding affinity for ~4-fold.31 Although it is not yet known whether this change in binding affinity is paralleled by a change in /H+ stoichiometry, a crystal structure of this ClC-ec1 mutant together with MD simulations might further illuminate mechanism of /Cl− selectivity in CLC anion/H+ coupling.
Early experimental studies showed a striking correlation between anion occupancy at Scen and Cl−/H+ coupling stoichiometry in ClC-ec1. Specifically, the halides Br− and Cl− are transported with a strict 2:1 exchange ratio to H+, whereas the pseudo halides, SCN− and SeCN−, are transported in a completely uncoupled manner.12 In ClC-ec1 crystal structures, Br− density is clearly observed at Scen, while SeCN− is not.12 Although difficult to interpret these data mechanistically, it was speculated that low occupancy at Scen could somehow promote occasional simultaneous opening of the inner and outer gates (which are in direct contact with Scen), thus creating an anion leak that degrades the coupling stoichiometry. Our simulation results provide an alternative explanation for uncoupling transport of nonhalides: the disruption of water wires that facilitate H+ transfer between Gluex and Gluin. The uncoupling ion SCN− remains bound in the vicinity of Scen (within the simulation time), but it is unable to sustain water wires. In this model, uncoupled SCN− transport need not necessarily involve simultaneous opening of both gates but could entail alternate opening of gates, similar to the alternate opening of gates that occurs in the coupled transporter but no longer coordinated with H+ transport. Such an uncoupling mechanism has precedent in ClC-ec1 Gluex mutations, where uncoupled Cl− transport involves protein conformational change similar to that observed in the coupled transporter.69 The idea that uncoupled Cl− and SCN− transport share some similarities is consistent with the observation that mutations at Gluin dramatically slow both Cl− and SCN− transport in ClC-5.14 We note also that our observation of SCN− binding to Scen is not inconsistent with the crystallo-graphic studies, in which the lack of anion density at Scen could reflect high disorder rather than lack of occupancy.
CONCLUSIONS
The coupling between the transport of different chemical species is at the heart of the mechanism of function of secondary transporters. In order to achieve stoichiometric transport of two ions, the transporter protein needs to be equipped with a precise molecular mechanism through which the translocation of the two (ionic) species are coupled. Despite the central role of this phenomenon in a large number of transporters, the underlying molecular mechanism is by and large lacking in the field of membrane transporters. Here, we have investigated this mode of operation in a representative member of the H+/Cl− antiporters. By systematically evaluating the differential binding modes of chemically diverse anions at the central anion binding site (Scen) and their effect on water wire formation between Gluex and Gluin in ClC-ec1 at an atomic level, we construct a qualitative explanation that connects the chemical nature of anionic substrates with their ability to assist in the coupled H+ transport. Our results infer that the difference in the anion/H+ coupling is due to their influence on the formation of water wires connecting Gluex and Gluin, which could be essential for H+ transport. This view adds a crucial layer of detail to our understanding of the Cl−/H+ coupling mechanism.
Supplementary Material
ACKNOWLEDGMENTS
This research is supported by National Institutes of Health grants R01-GM086749, U54-GM087519, and P41-GM104601. Simulations in this study have been performed using allocations at National Science Foundation Supercomputing Centers (XSEDE grant number MCA06N060).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b12062.
Figure S1, Interaction energy between the anions at Scen and ClC-ec1; Figure S2, anion-hydrogen RDF for Scen bound anions with protein or water hydrogen atoms; and Figure S3, coordination number of bound anions as a function of simulation time (PDF)
REFERENCES
- 1.Jentsch TJ. Crit. Rev. Biochem. Mol. Biol. 2008;43:3–36. doi: 10.1080/10409230701829110. [DOI] [PubMed] [Google Scholar]
- 2.Stauber T, Weinert S, Jentsch TJ. Compr. Physiol. 2012;2:1701–1744. doi: 10.1002/cphy.c110038. [DOI] [PubMed] [Google Scholar]
- 3.Jenntsch TJ, Stein V, Weinreich F, Zdebik A. Physiol. Rev. 2002;82:503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- 4.Picollo A, Pusch M. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- 5.Accardi A, Miller C. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 6.Scheel O, Zdebik AA, Lourdel S, Jentsch TJ. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 7.Rychkov GY, Pusch M, Roberts ML, Jentsch TJ, Bretag AH. J. Gen. Physiol. 1998;111:653–665. doi: 10.1085/jgp.111.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahlke C, Dürr C, George AL., Jr. J. Gen. Physiol. 1997;110:551–564. doi: 10.1085/jgp.110.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahlke C, Yu H, Beck CL, Rhodes TR, George AL., Jr. Nature. 1997;390:529–532. doi: 10.1038/37391. [DOI] [PubMed] [Google Scholar]
- 10.Ludewig U, Jentsch TJ, Pusch M. J. Physiol. 1997;498:691–702. doi: 10.1113/jphysiol.1997.sp021893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebeisen S, Heidtmann H, Cosmelli D, Gonzalez C, Poser B, Latorre R, Alvarez O, Fahlke C. Biophys. J. 2003;84:2306–2318. doi: 10.1016/S0006-3495(03)75036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguitragool W, Miller C. J. Mol. Biol. 2006;362:682–690. doi: 10.1016/j.jmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 13.De Stefano S, Pusch M, Zifarelli G. J. Biol. Chem. 2011;286:44134–44144. doi: 10.1074/jbc.M111.272815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zdebik AA, Zifarelli G, Bergsdorf E-Y, Soliani P, Scheel O, Jentsch TJ, Pusch M. J. Biol. Chem. 2008;283:4219–4227. doi: 10.1074/jbc.M708368200. [DOI] [PubMed] [Google Scholar]
- 15.Alekov AK, Fahlke C. J. Gen. Physiol. 2009;133:485–496. doi: 10.1085/jgp.200810155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaram H, Robertson JL, Wu F, Williams C, Miller C. Biochemistry. 2011;50:788–794. doi: 10.1021/bi1019258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng L, Campbell EB, Hsiung Y, MacKinnon R. Science. 2010;330:635–641. doi: 10.1126/science.1195230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leisle L, Ludwig CF, Wagner FA, Jentsch TJ, Stauber T. EMBO J. 2011;30:2140–2152. doi: 10.1038/emboj.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orhan G, Fahlke C, Alekov AK. Biophys. J. 2011;100:1233–1241. doi: 10.1016/j.bpj.2011.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maduke M, Pheasant DJ, Miller C. J. Gen. Physiol. 1999;114:713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahlke C. Am. J. Physiol.–Ren. Physiol. 2001;280:F748–F757. doi: 10.1152/ajprenal.2001.280.5.F748. [DOI] [PubMed] [Google Scholar]
- 22.Lim H-H, Stockbridge RB, Miller C. Nat. Chem. Biol. 2013;9:721–725. doi: 10.1038/nchembio.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brammer AE, Stockbridge RB, Miller C. J. Gen. Physiol. 2014;144:129–136. doi: 10.1085/jgp.201411225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stockbridge RB, Lim H-H, Otten R, Williams C, Shane T, Weinberg Z, Miller C. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15289–15294. doi: 10.1073/pnas.1210896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 27.Dutzler R, Campbell EB, MacKinnon R. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 28.Accardi A, Lobet S, Williams C, Miller C, Dutzler R. J. Mol. Biol. 2006;362:691–699. doi: 10.1016/j.jmb.2006.07.081. [DOI] [PubMed] [Google Scholar]
- 29.Han W, Cheng RC, Maduke MC, Tajkhorshid E. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1819–1824. doi: 10.1073/pnas.1317890111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walden M, Accardi A, Wu F, Xu C, Williams C, Miller C. J. Gen. Physiol. 2007;129:317–329. doi: 10.1085/jgp.200709756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picollo A, Malvezzi M, Houtman J, Accardi A. Nat. Struct. Mol. Biol. 2009;16:1294–1301. doi: 10.1038/nsmb.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Accardi A, Walden M, Nguitragool W, Jayaram H, Williams C, Miller C. J. Gen. Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim H-H, Miller C. J. Gen. Physiol. 2009;133:131–138. doi: 10.1085/jgp.200810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picollo A, Xu Y, Johner N, Berneche S, Accardi A. Nat. Struct. Mol. Biol. 2012;19:525–531. doi: 10.1038/nsmb.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smart OS, Goodfellow JM, Wallace BA. Biophys. J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garczarek F, Brown LS, Lanyi JK, Gerwert K. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Voth GA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6795–6800. doi: 10.1073/pnas.0408117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Sharpe MA, Qin L, Ferguson-Miller S, Voth GA. J. Am. Chem. Soc. 2007;129:2910–2913. doi: 10.1021/ja067360s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuang Z, Mahankali U, Beck TL. Proteins: Struct., Funct., Genet. 2007;68:26–33. doi: 10.1002/prot.21441. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Voth GA. Biophys. J. 2009;97:121–131. doi: 10.1016/j.bpj.2009.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freier E, Wolf S, Gerwert K. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11435–11439. doi: 10.1073/pnas.1104735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goyal P, Qian H-J, Irle S, Lu X, Roston D, Mori T, Elstner M, Cui Q. J. Phys. Chem. B. 2014;118:11007–11027. doi: 10.1021/jp503372v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaus M, Jin H, Demapan D, Christensen AS, Goyal P, Elstner M, Cui Q. J. Chem. Theory Comput. 2015;11:4205–4219. doi: 10.1021/acs.jctc.5b00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wraight CA. Biochim. Biophys. Acta, Bioenerg. 2006;1757:886–912. doi: 10.1016/j.bbabio.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Espinoza-Fonseca LM, Ramírez-Salinas GL. J. Am. Chem. Soc. 2015;137:7055–7058. doi: 10.1021/jacs.5b03814. [DOI] [PubMed] [Google Scholar]
- 46.Ko YJ, Jo WH. Biophys. J. 2010;98:2163–2169. doi: 10.1016/j.bpj.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng MH, Coalson RD. Biophys. J. 2012;102:1363–1371. doi: 10.1016/j.bpj.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Hermans J. Proteins: Struct., Funct., Genet. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Cohen J, Schulten K. Biophys. J. 2004;86:836–845. doi: 10.1016/S0006-3495(04)74159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bostick DL, Berkowitz ML. Biophys. J. 2004;87:1686–1696. doi: 10.1529/biophysj.104.042465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faraldo-Gómez JD, Roux B. J. Mol. Biol. 2004;339:981–1000. doi: 10.1016/j.jmb.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 52.Jorgensen W, Chandrasekhar J, Maudura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 53.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacKerell AD, Jr., Feig M, Brooks CL., III J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 55.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr., Pastor RW. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 57.Nosé S. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 58.Hoover WG. Phys. Rev. A: At., Mol., Opt. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 59.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP. J. Mol. Graphics. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 60.Robertson JL, Kolmakova-Partensky L, Miller C. Nature. 2010;468:844–846. doi: 10.1038/nature09556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwadate Y, Kawamura K, Igarashi K, Mochinaga J. J. Phys. Chem. 1982;86:5205–5208. [Google Scholar]
- 62.Ko YJ, Jo WH. J. Comput. Chem. 2010;31:603–611. doi: 10.1002/jcc.21432. [DOI] [PubMed] [Google Scholar]
- 63.Gervasio FL, Parrinello M, Ceccarelli M, Klein M. J. Mol. Biol. 2006;361:390–398. doi: 10.1016/j.jmb.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 64.Tansel B, Sager J, Rector T, Garland J, Strayer RF, Levine L, Roberts M, Hummerick M, Bauer J. Sep. Purif. Technol. 2006;51:40–47. [Google Scholar]
- 65.Richards LA, Schäfer AI, Richards BS, Corry B. Small. 2012;8:1701–1709. doi: 10.1002/smll.201102056. [DOI] [PubMed] [Google Scholar]
- 66.De Angeli A, Monachello D, Ephritikhine G, Frachisse JM, Thomine S, Gambale F, Barbier-Brygoo H. Nature. 2006;442:939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- 67.Zifarelli G, Pusch M. EMBO J. 2009;28:175–182. doi: 10.1038/emboj.2008.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bergsdorf E-Y, Zdebik AA, Jentsch TJ. J. Biol. Chem. 2009;284:11184–11193. doi: 10.1074/jbc.M901170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khantwal CM, Abraham SJ, Han W, Jiang T, Chavan TS, Cheng RC, Elvington SM, Liu CW, Mathews II, Stein RA, Mchaourab HS, Tajkhorshid E, Maduke M. eLife. 2016;5:e11189. doi: 10.7554/eLife.11189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.