Abstract
RAB5 GTPases are important regulators of endosomal membrane traffic. Among them Arabidopsis thaliana ARA7/RABF2b is highly conserved and homologues are present in fungal, animal and plant kingdoms. In land plants ARA7 and its homologues are involved in endocytosis and transport towards the vacuole. Here we report on the isolation of an ARA7 homologue (CaARA7/CaRABF2) in the highly evolved characean green alga Chara australis. It encodes a polypeptide of 202 amino acids with a calculated molecular mass of 22.2 kDa and intrinsic GTPase activity. Immunolabelling of internodal cells with a specific antibody reveals CaARA7 epitopes at multivesicular endosomes (MVEs) and at MVE-containing wortmannin (WM) compartments. When transiently expressed in epidermal cells of Nicotiana benthamiana leaves, fluorescently tagged CaARA7 localizes to small organelles (putative MVEs) and WM compartments, and partially colocalizes with AtARA7 and CaARA6, a plant specific RABF1 GTPase. Mutations in membrane anchoring and GTP binding sites alter localization of CaARA7 and affect GTPase activity, respectively. This first detailed study of a conventional RAB5 GTPase in green algae demonstrates that CaARA7 is similar to RAB5 GTPases from land plants and other organisms and shows conserved structure and localization.
Keywords: ARA7, Chara australis, endosomal trafficking, multivesicular endosome, RAB5 GTPase, RABF2
RAB GTPases are key regulators of membrane trafficking in eukaryotic cells (1,2). They belong to the family of small GTPases with a molecular mass of 20–25 kDa (1) and cycle between GTP and GDP conformations. In their activated, GTP-bound state RAB GTPases localize at membranes, where they recruit effector molecules and promote downstream reactions including tethering of transport vesicles or organelles to target membranes required for docking and fusion. Hydrolysis of GTP causes the release of RAB GTPases into the cytosol in a RAB GDP dissociation inhibitor-dependent manner. Activation and inactivation of GTPases are regulated by guanine nucleotide exchange factors (GEFs) and by GTPase activating proteins (GAPs) which drive the GTPase cycle of RAB proteins (3,4). Among RAB GTPases, members of the RAB5 family are responsible for endosomal trafficking in representatives of the fungal, animal and plant kingdom (1). In Arabidopsis thaliana the RAB5 group consists of RHA1 (RABF2a), ARA7 (RABF2b) and ARA6 (RABF1) (5,6). ARA6 is an enigmatic RAB5 GTPase, which differs from conventional RAB members in the sequence responsible for membrane anchoring (N-terminal myristoylation and palmitoylation instead of C-terminal prenylation) (5) and acts in the trafficking pathway from multivesicular endosomes (MVEs) to the plasma membrane especially under stress conditions (7–9). In addition to its role inexocytosis, a mutated form of ARA6 has been also shown to perturb vacuolar trafficking of soluble cargos in tobacco cells (10). ARA6 was long considered to be specific for land plants (11) but in recent studies we identified the first ARA6-like protein in green algae, CaARA6 from Chara australis (12) and a possible interaction partner, VAMP72 (13). ARA7 and RHA1 have a similar structure as mammalian RAB5 GTPases and are therefore considered to play similar roles. Indeed, several studies confirmed that ARA7 and RHA1 are involved in endocytosis and transport towards the vacuole in plant cells, e.g. (14,15). Conventional RAB5 is present on MVEs as shown by immunolabelling of electron microscope (EM) sections (16) and co-expression of AtARA7 with appropriate markers suggests that ARA7 is also localized at Golgi bodies (17). Fluorescent constructs of AtARA6 partially colocalize with organelles labelled by fluorescently tagged AtARA7 which either reflects maturation of endosomes or co-operation in membrane trafficking (6,8). RAB5 GTPases have been intensely studied in Arabidopsis and other higher plants (3,9). Far less is known about the structure and properties of RAB5 GTPases in algal cells.
In this study we investigated the structure, localization and the intrinsic GTPase activity of the ARA7 homologue of C. australis, CaARA7. Chara australis is a member of the characean green algae which are placed within the streptophytes indicating that they are close relatives of land plants (e.g. 18 and references therein). Within their multicellular thallus groups of small nodal cells regularly alternate with cylindrical internodes, which may become up to several cm long. The cytoplasm of the internodal cells consists of a stationary cortex including helical files of chloroplasts and a streaming endoplasm containing up to several thousand nuclei, Golgi bodies, trans-Golgi network (TGN) and MVEs. The central vacuole occupies more than 90% of the volume in mature cells. The rapid acto-myosin dependent cytoplasmic streaming, the huge size and the simple geometry make the internodal cells a useful model for studying various aspects of plant cell biology (19–21). Organelle behaviour during endo- and exocytosis has been studied in control and wounded cells (22) but little is known about the molecular players for membrane trafficking pathways in characean and other algae.
The results of this study confirm the presence of an ARA7 homologue in C. australis. CaARA7 has a similar structure as ARA7 members from A. thaliana and other organisms and localizes to MVEs. Immunolabelling and transient expression of fluorescently tagged CaARA7 revealed partial colocalization not only with AtARA7 but also with CaARA6. Mutations in the membrane anchoring and GTP-binding sites alter the localization of CaARA7 and affect GTPase activity, respectively. To our knowledge this is the first detailed study of a conventional RAB5 GTPase in green algae in spite of their wide distribution and their importance in membrane trafficking.
Results
Identification and molecular characterization of a conventional RAB5 GTPase in Chara
A cDNA library of C. australis was sequenced using a Roche GS FLX system (Eurofins MWG) and screened for sequences similar to A. thaliana ARA7, one of the conventional RAB5 sequences of plants (AtRABF2b; hereafter referred to as AtARA7). A highly similar cDNA sequence of 606 base pairs (CaARA7; accession number KM406482) was detected that resulted in a protein of 202 amino acids with a calculated molecular weight of 22.23 kDa.
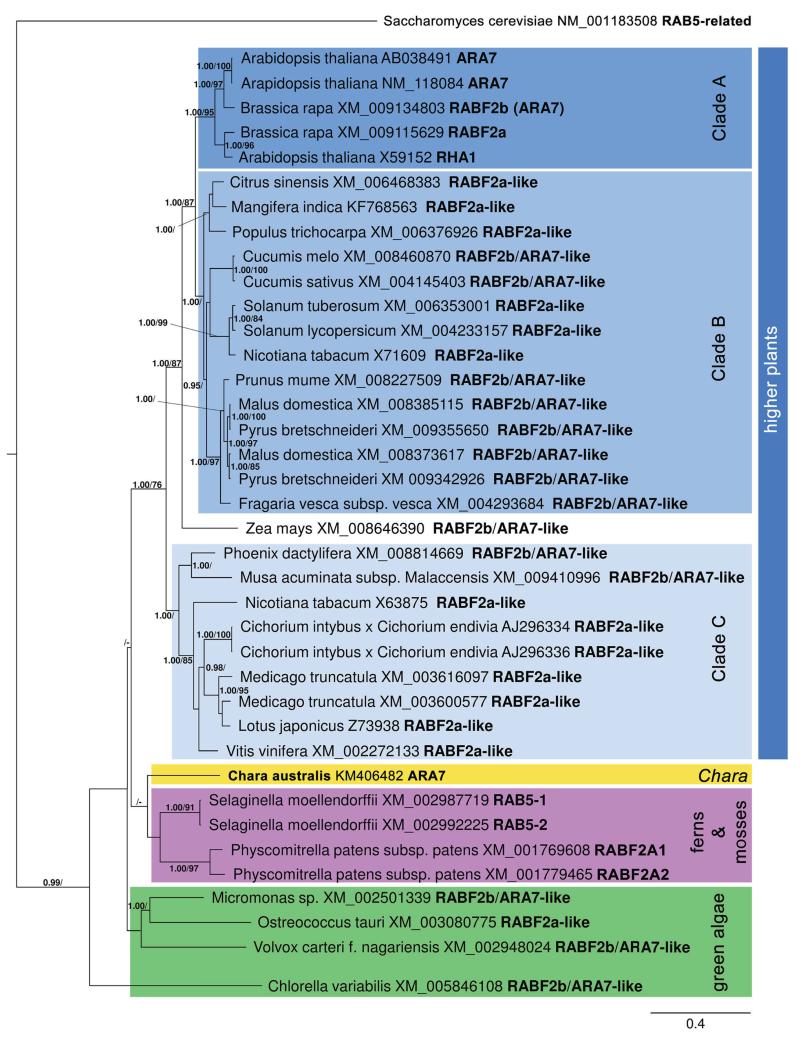
In order to classify CaARA7 in the family of conventional RAB5 GTPases phylogenetic analyses were performed. As shown in Figure 1, all aligned conventional RAB5 GTPases of higher plants are clustered together and were divided into three clades. Clade A comprises A. thaliana and Brassica rapa sequences, while clade B contains species of Anacardiaceae, Cucurbitaceae, Rosaceae, Rutaceae, Salicaceae and Solanaceae. Clade A represents a sister clade to B with support values of 1.00 PP and 87% MLB. The sequence of Zea mays (XM_008646390) is closely related (PP 1.00 and 87% MLB) to both clades. Clade C includes species of the families Arecaceae, Asteraceae, Fabaceae, Musaceae, Solanaceae and Vitaceae (PP 1.00 and < 70% MLB). Chara australis CaARA7 exhibits a close relationship to sequences of the genera Physcomitrella and Selaginella, but this arrangement is not supported. Sequences of other green algae (green box in Figure 1) are grouped together and are sister to all other aligned conventional RAB5 GTPase sequences.
Figure 1. Maximum clade credibility tree of different RAB5-GTPases calculated with BEAST.
Branches with posterior probabilities (PP)≥0.95%, which reflect the posterior median node heights for the clades, and ML bootstrap support (ML B)≥70% were considered as strongly supported. Saccharomyces cerevisiae (NM_001183508) constitutes as outgroup. Plant groups and higher plant clades are highlighted in different colours. The bar specifies substitutions per site.
Figure S1A, Supporting Information, shows the protein sequence of CaARA7 aligned with several conventional RAB5 protein sequences. CaARA7 protein sequence alignment with land plants such as A. thaliana (higher plant), Physcomitrella patens (moss) and Selaginella moellendorffii (spikemoss), as well as with other green algae (Micromonas sp. and Volvox carteri) revealed several highly conserved regions specific for RAB GTPases.
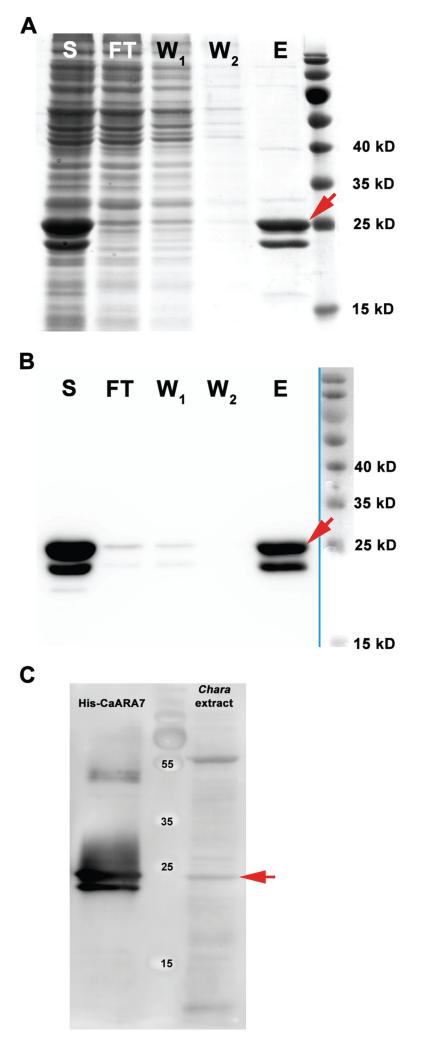
SDS-PAGE and western blot of recombinant CaARA7 and western blot of C. australis protein extract are shown in Figure 2. Figure 2A,B illustrates the purification of recombinant His-tagged CaARA7 from E. coli lysate. Both, SDS-PAGE and western blot His-tag detection revealed a CaARA7 double band at about 25 kDa. For detection of CaARA7 in Chara protein extracts a polyclonal antibody against CaARA7 antibody was used (Figure 2C). Recombinant His-tagged CaARA7 was used as a control. Interestingly, again two bands were detected in the recombinant His-CaARA7 sample, but only one band at about 25 kDa was detectable in Chara protein extract.
Figure 2. Purification of His-tagged CaARA7 and detection of CaARA7 in Chara protein extracts.
SDS-PAGE (A) and western blot (B) of different purification steps of recombinantly expressed Chara australis ARA7. Purification was performed by Ni-TED affinity column. Lanes are indicated as follows: S = soluble protein fraction of E. coli lysate; FT = flow through of affinity column; W1 and W2 = wash step one and two; E = eluted purified His-tagged CaARA7 protein (red arrows). Important molecular mass marker bands are indicated (right lane). C) Western blot of C. australis protein extract and purified His-tagged CaARA7. Recombinant His-CaARA7 was used for control. A polyconal antibody against ARA7 detected a prominent band with a molecular mass of about 25 kD in the protein extract of Chara (red arrow) and two bands in recombinant His-CaARA7. Important molecular mass marker bands are indicated (middle lane).
Intrinsic GTPase activity of CaARA7 and mutants
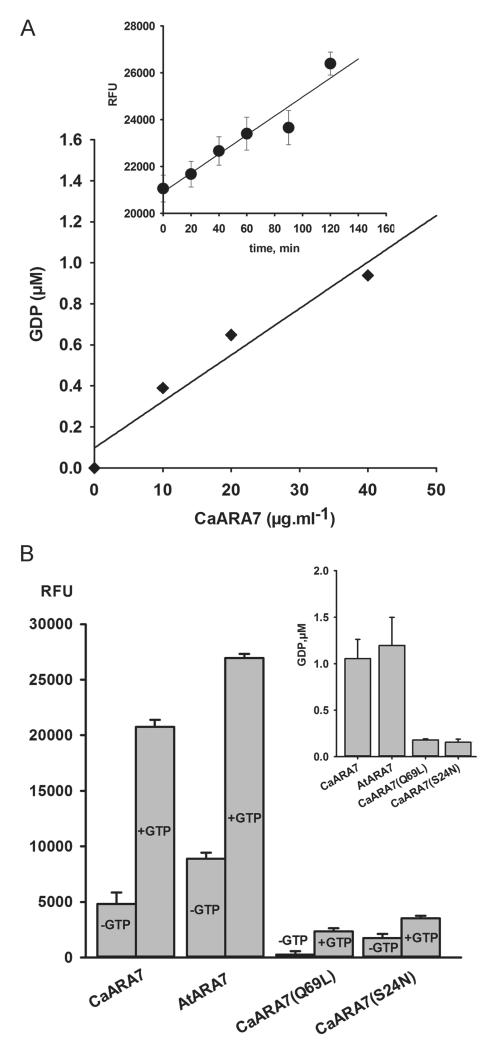
As RAB5 GTPases are known to display low intrinsic activity and we identified CaARA7 as being a member of this family, we were interested in investigating its ability to hydrolyse GTP in vitro. For this, we used a highly sensitive fluorescent GDP tracer-displacement assay (see Materials and Methods section) to measure GDP production upon incubation with GTP of recombinant wild-type and point mutated ARA7 proteins. Firstly, we detected an obvious correlation between the amount of released GDP and the concentration of CaARA7 after incubation with 10 μM GTP (Figure 3A). For a given CaARA7 concentration (40 mg/mL) the fluorescence signal increased proportionally to the incubation time, indicating continuous GDP production as a result of GTP hydrolysis (inset in Figure 3A). A further step was to compare the GTPase activity of recombinant CaARA7 and that of AtARA7 isolated and purified under the same conditions. As shown in Figure 3B (first two bar groups) the fluorescence signal increased significantly after 1 h incubation with 10 μM GTP, as compared to the control fluorescence values in the absence of the substrate. This clearly indicates that both CaARA7 and AtARA7 possess intrinsic GTPase activity. The extent of GDP production (inset in Figure 3B: 1.05 ± 0.20 μM for CaARA7 and 1.19 ± 0.30 μM for AtARA7) was similar when identical amounts of recombinant ARA7 proteins (40 mg/mL) were used (p = 0.436). A further approach in order to characterize the intrinsic GTPase activity of CaARA7 was to examine the effect of substitution mutations of particular residues which are known to play a key role in nucleotide binding and hydrolysis in RAB5 proteins and are situated within the first third from the N-terminus (Figure S1C) (23–26). A good candidate in this respect is the CaAra7Q69L mutant, where Gln at position 69 in the nucleotide binding G3 motif is substituted by Leu. In cognate RAB5 proteins this mutation is known to induce a very high affinity for GTP and to stabilize the GTP-bound, ‘constitutively active’ conformation, these RABs being defective in hydrolysing GTP (26,27). As expected, CaAra7Q69L displays a negligible GTPase activity (Figure 3B). Another interesting CaARA7 mutant with respect to the intrinsic GTPase activity is CaAra7S24N, where a Ser to Asn substitution at position 24 in the G1 motif dramatically lowers the affinity for GTP and stabilizes the RAB5 proteins in the GDP-bound ‘inactive’ form (Figure S1C) (26, 28). As can be seen in Figure 3B, this mutant has an approximately fivefold lower GTPase activity compared to the wild-type CaARA7. It has to be stressed that the method used only allows the measurement of a global, combined result of all reaction steps, i.e. GTP-binding and hydrolysis and GDP release so that any change in the rate of the afore mentioned steps will be observed as a change in the overall GTPase activity. In the case of the S24N mutant, the slow rate-limiting GDP-dissociation step (24, 27) together with the impaired GTP-binding ability (23) are most probably equally responsible for the low GDP-production.
Figure 3. Detection and analysis of the steady-state intrinsic GTPase activity of recombinant wild-type and point mutated ARA7 proteins.
A) GDP production by GTP hydrolysis in relation to CaARA7 concentration. Varying amounts of purified recombinant protein were incubated at room temperature in 96 well plates for 1 h with 10 μM GTP, then incubated with Transcreener®GDP stop-and-detection reagents and the fluorescent signal was measured with a fluorescence plate reader. GDP product formation was calculated by converting the fluorescent signal into concentration values using a standard curve (for details see 12). The inset shows GTP hydrolysis reaction progression for CaARA7 as relative fluorescence units (RFU) of displaced GDP-Alexa Fluor 594 tracer plotted against the incubation time. Samples containing 40 mg/mL were incubated with 10 μM GTP for different time periods (four replicates for each time point) and the fluorescent signal was measured as stated above. B) GTPase activity of CaARA7, AtARA7 and point mutated CaARA7 displayed as fluorescence signal in the absence and in the presence of 40 mg/mL protein, 10 μM substrate and after 1 h incubation. CaARA7 and AtARA7 hydrolyze GTP to GDP leading to a substantial increase of free GDP-fluorescent tracer as compared to the samples lacking substrate. For the point mutated CaAra7Q69L and CaAra7S24N the fluorescent signal (+GTP) is negligibly above the control signal (−GTP), indicating that these samples are GTPase deficient. In the inset the absolute GDP amounts produced within 1 h by the different ARA7 proteins are shown.
CaARA7 localization at MVEs in Chara internodal cells
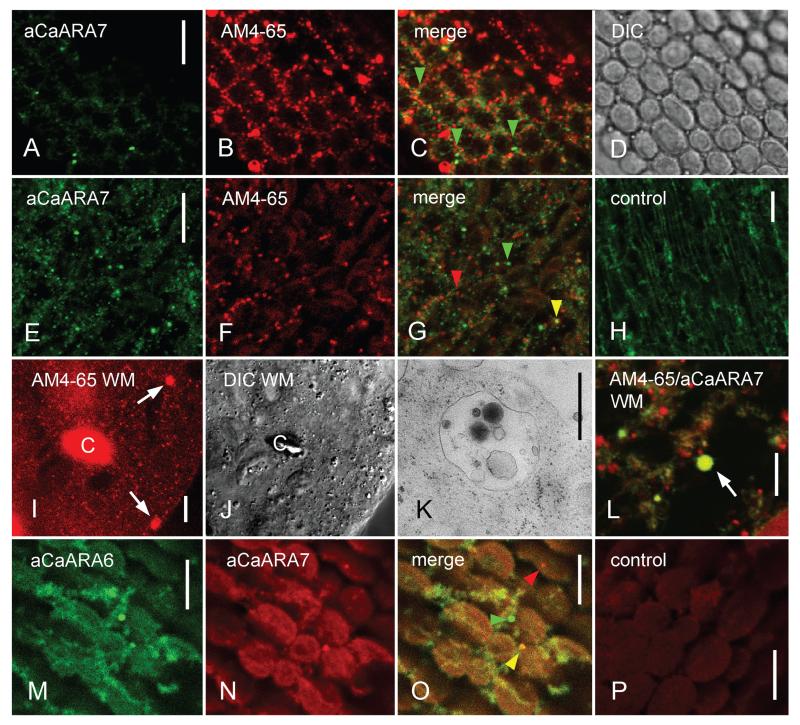
In cells of higher plants, ARA7 is found at MVEs which can be stained by endocytic tracers like the fluorescent styryl dyes FM4-64 and AM4-65 (e.g. 6, 17, 29). In order to compare the localization of CaARA7 with that of the FM-dyes we pulse-labelled internodal cells and fixed them for immunofluorescence after 1 h wash in AFW. During that period all putative endosomal organelles were stained by FM4-64 or AM4-65 and remained labelled for several hours without toxic effects (30). The cytoplasm of Chara internodal cells consists of a stationary cortex, containing files of helically aligned chloroplasts (Figure 4A–D) and the streaming endoplasm (Figure 4E–G). Immunofluorescence of internodal cells with the anti-CaARA7 revealed only few punctate structures with a size of up to one μm in the cortex and located between charasomes and the stationary chloroplasts (Figure 4A–D). Charasomes are convoluted regions of the plasma membrane and are required for efficient photosynthesis-dependent carbon uptake (31). Considerably more CaARA7-positive organelles were present in the endoplasm (Figure 4E) consistent with the distribution of MVEs (see below). Some of them colocalized with organelles stained by AM4-65 (Figure 4F) but organelles carrying only one fluorescence were also abundant (Figure 4G). A similar staining pattern could be obtained with an antibody against AtARA7 (5; Figure S2 Part 1).
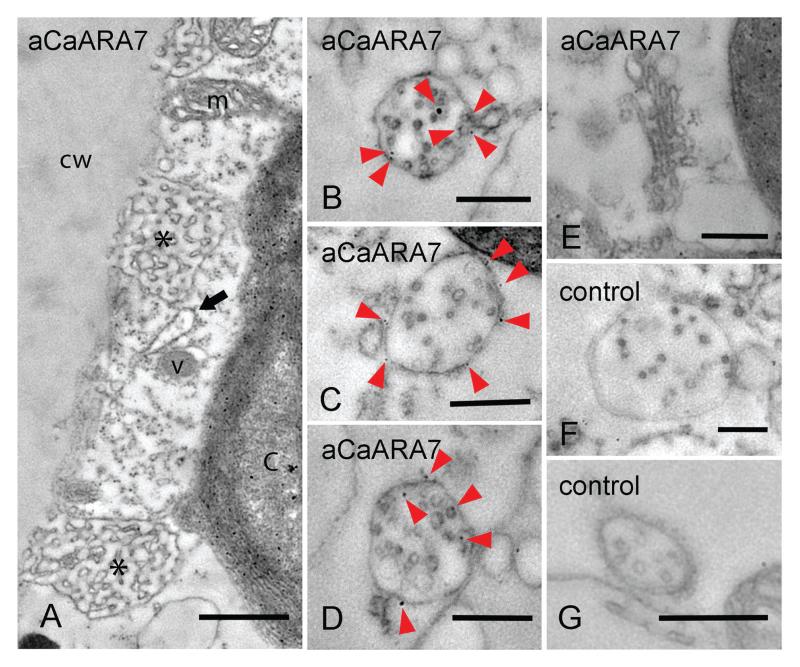
Figure 4. Immunofluorescence with anti-CaARA7 and anti-CaARA6 in control untreated internodal cells of Chara australis and in internodal cells treated with wortmannin (WM).
A–D) In the cortex few CaARA7 positive organelles (A) are present between AM4-65 labelled structures (mostly charasomes, B). Arrow heads in the merged image (C) indicate CaARA7 positive organelles; the corresponding differential interference contrast (DIC) image shows the stationary chloroplasts. E–G) Endoplasmic organelles labelled by anti-CaARA7 (E) partially colocalize with organelles stained with AM4-65 before immunolabelling (F). Yellow arrow heads in the merged image (G) indicate colocalizations, green and red arrow heads indicate organelles stained by anti-CaARA7 or AM4-65 only. H) Unspecific diffuse staining in the endoplasm after immunolabelling with PIS instead of primary antibody (control for E). I and J) Cytoplasmic droplet from an AM4-65 stained and WM-treated internodal cell. Arrows in (I) point to WM compartments scattered between smaller punctate organelles; (J) is the corresponding DIC image. ‘C’ = autofluorescent chloroplast. K) Electron micrograph of WM compartment. L) WM compartment labelled by AM4-65 (red) and by anti-CaARA7 (green) is indicated by an arrow. M–O) Endoplasmic organelles labelled by anti-CaARA6 (M) and by anti-CaARA7 (N). In the merged image (O) the yellow arrow head indicates an organelle labelled by both antibodies, green and red arrow heads indicate organelles labelled only by anti-CaARA6 or only by anti-CaARA7. P) Unspecific weak staining in the endoplasm after immunolabelling with PIS instead of primary antibody (control for N). Bars are 500 nm (K) and 10 μm.
In order to further characterize the CaARA7 carrying structures we used WM which causes homotypic fusion of MVEs into large compartments (32). For these experiments we used internodal cells which were pulse-labelled with AM4-65 and washed in artificial fresh water for 1 h as described above. When these internodal cells were treated with 25 μM WM for 2 h larger compartments up to 2330 nm in diameter appeared in the endoplasm. In contrast, the maximum size of AM4-65 labelled organelles in untreated cells was 1460 nm (for statistical analysis see Figure S2 Part 2). Because of the rapid streaming the WM compartments were difficult to visualize in the intact cell. They were, however, distinct in cytoplasmic droplets squeezed out from internodal cells (Figure 4I–J). The fine structure of Chara WM compartments is shown in Figure 4K and is similar to that described in higher plant cells (e.g. 33). Immunofluorescence of AM4-65-stained internodal cells proved that the large WM-induced compartments were recognized by anti-CaARA7 (Figure 4L) indicating that CaARA7 is present on MVEs.
In Arabidopsis root epidermal cells the fluorescence of AtARA7 constructs partially colocalize with those of the enigmatic RAB5 GTPase AtARA6 (6,8). Double immunofluorescence with antibodies against CaARA7 and CaARA6 likewise showed partial colocalization (Figure 4M–O). Specific labelling by the polyclonal antibodies was confirmed in negative controls using pre-immune serum instead of the first antibody (Figure 4H,P). Incubation with secondary antibody alone gave no signal (data not shown).
The antibody raised against CaARA7 was not suitable for the staining of EM sections but yielded satisfactory results after pre-embedment immunolabelling. Figure 5A shows an electron microscopical section from the periphery of an internodal cell which was fixed and treated with antibodies before embedment in acrylic resin. Smooth plasma membrane regions alternate with convoluted domains, the charasomes, which are often located in close proximity to cortical chloroplasts and mitochondria. Unspecific accumulation of gold beads was found at the chloroplasts but other organelles like vesicles, mitochondria or endoplasmic reticulum were not labelled. In the endoplasm, the CaARA7 antibody recognized epitopes at the surface, and occasionally within MVEs (Figure 5B–D). No or very few gold beads were found on other endoplasmic organelles such as Golgi bodies (Figure 5E). The specificity of MVE labelling by anti-CaARA7 was confirmed using cells incubated with pre-immune serum instead of the primary antibody (negative controls; Figure 5F,G).
Figure 5. Immunodetection of ARA7 at the fine structural level in internodal cells of Chara australis (A–E) and controls (F, G).
A) Slightly oblique section through the cortex of an internodal cell. Cell wall (cw), charasomes (asterisks), endoplasmic reticulum (arrow), vesicles (v) and mitochondria (m) are not labelled. Unspecific accumulation of gold particles is seen in the chloroplast (c). B–G) Longitudinal sections through the endoplasm. Numerous gold particles (arrow heads) are seen at multivesicular endosomes (B–D), but not at a Golgi body (E). Gold particles are absent from multivesicular endosomes treated with pre-immune serum instead of primary antibody (F, G; negative controls). Bars are 500 nm.
Transient expression of CaARA7 and organelle markers in tobacco leaf epidermal cells
Suitable methods for genetic transformation are not available for characean green algae. Therefore, we transiently expressed fluorescently tagged CaARA7 in epidermal cells of tobacco leaves (Nicotiana benthamiana) under the control of A. thaliana ubiquitin 10 promoter (34); compare (12). Either GFP or mCherry were fused to the N-terminus of ARA7 proteins and to the C-terminus of ARA6 proteins, respectively.
In vivo analysis of the localization of the GFP-CaARA7 fusion protein by confocal laser scanning microscopy (CLSM) revealed its location on small, punctate organelles with diameters ranging between 0.2 and 1.1 μm (mean ± SD: 0.65 ± 0.01; n = 280; Figure 6A,E, Figure S3 Part 1A and E, Part 2A). These organelles moved along linear or curved tracks, interrupted by oscillating motions (not shown). The GFP-CaARA7 fluorescence partially overlapped with that of FM4-65-stained endosomes (Figure S3 Part 1B–D). The Pearson’s and Spearman’s correlation coefficients took values between 0.476 and 0.784 and, respectively, 0.489–0.767 indicating a good colocalization of the fluorescent signals. GFP-CaARA7 was not detectable in organelles and membranes stained with the acidotropic LysoTracker Red which selectively accumulates in acidic compartments including lysosomes, autolysosomes and vacuoles (Figure S3 Part 1E–H) (35). This result is not surprising, since ARA7 proteins are considered to be basically found on MVB/LPVC, the lumen of which was recently shown to be neutral (pH 7.1) in tobacco epidermal cells (36).
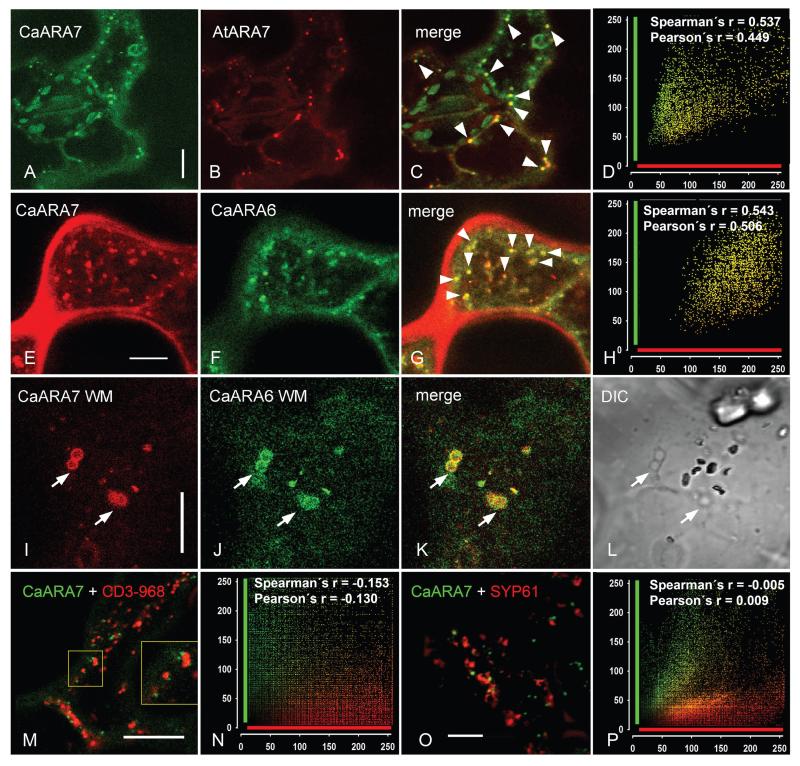
Figure 6. Transient co-expression of CaARA7 with organelle markers in epidermal cells of control untreated tobacco leaves and in epidermal cells of leaves infiltrated with wortmannin (WM).
A–D) In untreated cells the fluorescence of GFP-CaARA7 labelled organelles (A) frequently overlaps with the fluorescence of mCherry-AtARA7-labelled particles (B). Arrow heads in the merged image (C) indicate colocalizations; (D) is the corresponding scatter plot. E–H) Colocalization of mCherry-CaARA7 with CaARA6-GFP in control untreated epidermal cells of tobacco leaves (arrow heads in the merged image (G); (H) is the corresponding scatter plot). I–L) WM compartments (arrows) carry both mCherry-CaARA7 (I) and CaARA6-GFP (J). K) Is the merged image. Note characteristic WM compartments in the corresponding DIC image (L). M and N) Co-expression of wild-type GFP-CaARA7 (green) with the Golgi marker CD3-968-mCherry (red) (M, merged image) and cumulative scatterplot (N) show no colocalization. O and P) Co-expression of wild type green fluorescent GFP-CaARA7 with the red fluorescent TGN marker SYP61-mRFP (O, merged image) and cumulative scatterplot (P) illustrate lack of colocalization. Bars are 10 μm.
In order to get more information about the identity of the GFP-CaARA7 localizing organelles in tobacco leaf epidermal cells, we compared the distribution of CaARA7 with that of AtARA7 using pGI-AtUBQ10p∷GFP∷CaARA7 with pGI-AtUBQ10p∷mCherry∷AtARA7. When expressed alone, tagged AtARA7 localized to organelles which had a similar dynamic behaviour and a similar size as the fluorescent CaARA7 particles (data not shown). Co-expression of both proteins resulted in a high degree of overlap (Figure 6A–D) since Pearson’s coefficient ranged from 0.551 to 0.860 and Spearman’s coefficient was between 0.495 and 0.851 (n = 244).
We next compared the localization of CaARA7 with that of RAB5 GTPase ARA6 (5,12). When transiently expressed in tobacco mCherry-CaARA7 and CaARA6-GFP showed partial colocalization as illustrated in Figure 6E–H. Colocalization analysis performed on 881 punctate structures in 13 images produced a mean value for the Pearson’s coefficient of 0.541 and for the Spearman’s coefficient 0.523.
Interestingly, CaARA7 and CaARA6 completely colocalized on compartments induced by WM. The WM compartments (Figure 6I–L) had a mean size of 1.57 ± 0.02 μm (n = 142; Figure S3 Part 2B).
These data suggest not only that CaARA7 highly colocalizes with AtARA7 but also that there is a subpopulation of endosomal vesicles bearing both CaARA7 and CaARA6 when transiently expressed in tobacco epidermal cells. Similarly to these findings, only partial colocalization of fluorescently tagged AtARA6 and AtARA7 has been reported for Arabidopsis root epidermal cells (8). In order to prove that our transformation method yielded reliable results we co-expressed mCherry-AtARA7 and AtARA6-GFP in leaves of N. benthamiana. The results confirmed partial colocalization of the fluorescent constructs in untreated cells (Figure S3 Part 3A–D) and the WM compartments carried either AtARA7 (not shown), or AtARA6 or both proteins (Figure S3 Part 3E–H).
Kotzer et al. (17) using the same transient expression system found that AtARA7(AtRABF2b) localizes at least partially to the Golgi. Therefore we performed a co-expression with the red fluorescent Golgi marker CD3-968-mCherry (37) (Figure 6M). Obviously, CaARA7 labelled structures were distinct from the Golgi vesicles, which accounted for the low and negative values of both Pearson and Spearman correlation coefficients (Figure 6N). However, as can be seen from the inset in Figure 6M, there was sometimes a close association between the structures labelled green and red, even if no overlapping occurred. In a further attempt to determine the nature of the CaARA7 positive organelles, we used the co-expression with fluorescently tagged SYP61, a marker for TGN, frequently used in endosomal protein localization studies (38). As can be seen in Figure 6O,P the compartments labelled by SYP61-mRFP did not overlap with those bearing GFP-CaARA7, indicating that the TGN is not a target organelle for CaARA7.
CaARA7 localization affected by point mutations
The localization of RAB5 GTPases depends on their membrane anchoring domain and on proper nucleotide binding (e.g. 3). In order to characterize the membrane binding properties of CaARA7 we introduced point mutations corresponding to those used by Kotzer et al. (17) in AtARA7 (AtRABF2b) and transiently expressed the GFP-tagged mutants in tobacco leaf epidermal cells. The location of the different point mutations on the protein sequences of Arabidopsis and Chara ARA7 proteins is shown in Figure S1C.
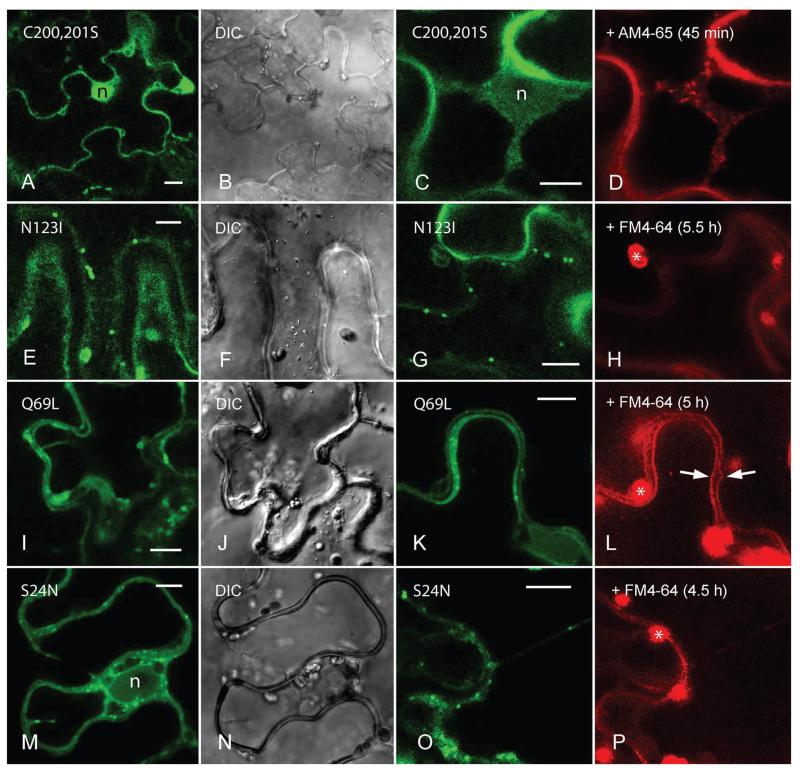
The GFP fusion of CaAra7C200,201S mutant lacking C-terminal isoprenylation was distributed throughout the cytosol and within the nucleus as expected for CaARA7 anchoring to membranes via isoprenylation (Figure 7A,B) (17). The guanine nucleotide-binding deficient mutant CaAra7N123I was mostly exclusively and strongly expressed in the cytosol and in the nucleus (not shown) or localized to organelles which were larger (about 1 μm in diameter) and less mobile than the organelles carrying the CaARA7 wild type (Figure 7E,F). The GTP-bound mutant CaAra7Q69L resided on slowly moving ring-like structures and on faster travelling dots with a size between 1 and 2 μm (Figure 7I,J). The GDP-bound mutant CaAra7S24N was found in the cytosol, as expected for inactive RAB GTPases, but also on mobile particles up to 1 μm in diameter and in the nucleus (Figure 7M,N).
Figure 7. Localization pattern of transiently expressed GFP-tagged CaARA7 mutants in leaf epidermal cells of Nicotiana benthamiana and comparison with organelles stained by endosomal dyes.
A and B) The CaAra7C200,201S mutant lacking C-terminal isoprenylation responsible for membrane anchoring is distributed throughout the cytoplasm and within the nucleus (n in A; B shows the corresponding DIC image). C and D) AM4-65-stained endosomes in the cytoplasm near the nucleus a cell expressing the CaAra7C200,201S mutant. E and F) The guanine nucleotide-binding deficient mutant CaAra7N123I localizes to approximately 1 μm sized mobile structures (E; F is the corresponding DIC image). G and H) FM4-64 is not internalized by cells expressing fluorescently tagged CaAra7N123I (autofluorescent chloroplasts are marked by asterisks). I and J) The GTP-bound mutant CaAra7Q69L resides on ring-like structures and dots with a size between 1 and 2 μm (I; J is the corresponding DIC image). K and L) The tonoplasts of cells expressing CaAra7Q69L become labelled by FM4-64 after 5 h staining (arrows). M and N) The GDP-bound mutant CaAra7S24N is found in the cytoplasm, on mobile particles sized up to 1 μm and in the nucleus (n in M; N is the corresponding DIC image). O and P) FM4-64 is not internalized in cells expressing the CaAra7S24N mutant. Bars are 10 μm.
In order to find out whether the presence of fluorescently tagged CaAra7 mutants affected endocytosis in leaf epidermal cells of Nicotiana bentamiana we applied the fluorescent endocytic tracers FM4-64 or AM4-65 and followed their internalization over a period of up to 22 h. In control cells expressing wild-type GFP-CaARA7, FM4-64-stained organelles could be detected already 30 min after dye addition (compare Figure 7C,D) and the tonoplast became labelled after about 5 h (compare Figure 7L). Similar results were obtained with cells expressing CaAra7C200,201S (Figure 7C,D) suggesting that this mutant – because of its exclusively cytosolic localization – cannot disturb proper functioning of the endogenous, membrane-bound tobacco ARA7. The rate of FM-internalization was also not affected by the constitutively active GTP-bound mutant CaAra7Q69L (Figure 7K,L) consistent with findings in HeLa cells (39). Internalization of the styryl dyes was, however, significantly delayed when fluorescently tagged CaAra7N123I (Figure 7G,H) or CaAra7S24N mutants (Figure 7O,P) were transiently expressed in tobacco leaves. In their epidermal cells AM4-65- or FM4-64-fluorescent organelles appeared in the cytoplasm only after several hours’ incubation if ever and the tonoplast never became labelled. Our observations are in line with results obtained with animal cell cultures in which overexpression of RAB5 GTPase mutants which were either GDP-bound or deficient in guanine nucleotide-binding had a severe inhibitory effect on the endocytic pathway (40–42).
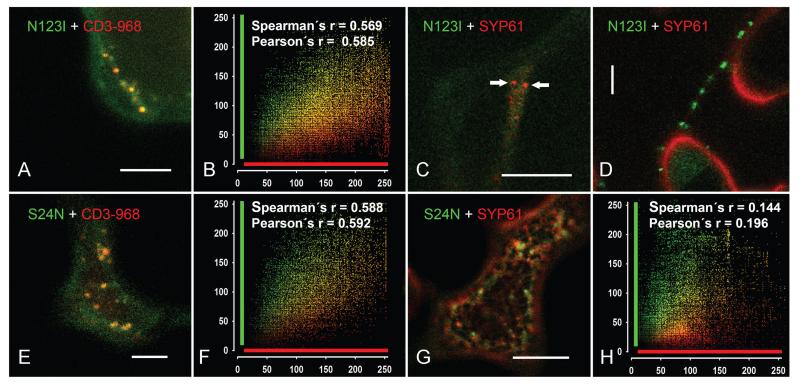
In order to clarify the nature of the organelles bearing the CaAra7N123I or the CaAra7S24N mutants, we performed co-expressions with either the Golgi marker CD3-968 or the TGN marker SYP61, both fluorescently tagged with mRFP. As illustrated in Figure 8A,B, the structures carrying the mutant CaAra7N123I significantly overlapped with the compartments labelled by the Golgi marker. Interestingly, in our hands, the expression of the TGN marker was impaired by the co-expression with the NI-mutant of CaARA7 (Figure 8C,D) and the fluorescent signal of SYP61 was mostly cytosolic. Punctate mobile organelles, probably corresponding to the TGN compartment, could be identified only in 17 out of approximately 4000 epidermal cells investigated (Figure 8C). In these very few cells with SYP61-positive organelles, however, the fluorescent signal of the NI-mutant was always cytosolic (Figure 8C). On the other hand, fluorescently tagged SYP61 was always cytosolic when cells expressed punctate CaAra7N123I-positive organelles. Therefore, colocalization studies were not possible.
Figure 8. Mutant CaARA7 localize to different compartments: Co-expression with Golgi and TGN markers in tobacco epidermal cells.
A and B) Merged image of a cell co-expressing GFP-CaAra7N123I with the Golgi marker CD3-968-mCherry (A) and cumulative scatterplot (B) indicate good colocalization. C and D) Co-expression of GFP-CaAra7N123I and the TGN marker SYP61-mRFP. Typical merged images show cells expressing either organelles carrying SYP61-mRFP (arrows in C) or organelles labelled with GFP-CaAra7N123I (D) whereas the other construct is cytosolic only. The number of particles occurring simultaneously with both constructs was not sufficient for colocalization analysis. E and F) Co-expression of GFP-CaAra7S24N with the Golgi marker CD3-968-mCherry (E) and cumulative scatterplot (F) show good colocalization (G and H) Co-expression of GFP-CaAra7S24N with the TGN marker SYP61-mRFP (G) and the cumulative scatterplot (H) show no colocalization. Bars are 10 μm.
The fluorescent particles with the SN-mutant of CaARA7 colocalized to a considerable extent with the Golgi marker CD3-968 (Figure 8E,F) but not with the compartments highlighted by the TGN marker SYP61-mRFP (Figure 8G,H).
Discussion
CaARA7 shows high protein sequence similarities to conventional RAB5 members
ARA7 is one of the conventional members of plant RAB5 GTPases. This study shows that characean green algae possess an ARA7-like protein with a high sequence similarity to other plant ARA7s.
Phylogenetic analyses of conventional RAB5 GTPase sequences of C. australis and various other plant and algae species were performed (see Figure 1). These analyses revealed that C. australis seems to be more closely related to ferns and mosses than to higher plants or other green algae. The green algae group (containing species of the genera Chlorella, Micromonas, Ostreococcus and Volvox) is located at the basis of the phylogenetic tree and thus shows a relationship to the characean as well as all other aligned plant sequences. However, as a result of poor support values the relationships within this group are not clarified. Similar results were indicated by a maximum likelihood analysis in a study of Ebine and coworkers performed in 2011 (8), where two algal sequences were included in tree calculation. Nonetheless, the positions of all algal sequences used in this study would most likely settle if more RAB5 GTPase sequences would be available. Especially sequences from other algal, fern and moss species would be important for more precise calculations. In our analysis higher plants are well separated of other groups and have no significance to establish a supported relationship with CaARA7. The group of higher plants investigated in our study is divided into three main clades. Interestingly, there are two similar Nicotiana tabacum RABF2a-like sequences: one in clade B and the other one in clade C. When aligned those sequences reveal only 70% similarity. Similar results were obtained by Ebine and coworkers (8), who also show such classification of RAB5 GTPases from the same species (e.g. Populus trichocarpa and Vitis vinifera). Calibrated phylogenetic analyses would be necessary to show the evolutionary development, in our case of both N. tabacum RABF2a-like proteins, which certainly could explain the sequence differences and origins.
Protein sequence alignment of the C. australis protein with RAB5s of land plants and other green algae showed several highly conserved regions that are specific for all members of the Ras superfamily of small GTPases (compare Figure S1A). First, there is a set of conserved G box GDP/GTP-binding motif elements (highlighted in green in Figure S1A) that comprise the following sequence patterns: G1, GxxxxGK[S/T]; G2, T; G3, DxxG; G4, [T/N][K/Q]xD; and G5, [C/S]A[K/L/T]; (where ‘x’ represents any amino acid). The N-terminally located G1 motif (also called Walker A motif) functions in positioning the triphosphate moiety of the bound nucleotide and forms the phosphate binding loop (P-loop). The Walker B motif contains a conserved glycine residue typically within the DxxG signature (G3 motif) and a distal [T/N][K/Q]xD G4 motif, which is responsible for guanine specificity (43,44). Amino acids involved in putative binding sites of different interacting proteins such as GDP-dissociation inhibitor (GDI), GEF, and different effector proteins are also marked in Figure S1A.
The C-terminus is mainly hypervariable, but usually ends with a conserved motif containing two cysteine residues. This motif consists of one of the following sequences: CC, CxC, CCxx and CCxxx (‘x’ represents any amino acid). In case of C. australis as well as in A. thaliana and P. patens the motif sequence is CCA (compare Figure S1A). The two cysteins are the sites of isoprenylation and therefore significant for membrane binding and GDI recognition (highlighted in red in Figure S1A) (45–48). The role of the C-terminal hypervariable domain (HVD) of RAB GTPases is controversially discussed. On the one hand HVD is supposed to act as a subcellular targeting signal (49,50), while on the other hand different interacting proteins (e.g. RAB effectors, GEFs) could also exhibit this function (51–54). Li and coworkers published a study in 2014 on different HVDs of human RAB GTPases (55). They showed that the HVD of RAB5a, which is closely related to ARA7 of plants, is not required for correct protein function as well as subcellular localization. They assumed that HVD of RAB5 serves more likely as an anchoring chain by connecting the GTPase domain physically with the target membrane (55).
Native CaARA7 was detected as a single band of about 25 kDa in protein extracts of C. australis (Figure 2C). On the other hand recombinantly expressed CaARA7 is always detected as a double band (Figure 2A–C). This can be explained by different states of post-translational modification in the excess of recombinant protein produced by E. coli. Thus, one band represents the mature and therefore fully processed form of CaARA7 while the second band shows an intermediate, not fully processed protein. This was confirmed by C-terminal deletions of RAB5s, which disabled post-translational isoprenylation. As a result only one band, the intermediate form of the protein, was visible (23,28,56).
Intrinsic GTPase activity of CaARA7 and mutants
Within cells the GTPase cycle of RAB proteins is controlled by various regulatory proteins: GTPase activating protein, GDP-dissociation inhibitor (GDI), GDI displacement factor (GDF) and GEF. However, RAB5 GTPases are also known to possess a low intrinsic GTPase activity as documented e. g. for AtARA6 and CaARA6 (5,12). In order to find out whether CaARA7 is able to hydrolyze GTP in vitro and in the absence of regulatory proteins we used a fluorescent assay to measure GDP production of recombinant AtARA7 and CaARA7 as well as that of mutated forms of CaARA7 in the presence and in the absence of GTP. These measurements confirmed that wild-type CaARA7 has a similar intrinsic GTPase activity as wild-type AtARA7. GTPase activity of the ‘constitutively active’ mutant CaAra7 Q69L which is defective in hydrolysing GTP was negligible as expected (26, 27) and GDP production is also significantly reduced in the ‘GDP-bound’ CaAra7S24N which has a very low affinity for GTP (26,28).
Localization of ARA7-like protein in Chara internodal cells and transient expression in tobacco leaf epidermal cells
The localization of CaARA7 in Chara internodal cells was studied with antibodies raised against CaARA7 (this study) and AtARA7 (5). Immunogold labelling of untreated cells and immunofluorescence of WM compartments strongly suggest that these antibodies recognize a CaARA7 epitope on MVEs as described for AtARA7 (16) and tobacco (17,29). The Golgi bodies, likewise reported as AtARA7 positive (17) were not labelled consistent with the absence of colocalization when fluorescently tagged CaARA7 was transiently expressed together with the fluorescently tagged Golgi marker (G-rb CD3-968 of Nelson) (37) in tobacco leaf epidermal cells. Furthermore, comparison of GFP- CaARA7 and the TGN marker SYP61-mRFP showed that they distributed to completely distinct compartments.
Immunofluorescence of internodal cells revealed partial colocalization of anti-CaARA7 and anti-CaARA6 consistent with the differential distribution of these proteins. CaARA7 appears to be exclusively present at MVEs (this study) whereas CaARA6 localizes not only to MVEs but also to the TGN and to the plasma membrane in Chara internodal cells (12).
When fluorescent constructs of CaARA7 and AtARA7 were transiently expressed in tobacco we frequently observed a high degree of colocalization as expected from the similar sequence and the EM data. There was also partial colocalization when mCherry-CaARA7 was simultaneously expressed with CaARA6-GFP similar to the partial overlapping of fluorescently tagged constructs of ARA7 and ARA6 reported for Arabidopsis plants and suspension-cultured cells (6,8). On the other hand, we found complete colocalization of CaARA7 and CaARA6 at WM compartments irrespective whether these proteins were immunolocalized in Chara internodal cells or transiently expressed as fluorescent constructs in tobacco leaf epidermal cells. In contrast, treatment of roots of Arabidopsis thaliana expressing fluorescent constructs of AtARA7 and AtARA6 with WM-induced swelling of both compartments but did not result in compartment mixing (8). We therefore co-expressed AtARA7 and AtARA6 in tobacco leaf epidermal cells and found that under these conditions WM compartments carried either the AtARA7 fluorescence or the AtARA6 fluorescence or both. The reasons for these different results remain elusive. They might reflect the properties of the proteins but artefacts due to the presence of fluorescent tags, the use of non-native promoters or the components of the ‘host’ cytoplasm are likely to influence protein localization, especially in cells stressed by infiltration or by an inhibitor. Apart from this, these findings illustrate how different conditions affect the fine tuning of membrane trafficking by these RAB5 GTPases.
Our data about the subcellular localization of GFP-tagged CaARA7 mutants were similar to those obtained with point mutations in AtARA7 (17). The GFP-tagged mutant CaAra7C200,201S lacks the C-terminal isoprenylation responsible for membrane anchorage and was therefore located in the cytosol. A cytosolic localization was also found for the nucleotide-free CaAra7N123I and the GDP-bound CaAra7S24N but the fluorescent constructs also localized to punctate organelles which, however, had a different size and behaviour as those labelled by wild-type GFP-CaARA7. Analysis of the co-expression of the nucleotide binding-defective fluorescently tagged N123I mutant of CaARA7 together with the fluorescent Golgi marker CD3-968-mCherry in tobacco epidermal cells prompted us to conclude that the organelles occasionally labelled by this mutant are to a substantial extent Golgi bodies. Likewise, the good colocalization of the S24N mutant with the same marker indicates that CaAra7S24N too is at least partially retained to the Golgi in accordance to Kotzer et al. (17) who reported that the S24N mutant of AtARA7 colocalizes with a Golgi marker. Taken together, these findings suggest the hypothesis that the ectopically expressed CaARA7 proteins reach their final destination (MVE/LPVC) in a Golgi-dependent manner. The involvement of CaARA7 in the late but not in early endosomal trafficking events is supported also by the total lack of colocalization of wild type as well as N123I and S24N mutants with the TGN-marker SYP61 under this experimental condition. The GTP-bound mutant GFP-CaAra7Q69L localized to ring-like structures which were similar to the enlarged MVEs described from transgenic Arabidopsis cells overexpressing AtAra7Q69L (57). The significantly delayed internalization of the endocytic tracer AM4-65 in cells expressing the nucleotide-free mutant protein CaAra7N123I and in cells expressing the GDP-bound mutant CaAra7S24N as compared to the wild-type CaARA7 is consistent with a role of ARA7 in endocytosis and highlights the importance of a functional GTPase cycle in membrane trafficking (4).
To summarize, the results of this study confirm the presence of a conventional RAB5/RABF2 GTPase in characean algae. The sequence, the intrinsic GTPase activity of CaARA7, and the localization at MVEs are similar to those of ARA7 from higher plants suggesting a similar function in endosomal trafficking towards the vacuole.
Materials and Methods
Plant material and culture conditions
Thalli of C. australis R.Br. were grown in 10–50 L aquaria filled with distilled water on a substrate of soil, peat and sand. Fluorescent lamps enabled a 16/8 h light/dark cycle during which the temperature was about 20°C. For microscopical studies, non-elongating, mature internodal cells of branchlets were harvested 1 day prior to experiments, trimmed of neighbouring internodal cells and incubated overnight in artificial fresh water (10−3 M NaCl, 10−4 M KCl, −4 M CaCl2). Whole thalli were used for biochemical and molecular biological analysis.
Nicotiana benthamiana plants were grown on standard fertilized soil (type ED73) in a growth chamber with following parameters: 16/8 h light/dark cycle, temperature adjusted to 23°C during light and to 22°C during dark phase, and relative humidity set to 60%. Four weeks old plants were used for Agrobacterium-mediated transient transfection as described (12). Protein expression was observed after 3–5 days.
Phylogenetic analyses
An alignment of the nucleotide sequences of C. australis and 27 diverse species of higher plants, ferns, mosses and green algae obtained from NCBI GenBank were generated using the program Geneious 6.1.2 created by Biomatters. The sequence Saccharomyces cerevisiae (NM_001183508) served as outgroup. The web-portal Alter (Alignment Transformation Environment 58) was applied to convert the alignment from FASTA format into a Nexus-file. Then, the dataset was analysed with the program JMODELTEST 2.1.1 using Akaiko Information Criterion (AIC) scores to achieve the optimal substitution model for phylogenetic analysis (59,60). The analysis was computed in BEAST 1.7.4 (61) on the basis of GTR+G model adjusted for each codon position with a fixed value for the gamma shape calculated by JMODELTEST. Three parallel analyses were run for 10 000 000 chains, which were performed under a lognormal relaxed clock and every 1000th tree was sampled. The three obtained log-files were controlled by the program TRACER v1.5 (62) if the analyses were suitable for the final phylogenetic tree configuration. The tree-files with branch lengths of each run were combined to one common tree-file using LOGCOMBINER 1.7.4 in BEAST. Finally, this file was used to summarize the sampled trees with a burn-in of 25% (7500 trees) to a maximum clade credibility tree by TREEANNOTATOR 1.7.4 in BEAST. The dataset was also analysed by ML bootstrapping in RAxML with the graphical front-end RAXMLGUI 1.3 using 1000 replicates (63,64). All trees were illustrated using the program FIGTREE v1.3.1. (65).
454 sequencing of Chara australis
Chara australis was collected from cultures described above. Thalli were rinsed with distilled water, gently blotted dry and frozen in liquid nitrogen and homogenized using mortar and pestle. Total RNA was extracted with TRI-Reagent according to manufacturer’s instructions (Sigma Aldrich). Transcriptomic data of C. australis were obtained from a normalized random primed cDNA library followed by 454 sequencing (Roche GS FLX system; Eurofins MWG). Different annotated ARA7 sequences were used for BLAST analyses (66) to reveal putative C. australis ARA7 cDNA. The accession number for CaARA7 is KM406482.
Vector construction
For vector construction PCRs were performed with Phusion High-Fidelity DNA polymerase (Thermo Scientific, #F530S) according to manufacturer’s instructions. Used primer sequences are listed in Table 1. For recombinant expression of His-tagged proteins ARA7 genes of A. thaliana and C. australis were cloned into Escherichia coli expression vector pQE30 (Qiagen). Therefore, total RNA was extracted either from thalli of C. australis or leaves of A. thaliana as described above and residual genomic DNA was digested using RNase-free DNase (Fermentas; #EN0521). First-strand cDNA was synthesized from 1 μg total RNA using M-MuLV Reverse Transcriptase (Fermentas, RevertAid #EP0441) and an anchored oligo(d)T primer-mix according to the supplier’s protocol. Obtained cDNA was used as template for PCR amplification in combination with either primer pair CaARA7_BamHI_fwd and CaARA7_c1903_rev (for C. australis), or AtARA7_BamHI_fwd and AtARA7_HindIII_rev (for A. thaliana). Both ARA7 genes were sub-cloned into pJet1.2 cloning vector (Fermentas, #K1231), digested with BamHI and HindIII and finally ligated into pQE30 expression vector.
Table 1.
List of primers used for cloning
| Primer name | Primer sequence |
|---|---|
| CaARA7_BamHI_fwd: | 5′-AAGGATCCATGGCGACTCTCGGGACCAG-3′ |
| CaARA7_c1903_rev: | 5′-CTCAAGCACAACAAGCACTTC-3′ |
| AtARA7_BamHI_fwd: | 5′-GGGGATCCATGGCTGCAGCTGGAAACAA-3′ |
| AtARA7_HindIII_rev: | 5′-GGGAAGCTTCTAAGCACAACAAGATGAGC-3′ |
| UBQ10_fwd (KpnI): | 5′-GGTACCCGACGAGTCAGTAATAAACG-3′ |
| UBQ10_rev (XhoI): | 5′-CTCGAGTGTTAATCAGAAAAACTCAG-3′ |
| GFP6_HindIII_fwd: | 5′-GGGAAGCTTATGAGTAAAGGAGAAGAACT-3′ |
| GFP6_SmaI_rev: | 5′-CCCCGGGGGCGCGCCTTTGTATAGTTC-3′ |
| mCherry_HindIII_fwd : | 5′-GGGAAGCTTATGGTGAGCAAGGGCGAGGA-3′ |
| mCherry_SmaI_rev: | 5′-CCCCGGGCTTGTACAGCTCGTCCATGC-3′ |
| AtARA7_SmaI_fwd: | 5′-CCCCGGGATGGCTGCAGCTGGAAACAA-3′ |
| AtARA7_NotI_rev: | 5′-CGGCGGCCGCCTAAGCACAACAAGATGAGC-3′ |
| CaARA7_SmaI_fwd: | 5′-CCCCGGGCATGGCGACTCTCGGGACCAG-3′ |
| CaARA7_NotI_rev: | 5′-CGGCGGCCGCTCAAGCACAACAAGCACTTC-3′ |
| CaAra7_S24N_fwd: | 5′-GGGTGCAGGGAAGAACAGCTTGGTGTTGCGC-3′ |
| CaAra7_ S24N_rev: | 5′-GCGCAACACCAAGCTGTTCTTCCCTGCACCC-3′ |
| CaAra7_Q69L_fwd: | 5′-GGGATACTGCTGGTCTCGAGAGGTATCATAGC-3′ |
| CaAra7_ Q69L_rev: | 5′-GCTATGATACCTCTCGAGACCAGCAGTATCCC-3′ |
| CaAra7_N123I_fwd: | 5′-CGCATTGGCGGGGATCAAAGTCGACTTGG-3′ |
| CaAra7_ N123I_rev: | 5′-CCAAGTCGACTTTGATCCCCGCCAATGCG-3′ |
List of all primers used for expression vector cloning and site directed mutagenesis as described in Materials and Methods section. Restriction sites are underlined.
For our localization experiments CaARA7 and AtARA7 were expressed using pGreenI-0029 (pGI; www.pgreen.ac.uk). The Arabidopsis thaliana ubiquitin 10 promoter (AtUBQ10p; At4g05320) was amplified using primers UBQ10_fwd and UBQ10_rev and A. thaliana genomic DNA as template. Genomic DNA was prepared by cetyltrimethylammonium bromide (CTAB) method according to (67). The promoter was cloned into pGI using restriction enzymes KpnI and XhoI. For construction of N-terminal fusions either mGFP6 or mCherry (GFP from pMDC107, see 68); mCherry from pCD3-960, see (55); both vectors obtained from NASC European Arabidopsis Stock Centre) were amplified using the following primers: GFP6_HindIII_fwd/GFP6_SmaI_rev and mCherry_HindIII_fwd/mCherry_SmaI_rev, respectively; GFP and mCherry genes were cloned downstream the AtUBQ10p (restriction sites: HindIII, SmaI). A. thaliana and C. australis cDNAs were prepared as described above and used as template for ARA7 amplification (AtARA7_SmaI_fwd and AtARA7_NotI_rev; CaARA7_SmaI_fwd and CaARA7_NotI_rev). Amplicons were cloned downstream of fluorescent protein encoding genes using restriction enzymes SmaI and NotI. Summarizing, following plasmids were obtained: pGI-AtUBQ10p∷GFP∷AtARA7, pGI-AtUBQ10p∷mCherry∷AtARA7, pGI-AtUBQ10p∷GFP∷CaARA7 and pGI-AtUBQ10p∷mCherry∷CaARA7.
Expression vectors carrying CaARA7 point mutations were constructed via site directed mutagenesis using pGI-AtUBQ10p∷GFP∷CaARA7 and pQE30-CaARA7 as templates. PfuUltra ™ High-Fidelity DNA Polymerase (Stratagene) and the following primers were used for plasmid amplification: CaAra7_S24N_fwd, CaAra7_S24N_rev, CaAra7_Q69L_fwd, CaAra7_Q69L_rev, CaAra7_N123I_fwd, CaAra7_N123I_rev. PCRs were performed as follows: 95°C for 30 seconds; 16 cycles (denaturation: 95°C for 30 seconds, annealing: 55°C for 1 min, extension: 68°C for 7 min for pGI template and 4 min 20 seconds for pQE template, respectively), followed by 2 min incubation on ice and DpnI template vector restriction (10 units; Fermentas, #ER1701) for 1 h at 37°C. Remaining mutagenized vectors were transformed into E. coli (strain XL-1; Stratagene) for amplification. Plasmids were verified by sequencing. For location of different point mutations see Figure S1C.
For information on ARA6 expression vector constructs (pGI-AtUBQ10p∷AtARA6∷GFP and pGI-AtUBQ10p∷CaARA6∷GFP) used in the colocalization experiments, please see (12). A protein sequence alignment of the conventional (ARA7) and plant-unique (ARA6) RAB5 GTPases of A. thaliana and C. australis used in this study can be seen in Figure S1B.
Expression and purification of recombinant CaARA7
The pQE30 constructs were transformed into E. coli (strain XL-1) and cells were grown to an absorbance (A600) of 0.6–1.0 in 250 mL LB medium containing 100 μg/mL ampicillin at 37°C under vigorous shaking. Cultures were cooled to 20°C and expression of proteins was induced by addition of 0.5 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG). After 20 h of vigorous shaking at 20°C cells were cooled to 4°C for 30 min prior to harvesting. All purification steps were performed at 4°C. E. coli were centrifuged and the resulting pellet was resuspended in equilibration buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0 adjusted with NaOH) in a ratio of 4 mL/g pellet. Lysozyme (1 mg/mL) and PMSF (phenylmethanesulfonylfluoride, 1 mM) were added to the suspension, which was shaken on ice for 30 min, followed by sonication and centrifugation (10 000 × g, 30 min, 4°C) in order to remove any residual insoluble residues. For purification of His-tagged recombinant ARA7 prepacked Protino ®Ni-TED 1000 columns (Macherey-Nagel, # 745110.50) were used according to manufacturer’s instructions. Recombinant protein was supplemented with 20% glycerol for storage at −20°C, analysed by SDS-PAGE and western blot, and used for GTPase assay. Purity of recombinant His-tagged CaARA7 was analysed by SDS-PAGE and colloidal Coomassie staining as well as by western blot analyses. For His-tag detection SuperSignal West His-Probe Kit (Thermo Scientific) was used according to the manufacturer’s instructions. Luminescence was detected by a LAS 3000 mini imaging system (Fujifilm).
GTPase activity
GTPase activity measurements were carried out using the Transcreener®GDP FI assay (BellBrook Labs) as previously described (12). This end-point assay employs a GDP Alexa594 red fluorescent tracer bound to a GDP antibody-quencher conjugate. GDP produced in the enzymatic reaction displaces the tracer bound in the complex which results in a corresponding fluorescence increase in samples which display GTPase activity.
The recombinant purified proteins were diluted at least 1:20 in order to achieve a working concentration in the reaction mixture of 40 μg/mL (unless otherwise stated) in a buffer containing 50 mM HEPES (pH 7.5), 4 mM MgCl2, 2 mM EGTA, 1% dimethyl sulfoxide (DMSO) and 0.01% Triton X-100. Basic control measurements were performed according to the manufacturer’s instructions as well as measurements without GTP added in order to determine the amount of residual GDP bound to the samples, since most of recombinant RAB GTPases are purified in the stable guanine nucleotide-bound form.
ARA7 antibody and western blot analyses
A polyclonal, affinity purified antibody against CaARA7 produced by Genscript was tested on C. australis protein extract and recombinant His-tagged CaARA7. The antibody was raised in rabbit against a mixture of peptides CNQGANDRYQRRGSA and CRKLPRANPAAQPTG located at the hypervariable region of the protein to ensure specific binding. For protein extracts whole C. australis thalli were homogenized in liquid nitrogen using mortar and pestle. 2.5 volumes of ice cooled extraction buffer (100 mM NaH2PO4 (pH 7.8), 100 mM KCl, 1 mM DTT, 0.04% Tween and protease inhibitor cocktail (P9599; Sigma) were added to the ground material and the suspension was shaken on ice for 15 min, followed by centrifugation (15 min, 7500 × g, 4°C). The resulting supernatant was used for SDS-PAGE and western blot analyses.
For western blot detection the anti-ARA7 antibody was diluted 1:5000 and the secondary antibody, an anti-rabbit IgG (alkaline phosphatase, Biocell), was used in a concentration of 1:20 000. Detection was carried out using CDP star detection kit (New England Biolabs) and a LAS 3000 mini imaging system (Fujifilm).
In vivo staining, inhibitor treatments and immunofluorescence of Chara internodal cells
Internodal cells were pulse labelled for 5 min with 10 μM of the green fluorescent endocytic tracer FM1-43FX [N-(3-triethylammoniumpropyl)-4-(dibutylamino)styryl pyridinium dibromide; Invitrogen; stock solution 0.5 mM in A. dest.] and the red fluorescent endocytic tracers FM4-64FX [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl)hexatrienyl) pyridinium dibromide; Invitrogen; 500 μM stock solution in A. dest.] and AM4-65 [(N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl)) hexatrienyl) pyridinium dibromide; Biotium; 1 mM stock solution in A. dest.], which is spectrally identical to the FM4-64. Staining with 10 μM of the acidotropic dye Lysotracker Red DND-99 (LTRed; Invitrogen; 1 mM stock solution in DMSO) required at least 1 h. Cells were treated with 25 μM wortmannin (WM; Enzo; 10 mM stock solution in DMSO). All dyes and inhibitors were diluted with artificial fresh water. Controls containing up to 2% DMSO had no visible effect on cytoarchitecture, cytoplasmic streaming or organelle dynamics.
The immunofluorescence procedure was as described in (31). For localization of ARA7-labelled organelles the polyclonal anti-CaARA7 (see above) was used at dilutions between 1:500 and 1:750 and combined with an Alexa Fluor 488-conjugated anti-rabbit IgG (Invitrogen; diluted 1:1000) or with an Alexa Fluor 546-conjugated anti-rabbit IgG (Thermo Fisher; diluted 3:1000). The monoclonal anti-CaARA6 (12) was used at a concentration of 1:10 and combined with anti-mouse CF488A (Sigma; diluted 3:1000). Double immunofluorescence was performed using a mixture of the primary antibodies against CaARA7 and CaARA6 diluted as described above and a mixture of the secondary antibodies anti-mouse CF488A and anti-rabbit IgG Alexa 546 diluted as described above. In separate experiments, an IgG antibody raised in rabbit against ARA7 from A. thaliana (5) was used at a dilution of 1:200 and combined with an Alexa Fluor 546-conjugated anti-rabbit IgG (Invitrogen; 1:1000). Rabbit pre-immune serum was used as negative control for the polyclonal antibodies.
For the comparison of anti-CaARA7-labelled organelles with structures stained by endocytic tracers, cells were pulse labelled with 10 μM of the fixable probes FM 1-43FX or AM4-65 and washed for 30 min in artificial fresh water before fixation.
Transient expression in Nicotiana benthamiana, in vivo staining and inhibitor treatments
For transient transfection of tobacco epidermal leaves Agrobacterium tumefaciens (strain GV3101) was transformed and infiltrated into 4 weeks old N. benthamiana plants as described in (12). For colocalization studies the mCherry-expressing vector G-rb CD3-968 of Nelson et al. (37), a marker of the Golgi apparatus was obtained from NASC European Arabidopsis Stock Centre. The SYP61-mRFP expressing pUBC-SYP61-mRFP, a TGN marker was kindly provided by Karin Schumacher (University of Heidelberg).
Tobacco leaf epidermal cells were stained by infiltration of leaves with 10 μM of FM4-64 (Invitrogen; 10 mM stock solution in DMSO) or with 10 μM LTRed. For inhibitor experiments leaves were infiltrated with 200 μM WM. This concentration caused the formation of WM compartments, within 2 h treatment. Lower concentrations had either no effect or required much longer incubation times with possible unspecific effects. Working solutions and controls were prepared with AFW ± DMSO as described above.
Confocal laser scanning microscopy
CLSM used in this study was a Leica TCS SP5 coupled to a DMI 6000B inverted microscope. Laser settings were as described in (31).
Live internodal cells were mounted in artificial fresh water ± dye or inhibitor. Leaf sections were either infiltrated with artificial fresh water (± dye or inhibitor; see above) or mounted in perfluorodecalin (69) in order to fill the intercellular air spaces.
Appropriate controls were made for all single and double labelling experiments in order to exclude bleed through and to distinguish signals from background fluorescence. Colocalizations were studied with sequential scan settings to avoid cross-talk between channels.
Observation times were usually restricted to 2 h in order to avoid toxic effects or redistribution of fluorescent dyes. Organelle dynamics were studied by analysing time series taken at minimum laser intensity and pixel time in order to avoid photo bleaching and stress response.
All images presented in this study are single sections unless otherwise stated. Images produced by the LSM software were further processed with Adobe Photoshop (Adobe Systems Inc.).
Image analysis, size measurements and statistics
IMAGEJ (http://rsbweb.nih.gov/ij) was used as described in (12) to count and measure ARA7-labelled organelles transiently expressed in tobacco epidermal cells. Colocalization analysis was performed with the IMAGEJ plug-in tool PSC Colocalization (70). For this purpose, 200–300 independent mobile fluorescent structures comprising between 30 000 and 70 000 pixel were manually masked with the brush tool of IMAGEJ and the background threshold was set to 10. Cumulative scatterplots and Pearson’s and Spearman’s correlation coefficients were calculated by examining images from several cells and using at least 130 fluorescent structures for each sample combination.
The size of AM4-65 stained organelles in the streaming endoplasm of control and WM-treated internodal cells was measured on randomly selected optical sections (40 × 40 μm) of video films taken at 370 milliseconds intervals. Parametric and non-parametric tests were performed with SIGMASTAT (Systat Software; version 3.11) in order to reveal significant differences (p ≤ 0.05) in size (given as largest cross-section or diameter). Graphs were generated with SIGMAPLOT (Systat Software; version 9.01).
Electron microscopy and immunolabelling
Chemical fixation of WM-treated internodal cells was as described in (71). Cells were embedded in Agar low viscosity resin (Agar Scientific) after dehydration in ethanol.
Immunolabelling of electron microscopical sections from chemically fixed or high pressure frozen cells did not yield satisfactory results. We therefore used pre-embedment immunolabelling to study the localization of CaARA7-positive epitopes at the fine structural level. Internodal cells were first fixed in 1% glutaraldehyde dissolved in 70% phosphate buffered saline (PBS; 1.4 M NaCl, 29.5 mM KCl, 23.8 mM KH2PO4, 76.1 mM Na2HPO4, pH7) for 20 min at room temperature. After a 15 min wash in PBS internodal cells were cut into small cylinders and treated as follows: blocking buffer (1% BSA and 50 mM glycine in PBS) for 30 min; polyclonal anti-CaARA7 diluted 1:500 in blocking buffer for 3 h; wash in PBS for at least 45 min; anti-rabbit IgG conjugated to 5 nm gold (BBI Solutions) 1:40 for 10 h at 4°C; 3× wash in PBS for 15 min each; postfixation in 1% glutaraldehyde in PBS for 30 min at room temperature and with 2% OsO4. After a short wash in distilled water cell fragments were dehydrated in an ethanol series at 4°C and finally embedded in LRGold (London Resin) as described in (72). Micrographs of ultrathin sections were taken with a LEO 912 transmission electron microscope (Zeiss) equipped with in-column energy filter and using elastic bright-field mode.
Supplementary Material
Acknowledgments
We are grateful to Raimund Tenhaken (University of Salzburg) for support and valuable discussion and to Karin Schumacher (University of Heidelberg) for SYP61-mRFP. This research was funded by the Austrian Science Fund (FWF project no. P 22957-B20 to IF).
Footnotes
The authors declare that they have no conflict of interest.
Additional Supporting Information may be found in the online version of this article:
References
- 1.Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci. 2007;120:3905–3910. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- 2.Fujimoto M, Ueda T. Conserved and plant-unique mechanisms regulating plant post-Golgi traffic. Front Plant Sci. 2012;3:197. doi: 10.3389/fpls.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito C, Ueda T. Chapter 4 Functions of RAB and SNARE proteins in plant life. In: Kwang WJ, editor. International Review of Cell and Molecular Biology. Academic Press; Burlington: 2009. pp. 183–233. [DOI] [PubMed] [Google Scholar]
- 4.Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A. VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell. 2007;19:3504–3515. doi: 10.1105/tpc.107.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueda T, Yamaguchi M, Uchimiya H, Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 2001;20:4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueda T, Uemura T, Sato MH, Nakano A. Functional differentiation of endosomes in Arabidopsis cells. Plant J. 2004;40:783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- 7.Bolte S, Schiene K, Dietz K-J. Characterization of a small GTP-binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol Biol. 2000;42:923–935. doi: 10.1023/a:1006449715236. [DOI] [PubMed] [Google Scholar]
- 8.Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, Thordal-Christensen H, Tsutsumi N, Nakano A, Ueda T. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 9.Ebine K, Miyakawa N, Fujimoto M, Uemura T, Nakano A, Ueda T. Endosomal trafficking pathway regulated by ARA6, a RAB5 GTPase unique to plants. Small GTPases. 2012;3:23–27. doi: 10.4161/sgtp.18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottanelli F, Foresti O, Hanton S, Denecke J. Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. Plant Cell. 2011;23:3007–3025. doi: 10.1105/tpc.111.085480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebine K, Ueda T. Unique mechanism of plant endocytic/vacuolar transport pathways. J Plant Res. 2009;122:21–30. doi: 10.1007/s10265-008-0200-x. [DOI] [PubMed] [Google Scholar]
- 12.Hoepflinger MC, Geretschlaeger A, Sommer A, Hoeftberger M, Nishiyama T, Sakayama H, Hammerl P, Tenhaken R, Ueda T, Foissner I. Molecular and biochemical analysis of the first ARA6 homologue, a RAB5 GTPase, from green algae. J Exp Bot. 2013;64:5553–5568. doi: 10.1093/jxb/ert322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoepflinger M, Hametner C, Ueda T, Foissner I. Vesicular trafficking in characean green algae and the possible involvement of a VAMP72-family protein. Plant Signal Behav. 2014;9:e28466. doi: 10.4161/psb.28466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhonukshe P, Baluska F, Schlicht M, Hlavacka A, Samaj J, Friml J, Gadella TWJ. Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev Cell. 2006;10:137–150. doi: 10.1016/j.devcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell. 2012;24:4205–4219. doi: 10.1105/tpc.112.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas TJ, Sliwinski MK, Martinez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell. 2007;19:1295–1312. doi: 10.1105/tpc.106.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci. 2004;117:6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- 18.Wodniok S, Brinkmann H, Glockner G, Heidel A, Philippe H, Melkonian M, Becker B. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun M, Foissner I, Lühring H, Schubert H, Thiel G. Characean algae: still a valid model system to examine fundamental principles in plants. Prog Bot. 2007;68:193–220. [Google Scholar]
- 20.Tazawa M, Shimmen T. How characean cells have contributed to the progress of plant membrane biophysics. Aust J Plant Physiol. 2001;28:523–539. [Google Scholar]
- 21.Foissner I, Wasteneys GO. Characean internodal cells as a model system for the study of cell organization. In: Kwang WJ, editor. International Review of Cell and Molecular Biology. Academic Press; Burlington: 2014. pp. 307–364. [DOI] [PubMed] [Google Scholar]
- 22.Foissner I, Wasteneys GO. The characean internodal cell as a model system for studying wound healing. J Microsc. 2012;247:10–22. doi: 10.1111/j.1365-2818.2011.03572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Barbieri A, Colombo MI, Stahl PD. Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J Biol Chem. 1994;269:14631–14635. [PubMed] [Google Scholar]
- 24.Hoffenberg S, Sanford JC, Liu S, Daniel DS, Tuvin M, Knoll BJ, Wessling-Resnick M, Dickey BF. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J Biol Chem. 1995;270:5048–5056. doi: 10.1074/jbc.270.10.5048. [DOI] [PubMed] [Google Scholar]
- 25.Anand B, Majumdar S, Prakash B. Structural basis unifying diverse GTP hydrolysis mechanisms. Biochemistry. 2013;52:1122–1130. doi: 10.1021/bi3014054. [DOI] [PubMed] [Google Scholar]
- 26.Olkkonen VM, Stenmark H. Role of rab GTPases in membrane traffic. In: Jeon KW, editor. International Review of Cytology – A Survey of Cell Biology. Academic Press Inc; Burlington: 1997. pp. 1–85. [DOI] [PubMed] [Google Scholar]
- 27.Kleuss C, Raw AS, Lee E, Sprang SR, Gilman AG. Mechanism of GTP hydrolysis by G-protein alpha subunits. Proc Natl Acad Sci USA. 1994;91:9828–9831. doi: 10.1073/pnas.91.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Stahl PD. Structure-function relationship of the small GTPase rab5. J Biol Chem. 1993;268:24475–24480. [PubMed] [Google Scholar]
- 29.Scheuring D, Viotti C, Kräger F, Känzl F, Sturm S, Bubeck J, Hillmer S, Frigerio L, Robinson DG, Pimpl P, Schumacher K. Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell. 2011;23:3463–3481. doi: 10.1105/tpc.111.086918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klima A, Foissner I. FM dyes label sterol-rich plasma membrane domains and are internalized independently of the cytoskeleton in characean internodal cells. Plant Cell Physiol. 2008;49:1508–1521. doi: 10.1093/pcp/pcn122. [DOI] [PubMed] [Google Scholar]
- 31.Schmoelzer PM, Hoeftberger M, Foissner I. Plasma membrane domains participate in pH banding of Chara internodal cells. Plant Cell Physiol. 2011;52:1274–1288. doi: 10.1093/pcp/pcr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang JQ, Cai Y, Miao YS, Lam SK, Jiang LW. Wortmannin induces homotypic fusion of plant prevacuolar compartments. J Exp Bot. 2009;60:3075–3083. doi: 10.1093/jxb/erp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam SK, Tse YC, Miao YS, Li H, Wang J, Lo SW, Jiang CJ. Molecular characterization of plant prevacuolar and endosomal compartments. J Integr Plant Biol. 2007;49:1119–1128. [Google Scholar]
- 34.Norris SR, Meyer SE, Callis J. The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol. 1993;21:895–906. doi: 10.1007/BF00027120. [DOI] [PubMed] [Google Scholar]
- 35.Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA. 2012;109:15942–15946. doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martiniere A, Bassil E, Jublanc E, Alcon C, Reguera M, Sentenac H, Blumwald E, Paris N. In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell. 2013;25:4028–4043. doi: 10.1105/tpc.113.116897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 38.Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct Funct. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- 39.Ceresa BP, Lotscher M, Schmid SL. Receptor and membrane recycling can occur with unaltered efficiency despite dramatic Rab5(q79l)-induced changes in endosome geometry. J Biol Chem. 2001;276:9649–9654. doi: 10.1074/jbc.M010387200. [DOI] [PubMed] [Google Scholar]
- 40.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 41.Bucci C, Lutcke A, Steele-Mortimer O, Olkkonen VM, Dupree P, Chiariello M, Bruni CB, Simons K, Zerial M. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 1995;366:65–71. doi: 10.1016/0014-5793(95)00477-q. [DOI] [PubMed] [Google Scholar]
- 42.Dinneen JL, Ceresa BP. Expression of dominant negative rab5 in HeLa cells regulates endocytic trafficking distal from the plasma membrane. Exp Cell Res. 2004;294:509–522. doi: 10.1016/j.yexcr.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 44.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 45.Araki S, Kaibuchi K, Sasaki T, Hata Y, Takai Y. Role of the C-terminal region of smg p25A in its interaction with membranes and the GDP/GTP exchange protein. Mol Cell Biol. 1991;11:1438–1447. doi: 10.1128/mcb.11.3.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musha T, Kawata M, Takai Y. The geranylgeranyl moiety but not the methyl moiety of the smg-25A/rab3A protein is essential for the interactions with membrane and its inhibitory GDP/GTP exchange protein. J Biol Chem. 1992;267:9821–9825. [PubMed] [Google Scholar]
- 47.Soldati T, Riederer MA, Pfeffer SR. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol Biol Cell. 1993;4:425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueda T, Matsuda N, Uchimiya H, Nakano A. Modes of interaction between the Arabidopsis Rab protein, Ara4, and its putative regulator molecules revealed by a yeast expression system. Plant J. 2000;21:341–349. doi: 10.1046/j.1365-313x.2000.00681.x. [DOI] [PubMed] [Google Scholar]
- 49.Chavrier P, Gorvel JP, Stelzer E, Simons K, Gruenberg J, Zerial M. Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature. 1991;353:769–772. doi: 10.1038/353769a0. [DOI] [PubMed] [Google Scholar]
- 50.Brennwald P, Novick P. Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature. 1993;362:560–563. doi: 10.1038/362560a0. [DOI] [PubMed] [Google Scholar]
- 51.Aivazian D, Serrano RL, Pfeffer S. TIP47 is a key effector for Rab9 localization. J Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y-W, Oesterlin LK, Tan K-T, Waldmann H, Alexandrov K, Goody RS. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol. 2010;6:534–540. doi: 10.1038/nchembio.386. [DOI] [PubMed] [Google Scholar]
- 53.Ali BR, Wasmeier C, Lamoreux L, Strom M, Seabra MC. Multiple regions contribute to membrane targeting of Rab GTPases. J Cell Sci. 2004;117(Pt 26):6401–6412. doi: 10.1242/jcs.01542. [DOI] [PubMed] [Google Scholar]
- 54.Beranger F, Paterson H, Powers S, de Gunzburg J, Hancock JF. The effector domain of Rab6, plus a highly hydrophobic C terminus, is required for Golgi apparatus localization. Mol Cell Biol. 1994;14:744–758. doi: 10.1128/mcb.14.1.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu Y. The role of the hypervariable C-terminal domain in Rab GTPases membrane targeting. Proc Natl Acad Sci USA. 2014;111:2572–2577. doi: 10.1073/pnas.1313655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li G, Barbieri MA, Stahl PD. Myristoylation cannot functionally replace the isoprenylation of Rab5. Arch Biochem Biophys. 1995;316:529–534. doi: 10.1006/abbi.1995.1070. [DOI] [PubMed] [Google Scholar]
- 57.Jia TR, Gao CJ, Cui Y, Wang JQ, Ding Y, Cai Y, Ueda T, Nakano A, Jiang LW. ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. J Exp Bot. 2013;64:2817–2829. doi: 10.1093/jxb/ert125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glez-Pena D, Gomez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:W14–W18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 61.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rambaut A, Drummond AJ. Tracer v1.4. 2007 Available from http://beast.bio.ed.ac.uk/Tracer.
- 63.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 64.Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12:335–337. [Google Scholar]
- 65.Rambaut A. FigTree: tree figure drawing tool. (Version 1.3.1) 2006–2009 http://tree.bio.ed.ac.uk/software/figtree/
- 66.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2002. [Google Scholar]
- 68.Curtis M, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Littlejohn GR, Gouveia JD, Edner C, Smirnoff N, Love J. Perfluorodecalin enhances in vivo confocal microscopy resolution of Arabidopsis thaliana mesophyll. New Phytol. 2010;186:1018–1025. doi: 10.1111/j.1469-8137.2010.03244.x. [DOI] [PubMed] [Google Scholar]
- 70.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protocols. 2008;3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 71.Foissner I. Induction of exocytosis in characean internodal cells by locally restricted application of chlortetracycline and the effect of cytochalasin B, depolarizing and hyperpolarizing agents. Plant Cell Environ. 1991;14:907–915. [Google Scholar]
- 72.Foissner I, Hoeftberger M. Immuno-gold labeling of resin-embedded electron microscopical sections. In: Žárský V, Cvrčková F, editors. Plant Cell Morphogenesis: Methods and Protocols. Springer Science Business Media; New York: 2014. pp. 183–193. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.