Abstract
Engineered protein therapeutics offer advantages, including strong target affinity, selectivity, and low toxicity, but like natural proteins can be susceptible to proteolytic degradation, thereby limiting their effectiveness. A compelling therapeutic target is mesotrypsin, a protease upregulated with tumor progression, associated with poor prognosis, and implicated in tumor growth and progression of many cancers. However, with its unique capability for cleavage and inactivation of proteinaceous inhibitors, mesotrypsin presents a formidable challenge to the development of biologic inhibitors. We used a powerful yeast display platform for directed evolution, employing a novel multi-modal library screening strategy, to engineer the human amyloid precursor protein Kunitz protease inhibitor domain (APPI) simultaneously for increased proteolytic stability, stronger binding affinity, and improved selectivity for mesotrypsin inhibition. We identified a triple mutant APPIM17G/I18F/F34V, with a mesotrypsin inhibition constant (Ki) of 89 pM, as the strongest mesotrypsin inhibitor yet reported; this variant displays 1459-fold improved affinity, up to 350,000-fold greater specificity, and 83-fold improved proteolytic stability vs wild-type APPI. We demonstrated that APPIM17G/I18F/F34V acts as a functional inhibitor in cell-based models of mesotrypsin-dependent prostate cancer cellular invasiveness. Additionally, by solving the crystal structure of the APPIM17G/I18F/F34V/mesotrypsin complex, we obtained new insights into the structural and mechanistic basis for improved binding and proteolytic resistance. Our study identifies a promising mesotrypsin inhibitor as a starting point for development of anticancer protein therapeutics and establishes proof-of-principle for a novel library screening approach that will be widely applicable for simultaneously evolving proteolytic stability in tandem with desired functionality for diverse protein scaffolds.
Keywords: protein engineering, directed evolution, proteolysis, protease inhibitor, enzyme inhibition, X-ray structure
INTRODUCTION
The human serine protease mesotrypsin, encoded by the PRSS3 gene, has recently emerged as a compelling new candidate drug target in cancer. Although its specific pathological roles are yet to be fully elucidated, the dysregulation and overexpression of mesotrypsin are associated with poor prognosis in many human tumors and with malignant behaviors in cancer models [1–9]. Mesotrypsin is particularly attractive as a potential target in the treatment of metastatic prostate cancer: it is associated with recurrence and metastasis and is upregulated in metastatic tumors. In cell culture and orthotopic mouse models, mesotrypsin drives invasive and metastatic phenotypes [2]. Likewise, in pancreatic cancer, higher mesotrypsin expression is associated with poorer patient survival, and in cell culture and animal models it promotes cancer cell proliferation, invasion, and metastasis [6]. Potent and selective inhibitors of mesotrypsin could thus offer both promise for the treatment of patients with aggressive metastatic cancers and tools to better dissect mesotrypsin function in cancer progression and metastasis.
Elucidating the mechanism of action of mesotrypsin and designing efficacious inhibitors are not trivial tasks [10–13]. A particular challenge in inhibitor development derives from the inability of mesotrypsin to form tight complexes with protein inhibitors, due to the presence of distinctive active site mutations [11, 13–15]. Furthermore, mesotrypsin also cleaves and inactivates many protein protease inhibitors as physiological substrates [12, 16, 17]. An additional challenge lies the inherent difficulty of obtaining selective inhibitors, since mesotrypsin shows high sequence homology and structural similarity with the major digestive trypsins (cationic and anionic trypsin) as well as with other serine proteases, including kallikreins and coagulation factors [18, 19]. It is thus not surprising that there are currently no effective inhibitory agents with high proteolytic stability, affinity and specificity for human mesotrypsin.
A conventional approach taken to developing inhibitors of other human serine proteases involves the engineering of natural serine protease inhibitors, such as those belonging to the Kunitz domain family [20–28]. These small proteins possess a protease-inhibitory binding loop of canonical conformation, supported by a compact scaffold with a three-dimensional structure that is maintained by hydrophobic packing and three disulfide bonds (Fig. S1C). From the perspective of developing selective drugs, the advantage of these inhibitors – over small molecules – is the availability of an extensive contact interface capable of interacting with multiple protease subsites. Initial attempts to engineer a potent polypeptide inhibitor of mesotrypsin by using bovine pancreatic trypsin inhibitor (BPTI) as a Kunitz domain scaffold that is relatively resistant to cleavage by mesotrypsin [29] revealed a number of drawbacks: BPTI exhibits a low affinity for mesotrypsin [15], a lack of target specificity [29], and a high potential for immune stimulation [30], and it may promote renal dysfunction [31]. Kunitz domains of human origin are likely to be less immunogenic, but they are much more susceptible than BPTI to cleavage and inactivation by mesotrypsin [16, 17]. However, if such liabilities could be overcome, an engineered human Kunitz domain possessing mesotrypsin affinity, enhanced proteolytic stability, and target selectivity could offer a promising avenue for cancer therapy.
We thus pursued a novel combinatorial strategy to engineer the human Kunitz domain amyloid precursor protein inhibitor (APPI) simultaneously for increased proteolytic stability, higher binding affinity, and improved selectivity for inhibition of mesotrypsin. In light of our previous studies showing that the proteolytic stability of Kunitz domains is modulated by residues within the scaffold [32], we constructed an all-gene random mutagenesis APPI library in which diversity was introduced throughout the binding loop and the scaffold. Employing a stepwise proteolytic stability/affinity maturation process, we used yeast surface display (YSD) as a directed evolution methodology; this technique has previously been used to engineer proteins for enhanced binding affinity [33–35], proper folding [36, 37], and improved thermal stability [38].
Here, the YSD selection platform was applied for the first time to simultaneously screen for both proteolytic stability and affinity. High-throughput screening of tens of millions of yeast-displayed mutants enabled the rapid isolation of multiple APPI mutants, each carrying one mutation (either in the active site or in the scaffold) that individually conferred a functional enhancement in mesotrypsin affinity and/or proteolytic stability. By combining three mutations, we generated APPIM17G/I18F/F34V, a triple mutant possessing the strongest affinity for mesotrypsin of any known inhibitor, with 1459-fold improved affinity and 83-fold improved proteolytic stability in comparison with wild-type APPI (APPIWT). Remarkably, this triple mutant displayed 350,000-fold enhanced specificity toward mesotrypsin. We then demonstrated that these optimized properties enabled APPIM17G/I18F/F34V to potently block mesotrypsin-dependent cancer cell invasiveness in cell-based assays, for which high binding affinities and exceptional protease stabilities are crucial. In addition, by solving the crystal structure of the APPIM17G/I18F/F34V/mesotrypsin complex, we revealed the molecular basis for the improved binding and proteolytic resistance of the engineered inhibitor as well as the basis for synergy between the mutations. Thus, our study establishes proof-of-principle for our novel multi-modal library screening approach. It also offers fundamental clues into the molecular basis of Kunitz domain stability and identifies a promising mesotrypsin inhibitor as a starting point for the development of anticancer protein therapeutics.
MATERIALS AND METHODS
Cells, reagents, and additional methods are described in SI Materials and Methods.
Generation of a combinatorial based APPI library
Synthesis and cloning of the DNA encoding APPIWT and the generation of the combinatorial APPI library are described in detail in SI Materials and Methods. In brief, the inhibitor domain of the amyloid precursor protein (APPIWT) gene was constructed on the basis of a published sequence (PDB id 1ZJD) by using codons optimized for both S. cerevisiae and Pichia pastoris usage and synthesized by PCR-assembly with six overlapping oligonucleotides. Next, a randomly mutated version of the APPI gene was constructed by error-prone PCR using nucleotide analogues and low-fidelity Taq polymerase with the APPIWT gene as a template. The resulting insert was amplified and transformed into yeast through homologous recombination, as previously described [39]. Random mutagenesis in the APPI sequence generated an APPI library with 0–3 mutations per clone, yielding an experimental library of about 9×106 clones.
Flow cytometry and cell sorting
The yeast-displayed APPI library and individual APPI variants were grown in SDCAA selective medium (as for SDCAA plates, but without agar; see SI Materials and Methods) and induced for expression with galactose medium (as for SDCAA, but with galactose instead of dextrose), according to established protocols [39]. Due to the different enzymatic turnover times of APPI and its variants by the target trypsins, i.e., bovine trypsin or mesotrypsin, two methods for trypsin-labeling were used, namely, ‘double staining’ and ‘triple staining,’ for the detection of proteolytically resistant clones, as described below. In the first step of labeling, approximately 1×106 cells were labeled with the appropriate catalytically active trypsin and a 1:50 dilution of mouse anti-c-Myc antibody in trypsin buffer (TB; 100 mM Tris-HCl, pH 8.0, 1 mM CaCl2) supplemented with 1% bovine serum albumin (BSA) for 30 min at room temperature. In this labeling step, the cells were exposed to biotinylated-bovine trypsin or mesotrypsin for ‘double staining.’ For ‘triple staining,’ the cells were first treated with non-biotinylated mesotrypsin, and then an additional labelling step was applied: the cells were washed with TB and incubated with 2 μM of biotinylated catalytically inactive mesotrypsin-S195A for 1 h at room temperature. Finally, for both ‘double staining’ and ‘triple staining,’ cells were washed with ice-cold TB, followed by incubation with a 1:800 dilution of fluorescein isothiocyanate (FITC)-conjugated streptavidin and a 1:50 dilution of phycoerythrin (PE)-conjugated anti mouse secondary antibody for 30 min on ice. Cells were washed again and analyzed by dual-color flow cytometry (Accuri C6; BD Biosciences).
Cell sorting of ‘triple-stained’ cells was carried out as described in Figure 2A with a iCyt Synergy FACS [Cytometry, Proteomics and Microscopy Unit (CPMU), National Institute of Biotechnology in the Negev (NIBN), Ben-Gurion University of the Negev (BGU)]. In brief, approximately 1×108 cells were first sorted to select for high expressing clones (c-Myc clear). Sorted cells were then grown in selective medium, and several colonies were sequenced [DNA Microarray and Sequencing Unit (DMSU), NIBN, BGU]. Following each triple staining sort, the number of yeast cells used for subsequent sorting was at least 10-fold in excess of the number of sorted cells. Several clones from each round of sorting were sequenced. The concentration of the target protein in each sort is shown in Figure 2A.
Prostate cancer cell assays
Matrigel invasion assays of PC3-M human prostate cancer cells were conducted as previously reported [2]. Cells with PRSS3 expression suppressed by lentiviral shRNA (NM_002771.2-454s1c1) served as a control in all experiments. Cells used for all other conditions were transduced with a control lentiviral shRNA that does not recognize any human genes. Prior to the invasion assays, cells were seeded at 1.5 × 106 cells per 10 cm dish (day 1). Thereafter, the medium was replaced with a mixture of 3.6 ml RPMI containing 10% fetal bovine serum (FBS), 10 μg/ml Polybrene® and 2.4 ml conditioned medium containing lentiviral particles (day 2). The medium was replaced again after 24 h, and cells were selected with 2 μg/ml puromycin (day 3). On day 4, cells were trypsinized and seeded into 24-well 8.0 μm cell culture inserts (BD) previously coated with 50 μg Matrigel (2 × 104 cells per insert in 400 μl of medium). APPIWT or APPIM17G/I18F/F34V proteins (10 nM–1 μM) were added to cell suspensions in some wells as indicated in Figure 4A,B. Quadruple biological replicates were performed. Cells were allowed to invade toward a chemoattractant medium (750 μl NIH/3T3 cell-conditioned serum free medium) for 18 h, and then non-invading cells were removed, and the filters were fixed, stained and analyzed as described previously [2]. Consistent results were obtained from five independent experiments.
PC3-M cells for 3D culture assays were similarly transduced with either PRSS3-targeted or control lentiviral shRNA according to the above schedule. On day 4, cells were seeded into 3D cultures in Matrigel using the ‘on-top’ protocol, essentially as described previously [2, 40]. Briefly, in 12-well plates, a base layer of 250 μl of 100% Matrigel was polymerized, PC3-M cells (2 × 104 cells/well) were seeded and allowed to adhere to the substratum, excess medium was aspirated, and cells were overlaid with 500 μl of medium supplemented with 10% Matrigel and 0.5% FBS plus 100 nM of APPIWT or APPIM17G/I18F/F34V where indicated in Figure 4C,D. Cultures were maintained at 37 °C in 5% CO2 for 3 d, photographed, and analyzed by counting and calculating the percentage of cells displaying spiky protrusions or branching morphology.
Protein complex crystallization, X-ray data collection, structure solution, and model refinement
APPIM17G/I18F/F34V was mixed with catalytically inactive mesotrypsin-S195A in an equimolar ratio at a total protein concentration of 4 mg/mL and then mixed 1:1 (v/v) with reservoir solution. The complex was crystallized at room temperature via the hanging drop method over a reservoir solution containing 0.1 M ammonium sulfate, 0.1 M Tris pH 7.5, and 20% PEG-1000. Crystals were harvested, cryoprotected, and flash cooled in liquid N2. X-ray diffraction data were collected at 100 K, at beamline 8.2.1 of the Advanced Light Source, Lawrence Berkeley National Laboratory, from one crystal that diffracted to 1.83 Å. Data were merged and scaled with XDS [41]. The crystal belonged to the space group P41212, with unit cell dimensions a = 78.15, b = 78.15, c = 243.73 and contained two copies of the complex in the asymmetric unit. The structure was solved by molecular replacement with MOLREP in CCP4 [42], using as the search model the complex of human mesotrypsin with APPIWT [Protein Data Bank (PDB) 3L33] [32]. Refinement employed alternating manual rebuilding in COOT [43] and automated refinement using REFMAC5 [44]. The quality of the final models was analyzed using wwPDB validation tools [45]. The coordinates and structure factors have been submitted to the Worldwide Protein Data Bank under the accession code 5C67.
RESULTS
Yeast-displayed APPIWT is rapidly cleaved by human mesotrypsin
The YSD system for directed evolution is based on the expression of a library of mutant proteins on the surface of yeast, followed by the selection of variants with improved affinity. However, this system has not been employed previously for identifying proteolytic cleavage or improving the proteolytic resistance of a displayed inhibitor. To test the compatibility of APPIWT with the YSD system, we cloned the coding region of APPIWT into a YSD plasmid for presentation on the Saccharomyces cerevisiae yeast surface as a fusion with the Aga2p agglutinin protein (Fig. S1). Correct folding of APPIWT was then verified by using flow activated cell sorting (FACS) for the detection of bound fluorescently labeled bovine trypsin, which is an established, tight-binding target of APPI [32]. We found that APPI displayed on the yeast surface was highly expressed and showed significant binding to bovine trypsin, thereby demonstrating proper folding of APPI (Figure 1A).
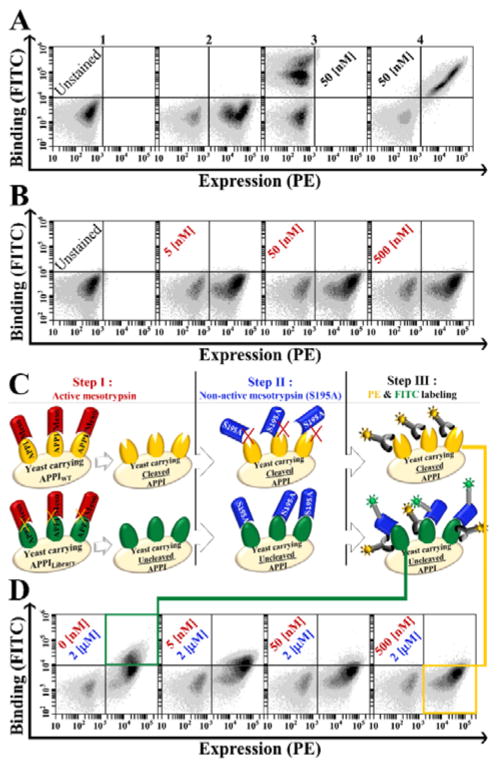
Figure 1. Yeast surface displayed APPIWT is cleaved by active mesotrypsin but detected by catalytically inactive mesotrypsin-S195A.
(A) Dual-color flow cytometric expression and folding analysis. APPI expression is shown on the X-axis and binding of APPI to bovine-trypsin (50 nM) on the Y-axis. Subpanels 1, 2, 3 and 4 represent unstained, PE-labeled expression, FITC-labeled binding and dual-labeled cells (demonstrating expression and binding, respectively). (B) APPIWT is not detected by active mesotrypsin. Panel B shows dual-labeled cells as in panel A, after exposure to different concentrations of FITC-labeled active mesotrypsin (in red). (C) General scheme of the ‘triple staining method’ for detection of residual uncleaved APPI by FITC-labeled catalytically inactive mesotrypsin-S195A after preincubation of cells with active mesotrypsin. (D) Intact APPI is detected by mesotrypsin-S195A. Panel D shows dual-labeled cells, as in panel B, after stepwise exposure to unlabeled active mesotrypsin (in red) followed by FITC-labeled mesotrypsin-S195A (in blue). Preincubation with increasing concentrations of active mesotrypsin correlated with the loss of the FITC signal, suggesting cleavage of surface-displayed APPIWT by active mesotrypsin. For all panels, the surface expression of APPI was detected by using a primary antibody against the C-terminal c-Myc tag and a PE-labeled secondary antibody, while binding to APPI was detected by a biotinylated target (bovine trypsin or mesotrypsin) and FITC-labeled streptavidin. Non-induced cells are shown in the bottom left quadrant of each plot. FITC- and PE-positive clones (green square) demonstrate resistant clones and FITC-negative PE-positive clones (yellow square) demonstrate cleaved clones.
Next, we assessed the ability of mesotrypsin to detect APPI displayed on the yeast cell surface. Even with a broad range of mesotrypsin concentrations, we were not able to detect mesotrypsin binding (Figure 1B). We hypothesized that surface-displayed APPI is rapidly proteolysed by mesotrypsin, preventing detection of the transient binding event. This explanation is consistent with the previously reported rapid cleavage of APPI by mesotrypsin in solution (enzymatic turnover time of 24 s [16]) and with the relatively long incubation time (at least 60 min) required for cell labeling prior to FACS. We thus employed a catalytically inactive form of mesotrypsin, in which the serine nucleophile is mutated to alanine (mesotrypsin-S195A) (Figure 1C). Unlike active mesotrypsin, mesotrypsin-S195A bound to surface-displayed APPI and produced a strong FACS signal (Figure 1D, left panel). Additionally, we found that preincubation of APPI-displaying yeast cells with active mesotrypsin prior to detection with mesotrypsin-S195A resulted in a concentration-dependent decrease in the FACS signal (Figure 1D, right panels), thereby confirming our hypothesis that surface-displayed APPI is rapidly proteolysed and depleted by mesotrypsin.
Simultaneous affinity and proteolytic stability maturation of an APPI library
We next postulated that active mesotrypsin and mesotrypsin-S195A could be used in a stepwise fashion to enrich an APPI diversity library for variants with proteolytic resistance. As a starting point, we generated a randomized library in which mutations were introduced throughout the entire APPI gene at a frequency of 0–3 mutations per clone, producing a library of about 9×106 independent variants. Diversity was introduced throughout the molecule, because while protease specificity and affinity are largely directed by the sequence of the canonical binding loop, proteolytic stability is a property strongly influenced by residues within the scaffold, as we showed previously [32].
Our unique screening strategy, designated ‘triple staining,’ consisted of three steps (Figure 1C). First, active mesotrypsin was incubated with the yeast-displayed APPI library and allowed to cleave the less-resistant APPI clones. Second, active mesotrypsin was washed out and replaced with biotinylated mesotrypsin-S195A, which bound selectively to the uncleaved (resistant) clones. Third, the bound mesotrypsin-S195A was visualized by staining with fluorescently labeled streptavidin, thereby facilitating detection. Elevated concentrations of active mesotrypsin were used as an evolutionary stimulus, with the fluorescently labeled mesotrypsin-S195A acting as a marker to facilitate identification of the most proteolytically resistant APPI variants (see ‘triple staining’ method, Figure 1C).
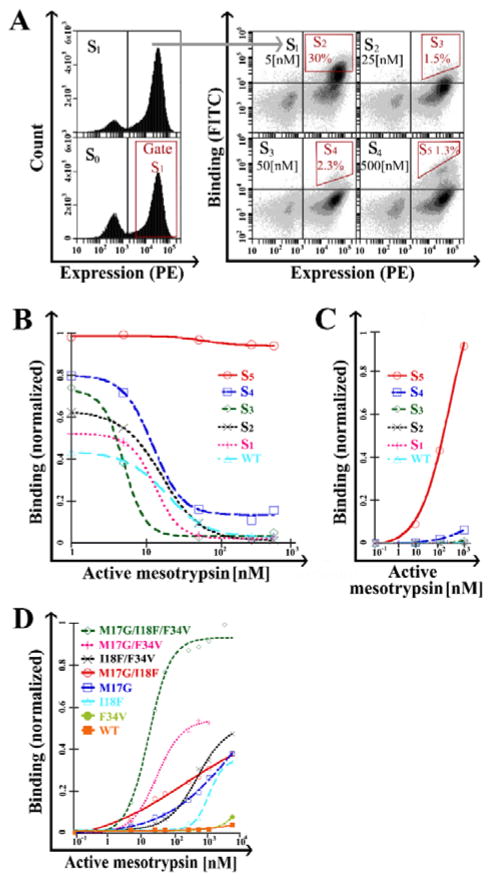
The initial library, termed S0 (where S stands for sort, and the number indicates the sort phase), was subjected to a round of expression enrichment based on c-Myc detection to yield the S1 library (Figure 2A, left plot). Subsequent rounds of affinity/stability maturation employed the ‘triple staining’ protocol. In sorting rounds S3, S4, and S5, diagonal sorting gates were used to select cell populations; this approach allowed binding normalization versus expression in real-time during the flow-cytometric sorting process, thereby dramatically decreasing bias of the expression level (i.e., the avidity effect) (Figure 2A).
Figure 2. Identification of APPI clones with improved resistance to cleavage.
(A) Stability maturation of the APPI library. FACS of single or dual-labeled cell populations for expression (S0 and S1) or both expression and binding (S1 to S5). The expressed population of APPI variants was sorted (S0), and the expression of the library was tested after enrichment (S1). Next, each cycle of stability maturation (S2 to S5) was performed with elevated concentrations of active mesotrypsin (as noted in the upper right quadrant of each plot) and a fixed concentration of inactive mesotrypsin (2 μM). Sorting gates are marked in red. (B) ‘Triple staining’ and (C) ‘double staining’ analysis of cell populations from library maturation cycles. (D) ‘Double staining’ analysis of cells expressing M17G, I18F, F34V and combination variants. A leftward shift in the sigmoid shape indicates a higher affinity, whereas a higher binding signal under saturating conditions indicates higher proteolytic stability. In panels B–D, the Y-axis represents mean fluorescence intensity normalization of binding to expression. Data was analyzed using KaleidaGraph software, with a sigmoidal curve fit. For all panels, the surface expression of APPI was detected by using a primary antibody against the C-terminal c-Myc tag and a PE-labeled secondary antibody, while binding to APPI was detected by biotinylated mesotrypsin and FITC-labeled streptavidin.
‘Triple staining’ analysis of cells displaying APPIWT and cell populations from the library maturation cycles (S1 to S5) showed that the more advanced the sort, the higher the stability and affinity of the mutant library for mesotrypsin (Figure 2B). Remarkably, the S5 pool showed high tolerance to the proteolytic activity of mesotrypsin at all enzyme concentrations used. We then determined whether it would be possible to detect the binding interaction between active mesotrypsin and each of the sort generation clone pools (as was done with bovine trypsin). Indeed, ‘double staining’ analysis with FITC-labeled active mesotrypsin showed high binding in pools derived from the advanced sorting rounds (S4 and S5; Figure 2C). These results suggest a high representation of proteolytically resistant APPI variants in the S5 pool of stability-matured variants.
Identification of surface displayed APPI clones with improved mesotrypsin affinity and resistance to cleavage
DNA sequencing of 37 randomly selected APPI clones from S5 showed three repeating mutations, M17A, I18F, and F34V, along with a number of unique mutations (Table S1). In a previous rational mutagenesis study, we had identified M17A as a mutation mediating improved mesotrypsin affinity and resistance to cleavage and had also found that Gly at the same position conferred an even greater improvement [29]. Accordingly, we chose M17G to replace M17A for subsequent analysis of single and composite mutants.
Staining of the individual YSD clones with active mesotrypsin showed that M17G and I18F exhibited high binding affinity and proteolytic stability, whereas F34V had only marginally enhanced binding affinity and stability vs. APPIWT (Figure 2D). The three mutations are spatially close to each other in the three-dimensional structure of APPI and may be expected to interact physically. To better understand the potential functional interactions between the mutations, we investigated the effect of all possible combinations (Figure 2D), which allowed us to assess additive, cooperative (beneficial dependence), or uncooperative (harmful dependence) interactions between mutations with respect to affinity and proteolytic resistance. Importantly, the results imply an additive or cooperative effect, in which the triple mutant showed remarkably higher binding affinity and proteolytic stability than the other combinations.
Affinity/stability-matured APPI variants show improved mesotrypsin inhibition in solution
To accurately assess mesotrypsin affinity and proteolytic stability independently, we expressed and purified soluble forms of the mutant proteins (Fig. S2, S3 and S4). Correct folding (Fig. S5) and high thermostability (Fig. S5 and S6) were observed for all proteins [using circular dichroism (CD)]. Thereafter, we determined the inhibition constants (Ki) (see SI; eqn 1), approximating the enzyme-inhibitor dissociation constants, by testing APPIWT and mutated variants as inhibitors of mesotrypsin catalytic activity against the small chromogenic peptide substrate Z-GPR-pNA.
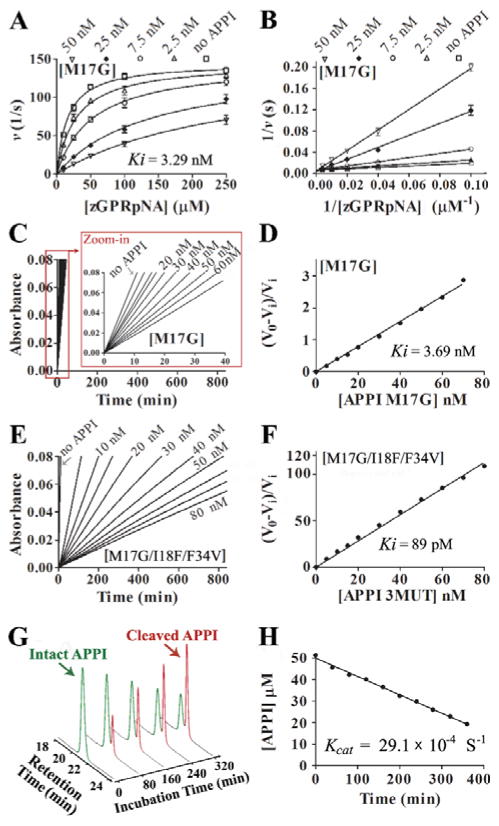
We observed a classic competition pattern of inhibition for all inhibitors (Figure 3A and B and Fig. S7A–D) and obtained a Ki value for APPIWT of 131±17 nM (Table 1), consistent with previous work [16]. APPIM17G showed a ~40-fold improvement in Ki, as previously reported [29], and APPII18F showed a similar improvement, whereas APPIF34V showed a ~threefold improvement in mesotrypsin affinity (Table 1).
Figure 3. Kinetics of mesotrypsin inhibition by APPI and hydrolysis of APPI by mesotrypsin.
(A) Competitive patterns of mesotrypsin inhibition by APPIM17G. Mesotrypsin cleavage of peptide substrate Z-GPR-pNA is competitively inhibited by APPIM17G. (B) The Lineweaver-Burk double reciprocal transform of the data used in panel A. The APPI (inhibitor) concentration is given at the top of each plot; the mesotrypsin concentration was 0.25 nM. Data was fitted globally to the competitive inhibition equation using Prism, GraphPad Software. (C, E) Slow, tight binding inhibition of mesotrypsin by APPI variants. Steady-state equilibrium for the reactions of APPIM17G (C) and APPIM17G/I18F/F34V (E) with various concentrations of APPI and 145 μM of peptide substrate Z-GPR-pNA. (D, F) A re-plot of slopes (V0 and Vi) calculated from the binding curves shown in panels C and E, respectively, where V0 is the uninhibited rate and Vi is the rate in the presence of inhibitor, which allows calculation of Ki using eq. 2 (as described in SI Materials and Methods under “ Inhibition studies”). (G) Kinetics of APPIM17G/I18F/F34V hydrolysis by mesotrypsin. Representative HPLC chromatograms are shown from a time course of APPIM17G/I18F/F34V hydrolysis by mesotrypsin. Green and red peaks represent intact and cleaved inhibitor, respectively. (H) Initial rate of hydrolysis, from which kcat is calculated. Disappearance of intact APPIM17G/I18F/F34V was quantified by integration of the HPLC peak in a time course that is illustrated in panel G. Hydrolysis reaction contained 50 μM of APPIM17G/I18F/F34V and 2.5 μM of enzyme.
Table 1.
Kinetic constants of mesotrvpsin with APPI variants
| Inhibitor | Ki (M)‡ | Ki (fold) | Kcat(s−1)‡ | Turnover time (s)‡ | Turnover time (fold) |
|---|---|---|---|---|---|
| APPI-WT | *(1.31±0.17)×10−7 | 1 | (35.6±2.3)×10−3 | 28.1±1.8 | 1 |
| APPI-F34V | *(5.01±0.46)×10−8 | 2.6 | (16.1±1.6)× 10−3 | 62.4±4.3 | 2.2 |
| APPI-M17G | **3.69×10−9 | 34.8 | |||
| APPI-M17G | *(3.29±0.25 )×10−9 | 39.8 | (15.6±1.5)×10−3 | 64.3±6.4 | 2.3 |
| APPI-118F | *(3.29±0.21)×10−9 | 39.8 | (10.4±0.9)×10−3 | 96.5±8.2 | 3.4 |
| APPI-118F/F34V | **(3.3±0.07)×10−9 | 39.8 | (5.35±0.2)×10−3 | 187.1±7.7 | 6.6 |
| APPI-M17G/F34V | **(1.40±0.11)×10−9 | 93.6 | (37.1±1.4)×10−4 | 270.0±10.1 | 9.6 |
| APPI-M17G/118F | **(45.25±0.36)×10−11 | 290 | (29.1±0.6)×10−4 | 344.0±7.6 | 12.2 |
| APPI-M17G/118F/F34V | **(89.8±0.23)×10−12 | 1459 | (4.29±0.3)×10−4 | 2336.7±140.0 | 83 |
represent fitted to Equations 1 and 2 (see SI), respectively.
Values arc means ± SD.
Since the lowest Ki values of our single-mutation APPI variants were in the lower nanomolar range, close to the practical limit of determination using the classical competitive inhibition equation in our assay, it was not possible to apply this method for combination variants with lower Ki values. Therefore, an alternative kinetic treatment, suitable for the quantification of slow, tight binding behavior (see SI; Eq. 2) was thus used, as shown in Figure 3C–F and Fig. S7F–J and summarized in Table 1.
To compare the results obtained from the slow-tight binding vs. the classical competitive inhibition studies, APPIM17G inhibition was evaluated using both approaches, with the results showing a high correlation between the two methods (Figure 3A–D and Table 1). As anticipated, the Ki values for double and triple mutants were, for the most part, significantly enhanced vis-à-vis those for the single mutants (Table 1). In particular, we observed an outstanding improvement in binding – of more than three orders of magnitude – of our triple mutant variant (Ki = 89.8 pM) vs. the wild type (Ki = 131,000 pM) (Figure 3E and F and Table 1).
Affinity/stability-matured APPI variants show marked enhancement in proteolytic resistance in solution
To evaluate whether our ‘stability maturation’ strategy did indeed produce variants with improved proteolytic resistance towards mesotrypsin, we directly measured rates of inhibitor cleavage by mesotrypsin in HPLC-based time course hydrolysis assays. Because the proteolysis reactions were conducted at APPI variant concentrations far above their measured Ki values (which also represent Km values for cleavage of the APPI variants as mesotrypsin substrates), the observed hydrolysis rates represent the catalytic rate constants (kcat). The individual mutations resulting from the stability maturation process gave modestly improved (two- to fourfold) proteolytic stability (Table 1). Pairs of mutations resulted in greater improvements (6- to 12-fold), while the APPIM17G/I18F/F34V triple mutant displayed a remarkable 83-fold improvement in proteolytic resistance towards mesotrypsin (Figure 3G,H, Fig. S8 and Table 1).
Most importantly, when combined to form the APPIM17G/I18F/F34V triple mutant, the three mutations (M17G, I18F, and F34V) act cooperatively in most cases with respect to affinity and in all cases with respect to proteolytic stability (See SI).
To test the specificity of the APPIM17G/I18F/F34V triple mutant, we selected cationic trypsin, anionic trypsin, factor XIa (FXIa) and kallikrein-6 [46] as targets that bind tightly to APPIWT and therefore serve as competitors for in vivo mesotrypsin binding. Importantly, we found that while APPIM17G/I18F/F34V showed greatly improved binding affinity toward mesotrypsin vis-à-vis APPIWT, affinity improvements toward kallikrein-6 and cationic and anionic trypsins were negligible, and affinity was substantially weakened toward the APPI physiological target FXIa. Thus, the mutations present in APPIM17G/I18F/F34V result in enhancement of specificity toward mesotrypsin over other proteases by three to five orders of magnitude (Table 2). These results suggest that APPIM17G/I18F/F34V may indeed constitute a suitable candidate for in vivo applications targeting mesotrypsin.
Table 2.
The inhibitor specificity of APPI-M17G/118F/F34V towards a range of human serine proteases
| Inhibitor | Ki for mesotrypsin (M)‡ | Ki for kallikrein-6(M)‡ | Ki for cationic trypsin(M)‡ | Ki for anionic trypsin (M)‡ | Ki for FXla (M)‡ | |
|---|---|---|---|---|---|---|
| APPI-WT | (1.31±0.17)×10−7 | (2.23±0.18)×10−9 | (6.27±1.01)×10−12 | (1.74±0.05)×10−12 | (4.1±0.14)×10−10 | |
| APPI-M17G/118F/F34V | (89.8±0.23)×10−12 | (1.09±0.12) ×10−9 | (4.96±0.25) ×10−12 | (1.47±0.02×10−12 | (9.84±0.32)×10−8 | |
|
| ||||||
| Ki (fold) | 1459 | 2.04 | 1.3 | 1.18 | 4.16×10−3 | |
|
|
1 | 715 | 1122 | 1236 | 350000 | |
Values are means ± SD.
APPIM17G/I18F/F34V variant reveals enhanced potency for inhibition of mesotrypsin-dependent cancer cell invasiveness
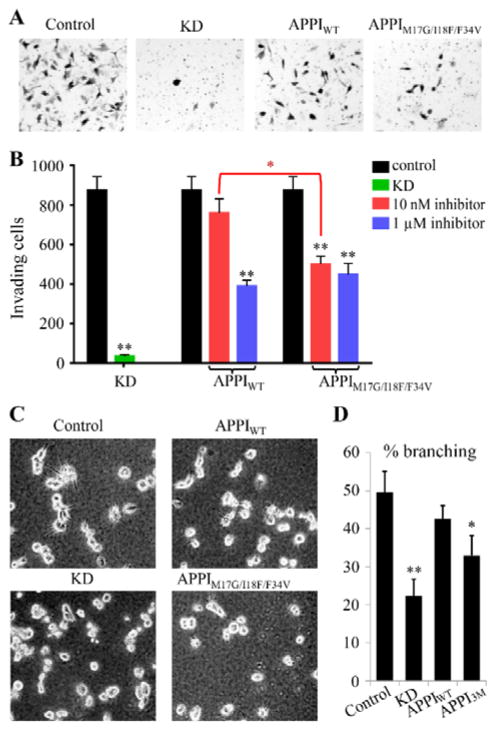
To evaluate the ability of the APPIM17G/I18F/F34V triple mutant to inhibit invasive behavior, we used human PC3-M cells, a hormone-independent, highly aggressive and metastatic prostate cancer cell line [47]. Matrigel invasion assays confirmed that mesotrypsin expression is essential for the invasiveness of these cells, since transduction with a lentiviral shRNA construct targeting the PRSS3 gene resulted in profound inhibition of cells passing through the membrane barrier (Figure 4A,B, KD control), as previously reported [2]. When control cells with endogenous PRSS3 expression were treated with 10 nM APPIM17G/I18F/F34V, significant inhibition of invasion was observed, whereas 10 nM APPIWT did not produce a significant effect. At much higher inhibitor concentrations (1 μM), both inhibitors produced similar maximum inhibitory effects of ~50%.
Figure 4. Enhanced potency of APPIM17G/I18F/F34V for inhibition of prostate cancer cell invasiveness.
(A,B) In Matrigel invasion assays, shRNA knockdown (KD) of PRSS3 or treatment with inhibitors APPIWT or APPIM17G/I18F/F34V led to reductions in PC3-M cellular invasion compared to control cells. (A) Images are shown for representative fields from stained invasion filters for (left to right) control cells, cells with PRSS3 knockdown (KD), cells treated with 10 nM APPIWT, and cells treated with 10 nM APPIM17G/I18F/F34V. (B) Bar graph shows means and S.E.M. for quadruplicate biological replicates. Black bars represent control cell samples, green bar represents cells with PRSS3 knockdown (KD), red bars represent cells treated with 10 nM inhibitor (APPIWT or APPIM17G/I18F/F34V as indicated), and blue bars represent cells treated with 1 μM inhibitor (APPIWT or APPIM17G/I18F/F34V as indicated). (C,D) In Matrigel 3D cell culture assays, shRNA knockdown of PRSS3 (KD) or treatment with 100 nM APPIM17G/I18F/F34V (indicated as APPI3M in panel D), but not with APPIWT, led to a significant reduction in the invasive branching morphology of PC3-M cells compared to control cells. (C) Images are shown for representative fields in 3D cultures for control cells, cells with PRSS3 knockdown (KD), cells treated with 100 nM APPIWT, and cells treated with 100 nM APPIM17G/I18F/F34V, as indicated. (D) Bar graphs shows means and S.E.M. for quadruplicate biological replicates. **P<0.005 for t-test comparisons of indicated conditions versus control. *P = 0.02 for t-test comparison of 10 nM treatment conditions for APPIWT vs. APPIM17G/I18F/F34V in panel B; P = 0.034 for t-test comparison of APPIM17G/I18F/F34V (APPI3M) vs. control in panel D.
To further explore the impact of APPI-based inhibitors on prostate cancer cells in a physiologically relevant environment, we grew PC3-M cells in 3D Matrigel culture. As reported previously [2], these cells displayed a native growth morphology characterized by spiky protrusions when grown in 3D (Figure 4C, upper left panel). Lentiviral shRNA silencing of PRSS3 significantly reduced the proportion of cells displaying this invasive morphology, as did treatment of the cultures with 100 nM APPIM17G/I18F/F34V (Figure 4C,D). In contrast, treatment of cells with APPIWT at the same concentration did not produce a comparable effect (Figure 4C,D). Taken together, these experiments demonstrate the enhanced potency of APPIM17G/I18F/F34V compared to APPIWT for suppression of cellular invasiveness. The inability of either APPIWT or APPIM17G/I18F/F34V to suppress invasion or invasive morphology to the same extent as mesotrypsin knockdown may result from inadequate selectivity or from competition for binding from other proteases in the cellular milieu; this possibility reinforces the necessity for continued engineering efforts to further enhance the selectivity of mesotrypsin-targeted APPI variants.
High-resolution crystal structure of APPIM17G/I18F/F34V bound to mesotrypsin provides insight into functional improvements
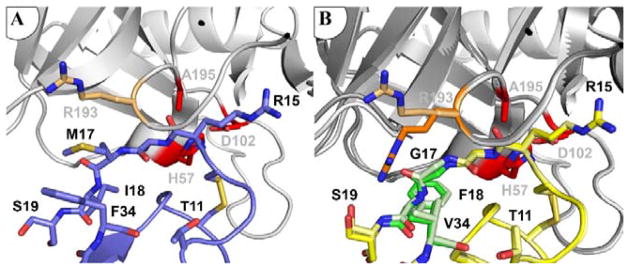
To gain insight into the mechanistic basis for improved mesotrypsin affinity and resistance to proteolysis of APPIM17G/I18F/F34V, we co-crystallized the inhibitor with mesotrypsin and solved and refined the structure of the complex at 1.83 Å resolution (Table S2). Overall, the structure is highly similar to the previously-solved APPIWT complex with mesotrypsin [PDB ID: 3L33; [32]], including similar disulfide bond topology of the inhibitor. Mutations at APPI residues 17 and 18, located on the primed side of the canonical binding loop, altered enzyme-inhibitor interactions in the S2′ and S3′ subsites. The mesotrypsin S2′ subsite is defined by the atypical Arg-193 residue (a highly conserved Gly in other trypsins), responsible for steric interactions that disfavor bulky P2′ substrate or inhibitor residues [29]. Arg-193 has been found to adopt multiple conformations depending on the P2′ residue of a bound inhibitor [17, 29]. In the structure of mesotrypsin bound to APPIWT [PDB ID: 3L33; [32]], Arg-193 is pushed upward by the inhibitor P2′ residue Met-17 into a cleft between the two beta-barrels of mesotrypsin (Figure 5A). The mutation of APPI Met-17 to Gly removes constraints on the conformation of Arg-193 in the complex, and, intriguingly, Arg-193 adopts different conformations in the two copies of the complex in our new crystal structure (Figure 5B). One conformation is similar to the ‘up’ conformation observed in the APPIWT complex structure, while the other is similar to the ‘down’ conformation previously seen in structures of mesotrypsin bound to the small molecule inhibitor benzamidine [PDB ID:1H4W; [13]] or to mutant BPTI possessing Gly at the P2′ position [PDB ID:3P92; [29]]. The observation of both Arg-193 conformations within the same crystal reveals that these conformations are roughly energetically equivalent and suggests that the improved affinity attributable to Gly-17 may result, at least in part, from preservation of conformational entropy in the vicinity of Arg-193 upon binding. The elimination of unfavorable steric interactions between Arg-193 and Met-17 (as found in APPIWT) [29, 32] may additionally contribute to the improved mesotrypsin affinity of variants possessing Gly-17.
Figure 5. Mesotrypsin Arg-193 adopts multiple conformations in the complex with APPIM17G/I18F/F34V.
(A) Arg-193 conformation is constrained by interaction with APPI Met-17 in the mesotrypsin complex with APPIWT. Mesotrypsin is shown in gray with catalytic triad residues in red, and the Arg-193 side chain rendered as light orange sticks; APPIWT is shown in blue (PDB entry: 3L33). (B) In complex with APPIM17G/I18F/F34V, mesotrypsin Arg-193 is found in different conformations in the two copies of the structure in the asymmetric unit of the crystal, which here are shown superposed. Mesotrypsin is shown in gray, with catalytic triad residues in red; APPIM17G/I18F/F34V is shown in yellow, with mutated residues in green. The distinct ‘up’ and ‘down’ conformations of Arg-193 are illustrated in light and dark orange, respectively.
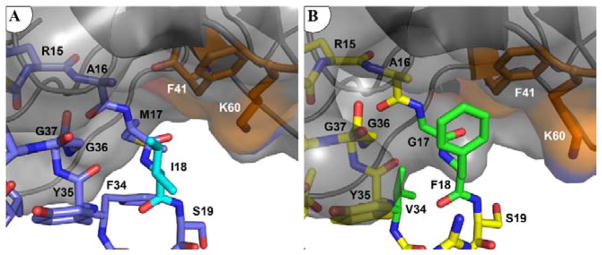
The mutation of APPI Ile-18 to Phe optimizes intermolecular packing in the S3′ subsite, resulting in the formation of new van der Waals forces, ring-stacking and hydrophobic contacts with Phe-41 and Lys-60 of mesotrypsin (Figure 6). The closest interatomic distance between the aromatic rings of APPI Phe-18 and mesotrypsin Phe-41 is 3.5 Å, which is consistent with the 3.8 Å that is typical of stabilizing aromatic pairs within proteins [48, 49]. The APPI Phe-18 aromatic ring is also located within 5 Å of the Nζ atom of mesotrypsin Lys-60, suggesting that favorable amino-aromatic interactions [50] may also potentially contribute to the enhancement of binding affinity. Taken together, these interactions enhance affinity by providing energetically favorable contacts between the enzyme and inhibitor, which are absent in the wild-type APPI-mesotrypsin complex.
Figure 6. APPIM17G/I18F/F34V displays improved complementarity with the mesotrypsin S3′ subsite.
(A) APPIWT shows limited interactions with the mesotrypsin S3′ subsite. Mesotrypsin cartoon and surface are shown in gray, with Phe-41 and Lys-60 in orange; APPIWT is shown in blue, with Ile-18 in cyan (PDB entry: 3L33). (B) Phe-18 of APPIM17G/I18F/F34V forms hydrophobic, ring-stacking, and amino-aromatic interactions with mesotrypsin Phe-41 and Lys-60 residues. Mesotrypsin is rendered and colored as above, with APPIM17G/I18F/F34V in yellow and mutated residues in green.
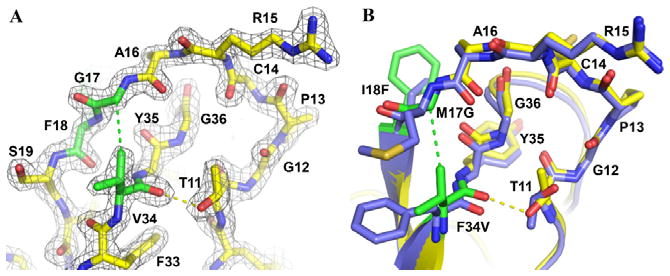
The mutation of Phe-34 to Val in APPI is located within the inhibitor scaffold, rather than in the canonical binding loop (Figure 7A). The Val-34 side chain approaches within 3.7 Å of the hydroxyl group of mesotrypsin Tyr-151 but otherwise does not make close contact with the enzyme. Residues 34 and 17 lie adjacent in the Kunitz domain structure, and the side chain of residue 34 constrains the conformation and mobility of residue 17. Val-34 Cγ1 forms a close contact of 3.5 Å with Gly-17 Cα, which is not present in APPIWT (Figure 7A,B); the resulting tighter intramolecular packing within APPIM17G/I18F/F34V potentially imposes greater constraints on the backbone mobility of residue 17. Consistent with this observation, the positive cooperativity of M17G and F34V seen in analyses of affinity improvements, proteolytic resistance, and energetic additivity (Table 1 and Fig. S9) suggests the possibility that the major importance of Val-34 may lie in its influence on the conformation and dynamics of Gly-17. Comparison of the structures of APPIWT and APPIM17G/I18F/F34V additionally reveals subtle backbone shifts of up to ~1 Å that enable the formation in the triple mutant of a new hydrogen bond between the carbonyl oxygen of Val-34 and the hydroxyl of Thr-11 (Figure 7A,B). This H bond, absent in most structures of APPIWT but present in all structures of BPTI, may confer additional rigidity on the inhibitor scaffold and hence contribute to proteolytic stability.
Figure 7. Enhanced intramolecular stabilizing features of APPIM17G/I18F/F34V.
The structure of APPIM17G/I18F/F34V bound to mesotrypsin reveals a new hydrophobic contact between Val-34 Cγ1 and Gly-17 Cα (green dashed line) and a new H-bond between Val-34 O and Thr-11 OH (yellow dashed line). (A) 2Fo-Fc density map is shown contoured at 2.0σ. APPIM17G/I18F/F34V is shown in yellow, with mutated residues in green. (B) APPIM17G/I18F/F34V is colored as above, superimposed upon APPIWT in blue to highlight small shifts in backbone positions. Thr-11 adopts a different rotamer conformation to facilitate formation of the H-bond.
DISCUSSION
During the past decade, mesotrypsin has emerged as a significant player in different stages of cancer development, being associated with cell malignancy in multiple cancers, including lung, colon, breast, pancreas and prostate cancers [1–6]. These studies have implicated mesotrypsin as a potential new therapeutic target. In the present study, we have identified a novel inhibitor with an unprecedented combination of mesotrypsin affinity, proteolytic stability, and biological potency, offering promise for development of new therapeutics.
Early studies of transendothelial migration in non-small cell lung cancer (NSCLC) cultures showed mesotrypsin overexpression to be associated with invasion and metastasis, while comparative microarray assays of cells taken from NSCLC patients showed mesotrypsin overexpression to be predictive of poor survival [1]. The first report to offer some mechanistic insight into the putative role of mesotrypsin in cancer progression showed a correlation between the upregulation of mesotrypsin and malignant progression in a breast cancer cell line. In that study, it was found that CD109, a cell-surface glycoprotein, acts as a potential mesotrypsin substrate involved in driving malignancy [5]. An indirect tumorigenic effect (in pancreatic cancer cells) – mediated by upregulation of VEGF expression – via the protease-activated receptor1 (PAR1)-mediated ERK pathway has also been suggested for mesotrypsin [6]. A more direct, and specific, role for the proteolytic activity of mesotrypsin in cancer cell invasiveness was recently described for metastatic prostate tumors [2]. While treatment with mesotrypsin directly promoted an invasive cellular phenotype, neither cationic trypsin nor a non-catalytic mesotrypsin variant could similarly drive this invasive phenotype, suggesting that the promotion of invasion depends on the specific proteolytic activity of mesotrypsin [2]. Indeed, a clue to the role played by mesotrypsin in metastasis may be found in the enhanced catalytic capability of mesotrypsin to hydrolyze canonical trypsin inhibitors that are highly abundant in the tumor microenvironment, such as human Kunitz protease inhibitor domains from amyloid precursor-like protein 2 (APLP2), bikunin, hepatocyte growth factor activator inhibitor type 2 (HAI2) and others [16, 17]. Cleavage and inactivation of these inhibitors as physiological substrates by mesotrypsin may plausibly contribute to its significant role in the mechanism of metastasis enhancement [16, 17]. In the present work, we have engineered a human Kunitz domain inhibitor that is remarkably resistant to mesotrypsin proteolysis, which we expect will block mesotrypsin’s metastasis-promoting activity.
Although mesotrypsin and other trypsins share many residues that contribute to similar primary specificity, mesotrypsin exhibits unique sequence and structural features that are responsible for its enhanced catalytic capability and distinct resistance towards canonical trypsin inhibitors. This resistance is most notably the result of two evolutionary mutations in mesotrypsin: the substitution of Gly-193 by Arg, which clashes sterically with the inhibitors, and the substitution of Tyr-39 by Ser, which prevents the formation of a hydrogen bond within the mesotrypsin/inhibitor complexes [11, 14, 15]. These mutations are thus responsible for the unusually low affinity of mesotrypsin (relative to typical trypsins) for polypeptide trypsin inhibitors, and also contribute to the more surprising ability of mesotrypsin to cleave several canonical trypsin inhibitors as substrates [12, 16, 17]. Additional unique mesotrypsin residues, Lys-74 and Asp-97, located on the periphery of the active site, have further evolutionarily adapted mesotrypsin for efficient cleavage of trypsin inhibitors [14]. The net effect of these evolutionary adaptations is to weaken favorable interactions, promote unfavorable interactions, and enhance protein dynamics at the interface between mesotrypsin and a bound canonical trypsin inhibitor [11, 14, 15]. Since the rate-limiting step in canonical inhibitor proteolysis is attributed to the hydrolysis of the acyl enzyme (requiring dissociation of the primed side residues of the canonical binding loop from the enzyme’s active site) [51], the weakened association and enhanced dynamics present in mesotrypsin-inhibitor complexes are expected to result in more rapid deacylation and expulsion of the cleaved inhibitor from the active site.
Notably, we found that the combination of three mutations in APPIM17G/I18F/F34V produces a mesotrypsin-inhibitor complex featuring a much more complementary binding interface, at which new favorable interactions of the inhibitor with mesotrypsin S2′ and S3′ subsites are exhibited on the primed side of the active site (Figures 6, 7); this explains the improved affinity of APPIM17G/I18F/F34V for mesotrypsin. To the extent that these favorable interactions are maintained during catalysis and following inhibitor cleavage, they may be expected to retard the dissociation of the cleaved inhibitor from the active site, thus contributing to the markedly improved stability of APPIM17G/I18F/F34V to proteolysis by mesotrypsin. The APPIM17G/I18F/F34V structure also revealed new stabilizing intramolecular features within the inhibitor scaffold (Figure 7); these may dampen the conformational dynamics of the molecule, thus further retarding proteolysis. The functional cooperativity between the three mutated residues, evident in our analyses of free energy additivity (Fig. S9), appears to be quite complex, and perhaps cannot be fully understood from the crystal structure alone, in the absence of data to describe the impact of the mutations on protein dynamics. Nonetheless, based on the crystal structure, we suggest that the intimate packing between Val-34 and Gly-17 (Figure 7) may synergistically stabilize inhibitor scaffold-binding loop interactions and thus slow proteolysis. Gly-17 and Phe-18 each independently enhance affinity for the enzyme and, by slowing dissociation of primed-side residues from the enzyme, also retard proteolysis. However, it appears that only in the triple mutant is there full optimization of both intramolecular packing within the inhibitor and intermolecular packing with the enzyme, resulting in greater cooperative enhancements in both affinity and proteolytic resistance.
While the capacity of mesotrypsin to cleave inhibitors presents a challenge from the perspective of inhibitor development, once a protein scaffold has been sufficiently stabilized to resist mesotrypsin cleavage, these very same distinctive active site features may provide an opportunity to develop mesotrypsin-selective inhibitors that can be exploited for therapeutic and diagnostic purposes. Our prior attempts to develop peptide-based mesotrypsin inhibitors using a structure-guided rational mutagenesis approach [29, 32] have demonstrated that mesotrypsin affinity is largely modulated by specific residues within the inhibitor binding loop, whereas inhibitor stability to mesotrypsin hydrolysis is highly influenced by scaffold residues. While the contributions of several binding loop residues to affinity have been previously identified [29, 32], the specific scaffold residues that contribute to proteolytic resistance have remained obscure until now. In this study, we have taken an unbiased combinatorial directed evolution approach – incorporating diversity throughout both the binding loop and scaffold – enhanced by a unique “triple staining” screening strategy, to enable, for the first time, simultaneous evolution of proteolytic stability and affinity. Intriguingly, our screening identified both binding loop and scaffold residues that acted cooperatively to enhance both mesotrypsin affinity and proteolytic stability. The best mesotrypsin inhibitor that we identified using this approach exhibited both 1459-fold higher affinity and 83-fold greater proteolytic stability than the parent molecule, making it by far the most potent inhibitor of mesotrypsin reported to date. Importantly, this novel inhibitor also reduced cellular invasion at nanomolar concentrations in cell-based assays (Figure 4), thus showing promise for development as a therapeutic or diagnostic agent. We anticipate that continued engineering efforts using this platform and screening strategy may further enhance the selectivity of mesotrypsin-targeted APPI variants, providing improved efficacy in the cellular context.
In conclusion, mesotrypsin has recently been identified as a protease that is upregulated in concert with tumor progression in many different high incidence and poor prognosis cancers. It has been found that silencing of mesotrypsin expression can inhibit cancer growth, invasion, and metastasis in cell culture and in mouse models, thus implicating mesotrypsin as a putative therapeutic target. However, it has proved difficult to engineer mesotrypsin inhibitors that possess high affinity and proteolytic stability. This study demonstrates that APPI provides a suitable protein scaffold for developing mesotrypsin inhibitors and that the YSD platform in concert with our unique screening approach offers a powerful method to simultaneously engineer proteolytic resistance, affinity and specificity. Such novel inhibitors will offer promising avenues for cancer therapy and imaging applications; they will also constitute extremely valuable laboratory reagents for deciphering the specific mechanisms by which mesotrypsin drives cancer progression. Finally, as proteolytic degradation is a general challenge faced in development of protein therapeutics, we suggest that similar YSD screening strategies employing active protease exposure as evolutionary stimuli may have wide applicability to the development of diverse protein-based drugs and diagnostics for many applications.
Supplementary Material
Summary statement.
Cancer is a leading cause of morbidity and mortality worldwide. The results presented here pave the way to develop new therapies targeting mesotrypsin, an enzyme that contributes to progression and metastasis of lung, prostate, breast, and pancreatic cancers.
Acknowledgments
The authors thank Professor Amnon Horovitz (Weizmann Institute of Science) for helpful discussions. We thank Dr. Alon Zilka for his technical assistance. FACS experiments were performed at the NIBN proteomics unit. This work was supported by the European Research Council “Ideas program” ERC-2013-StG (contract grant number: 336041) and the Prostate Cancer Foundation (PCF) to N.P. and by NIH grant R01CA154387 to E.S.R. X-ray diffraction data were measured at the Berkeley Center for Structural Biology (supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute) at the Advanced Light Source (supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231).
Abbreviations
- APPI
amyloid precursor protein Kunitz protease inhibitor domain
- BGU
Ben-Gurion University of the Negev
- BPTI
bovine pancreatic trypsin inhibitor
- BSA
bovine serum albumin
- CD
circular dichroism
- CPMU
Cytometry, Proteomics and Microscopy Unit
- DMSU
DNA Microarray and Sequencing Unit
- FACS
flow activated cell sorting
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- FXIa
factor XIa
- KD
knockdown
- NIBN
National Institute of Biotechnology in the Negev
- NSCLC
non-small cell lung cancer
- PE
phycoerythrin
- TB
trypsin buffer
- YSD
yeast surface display
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest with respect to publication of this paper.
References
- 1.Diederichs S, Bulk E, Steffen B, Ji P, Tickenbrock L, Lang K, Zanker KS, Metzger R, Schneider PM, Gerke V, et al. S100 family members and trypsinogens are predictors of distant metastasis and survival in early-stage non-small cell lung cancer. Cancer Res. 2004;64:5564–5569. doi: 10.1158/0008-5472.CAN-04-2004. [DOI] [PubMed] [Google Scholar]
- 2.Hockla A, Miller E, Salameh MA, Copland JA, Radisky DC, Radisky ES. PRSS3/mesotrypsin is a therapeutic target for metastatic prostate cancer. Mol Cancer Res. 2012;10:1555–1566. doi: 10.1158/1541-7786.MCR-12-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Zhang L, Wu Q, Boyd DD. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J Biol Chem. 2008;283:35295–35304. doi: 10.1074/jbc.M806965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dozmorov MG, Hurst RE, Culkin DJ, Kropp BP, Frank MB, Osban J, Penning TM, Lin HK. Unique patterns of molecular profiling between human prostate cancer LNCaP and PC-3 cells. Prostate. 2009;69:1077–1090. doi: 10.1002/pros.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hockla A, Radisky DC, Radisky ES. Mesotrypsin promotes malignant growth of breast cancer cells through shedding of CD109. Breast Cancer Res Treat. 2010;124:27–38. doi: 10.1007/s10549-009-0699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang G, Cao F, Ren G, Gao D, Bhakta V, Zhang Y, Cao H, Dong Z, Zang W, Zhang S, et al. PRSS3 promotes tumour growth and metastasis of human pancreatic cancer. Gut. 2010;59:1535–1544. doi: 10.1136/gut.2009.200105. [DOI] [PubMed] [Google Scholar]
- 7.Ghilardi C, Silini A, Figini S, Anastasia A, Lupi M, Fruscio R, Giavazzi R, Bani M. Trypsinogen 4 boosts tumor endothelial cells migration through proteolysis of tissue factor pathway inhibitor-2. Oncotarget. 2015 doi: 10.18632/oncotarget.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma R, Ye X, Cheng H, Ma Y, Cui H, Chang X. PRSS3 expression is associated with tumor progression and poor prognosis in epithelial ovarian cancer. Gynecologic oncology. 2015;137:546–552. doi: 10.1016/j.ygyno.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 9.Han S, Lee CW, Trevino JG, Hughes SJ, Sarosi GA., Jr Autocrine extra-pancreatic trypsin 3 secretion promotes cell proliferation and survival in esophageal adenocarcinoma. PloS one. 2013;8:e76667. doi: 10.1371/journal.pone.0076667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salameh MA, Radisky ES. Biochemical and structural insights into mesotrypsin: an unusual human trypsin. Int J Biochem Mol Biol. 2013;4:129–139. [PMC free article] [PubMed] [Google Scholar]
- 11.Salameh MA, Soares AS, Alloy A, Radisky ES. Presence versus absence of hydrogen bond donor Tyr-39 influences interactions of cationic trypsin and mesotrypsin with protein protease inhibitors. Protein Sci. 2012;21:1103–1112. doi: 10.1002/pro.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szmola R, Kukor Z, Sahin-Toth M. Human mesotrypsin is a unique digestive protease specialized for the degradation of trypsin inhibitors. J Biol Chem. 2003;278:48580–48589. doi: 10.1074/jbc.M310301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katona G, Berglund GI, Hajdu J, Graf L, Szilagyi L. Crystal structure reveals basis for the inhibitor resistance of human brain trypsin. J Mol Biol. 2002;315:1209–1218. doi: 10.1006/jmbi.2001.5305. [DOI] [PubMed] [Google Scholar]
- 14.Alloy AP, Kayode O, Wang R, Hockla A, Soares AS, Radisky ES. Mesotrypsin Has Evolved Four Unique Residues to Cleave Trypsin Inhibitors as Substrates. J Biol Chem. 2015;290:21523–21535. doi: 10.1074/jbc.M115.662429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salameh MA, Soares AS, Hockla A, Radisky ES. Structural basis for accelerated cleavage of bovine pancreatic trypsin inhibitor (BPTI) by human mesotrypsin. J Biol Chem. 2008;283:4115–4123. doi: 10.1074/jbc.M708268200. [DOI] [PubMed] [Google Scholar]
- 16.Salameh MA, Robinson JL, Navaneetham D, Sinha D, Madden BJ, Walsh PN, Radisky ES. The amyloid precursor protein/protease nexin 2 Kunitz inhibitor domain is a highly specific substrate of mesotrypsin. J Biol Chem. 2010;285:1939–1949. doi: 10.1074/jbc.M109.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pendlebury D, Wang R, Henin RD, Hockla A, Soares AS, Madden BJ, Kazanov MD, Radisky ES. Sequence and conformational specificity in substrate recognition: several human Kunitz protease inhibitor domains are specific substrates of mesotrypsin. J Biol Chem. 2014;289:32783–32797. doi: 10.1074/jbc.M114.609560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emi M, Nakamura Y, Ogawa M, Yamamoto T, Nishide T, Mori T, Matsubara K. Cloning, characterization and nucleotide sequences of two cDNAs encoding human pancreatic trypsinogens. Gene. 1986;41:305–310. doi: 10.1016/0378-1119(86)90111-3. [DOI] [PubMed] [Google Scholar]
- 19.Wiegand U, Corbach S, Minn A, Kang J, Muller-Hill B. Cloning of the cDNA encoding human brain trypsinogen and characterization of its product. Gene. 1993;136:167–175. doi: 10.1016/0378-1119(93)90460-k. [DOI] [PubMed] [Google Scholar]
- 20.Dennis MS, Herzka A, Lazarus RA. Potent and selective Kunitz domain inhibitors of plasma kallikrein designed by phage display. J Biol Chem. 1995;270:25411–25417. doi: 10.1074/jbc.270.43.25411. [DOI] [PubMed] [Google Scholar]
- 21.Dennis MS, Lazarus RA. Kunitz domain inhibitors of tissue factor-factor VIIa. II. Potent and specific inhibitors by competitive phage selection. J Biol Chem. 1994;269:22137–22144. [PubMed] [Google Scholar]
- 22.Dennis MS, Lazarus RA. Kunitz domain inhibitors of tissue factor-factor VIIa. I. Potent inhibitors selected from libraries by phage display. J Biol Chem. 1994;269:22129–22136. [PubMed] [Google Scholar]
- 23.Markland W, Ley AC, Ladner RC. Iterative optimization of high-affinity protease inhibitors using phage display. 2. Plasma kallikrein and thrombin. Biochemistry. 1996;35:8058–8067. doi: 10.1021/bi952629y. [DOI] [PubMed] [Google Scholar]
- 24.Markland W, Ley AC, Lee SW, Ladner RC. Iterative optimization of high-affinity proteases inhibitors using phage display. 1. Plasmin. Biochemistry. 1996;35:8045–8057. doi: 10.1021/bi9526286. [DOI] [PubMed] [Google Scholar]
- 25.Roberts BL, Markland W, Ley AC, Kent RB, White DW, Guterman SK, Ladner RC. Directed evolution of a protein: selection of potent neutrophil elastase inhibitors displayed on M13 fusion phage. Proc Natl Acad Sci U S A. 1992;89:2429–2433. doi: 10.1073/pnas.89.6.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Wang BE, Moran P, Lipari T, Ganesan R, Corpuz R, Ludlam MJ, Gogineni A, Koeppen H, Bunting S, et al. Pegylated kunitz domain inhibitor suppresses hepsin-mediated invasive tumor growth and metastasis. Cancer Res. 2009;69:8395–8402. doi: 10.1158/0008-5472.CAN-09-1995. [DOI] [PubMed] [Google Scholar]
- 27.Devy L, Rabbani SA, Stochl M, Ruskowski M, Mackie I, Naa L, Toews M, van Gool R, Chen J, Ley A, et al. PEGylated DX-1000: pharmacokinetics and antineoplastic activity of a specific plasmin inhibitor. Neoplasia. 2007;9:927–937. doi: 10.1593/neo.07544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams A, Baird LG. DX-88 and HAE: a developmental perspective. Transfusion and apheresis science : official journal of the World Apheresis Association : official journal of the European Society for Haemapheresis. 2003;29:255–258. doi: 10.1016/S1473-0502(03)00170-8. [DOI] [PubMed] [Google Scholar]
- 29.Salameh MA, Soares AS, Hockla A, Radisky DC, Radisky ES. The P(2)′ residue is a key determinant of mesotrypsin specificity: engineering a high-affinity inhibitor with anticancer activity. Biochem J. 2011;440:95–105. doi: 10.1042/BJ20110788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fioretti E, Angeletti M, Citro G, Barra D, Ascoli F. Kunitz-type inhibitors in human serum. Identification and characterization. J Biol Chem. 1987;262:3586–3589. [PubMed] [Google Scholar]
- 31.Bajaj MS, Ogueli GI, Kumar Y, Vadivel K, Lawson G, Shanker S, Schmidt AE, Bajaj SP. Engineering kunitz domain 1 (KD1) of human tissue factor pathway inhibitor-2 to selectively inhibit fibrinolysis: properties of KD1-L17R variant. J Biol Chem. 2011;286:4329–4340. doi: 10.1074/jbc.M110.191163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salameh MA, Soares AS, Navaneetham D, Sinha D, Walsh PN, Radisky ES. Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin. J Biol Chem. 2010;285:36884–36896. doi: 10.1074/jbc.M110.171348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graff CP, Chester K, Begent R, Wittrup KD. Directed evolution of an anti-carcinoembryonic antigen scFv with a 4-day monovalent dissociation half-time at 37 degrees C. Protein Eng Des Sel. 2004;17:293–304. doi: 10.1093/protein/gzh038. [DOI] [PubMed] [Google Scholar]
- 34.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 35.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci U S A. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YS, Bhandari R, Cochran JR, Kuriyan J, Wittrup KD. Directed evolution of the epidermal growth factor receptor extracellular domain for expression in yeast. Proteins. 2006;62:1026–1035. doi: 10.1002/prot.20618. [DOI] [PubMed] [Google Scholar]
- 37.Kieke MC, Shusta EV, Boder ET, Teyton L, Wittrup KD, Kranz DM. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc Natl Acad Sci U S A. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 39.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 40.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nature methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D: Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Read RJ, Adams PD, Arendall WB, 3rd, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, et al. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navaneetham D, Sinha D, Walsh PN. Mechanisms and specificity of factor XIa and trypsin inhibition by protease nexin 2 and basic pancreatic trypsin inhibitor. J Biochem. 2010;148:467–479. doi: 10.1093/jb/mvq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozlowski JM, Fidler IJ, Campbell D, Xu ZL, Kaighn ME, Hart IR. Metastatic behavior of human tumor cell lines grown in the nude mouse. Cancer Res. 1984;44:3522–3529. [PubMed] [Google Scholar]
- 48.McGaughey GB, Gagne M, Rappe AK. pi-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 49.Thomas A, Meurisse R, Charloteaux B, Brasseur R. Aromatic side-chain interactions in proteins. I. Main structural features. Proteins. 2002;48:628–634. doi: 10.1002/prot.10190. [DOI] [PubMed] [Google Scholar]
- 50.Burley SK, Petsko GA. Amino-aromatic interactions in proteins. FEBS Lett. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- 51.Radisky ES, Koshland DE., Jr A clogged gutter mechanism for protease inhibitors. Proc Natl Acad Sci U S A. 2002;99:10316–10321. doi: 10.1073/pnas.112332899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson DS, Keefe AD. Random mutagenesis by PCR. Curr Protoc Mol Biol. 2001;Chapter 8(Unit 8):3. doi: 10.1002/0471142727.mb0803s51. [DOI] [PubMed] [Google Scholar]
- 53.Looke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011;50:325–328. doi: 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chase T, Jr, Shaw E. p-Nitrophenyl-p′-guanidinobenzoate HCl: a new active site titrant for trypsin. Biochem Biophys Res Commun. 1967;29:508–514. doi: 10.1016/0006-291x(67)90513-x. [DOI] [PubMed] [Google Scholar]
- 55.Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 56.Horovitz A, Rigbi M. Protein-protein interactions: additivity of the free energies of association of amino acid residues. J Theor Biol. 1985;116:149–159. doi: 10.1016/s0022-5193(85)80135-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.