Abstract
Translesion synthesis (TLS) is a mutagenic branch of cellular DNA damage tolerance that enables bypass replication over DNA lesions carried out by specialized low-fidelity DNA polymerases. The replicative bypass of most types of DNA damage is performed in a two-step process of Rev1/Polζ-dependent TLS. In the first step, a Y-family TLS enzyme, typically Polη, Polι or Polκ, inserts a nucleotide across DNA lesion. In the second step, a four-subunit B-family DNA polymerase Polζ (Rev3/Rev7/PolD2/PolD3 complex) extends the distorted DNA primer-template. The coordinated action of error-prone TLS enzymes is regulated through their interactions with the two scaffold proteins, the sliding clamp PCNA and the TLS polymerase Rev1. Rev1 interactions with all other TLS enzymes are mediated by its C-terminal domain (Rev1-CT), which can simultaneously bind the Rev7 subunit of Polζ and Rev1-interacting regions (RIRs) from Polη, Polι or Polκ. In this work, we identified a previously unknown RIR motif in the C-terminal part of PolD3 subunit of Polζ whose interaction with the Rev1-CT is among the tightest mediated by RIR motifs. Three-dimensional structure of the Rev1-CT/PolD3-RIR complex determined by NMR spectroscopy revealed a structural basis for the relatively high affinity of this interaction. The unexpected discovery of PolD3-RIR motif suggests a mechanism of 'inserter' to 'extender' DNA polymerase switch upon Rev1/Polζ-dependent TLS, in which the PolD3-RIR binding to the Rev1-CT (i) helps displace the 'inserter' Polη, Polι or Polκ from its complex with Rev1, and (ii) facilitates assembly of the four-subunit 'extender' Polζ through simultaneous interaction of Rev1-CT with Rev7 and PolD3 subunits.
Introduction
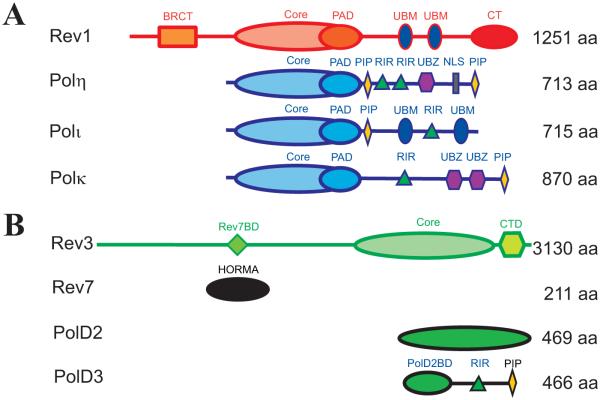
Genomic DNA undergoes constant modification by endogenous and environmental genotoxic agents1. If unrepaired, resulting lesions can block DNA replication by the high-fidelity DNA polymerases, Polδ and Polε, and trigger a cascade of events leading to cell death1,2. To evade this catastrophic scenario, cells use specialized translesion synthesis (TLS) DNA polymerases that can copy over DNA lesions encountered at the replication fork or fill damage-containing single-stranded DNA gaps left after replication, while leaving the DNA damage unrepaired3-7. In humans, the major TLS enzymes include the Y-family DNA polymerases Rev1, Polη, Polι and Polκ, and a B-family DNA polymerase Polζ3-7 (Figure 1).
Figure 1.
A. Structural domains of human Y-family TLS DNA polymerases Rev1, Polη, Polι, Polκ3-7. The core polymerase domain consists of the palm, finger and thumb domains, and a polymerase associated domain (PAD) unique to Y-family TLS enzymes. In addition, Y-family TLS polymerases possess accessory domains and motifs that mediate a range of protein-protein interactions. B. Subunits of human B-family TLS DNA polymerase Polζ and their domain arrangement42 (see text for details).
Certain DNA lesions can be accurately and efficiently bypassed by a single TLS enzyme. For example, Polη can copy over one of the most common UV-induced DNA lesions, T-T cyclobutane pyrimidine dimers (T-T CPD), in an error-free manner8. However, the bypass of most types of DNA damage is performed in a two-step process of Rev1/Polζ-dependent TLS9,10. In the first step of this process, an 'inserter' Y-family TLS DNA polymerase (typically one of Polη, Polι or Polκ) incorporates a nucleotide across the lesion. In the second step, an 'extender' TLS polymerase (typically Polζ) extends the aberrant DNA primer terminus before the replicative polymerase can take over DNA synthesis. Certain TLS enzymes are specialized for a bypass of different types of DNA lesions8,11,12. Thus, in addition to the bypass of TT-CPDs by Polη8, another Y-family TLS enzyme Polκ can efficiently replicate over N2-dG adducts such as the N2-benzo[a]pyrene-dG (BaP-dG) adducts resulting from exposure to smoke11,12. Therefore, the insertion step of the Rev1/Polζ-dependent TLS may also include selection of an appropriate Y-family enzyme for the bypass of a given DNA lesion. Despite recognition that the two-step Rev1/polζ-dependent TLS is a general mechanism for DNA lesion bypass that accounts for the majority of mutagenesis in eukaryotes3-7,10, the molecular mechanisms of TLS enzyme selection and polymerase switching events still remain elusive.
The exchange of replicative to TLS DNA polymerases at replication forks stalled by DNA damage is triggered by Rad6/Rad18-dependent mono-ubiquitination of the sliding clamp PCNA13-15, which serves as a binding platform for DNA replication and damage response proteins16. Like replicative DNA polymerases (Polδ, Polε), Y-family TLS enzymes (Polη, Polι, Polκ) bind PCNA via a consensus PCNA-interacting protein (PIP) box motif4,6,16 (Figure 1A). Rev1 is unique among Y-family TLS enzymes since it lacks the PIP-box motif and instead binds PCNA via its N-terminal BRCA1 C-Terminus (BRCT) domain17,18 and/or Polymerase Associated Domain (PAD)19 (Figure 1A). On the other hand, unlike replicative DNA polymerases, all eukaryotic Y-family TLS enzymes have ubiquitin-binding motifs (UBM) or ubiquitin-binding motifs zinc finger (UBZ) domains20 (Figure 1A) that augment their interaction with ubiquitinated PCNA and facilitate the replicative to TLS DNA polymerase switch, presumably through an affinity driven competition21.
In humans, selection of an appropriate 'inserter' TLS polymerase and an 'inserter' to 'extender' polymerase switch during Rev1/Polζ-dependent TLS likely involve a rearrangement of protein-protein interactions that are mediated by Rev13-7. Rev1 is a Y-family DNA polymerase whose catalytic activity is limited to incorporation of dCMP opposite to a G-template and bypass of certain DNA lesions such as abasic sites or G-adducts4,22-26. However, Rev1’s major role in TLS is not as a catalytic DNA polymerase27, but as a scaffold that recruits other TLS enzymes to DNA lesions and mediates assembly of the multi-polymerase complexes acting in Rev1/polζ-dependent TLS28-32. Rev1 interactions with other proteins are mediated by two unique modular domains missing in other Y-family TLS enzymes: the N-terminal Rev1-BRCT domain that interacts with PCNA17,18 and the C-terminal Rev1-CT domain that can simultaneously bind Rev1-interacting regions (RIR) of one of the 'inserter' Y-family polymerases (Polη, Polι or Polκ) and a Rev7 subunit of the 'extender' Polζ32-39 (Figure 1A). In addition, the PAD domain of yeast Rev1 was implicated in binding to PCNA19, Polη40 and Rev741; these interactions, however, have not been confirmed in vertebrates. In spite of the increasing knowledge on the structure and interactions of the Rev1 modular domains, their precise roles in mediating TLS polymerase selection and exchange are yet to be established.
The B-family TLS polymerase, Polζ, is a master 'extender' enzyme involved in Rev1/polζ-dependent TLS across a wide range of DNA lesions10,42,43, although the Y-family TLS polymerase Polκ has also demonstrated ability to extend mismatched base pairs44,45. In addition, it was shown that Polζ can also perform a nucleotide insertion step for some DNA lesions46-49. The minimal functional unit of Polζ is a complex of the catalytic Rev3 and accessory Rev7 subunits46 (Figure 1B). Recently, several groups have shown that a more efficient and processive form of Polζ includes two additional subunits, human PolD2 (p50) and PolD3 (p66) or their yeast homologues Pol31 and Pol32 (Figure 1B), that are also accessory subunits of the lagging strand replicative DNA polymerase Polδ42,50-53. Assembly of the four-subunit Polζ (below called Polζ4) is mediated by interaction of the PolD2 (Pol31) subunit with the Rev3 C-terminus, which contains a zinc finger domain and an iron-sulfur 4Fe-4S cluster50,54. Additionally, PolD2 (Pol31) forms a stable complex with the N-terminal domain of PolD3 (Pol32)55. The finding that Polζ may function as a four-subunit enzyme has led to the suggestion that the polymerase switches upon Rev1/Polζ-dependent TLS involve dissociation of the Polδ catalytic PolD1 and PolD2/PolD3 subunits, with the latter becoming a part of Polζ442,50. Consistent with this, it was shown that DNA damage causes Def1-dependent degradation of the catalytic subunit of yeast Polδ56 and degradation of the small PolD4 (p12) subunit from human Polδ resulting in its destabilization57. On the other hand, it was demonstrated that yeast Polζ4 is stable in all phases of cell cycle irrespective of DNA damage, arguing against direct sharing of subunits between Polδ and Polζ51. It also remains unclear how Polζ4 assembly and subunit exchange occurs on the leading DNA strand replicated by Polε58 (although recent reports suggest that Polδ may play a role in replication of both strands59,60), as well as what molecular events lead to the insertion step performed by Y-family TLS enzymes that precedes primer-template extension by Polζ.
The four-subunit Polζ4 has enhanced interaction with PCNA via the PIP-box motif in the PolD3 (Pol32) C-terminus61,62, which can be further strengthened in the Rev1/Polζ complex through the PCNA-binding domains of Rev117-19 and the Rev1-UBM motifs that can bind a ubiquitin moiety attached to PCNA20,63. Therefore, irrespective of whether Polζ4 assembles directly before the extension step or is recruited as pre-assembled complex, it is likely that the 'extender' Polζ4 can displace the 'inserter' Polη, Polι or Polκ after the completion of insertion step via an affinity driven competition. In this work, we show a direct physical interaction between the human Rev1-CT domain and the accessory PolD3 subunit of Polζ, which provides additional evidence in favor of this hypothesis. The Rev1-CT - PolD3 interaction is mediated by a Rev1-interacting region (RIR) in the unstructured C-terminal part of PolD3 (residues 144-466) following the N-terminal domain (residues 1-144) that forms a tight complex with PolD255 (Figure 1B). This newly discovered PolD3-RIR motif competes for the Rev1-CT binding with similar RIR motifs found in Polη, Polι and Polκ32,33. Thus, in the 'extender' Rev1/Polζ4 complex, the two independent binding sites on the Rev1-CT (one for RIR motifs and another for Rev7) can be simultaneously occupied by the Rev7 and PolD3 subunits of Polζ. This finding suggests a new role for the Rev1-CT - PolD3 interaction in stabilization of the 'extender' Rev1/Polζ4 assembly and in facilitating the 'inserter' to 'extender' polymerase switch by displacing the 'inserter' Polη, Polι or Polκ from their complex with Rev1.
Materials and Methods
Protein sample preparation
Human Rev1 C-terminal domain (Rev1-CT; residues 1158-1251) was expressed and purified as described previously33. The final sample of the domain used for NMR structure determination of the Rev1-CT/PolD3-RIR complex contained 0.7 mM 15N/13C protein, 50 mM sodium phosphate buffer, 100 mM NaCl, 0.5 mM EDTA, 2 mM DTT, 0.05% NaN3, 10% D2O, pH 7.0. A custom-synthesized peptide (GenScript) including the predicted RIR motif from human PolD3 (PolD3-RIR; residues 231-246; KGNMMSNFFGKAAMNK) has low solubility at the conditions used in our NMR experiments. Therefore, the Rev1-CT/PolD3-RIR complex was prepared by dissolving lyophilized unlabeled PolD3-RIR peptide directly in the 15N/13C Rev1-CT domain solution and incubating the sample at room temperature for several hours. The excess undissolved peptide was removed by centrifugation and formation of the complex was monitored by recording 1H-15N HSQC spectrum of the domain. The procedure was repeated until only one set of the Rev1-CT domain peaks corresponding to the peptide-bound form of the domain was left in the spectrum. Similar to the Rev1-CT/Polη-RIR interaction33, the Rev1-CT/PolD3-RIR binding is slow on the NMR time scale, so that two sets of peaks were visible corresponding to free and bound forms until the protein is fully saturated with peptide.
NMR resonance assignment and structure calculation of the complex
All NMR spectra were collected at 15 ºC on Agilent VNMRS spectrometers operating at 11.7 and 18.8 T magnetic fields equipped with cold probes; spectra were processed in NMRPipe64 and analyzed in CARA65. 15N, 13C and 1H NMR resonance assignments for the 15N/13C Rev1-CT domain in complex with the unlabeled PolD3-RIR peptide were obtained from two-dimensional 1H-15N HSQC and 1H-13C HSQC and three-dimensional HNCO, HNCACB, HBHA(CO)HN, HC(C)H-TOCSY and (H)CCH-TOCSY spectra66. 1H resonance assignments for the unlabeled PolD3-RIR peptide bound to the Rev1-CT domain were derived from two-dimensional 15N,13C-filtered TOCSY and NOESY (250 ms mixing time) spectra67. Spatial structure calculation for the complex was based on 1H-1H distance restraints derived from three types of NOESY spectra : (i) intramolecular distance restraints for the Rev1-CT domain were obtained from three-dimensional 15N- and 13C-edited NOESY-HSQC spectra66, (ii) intermolecular protein-peptide distance restraints were obtained from three-dimensional 13C- and 15N-edited 15N,13C-filtered NOESY-HSQC spectra67 (see Supplementary Figure S3), and (iii) intramolecular distance restraints for the PolD3-RIR peptide were obtained from a two-dimensional 15N,13C-filtered NOESY spectrum67.
The structure of the Rev1-CT/PolD3-RIR complex was calculated in CYANA68 based on (i) inter- and intramolecular 1H-1H distance restraints obtained from the NOESY spectra described above, (ii) backbone dihedral φ and ψ angle restraints derived from the backbone 1H, 15N, and 13C chemical shifts using the program TALOS+69, and (iii) hydrogen bond restraints added based on dihedral φ/ψ angle and NOE analysis. Intramolecular NOE correlations for the Rev1-CT domain were automatically assigned using CYANA68, while intermolecular protein-peptide and intra-peptide NOE correlations were assigned manually. A total of 200 structures of the Rev1-CT/PolD3-RIR complex were generated; the 20 lowest-energy structures were subsequently refined by constrained molecular dynamic simulations in explicit solvent in the software CNS70. NMR-based restraints used for the Rev1-CT/PolD3-RIR structure calculation and structure refinement statistics are summarized in Table 1.
Table 1.
NMR-based restraints used for structure calculation of the Rev1-CT domain complex with the PolD3-RIR peptide (residues 231-246).
| Total | Rev1-CT | PolD3 | |
|---|---|---|---|
| Summary of restraints | |||
|
| |||
| Total NOE distance restraints | 2580 | 2411 | 86 |
| Short range (|i-j|≤1) | 1074 | 989 | 85 |
| Medium range (1<|i-j|≤4) | 861 | 860 | 1 |
| Long range (|i-j|>4) | 562 | 562 | |
| Intermolecular | 83 | ||
| Dihedral angles restraints (φ/ ψ) | 85/80 | 78/78 | 7/2 |
| Hydrogen bonds | 57 | 57 | |
| Structure refinement statistics | |||
|
| |||
| Deviation from NMR-based restraints | |||
| NOE (Å) | 1.04 | ||
| Dihedral restraints (degrees) | 0.82 | ||
| Deviation from idealized geometry | |||
| Bonds (Å) | 0.02 | ||
| Angles (degrees) | 1.3 | ||
| Ramachandran plot favorable for selected residues a | 97.5% | ||
| RMSD, pairwise (Å) | |||
|
| |||
| All residues | |||
| Backbone atoms | 0.94±0.16 | ||
| Heavy atoms | 1.56±0.16 | ||
| Residues from regular secondary structure elements a | |||
| Backbone atoms | 0.68±0.14 | 1.63±0.51 | |
| Heavy atoms | 1.38±0.15 | 2.80±0.79 | |
- Residues 1165-1178, 1184-1199, 1203-1219, 1224-1243 of the Rev1-CT domain and residues 235-243 of the PolD3-RIR peptide involved in interaction with the domain.
SPR binding assays
Surface plasmon resonance (SPR) interaction assays were performed on a Biacore T100 instrument (GE Healthcare). PolD3-RIR (residues 231-246; CKGNMMSNFFGKAAMNK) and Polκ-RIR (residues 560-575; CEMSHKKSFFDKKRSER) peptides modified with the N-terminal cysteine residue were immobilized on CM5 chips using a thiol coupling kit. All SPR experiments were performed in 10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% (v/v) surfactant P20 buffer at 25 °C. Varying concentrations of the Rev1-CT domain, expressed and purified as described previously33, were injected over the chip surface for 300 to 500 s at a flow rate of 20 µL/min, followed by dissociation of the complex. Blank buffer injections were performed every 2 to 3 cycles during the concentration series and were used to monitor the stability of the experiment. Analyte responses were corrected for signals resulting from blank buffer injections both on a reference channel with no peptide immobilized and on channels with immobilized peptides. Dissociation constants (Kd) for the Rev1-CT/PolD3-RIR and Rev1-CT/Polκ-RIR complexes were determined by equilibrium analysis of SPR sensorgrams using the Biacore T200 evaluation software (v.2.0).
Results
PolD3 subunit of human DNA polymerases δ and ζ contains a Rev1 interacting region
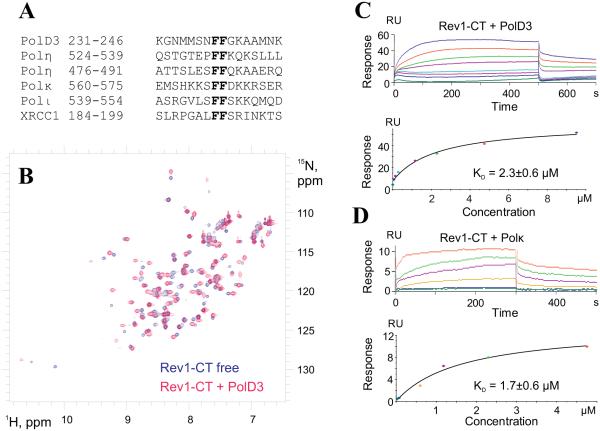
In vertebrates, the Rev1-CT domain has independent binding sites for (i) the RIR motifs of the 'inserter' Y-family TLS DNA polymerases Polη, Polι and Polκ and (ii) the accessory Rev7 subunit of the 'extender' B-family TLS polymerase Polζ32-39. The consensus RIR motif, which can be defined as 'nFFhhhh'32,33, consists of two consecutive phenylalanine residues preceded by an N-cap residue ('n': N, D, S, T, C or P)71,72 and followed by at least four residues that can form an α-helix ('h': all residues but P) (Figure 2A); lysine residue in the position +4 from the first phenylalanine is optional (found in 3 of 6 motifs shown in Figure 2A). Initially, RIR motifs were found in vertebrate Y-family TLS DNA polymerases only and were proposed to facilitate Rev1-mediated selection of the most appropriate 'inserter' polymerase to bypass a given type of DNA lesion32. Recent discovery of the RIR motif in XRCC1, a protein involved in single-strand break and base excision repair (Figure 2A), suggests a broader role for Rev1-CT mediated interactions in regulating cross-talk of TLS and other DNA damage response pathways73; the Rev1 - XRCC1 interaction may also help recruit DNA polymerase Polβ that can bypass some lesions74,75.
Figure 2.
A. Primary sequence alignment of the Rev1-interacting regions (RIR) from human PolD3 subunit of Polδ and Polζ, Y-family TLS DNA polymerases Polη, Polι and Polκ, and a single-strand break and base excision repair protein, XRCC173. B. Overlay of 1H-15N HSQC spectra of the free 15N/13C Rev1-CT domain (blue) and its complex with the unlabeled PolD3 RIR-motif (magenta). B, C. Surface plasmon resonance (SPR) sensorgrams for the Rev1-CT domain injected over the PolD3-RIR (plot C) and Polκ-RIR (plot D) peptides immobilized on a sensor chip surface (top panels) and best fits of the steady state response values to a two-state binding model (bottom panels).
The Rev1-CT domain in all eukaryotes interacts with Polζ via its accessory Rev7 subunit4. Electron microscopy reconstruction revealed that the four subunits of yeast Polζ4 (Rev3/Rev7/Pol31/Pol32) are well organized relative to each other, with Pol32 (analogue of human PolD3) located in close proximity to Rev776. The distinct spatial arrangement of yeast Polζ4 in this reconstruction is consistent with Polζ4 assembly stabilized by a number of intersubunit interactions, including those between Rev3 and Rev7, between Pol31 (analogue of human PolD2) and Rev3 C-terminus, and between Pol31 (PolD2) and the N-terminal domain of Pol32 (PolD3)55. In addition, spatial proximity of the Rev7 and Pol32 subunits has led to identification of a direct interaction between yeast Rev7 and the unstructured C-terminal part of Pol32, which has been demonstrated by pull-down assay with purified proteins76.
Interestingly, the electron microscopy-derived model of yeast Polζ4 suggests that, in the context of four-subunit assembly, the Rev7 subunit is accessible for interaction with the Rev1-CT domain76. This implies that, in the Rev1/Polζ4 complex, the Rev1-CT domain likely associates with Rev7 and is located in the proximity to Pol32 (homologue of human PolD3). Therefore, keeping in mind that human Rev1-CT has independent binding sites for Rev7 and RIR motifs33-39, we have examined the primary sequence of the unstructured C-terminal part of human PolD3 and identified a RIR motif centered at residues F238 - F239 (Figure 2A). The discovery of a RIR motif in human PolD3 brings up the intriguing possibility that the two independent binding sites on Rev1-CT can be simultaneously occupied by two distinct subunits of Polζ4, Rev7 and PolD3. This suggests a new role for Rev1-CT in stabilization of Polζ4 assembly and implies a possible mechanism of 'inserter' to 'extender' TLS polymerase switch (see below).
Rev1-CT/PolD3 binding is among the strongest interactions mediated by RIR-motifs
To confirm the Rev1-CT/PolD3-RIR interaction experimentally, we studied the formation of their complex by NMR spectroscopy. Figure 2B shows a comparison of 1H-15N HSQC spectra of the free 15N-labeled Rev1-CT domain (blue) and its complex with a 16 amino acid peptide corresponding to PolD3 residues 231-246 encompassing the PolD3-RIR motif (magenta). A number of peaks in the Rev1-CT domain have shifted to new positions after mixing the domain with the peptide (see Materials and Methods) consistent with specific binding. The pattern of chemical shift changes for the Rev1-CT domain bound to PolD3-RIR is similar to that we previously reported for the Rev1-CT/Polη-RIR complex formation33, suggesting that the two RIR motifs interact with the domain in a similar configuration.
In order to verify the Rev1-CT/PolD3-RIR interaction by another method and determine dissociation constant Kd for the complex, we have also monitored complex formation using a surface plasmon resonance (SPR) binding assay. In this assay, PolD3-RIR peptide (residues 231-246) modified with an N-terminal cysteine residue was immobilized on a sensor chip surface using thiol coupling chemistry, followed by monitoring complex formation and dissociation after injection of various concentrations of the Rev1-CT domain. To enable a comparison with previously published results32,73, we also performed the SPR assay to probe the Rev1-CT domain binding to the peptide that includes the RIR-motif from TLS polymerase Polκ (residues 560-575). Figures 2C and 2D, respectively, show SPR sensorgrams for the Rev1-CT domain interaction with the immobilized PolD3-RIR and Polκ-RIR peptides, and best fits of the steady state response values to a two-state binding model. The data analysis resulted in dissociation constants Kd of 2.3±0.6 µM for the Rev1-CT/PolD3-RIR and 1.7±0.6 µM Rev1-CT/Polκ-RIR complexes indicative of medium affinity binding.
The Kd value of 7.6 µM for the Rev1-CT/Polκ-RIR complex has been previously reported by Ohashi et al.32 based on an SPR assay similar to that used in our work. Another group73 reported a much stronger affinity for the Rev1-CT/Polκ-RIR complex of about 0.3 µM based on (i) binding assays that used changes in intrinsic tryptophan fluorescence of the Rev1-CT domain upon its titration with RIR peptides, and (ii) an NMR binding assay that monitored changes in peak intensities in 19F NMR spectrum of the 4-fluorophenylalanine-labeled Polκ-RIR peptide upon its titration into the Rev1-CT solution. In spite of significant differences in the reported absolute affinities determined using different methods, the two previous studies32,73 were consistent in concluding that the Polκ-RIR motif binds the Rev1-CT domain about an order of magnitude stronger than RIR motifs from Polη, Polι or XRCC1. Our SPR binding data, on the other hand, suggest that PolD3-RIR affinity for Rev1-CT (2.3 µM; Figure 2C) is about as high as that of Polκ-RIR (1.7 µM; Figure 2D), which is the tightest among RIR motifs.
In addition to showing the Rev1-CT/PolD3-RIR binding by NMR and SPR, we also used yeast two-hybrid assays in an attempt to investigate the Rev1-CT domain interaction with longer constructs encoding the C-terminal part of PolD3, as we had previously done when characterizing the Rev1-CT/Polη interaction33 (see Supplementary Materials). In contrast to what has been reported for Polη, Polι and Polκ32,33,36, the yeast two-hybrid assays were unable to detect the interaction between the Rev1-CT domain and any of the considered PolD3 fragments, possibly due to low expression level or low stability/solubility of the PolD3 constructs used for these assays. In this respect, we note that the PolD3-RIR peptide used for our NMR and SPR experiments is significantly more hydrophobic and less soluble than the corresponding RIR-peptides from Polη, Polι and Polκ (Figure 2A), and that the RIR motif centered at F238 - F239 is located in a relatively hydrophobic region of the PolD3 C-terminus.
Structure of the Rev1-CT/PolD3-RIR complex
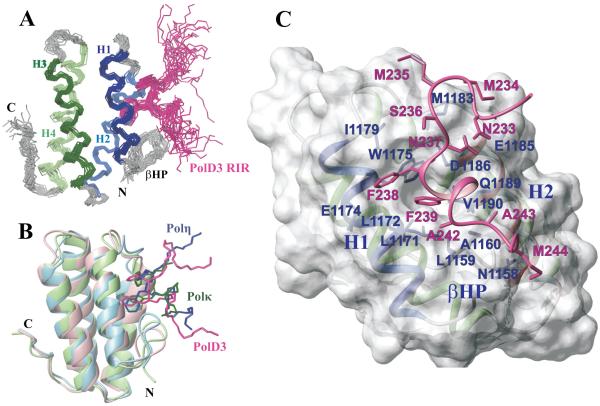
To characterize the Rev1-CT/PolD3 interaction in more detail, we have determined the three-dimensional structure of the Rev1-CT domain in complex with a 16 amino acid synthesized peptide containing the PolD3-RIR motif (residues 231-246) by NMR spectroscopy. Figure 3A shows the ensemble of 20 lowest energy structures of the complex. The generated ensemble agrees well with the input experimental data and displays the backbone root-mean-square deviation (RMSD) of 0.68 Å for regular secondary structure elements of the Rev1-CT domain and 1.63 Å for the PolD3 fragment 235-243 that directly interacts with the domain (Table 1).
Figure 3.
A. Superposition of 20 lowest-energy structures of the human Rev1-CT/PolD3-RIR complex. The Rev1-CT domain α-helices are shown in different colors; PolD3 peptide (residues 231-246) is shown in magenta with F238 and F239 side-chains highlighted. B. Comparison of known structures of the human Rev1-CT domain in complexes with RIR motifs from different partners: Polκ-RIR (green; PDB# 2LSI), Polη-RIR (blue; PDB# 2LSK) and PolD3-RIR (magenta; PDB# 2N1G, this work). B. Close up view of the PolD3-RIR interaction with the Rev1-CT domain surface. Side-chains of key residues of the Rev1-CT domain and PolD3-RIR motif that mediate intermolecular interactions are labeled in blue and magenta, respectively.
The structure of human Rev1-CT/PolD3-RIR complex includes key elements that are similar to previously reported structures of the Rev1-CT complexes with RIR motifs from Polη and Polκ33,36,39. The Rev1-CT domain forms a 4-helix bundle with α-helices spanning residues 1165-1178 (H1), 1184-1199 (H2), 1203-1219 (H3) and 1224-1243 (H4) (Figure 3A,B). The N-terminal β-hairpin (βHP) stabilized by the two backbone hydrogen bonds between L1159 and A1162 (NH1159-CO1162, CO1159-NH1162) docks against helices H1 and H2 and creates a hydrophobic patch on the domain surface with two binding pockets that can accommodate the sequential F238 and F239 of the PolD3-RIR motif (Figure 3C). The first hydrophobic pocket is formed by side-chains of L1159 from βHP, L1171, L1172, W1175 from H1 and D1186, Q1189, V1190 from H2 of the Rev1-CT domain and is deep enough to allow full insertion of the aromatic ring of F239 from the PolD3-RIR motif. The side chain of F238 from the PolD3-RIR fits into a shallower pocket where it interacts with E1174, W1175 and I1179 from helix H1 of the Rev1-CT domain. Similar to what has been described for the Rev1-CT/Polη-RIR complex33, the negatively charged side-chain of D1186 of the Rev1-CT points towards the peptide backbone and interacts with HN of F238 and F239 via a charge-dipole interaction (Figure 3C). Notably, the hydrophobic patch on the Rev1-CT surface that mediates its interaction with the PolD3-RIR includes three extra residues: A1160 and G1161 from the βHP that interact with A242, A243 and M244 from the C-terminal part of the PolD3-RIR peptide, and M1183 from the H1-H2 loop that interacts with M234 - N237 N-terminal to the FF pair of the RIR motif (Figure 3C). In addition, the negatively charged E1185 from helix H2 interacts with the backbone HN of N233 from the N-terminal part of the PolD3-RIR peptide (Figure 3C).
Figure 3B shows a superposition of the Rev1-CT/PolD3-RIR structure reported in this work (magenta) and the structures of Rev1-CT complexes with RIR motifs from Polη (blue; PDB ID: 2LSK)33 and Polκ (green; PDB ID: 2LSI)39. Overall, the mode of RIR motif interaction with the Rev1-CT domain is conserved among all complexes whose structures are available in the Protein Data Bank (PDB). In all structures, (i) aromatic side-chains of the two sequential phenylalanine residues of RIR motifs interact with pre-formed binding pocket on the domain surface, and (ii) the FF pair and the following several residues of RIR motifs are found in α-helical conformation. In all cases, the Rev1-CT - RIR complex is stabilized by polar interactions between the backbone amides of the two phenylalanine residues of the RIR motif and Oδ of D1186 of the Rev1-CT domain. However, in contrast to the RIR motifs from Polη and Polκ that upon binding form 2 to 3 turns of an α-helix33,36,39, only 3 residues of the PolD3-RIR motif bound to Rev1-CT were found in an α-helical conformation (Figure 3). A possible reason is the presence of G240 immediately following the FF pair, which has the second lowest (after P) propensity to form an α-helix among amino acids77. This configuration of the PolD3-RIR peptide, however, is sufficient to maintain the correct orientation of the sequential F238 and F239 towards the domain surface. Interestingly, a relatively short α-helix has been also reported for the XRCC1-RIR motif bound to the Rev1-CT domain73. On the other hand, the structure of the Rev1-CT domain does not undergo significant changes upon binding different RIR motifs. Mean pairwise RMSD for the Rev1-CT domain bound to RIR motifs from Polη (2LSK)33, Polκ (2LSI)39 and PolD3 (this work) is 1.1 Å for the backbone and 2.1 Å for all heavy atoms (calculated for residues from regularly structure elements), while RMSD between the free (2LSY)33 and PolD3-bound Rev1-CT is 1.4 Å for the backbone and 2.3 Å for all heavy atoms.
The lack of an extended α-helix in the bound form of the PolD3-RIR peptide poses a question of why the Rev1-CT/PolD3-RIR interaction is among the strongest mediated by RIR motifs. In the Rev1-CT complexes with RIR motifs from Polη and Polκ33,36,39, most intermolecular contacts are mediated by the six residues on the side of the RIR α-helix facing the domain (underlined in 'nFFhhhh', where 'n' is the N-cap residue, and 'h' is a helix-forming residue). In contrast, the PolD3-RIR motif adopts a more extended conformation with several additional residues involved in direct interaction with the Rev1-CT domain. Thus, protein-peptide NOEs were observed for the PolD3 fragment spanning residues 233-244, resulting in 83 intermolecular distance restraints that were used for structure calculation of the Rev1-CT/PolD3- RIR complex (Table 1). This number is greater than the number of protein-peptide distance restraints previously used for NMR structure calculation of the Rev1-CT/Polη-RIR complex, in which the RIR motif forms a 9-residue α-helix33. In the Rev1-CT/PolD3-RIR complex, intermolecular NOEs were observed for the four residues N-terminal to the FF pair (N233, M235, S236 and N237), which interact with the H1-H2 loop and the beginning of α-helix H2 (Figure 3C). Furthermore, intermolecular NOEs were observed for G240, A242, A243 and M244 (following the FF pair) that form an extended tail in the PolD3-RIR, which interacts with the tip of the Rev1-CT N-terminal β-hairpin formed by A1160 and G1161 (Figure 3C). While opposing the PolD3-RIR α-helix formation, interactions mediated by the above PolD3 residues contribute to an extensive Rev1-CT/PolD3-RIR interface, likely resulting in additional stabilization of the complex.
Discussion
Regulation of translesion synthesis (TLS) in eukaryotes occurs on multiple levels, including transcriptional control of protein expression, protein degradation, protein post-translational modifications and, most importantly, protein interactions3-7. The access of TLS enzymes to DNA, assembly of multi-polymerase complexes and polymerase switching events are regulated via a range of protein-protein and protein-DNA interactions mediated by interaction motifs, modular domains and subunits of TLS DNA polymerases3-7 (Figure 1). Some of these interactions are fairly tight, while many others such as Rev1-CT interactions with RIR motifs from human Polη, Polι, Polκ and PolD3 are weaker (µM to mM affinities) and are often competitive with one another32,33,36,39. One can expect that, owing to these relatively weak and competitive interactions, the TLS assembly is configured differently at each step of DNA lesion bypass, at different types of lesions and at replication forks vs. postreplication gaps, with only a subset of possible interactions formed at a given time.
In this work, we discovered a previously unknown RIR motif in the unstructured C-terminal part of PolD3 subunit of human Polδ and Polζ (Figure 2A). We studied the Rev1-CT - PolD3-RIR interaction by NMR and SPR, and demonstrated that the Rev1-CT/PolD3-RIR and Rev1-CT/Polκ-RIR complexes are among the tightest mediated by RIR motifs (dissociation constants Kd of 2.3±0.6 and 1.7±0.6 µM, respectively; Figures 2B-D). Furthermore, we used NMR spectroscopy to determine the three-dimensional structure of the Rev1-CT/PolD3-RIR complex, revealing a structural basis for the relatively high affinity of this interaction (Figure 3). The unexpected finding of a RIR motif in the PolD3 subunit of Polζ that can bind the Rev1-CT domain suggests a new role for Rev1-CT in stabilization of the four-subunit Polζ4 and offers potential new insights into the mechanism of 'inserter' to 'extender' polymerase switch upon mutagenic Rev1/Polζ-dependent TLS. This finding fits well into a model of reorganization of a multi-polymerase complex upon Rev1/Polζ-dependent TLS discussed below, in which the assembly of the 'extender' four-subunit Polζ4 results in Polζ acquiring Rev1 and PCNA interacting motifs found in the PolD3 subunit (Figure 1) that help Polζ displace an 'inserter' enzyme (one of Polη, Polι and Polκ) via an affinity driven competition.
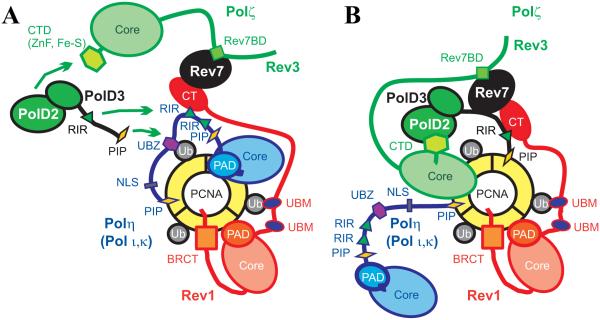
Figure 4 illustrates a possible model of reorganization of protein-protein interactions in a multi-polymerase complex that carries out two-step Rev1/Polζ-dependent TLS. Following DNA damage, the Rad6/Rad18 hetero-dimer is recruited to stretches of single-stranded DNA formed at stalled replication forks78 and mono-ubiquitinates the sliding clamp PCNA13-15. This event signals switching from normal DNA replication to the insertion step of TLS performed by Y-family DNA polymerases Polη, Polι or Polκ13-15 (Figures 4A); Rev14,22-26 and Polζ46-49 can also insert opposite particular lesions. Y-family TLS enzymes can interact with ubiquitinated PCNA via their PCNA-binding domains (PIP-box motifs in Polη, Polι, Polκ or BRCT domain in Rev1) and ubiquitin-binding UBM (Rev1, Polι) or UBZ (Polη, Polκ) domains that are not present in replicative DNA polymerases (Polδ, Polε)20. Ubiquitin-binding domains strengthen interactions of TLS enzymes with PCNA and mediate the replicative to TLS DNA polymerase switch, which presumably occurs via affinity-driven competition21. Additionally, the replicative to TLS DNA polymerase switch may involve destabilization and disassembly of the multi-subunit replicative polymerase Polδ56,57, resulting in release of a complex of its accessory PolD2/PolD3 subunits.
Figure 4.
A model or reorganization of protein-protein interactions and 'inserter' to 'extender' DNA polymerase switch upon a two-step Rev1/Polζ-dependent TLS. A. A possible configuration of the multi-polymerase TLS complex, including TLS DNA polymerases Rev1, and Polη (Polι, Polκ) and Polζ, assembled on mono-ubiquitinated PCNA at the nucleotide 'insertion' step of Rev1/Polζ-dependent TLS. In this configuration, a catalytic domain of the 'inserter' Y-family TLS polymerase Polη has access to DNA. Polη interactions with mono-ubiquitinated PCNA are mediated by Polη-UBZ domain and Polη-PIP motif. Rev1 binding to mono-ubiquitinated PCNA is mediated by Rev1-BRCT, Rev1-PAD, Rev1-UBM domains and to Polη-RIR by the Rev1-CT domain. In addition, the Rev1-CT domain mediates recruitment of Polζ through interaction with its Rev7 subunit. Presumably, Polη is recruited to stalled replication forks independently of other TLS enzymes via interaction with Rad18, which mono-ubiquitinates PCNA85. Rev1 is reportedly recruited to replication foci independently of Polη31,79, while Polζ is recruited via interaction with Rev179 either as two- or four-subunit (Polζ4) complex. In case Polη fails to insert a nucleotide across DNA lesion, a more appropriate 'inserter' Y-family enzyme (Polι, Polκ) may take over Polη by interacting with mono-ubiquitinated PCNA via its UBM/UBZ/PIP-box and trading its RIR motif with Polη-RIR bound to the Rev1-CT. B. A possible configuration of the multi-polymerase TLS complex at the primer-templete 'extension' step of Rev1/Polζ-dependent TLS. In this configuration, a catalytic domain of the 'extender' B-family polymerase Polζ has access to DNA. The 'extender' Polζ4 may take over the 'inserter' Polη (Polι or Polκ) by displacing Polη-RIR motif bound to the Rev1-CT with the PolD3-RIR and, in this way, disrupting Polη/Rev1 interaction. The Rev1-CT domain interactions with Rev7 and PolD3 subunits of Polζ stabilize the 'extender' Rev1/Polζ4 assembly that can interact with ubiquitinated PCNA via Rev1-BRCT, Rev1-PAD, Rev1-UBM domains and PolD3-PIP motif.
The formation of Polη foci and one-polymerase bypass of the Polη cognate lesion, UV-induced TT-CPD, depends on Polη interactions with PCNA and ubiquitin mediated by PIP-box and UBZ domains20,21, but not on the Polη interaction with Rev1 mediated by a RIR motif79. Thus, a Polη variant with F to A mutations in both RIR motifs (see Figure 2A) was able to suppress UV-induced mutagenesis and complement UV sensitivity of XPV cells to a similar extent as the wild-type Polη, suggesting that the Polη mutant effectively promotes single-polymerase bypass of the UV-induced TT-CPDs80. On the other hand, this Polη variant resulted in increased rate of spontaneous mutations as compared to wild-type Polη, consistent with a model where the Polη-RIR - Rev1 interaction is required for TLS across other types of DNA damage that Polη alone is unable to efficiently bypass80. A Polκ variant with F to A mutations in its RIR motif, which is defective in Rev1 binding, was unable to complement Polκ−/− mouse embryonic fibroblasts after exposure to benzo[a]pyrene in a DNA damage sensitivity assay, showing that the Polκ-RIR - Rev1 interaction is required for the bypass of the Polκ cognate DNA lesion BaP-dG32. Taken together, these findings suggest that if one Y-family TLS enzyme cannot efficiently copy over a DNA lesion, the Rev1-CT interactions with RIR motifs from this and other TLS enzymes facilitate selection of a more appropriate TLS DNA polymerase capable of completing the task. For example, if Polη is not proficient in incorporating a nucleotide across a DNA lesion, it may be replaced by another TLS enzyme such as Polκ whose RIR motif binds the Rev1-CT domain an order of magnitude stronger than RIR motifs from Polη32,73.
Previous studies of the Rev1-CT domain interactions with RIR motifs from Polη, Polι and Polκ did not address whether the Rev1-CT - RIR binding is only required for the selection of an appropriate 'inserter' TLS enzyme, or whether it also plays a role in the 'inserter' to 'extender' polymerase switch. The 'extender' Polζ is recruited to replication foci by Rev1 presumably through the Rev1-CT - Rev7 interaction79. The discovery of RIR motif in the C-terminal part of the PolD3 subunit of Polζ reported in this work suggests a possible key role for the Rev1-CT - PolD3-RIR interaction in facilitating the 'inserter' (Polη, Polι or Polκ) to 'extender' (Polζ) TLS polymerase switch (Figures 4). First, PolD3-RIR competes with RIR motifs from Y-family 'inserter' TLS enzymes for Rev1-CT binding and thus can help displace an 'inserter' enzyme from its complex with Rev1, weakening the 'inserter' enzyme interaction with the TLS complex assembled on mono-ubiquitinated PCNA. Second, the Rev1-CT - PolD3-RIR interaction can facilitate the assembly of 'extender' Polζ4 (Rev3/Rev7/PolD2/PolD3 complex)50,53. In the Rev1/Polζ4 complex, the two independent binding sites on the Rev1-CT domain34-39 can simultaneously interact with the Rev7 and PolD3 subunits of Polζ (Figure 4B). These interactions may play a role in stabilizing the Polζ4 complex in addition to interactions between the Rev7-binding motif of Rev3 (Rev7BD) and Rev781, between the Rev3 C-terminal domain (CTD) and PolD250,54, between PolD2 and the N-terminal domain of PolD355 and, possibly, between the PolD3 C-terminus and Rev7 (as shown in yeast76) (Figure 4B). The resulting 'extender' Rev1/Polζ4 complex would have an enhanced interaction with mono-ubiquitinated PCNA mediated by the Rev1-UBM20,63, Rev1-BRCT17,18 and presumably Rev1-PAD19 domains as well as by a PIP-box motif in the PolD3 C-terminus61,62. On the other hand, the 'inserter' Y-family TLS enzyme, which no longer interacts with Rev1, would have a decreased interaction with the TLS complex assembled on mono-ubiquitinated PCNA, and can be displaced from the DNA primer-template by an affinity driven competition (Figure 4B).
Interestingly, the only 'inserter' Y-family TLS polymerase in S. cerevisiae yeast, Polη, lacks a RIR motif and arguably interacts with the Rev1-PAD, but not with the Rev1-CT domain40. Consistent with this, inspection of the primary sequence of Pol32 subunit of yeast Polζ (homologue of human PolD3) did not reveal obvious candidate RIR motifs capable of Rev1-CT binding except 338-QGTLESFFKRKAK-350 at the very C-terminus, which serves as PCNA-interacting PIP-box motif. In addition, Rev1-CT – Pol32 binding was not detected in pull-down assays with purified proteins82. These data may suggest that regulation of TLS polymerase selection and exchange via the Rev1-CT – RIR motif interaction is a mechanism evolved in higher eukaryotes together with the emergence of additional Y-family TLS DNA polymerases, Polι and Polκ. Furthermore, higher eukaryotes acquired additional levels of TLS control such as negative regulation of Rev1/Polζ-dependent TLS by a putative human metalloprotease Spartan83, which is not found in yeast42. Remarkably, the primary sequence of human Spartan also contains a previously unnoticed RIR motif centered at F420 - F421 (whose interaction with Rev1-CT has been confirmed by 15N NMR; Supplementary Figure S2) that might help explain why Spartan depletion promotes complex formation between Rev1 and the PolD3 subunit of Polζ83. One should note, however, that the N-terminal part of human Rev1-CT responsible for interaction with RIR motifs exhibits higher sequence conservation between human and yeast than the C-terminal part that binds the Rev7 subunit of Polζ84, the interaction conserved between human and yeast4. Therefore, one cannot exclude a possibility that the yeast Rev1-CT domain might weakly interact with a RIR sequence somewhat different from that described in higher eukaryotes (e.g. 214-NLFVEDD-220 or 290-SFIDEDG-296 found in the C-terminal part of yeast Pol32 where the two residues with bulky hydrophobic side-chains follow an N-capping residue).
In summary, we have described interaction between the Rev1-CT domain and a newly discovered RIR motif from the C-terminal part of the PolD3 subunit of human Polδ and Polζ. The structure of the Rev1-CT/PolD3-RIR complex shows that PolD3-RIR binds Rev1-CT in a manner similar to RIR motifs from Polη and Polκ33,36,39, with the two sequential phenylalanine residues of the RIR motif mediating critical intermolecular contacts (Figure 3). However, in contrast to RIR motifs from Polη and Polκ, which upon binding form extended α-helices, in the Rev1-CT/PolD3-RIR complex only three residues of the PolD3-RIR were found in α-helical conformation. And yet, because of extensive contacts between Rev1-CT and RIR residues preceding and following FF pair, the Rev1-CT - PolD3-RIR interaction is among the strongest mediated by RIR motifs (Figures 2,3). The unexpected finding of RIR motif in the PolD3 subunit of Polδ and Polζ suggests a new role for Rev1-CT in stabilization of the functional four-subunit Polζ4 (Rev3/Rev7/PolD2/PolD3 complex)4, in which the Rev1-CT domain can simultaneously bind Rev7 and PolD3 subunits. Furthermore, this finding, along with previously published interaction data, provides new insights into the mechanism of 'inserter' (Polη, Polι, Polκ) and 'extender' (Polζ) polymerase switch upon mutagenic Rev1/Polζ-dependent TLS. In the proposed model of polymerase exchange (Figure 4), the relatively high affinity binding of PolD3-RIR to Rev1-CT helps displace the 'inserter' Polη, Polι or Polκ from its complex with Rev1 and promotes the formation the 'extender' Rev1/Polζ4 assembly, thus modulating affinities of 'inserter' and 'extender' complexes to mono-ubiquitinated PCNA.
Supplementary Material
Acknowledgements
The authors thank Dr. Irina Bezsonova for helpful discussion and assistance with preparation of the manuscript graphics. G.C.W. is an American Cancer Society Professor.
Funding Information
This work was supported by UCHC startup funds, a CICATS pilot grant and a CNPq Science without Boarders grant to D.M.K., and National Institute of Environmental Health Sciences Grant ES015818 to G.C.W.
Footnotes
Data Deposition
Atomic coordinates for the Rev1-CT/PolD3-RIR complex were deposited in the Protein Data Bank with PDB ID 2N1G. Backbone and side-chain chemical shifts of the complex were deposited in the BioMagResBank (BMRB) with entry number 25559.
Supplementary Materials
A description of yeast two-hybrid assays that probe Rev1-CT - PolD3 interaction; Figure S1 summarizing the results of yeast two-hybrid assay; Figure S2 showing 1H-15N HSQC spectrum of the Rev1-CT domain in the absence and presence of Spartan RIR motif; Figure S3 showing (A) selected strips from 13C-edited, 13C/15N-filtered NOESY-HSQC spectrum with intermolecular NOE correlations between the Rev1-CT domain and PolD3-RIR peptide, and (B) mapping of the NOE-derived intermolecular distance restraints onto the structure of the Rev1-CT/PolD3-RIR complex.
References
- 1.Friedberg EC, Friedberg EC. DNA repair and mutagenesis. 2nd ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 2.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo CX, Kosarek-Stancel J, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase Pol η. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 9.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 10.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 11.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells - The role of DNA polymerase κ. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 12.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 13.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg EC, Lehmann AR, Fuchs RPP. Trading places: How do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang ZH, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Pol η and Pol δ by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Guo CX, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: Significance of the REV1 BRCT domain in vitro and in vivo. Mol. Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 18.Pustovalova Y, Maciejewski MW, Korzhnev DM. NMR mapping of PCNA interaction with translesion synthesis DNA polymerase Rev1 mediated by Rev1-BRCT domain. J. Mol. Biol. 2013;425:3091–3105. doi: 10.1016/j.jmb.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 19.Sharma NM, Kochenova OV, Shcherbakova PV. The non-canonical protein binding site at the monomer-monomer Interface of yeast proliferating cell nuclear antigen (PCNA) regulates the Rev1-PCNA Interaction and Polζ/Rev1-dependent translesion DNA synthesis. J. Biol. Chem. 2012;286:33557–33566. doi: 10.1074/jbc.M110.206680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 21.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 22.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 23.Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J. Biol. Chem. 2001;276:15051–15058. doi: 10.1074/jbc.M008082200. [DOI] [PubMed] [Google Scholar]
- 24.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 25.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiltrout ME, Walker GC. The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics. 2011;187:21–35. doi: 10.1534/genetics.110.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson JR, Gibbs PEM, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 29.Guo CX, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 31.Tissier A, Kannouche P, Reck M-P, Lehmann AR, Fuchs RPP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase polη and REV1 protein. DNA Rep. 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi E, Hanafusa T, Kamei K, Song I, Tomida J, Hashimoto H, Vaziri C, Ohmori H. Identification of a novel REV1-interacting motif necessary for DNA polymerase κ function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pozhidaeva A, Pustovalova Y, D'Souza S, Bezsonova I, Walker GC, Korzhnev DM. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase η. Biochemistry. 2012;51:5506–5520. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pustovalova Y, Bezsonova I, Korzhnev DM. The C-terminal domain of human Rev1 contains independent binding sites for DNA polymerase η and Rev7 subunit of polymerase ζ. FEBS Lett. 2012;586:3051–3056. doi: 10.1016/j.febslet.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, Kim H, D'Andrea AD, Walker GC, Zhou P. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol ζ and Pol κ. J. Biol. Chem. 2012;287:33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojtaszek J, Liu JX, D'Souza S, Wang S, Xue YH, Walker GC, Zhou P. Multifaceted recognition of vertebrate Rev1 by translesion polymerases ζ and κ. J. Biol. Chem. 2012;287:26400–26408. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7. J. Biol. Chem. 2012;287:33847–33852. doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie W, Yang X, Xu M, Jiang T. Structural insights into the assembly of human translesion polymerase complexes. Protein Cell. 2012;3:864–874. doi: 10.1007/s13238-012-2102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu D, Ryu K-S, Ko J, Sun D, Lim K, Lee J-O, Hwang J. m., Lee Z.-w., Choi B-S. Insights into the regulation of human Rev1 for translesion synthesis polymerases revealed by the structural studies on its polymerase-interacting domain. J. Mol. Cell. Biol. 2013;5:204–206. doi: 10.1093/jmcb/mjs061. [DOI] [PubMed] [Google Scholar]
- 40.Acharya N, Haracska L, Prakash S, Prakash L. Complex formation of yeast Rev1 with DNA polymerase η. Mol. Cell. Biol. 2007;27:8401–8408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol. Cell. Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova AV, Burgers PM. Eukaryotic DNA polymerase ζ. DNA Rep. 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol ζ) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 44.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol.Cell. Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 47.Johnson RE, Yu S-L, Prakash S, Prakash L. Yeast DNA polymerase ζ is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y-C, Li L, Makarova AV, Burgers PM, Stone MP, Lloyd RS. Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis. 2014;35:1461–1468. doi: 10.1093/carcin/bgu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Y-C, Li L, Makarova AV, Burgers PM, Stone MP, Lloyd RS. Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB(1)-N7-Gua. J. Biol. Chem. 2014;289:18497–18506. doi: 10.1074/jbc.M114.561563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA Polymerase δ and ζ Switch by Sharing Accessory Subunits of DNA Polymerase δ. J. Biol. Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee Y-S, Gregory MT, Yang W. Human Polζ purified with accessory subunits is active in translesion DNA synthesis and complements Polη in cisplatin bypass. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Netz DJA, Stith CM, Stuempfig M, Koepf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PMJ, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nature Chem. Biol. 2012;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH. X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle. 2008;7:3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daraba A, Gali VK, Halmai M, Haracska L, Unk I. Def1 Promotes the Degradation of Pol3 for Polymerase Exchange to Occur During DNA-Damage-Induced Mutagenesis in Saccharomyces cerevisiae. PLoS Biol. 2014;12:e1001771. doi: 10.1371/journal.pbio.1001771. Article No.: e1001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, Zhou Y, Trusa S, Meng X, Lee EYC, Lee MYWT. A novel DNA damage response - Rapid degradation of the p12 subunit of DNA polymerase δ. J. Biol. Chem. 2007;282:15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- 58.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol. Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyabe I, Mizuno K, Keszthelyi A, Daigaku Y, Skouteri M, Mohebi S, Kunkel TA, Murray JM, Carr AM. Polymerase δ replicates both strands after homologous recombination-dependent fork restart. Nat. Struct. Mol. Biol. 2015;22:932–938. doi: 10.1038/nsmb.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johansson E, Garg P, Burgers PMJ. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 62.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo C, Tang T-S, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 65.Keller RLJ. The Computer Aided Resonance Assignment Tutorial. Cantina Verlag; Switzerland: 2004. [Google Scholar]
- 66.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999;34:93–158. [Google Scholar]
- 67.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage lambda N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997;119:6711–6721. [Google Scholar]
- 68.Guntert P. Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:105–125. [Google Scholar]
- 69.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 71.Doig AJ, Baldwin RL. N- and C-capping preferences for all 20 amino acids in alpha-helical peptides. Protein Sci. 1995;4:1325–1336. doi: 10.1002/pro.5560040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viguera AR, Serrano L. Stable proline box motif at the N-terminal end of alpha-helices. Protein Sci. 1999;8:1733–1742. doi: 10.1110/ps.8.9.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gabel SA, DeRose EF, London RE. XRCC1 interaction with the REV1 C-terminal domain suggests a role in post replication repair. DNA Rep. 2013;12:1105–1113. doi: 10.1016/j.dnarep.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.London RE. The structural basis of XRCC1-mediated DNA repair. DNA Rep. 2015;30:90–103. doi: 10.1016/j.dnarep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith LA, Makarova AV, Samson L, Thiesen KE, Dhar A, Bessho T. Bypass of a psoralen DNA interstrand cross-link by DNA polymerases β, ι, and κ in vitro. Biochemistry. 2012;51:8931–8938. doi: 10.1021/bi3008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gomez-Llorente Y, Malik R, Jain R, Choudhury JR, Johnson RE, Prakash L, Prakash S, Ubarretxena-Belandia I, Aggarwal AK. The architecture of yeast DNA polymerase ζ. Cell Reports. 2013;5:79–86. doi: 10.1016/j.celrep.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chakrabartty A, Baldwin RL. Stability of alpha-helices. Adv. Protein Chem. 1995;46:141–76. [PubMed] [Google Scholar]
- 78.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: A potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 79.Andersen PL, Xu F, Ziola B, McGregor WG, Xiao W. Sequential assembly of translesion DNA polymerases at UV-induced DNA damage sites. Mol. Biol. Cell. 2011;22:2373–2383. doi: 10.1091/mbc.E10-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akagi J-I, Masutani C, Kataoka Y, Kan T, Ohashi E, Mori T, Ohmori H, Hanaoka F. Interaction with DNA polymerase η is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Rep. 2009;8:585–599. doi: 10.1016/j.dnarep.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 81.Hara K, Hashimoto H, Murakumo Y, Kobayashi S, Kogame T, Unzai S, Akashi S, Takeda S, Shimizu T, Sato M. Crystal structure of human Rev7 in complex with a human Rev3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J. Biol. Chem. 2010;285:12299–12307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Acharya N, Johnson RE, Pages V, Prakash L, Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim MS, Machida Y, Vashisht AA, Wohlschlegel JA, Pang Y-P, Machida YJ. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res. 2012;41:1661–1668. doi: 10.1093/nar/gks1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D'Souza S, Waters LS, Walker GC. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair. 2008;7:1455–1470. doi: 10.1016/j.dnarep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.