Abstract
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) systems mediate adaptive immunity in diverse prokaryotes. CRISPR-associated Cas1 and Cas2 proteins have been shown to enable adaptation to new threats in Type I and II CRISPR systems by the acquisition of short segments of DNA (“spacers”) from invasive elements. In several Type III CRISPR systems, Cas1 is naturally fused to a reverse transcriptase (RT). In the marine bacterium Marinomonas mediterranea (MMB-1), we show that an RT-Cas1 fusion enables the acquisition of RNA spacers in vivo in an RT-dependent manner. In vitro, the MMB-1 RT-Cas1 and Cas2 proteins catalyze ligation of RNA segments into the CRISPR array, followed by reverse transcription. These observations outline a host-mediated mechanism for reverse information flow from RNA to DNA.
RNA-guided host defense mechanisms associated with Clustered Regularly Interspersed Short Palindromic Repeat (CRISPR) arrays exist in most bacteria and archaea (1, 2). Their target specificity derives from a series of unique “spacers” – many identical to DNA sequences from phage, transposon, and plasmid mobilomes – interspersed within CRISPR arrays (3-5). Transcripts from these CRISPR arrays are processed into short, structured RNAs, which form a complex with CRISPR associated (Cas) endonucleases and target invasive nucleic acids, thereby conferring immunity (6, 7). CRISPR-Cas systems have been phylogenetically grouped into five types (8, 9). Homologs of the cas1 and cas2 genes are conserved across diverse CRISPR types (9, 10), with direct evidence for a role in the physical integration of new spacers from invasive DNA into CRISPR arrays in a few Type I and II systems (11-14). Spacer acquisition allows the host to adapt to new threats.
The ability of Type III systems to target RNA in addition to DNA (15-21) raises the possibility of natural spacer acquisition from RNA species. Direct acquisition of RNA spacers would add to the handful of known mechanisms for the reverse flow of genetic information from RNA into DNA genomes (22-27).
Examination of bacterial genomes has revealed a class of CRISPR-associated coding regions with Cas1 fused to a putative reverse transcriptase (RT) (10, 28-30). These RT-Cas1 fusions raise the possibility of a concerted mechanism of spacer acquisition involving reverse transcription of RNA to DNA: a potentially host-beneficial mechanism for RNA-to-DNA information flow.
Common features of RT-Cas1 fusions
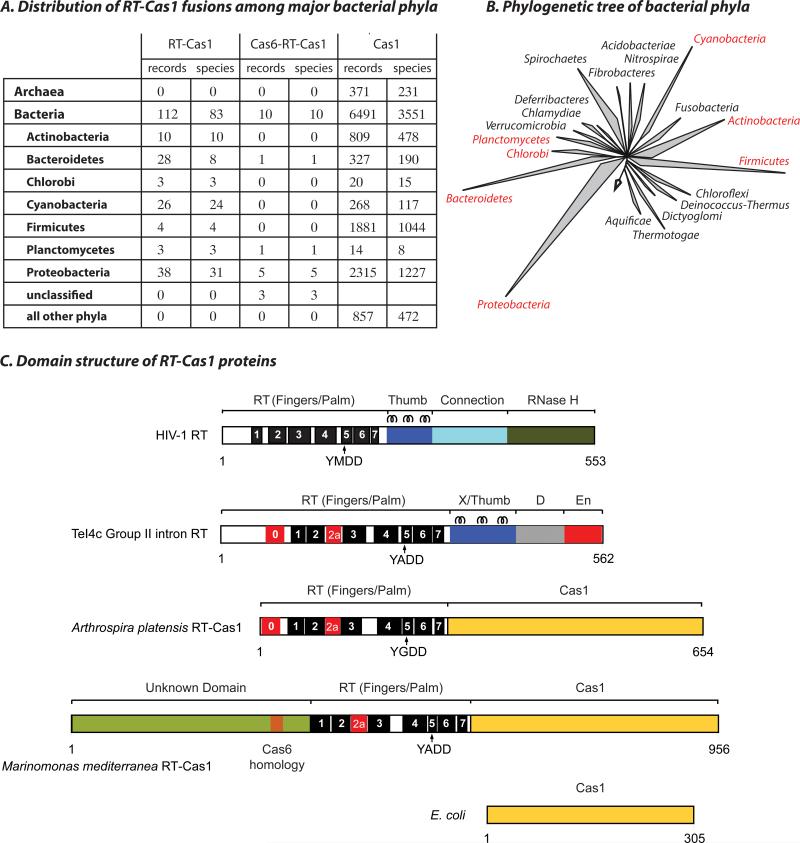
To examine the phylogenetic distribution of fused RT-Cas1 genes, we used the NCBI Conserved Domain Architecture Retrieval Tool (CDART) to retrieve protein records containing both a Cas1 domain (PF01867) and an RT domain of any origin (PF00078). Of 93 RT-Cas1 bearing species, all were from bacteria and none from archaea. RT-Cas1 fusions were most prevalent among cyanobacteria, with 21% of cas1-bearing cyanobacteria carrying such fusions (Fig. 1A,B). RT-Cas1 fusions with sufficient flanking sequence for type classification were exclusively associated with Type III CRISPR systems (Table S1); conversely, ~8% of bacterial Type III CRISPR systems carry RT-Cas1 fusions.
Figure 1. Phylogenetic distribution and domain structure of RT-Cas1 fusions.
(A) Taxonomic summary of unique RT-Cas1 protein records obtained from the NCBI CDART engine (current as of 05/2015). Numbers of Cas1 protein records and bacterial species are shown with (left) a fused RT domain; (center) RT and an additional N-terminal extension containing a Cas6-like motif; and (right) Cas1 with no additional annotated domain. Only phyla containing RT-Cas1 fusions are listed. (B) 16S rRNA-based tree showing major bacterial phyla, with RT-Cas1 containing phyla in red (adapted from (47)). (C) Schematic showing the domain organization of HIV RT (P03366), a group II intron RT (TeI4c from Thermosynechococcus elongatus BP-1; WP_011056164), Arthrospira platensis RT-Cas1 (WP_006620498), Marinomonas mediterranea RT-Cas1 (WP_013659858), and E. coli Cas1 (NP_417235). Conserved RT motifs as defined in (48) are labeled 1 to 7. Motifs 0 and 2a are conserved in mobile group II intron and non-LTR-retrotransposon RTs (32). The YxDD sequence found in motif 5 contains two Asp residues at the RT active site. D: DNA binding domain, En: Endonuclease domain. Three α-helices found in the Thumb/X domain of HIV and group II intron RTs are indicated.
The Cas1-fused RT domains were most closely related to RTs encoded by mobile genetic elements (retrotranspons) known as mobile group II introns (29, 30). We identified two related structural families of RT-Cas1 proteins. The more abundant family carries a canonical N-terminal RT domain with a conserved ‘RT-0’ motif characteristic of group II intron and non-LTR-retrotransposon RTs (31, 32). The other lacks the RT-0 motif, starting instead with an additional N-terminal domain containing a putative Cas6-like RNA recognition motif (RRM) of the RAMP (Repeat-Associated-Mysterious Protein; (10)) superfamily. Alignments of the retrovirus HIV-1 RT and a group II intron RT (Thermosynechococcus elongatus TeI4c RT; (33)) with representatives of the two RT-Cas1 fusion families (from Arthrospira platensis and Marinomonas mediterranea) revealed that both Cas1-fused RTs contain the seven conserved sequence motifs characteristic of the fingers and palm regions of retroviral RTs. Each also shares the RT-2a motif, which is conserved in group II intron RTs and related proteins but not present in retroviral RTs, such as HIV-1 RT (31, 32). The thumb/X domain, which is found in retroviral and group II intron RTs just downstream of the RT domain, appears to be missing in the Cas1-associated RTs (Fig. 1C).
The structural subcategories, limited phylogenetic distribution, and exclusive association with a subset of CRISPR types are consistent with a small number of common origins of RT-Cas1 fusions (10, 29).
Spacer acquisition by the M. mediterranea MMB-1 Type III-B machinery in an E. coli host
To test whether RT-Cas1 proteins could facilitate the acquisition of new spacers and to determine whether such spacers might be acquired from RNA, we chose the Type III-B CRISPR locus in M. mediterranea (MMB-1) (34), as this was the only known, easily cultured, non-pathogenic member of the well-studied γ-proteobacterium class that contains an RT-Cas1 gene.
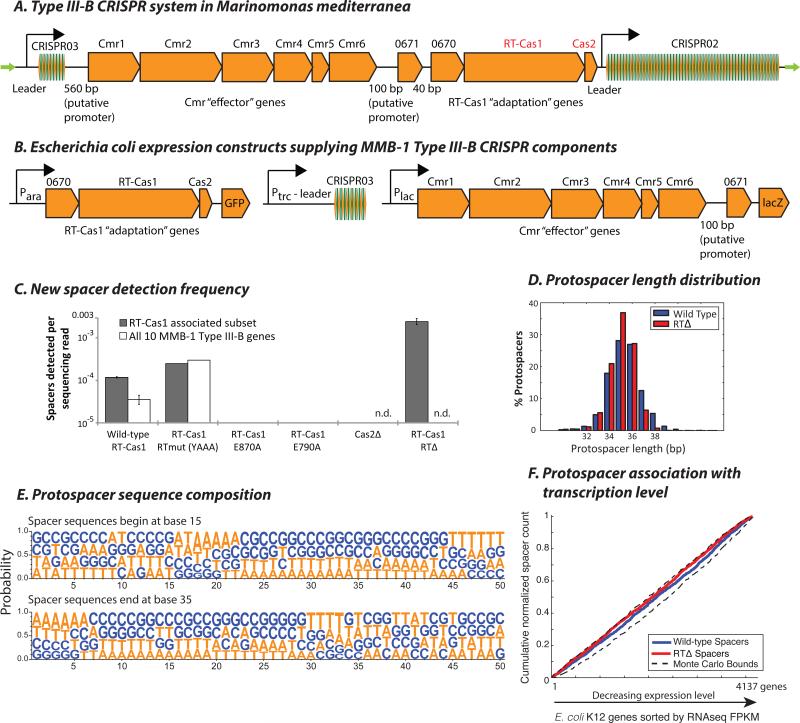
We first assessed spacer acquisition following transplantation of the locus into the canonical γ-proteobacterium experimental model, E. coli. We constructed expression vectors carrying the Type III-B operon of MMB-1 in two configurations (either as a single cassette consisting of the CRISPR03 array (35), the genes encoding RT-Cas1 and Cas2, and an adjacent gene (“Marme_0670”) with limited homology to the “NERD” family (36), or together with a second cassette additionally encoding the remaining CRISPR-associated factors, Cmr1-Cmr6 and Marme_0671) (Fig. 2A,B). Acquisition of new spacers into CRISPR03 was evident from PCR amplification of the region between the leader sequence and the first native spacer, followed by high-throughput sequencing. We identified newly acquired spacers in transformants expressing either the full complement of Cas genes, or the subset containing only the potential “adaptation” genes (RT-Cas1, Cas2, Marme_0670). Bona fide spacer acquisition is evidenced by the precise junctions between the inserted spacer DNA and CRISPR repeats (Fig. S1A), and by the diversity of acquired spacers (Fig. S1B,D).
Figure 2. Spacer acquisition in E. coli by ectopic expression of MMB-1 Type III-B CRISPR components.
(A) The MMB-1 Type III-B CRISPR operon consists of an 8-spacer CRISPR array (CRISPR03), followed by a canonical 6-gene cassette putatively encoding the Type III-B Cmr effector complex, two genes of unknown function (Marme_0671 and Marme_0670), then the RT-Cas1 and Cas2 genes, and finally a larger 58-spacer CRISPR array (CRISPR02). The locus is flanked by two ~200 bp direct repeats (green arrows). (B) Arrangement of MMB-1 Type III-B CRISPR components under inducible promoters on pBAD vectors for ectopic expression in E. coli. (C) Spacer detection frequency after overnight induction of E. coli carrying pBAD expression vectors with arabinose and IPTG. Wild-type RT-Cas1, RT active site mutant (YAAA), and Cas1 domain mutants E790A and E870A were tested with or without the Plac driven Cmr “effector” gene cassette. Cas2 Δ32-* and RT domain Δ299-588 deletion mutants were tested without the Cmr cassette. Where shown, bars indicate values for two biological replicates (n.d.: not determined). (D) Histogram showing normalized counts of E. coli genomic protospacers from the wild-type RTCas1 and RTΔ spacer acquisition experiments, distributed by mappable length. Pooled data from several experiments are presented. (E) Nucleotide probabilities at each position along the wild-type RT-Cas1-acquired protospacers in (D) including 15 bp of flanking sequence on each side. Due to varying protospacer lengths, two panels are shown with spacer 5’ and 3’ ends anchored at positions 15 and 35, respectively. (F) Cumulative normalized distribution of spacers in (D) among E. coli protein-coding ORFs sorted by expression level (normalized RNAseq read counts from (49); FPKM: fragments per kb per million reads), with most highly expressed genes listed first. 2,470 wild-type RT-Cas1, and 5,569 RTΔ-acquired spacers mapping to E. coli genes are included. Dashed black lines show the range of values from a Monte-Carlo simulation with random assortment (no transcription-related bias).
Specificity was further tested by evaluating requirements for RT-Cas1 and Cas2 for spacer acquisition. We constructed two point mutations, E870A and E790A, in the putative Cas1 active site of MMB-1 RT-Cas1 based on a 3-D homology model computed using the Archaeoglobus fulgidus Cas1 crystal structure (37). Each point mutation abolished spacer acquisition, as did a 60 aa C-terminal deletion in Cas2 (Fig. 2C).
The majority (~85%) of newly acquired spacers mapped to the E. coli genome, with the rest being derived from plasmid DNA (Fig. S1D). Over 70% of the spacers were 34-36 bp in length (Fig. 2D). Consistent with observations of interference mechanisms in other Type III CRISPR systems (7), we found no evidence for a conserved protospacer-adjacent motif (PAM) or other sequence signature associated with protospacer choice (Fig. 2E). We observed no bias for the sense strand among spacers acquired from annotated E. coli genes (Fig. S2A), and no enrichment of spacers derived from highly transcribed genes (Fig. 2F). Spacer acquisition was unhindered when the RT domain of RT-Cas1 was mutated or deleted (Fig. 2C), consistent with a DNA-based mechanism under these conditions. Indeed, deletion of the entire 290 aa conserved region of the RT domain resulted in a ~20 fold increase in spacer acquisition frequency (see note (38)), with no apparent differences in the characteristics of the pool of acquired spacers (Figs. 2C-F, S2A, S3A).
Transcription-associated spacer acquisition in M. mediterranea is RT-dependent
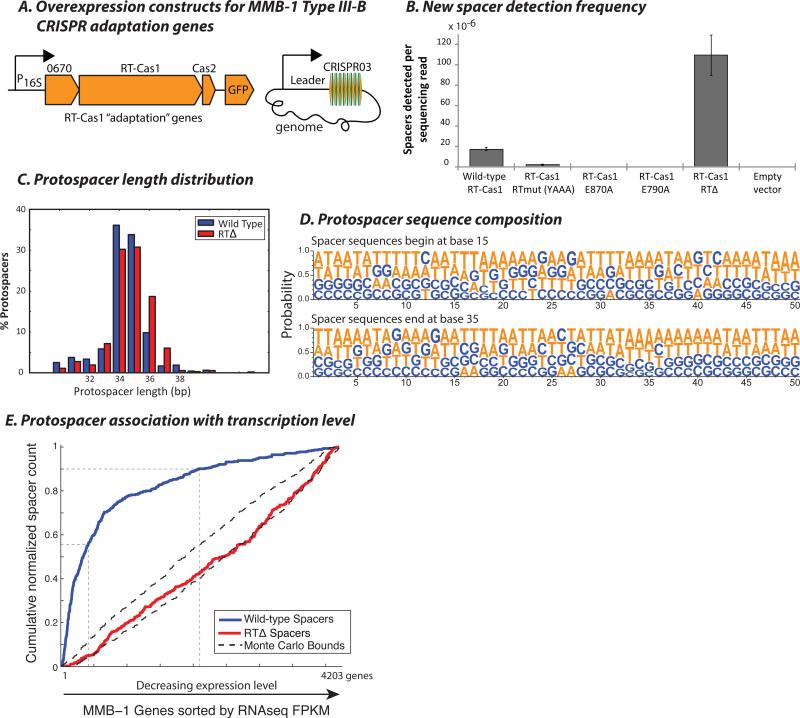
Our inability to detect RNA spacer acquisition in the ectopic E. coli assay could reflect the absence of required factors or conditions that are present in the native host M. mediterranea MMB-1. To assay spacer acquisition in MMB-1, we overexpressed the RT-Cas1 and Cas2 ORFs along with Marme_0670 from a broad-host-range plasmid (pKT230), using the 100 bp sequence upstream of the MMB-1 16S rRNA gene as a promoter (Fig. 3A). We recovered newly acquired spacers from the genomic copy of the CRISPR03 array and found that the vast majority (~95%) mapped to the MMB-1 genome, with an expected proportion mapping to the expression vector (Figs. S1C,D, S4). Although the endogenous Type III-B CRISPR operon was still present in these strains, we found that plasmid-driven over-expression of adaptation genes was critical for detectable acquisition of new spacers: parallel analysis of transconjugants where plasmid-driven RT-Cas1 had the mutation E870A or E790A at the putative Cas1 active site, or transconjugants carrying an empty vector failed to identify any new spacers (Fig. 3B). As in E. coli, most (>75%) of the new protospacers were 34-36 bp in length (Fig. 3C), and we did not observe a PAM-like sequence at either the 5’ or 3’ ends of the acquired spacers (Fig. 3D).
Figure 3. RT-Cas1 mediated spacer acquisition in Marinomonas mediterranea.
(A) Arrangement of Marme_0670, RT-Cas1, and Cas2 genes on pKT230 broad-host-range vectors under control of the putative 16S rRNA promoter (100 bp sequence upstream of the M. mediterranea 16S rRNA gene) for over-expression in MMB-1. New spacers were amplified from the genomic CRISPR03 array. (B) Spacer detection frequency after overnight growth of MMB-1 transconjugants carrying pKT230 over-expression vectors. Two clones each from two independent conjugations carrying either wild-type RT-Cas1, Cas1 domain mutants E790A or E870A, RT domain Δ299-588 deletion mutants, or an empty pKT230 vector were tested. Bars depict spacer acquisition frequencies for two transconjugants. (C) Histogram showing normalized counts of MMB-1 genomic protospacers from the wild-type RT-Cas1 and RTΔ spacer acquisition experiments, distributed by mappable length. Pooled data from several experiments are presented. (D) Nucleotide probabilities at each position along the wild-type RT-Cas1-acquired protospacers in (C) including 15 bp of flanking sequence on each side. Due to varying protospacer lengths, two panels are shown with spacer 5’ and 3’ ends anchored at positions 15 and 35, respectively. (E) Cumulative distribution of spacers in (C) among MMB-1 genes sorted by RNAseq FPKM, with most highly expressed genes listed first. 455 wild-type RT-Cas1, and 341 RTΔ-acquired spacers mapping to MMB-1 genes are included. Guides are drawn along the x-axis at top 10% and top 50% genes by expression level. Monte Carlo bounds were calculated as in Figure 2F. rRNA genes have been excluded from this analysis as spacers were rarely acquired from rRNA.
In contrast to the E. coli dataset, the genomic regions most frequently sampled by the RT-Cas1 spacer acquisition machinery in MMB-1 appeared to be genes that are typically highly expressed in bacteria. We further investigated this association between expression and spacer-capture by obtaining RNAseq expression profiles of two independent MMB-1 transconjugants carrying the RT-Cas1 expression vector. The 10% most highly expressed genes accounted for over 50% of newly acquired spacers, with the top 50% of expressed genes accounting for 90% of newly acquired spacers (Fig. 3E). Next, we tested whether this transcriptional association was dependent on the RT domain of RT-Cas1. Deletion of the conserved RT domain of RT-Cas1 abolished the preference for highly transcribed genes (Fig. 3E; also see Fig. S5), while maintaining a comparable length and sequence distribution for the acquired spacer repertoire (Figs. 3B,C, S2B, S3B, S4). Together, these data demonstrate an RT-dependent bias toward acquisition of spacers from highly transcribed regions.
Spacers acquired from transcribed regions could conceivably be integrated into the CRISPR array in either negative or positive orientation. Among spacers that mapped to MMB-1 transcripts, we observed at most a limited preference for the sense strand (Fig. S2B,C). The lack of a strong bias implies a degree of directional flexibility in the integration mechanism, potentially yielding a system in which only a fraction of spacers is able to protect against a single-stranded DNA or RNA target.
RT-Cas1 mediated spacer acquisition from RNA
The observed association between the gene expression level and frequency of spacer acquisition in MMB-1, combined with the requirement for the RT domain for this association, is consistent with an acquisition process involving reverse transcription of an RNA. Nonetheless, an alternative hypothesis is that acquisition of DNA spacers could result from increased accessibility of DNA in regions of high transcriptional activity.
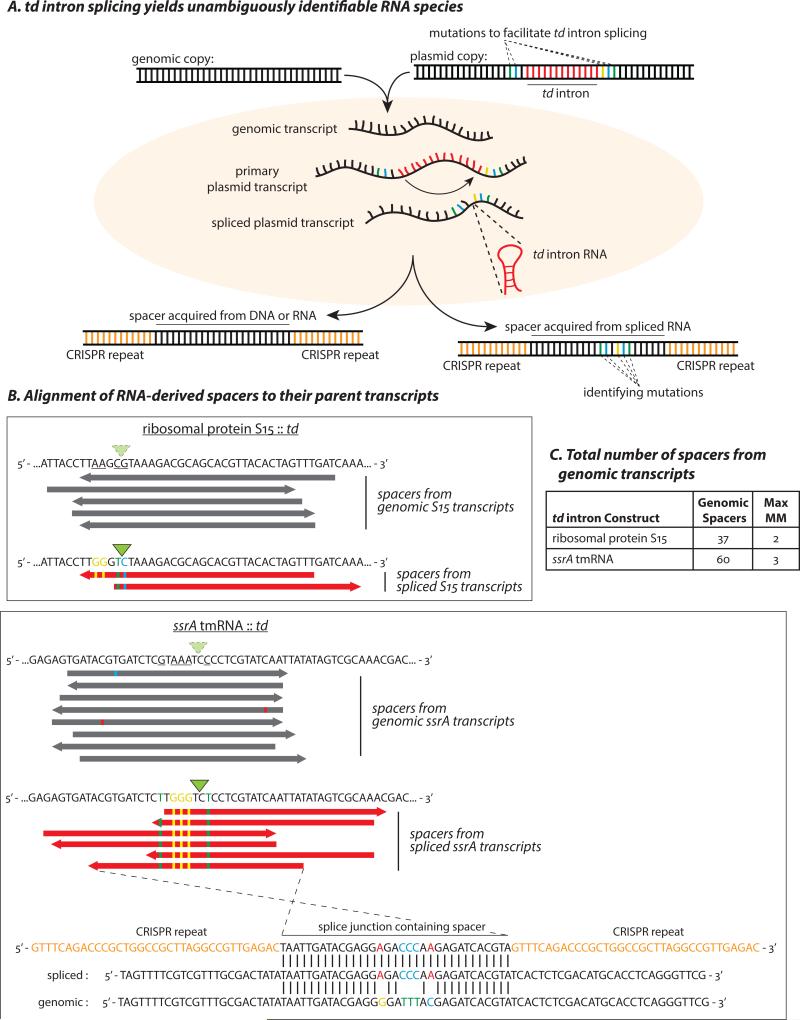
Acquisition of DNA spacer sequences from an RNA can be tested by placing a functional intron into a transcript, which is spliced to yield a ligated-exon junction sequence that is then captured as DNA (25). To test whether the RT-Cas1 complex could acquire spacers directly from RNA, we used the self-splicing td group I intron, a ribozyme that catalyzes its own excision from its parent transcript, leaving behind a splice junction that was not present as a DNA sequence (39). We produced intron-interrupted versions of two MMB-1 genes – the ssrA gene, encoding a small non-coding RNA (tmRNA; (40)) and Marme_0982, encoding ribosomal protein S15 – in both cases inserting the intron at sites that were well sampled in our spacer libraries. Each construct was designed with 4-5 mutations to optimize the flanking exon-sequences for td intron splicing. These mutations allow us to unambiguously distinguish between spliced (plasmid expressed) and native (genomic) ssrA and ribosomal protein S15 transcripts (Fig. 4A). After confirming self-splicing in vitro (Fig. S6A), we placed the td intron-containing genes on our RT-Cas1 over-expression plasmids, and expressed them in MMB-1 from their native promoters. To assess the transcription level of the engineered coding regions relative to their endogenous counterparts in vivo, we performed high-throughput sequencing of RT-PCR amplicons spanning the splice junctions, finding that ~30% of all ribosomal protein S15 transcripts, and ~16% of all ssrA tmRNA transcripts were produced by splicing in the respective transconjugants (Fig. S6B).
Figure 4. Spacer acquisition from RNA in the MMB-1 Type III-B system.
(A) Spacers acquired from a host genome could conceivably originate from either RNA or DNA. To test for an RNA origin, we used an engineered self-splicing transcript, which produces an RNA sequence junction that is not encoded by DNA. Bases that were mutated to provide flanking exon sequences favorable for td intron splicing are separated by the 393 bp intron in the DNA template. Following transcription and splicing, the two exons are brought together to form a novel junction containing the “identifying mutations”. Newly acquired spacers that contain this exon-junction indicate spacer acquisition from an RNA target. (B) Alignments of some of the genome-contiguous spacers (gray) and several newly acquired exon-junction spanning spacers (red) to the genomic and split-gene sequences, respectively. Bases mutated to facilitate td intron splicing are underlined in the genomic sequences. Identifying mutations are depicted as colored bases, and the splice sites are indicated by green triangles. The highlighted ssrA exon-junction spanning spacer (bottom) is antisense to the spliced tmRNA and differs from a putative DNA template by the 5 expected mutations. (C) All unique spacers spanning the td-intron splice site that did not carry the engineered mutations. The maximum number of mismatches when these spacers were mapped to the wild-type genomic locus is indicated. None of the identifying mutations were observed among these sporadic mismatches. The spacers in (B) were in addition to four spacers (1 for the S15 and 3 for the ssrA construct) that align to the unspliced exon-intron junction and could have been derived from either DNA or (nascent) RNA.
We assayed for newly integrated spacers in plasmid copies of CRISPR03, recovering 80,136 new spacers mapping to the MMB-1 genome. The protospacer length, sequence composition, and bias for highly expressed genes remained consistent with our previous results in MMB-1 (Fig. S7). We found 2 spacers spanning the ribosomal protein S15 splice junction, and 6 spacers spanning the tmRNA splice junction from two independent cultures of two independent transconjugants, thereby confirming that the RT-Cas1 spacer acquisition machinery is capable of acquiring spacers from RNA molecules (Fig. 4B,C). We observed both sense and antisense spacers spanning the synthetic splice junctions from both the ssrA and ribosomal protein S15 constructs (Fig. 4B), further indicating flexibility in orientation of spacer acquisition relative to the leader. We considered the possibility that these spacers might have been acquired from an extended cDNA copy of the spliced transcripts generated through indiscriminate RT activity. Such cDNA sequences would have been detectable by highly sensitive targeted sequencing assays and were not seen (Fig. S6C).
While these experiments demonstrate the ability of this system to acquire spacers from RNA, the RT-domain deletion experiments in which spacer acquisition was not biased toward transcribed regions (Fig. 3E) indicate that the system can also acquire spacers from DNA. Nonetheless, the strong transcriptional bias seen with wild-type RT-Cas1 in MMB-1 indicates that most spacer acquisitions driven by the intact RT-Cas1 fusion protein under our conditions are from RNA.
Ligation of RNA and DNA oligonucleotides directly into CRISPR repeats by an RT-Cas1/Cas2 complex
The E. coli Cas1/Cas2 complex has been shown to ligate dsDNA directly into a supercoiled plasmid containing a CRISPR array by a concerted cleavage-ligation (transesterification) mechanism analogous to that of retroviral integrases (41). To investigate how MMB-1 RT-Cas1 functions in spacer acquisition, we reconstituted this activity in vitro using purified RT-Cas1 and Cas2 proteins. We confirmed that wild-type RT-Cas1 protein has RT activity that is abolished by deletion of the RT domain (RTΔ) or mutations at the RT active site (YADD → YAAA at aa pos. 530-533) (Fig. S8). To assay spacer acquisition, the purified RT-Cas1 and Cas2 proteins were incubated with putative spacer precursors (“protospacers”) corresponding to DNA or RNA oligonucleotides of different lengths, and a linear 268 bp internally labeled CRISPR DNA substrate containing the leader, the first two repeats, and interspersed spacer sequences from the MMB-1 CRISPR03 array (Fig. 5A). The reactions also included dNTPs to enable reverse transcription of a ligated RNA oligonucleotide.
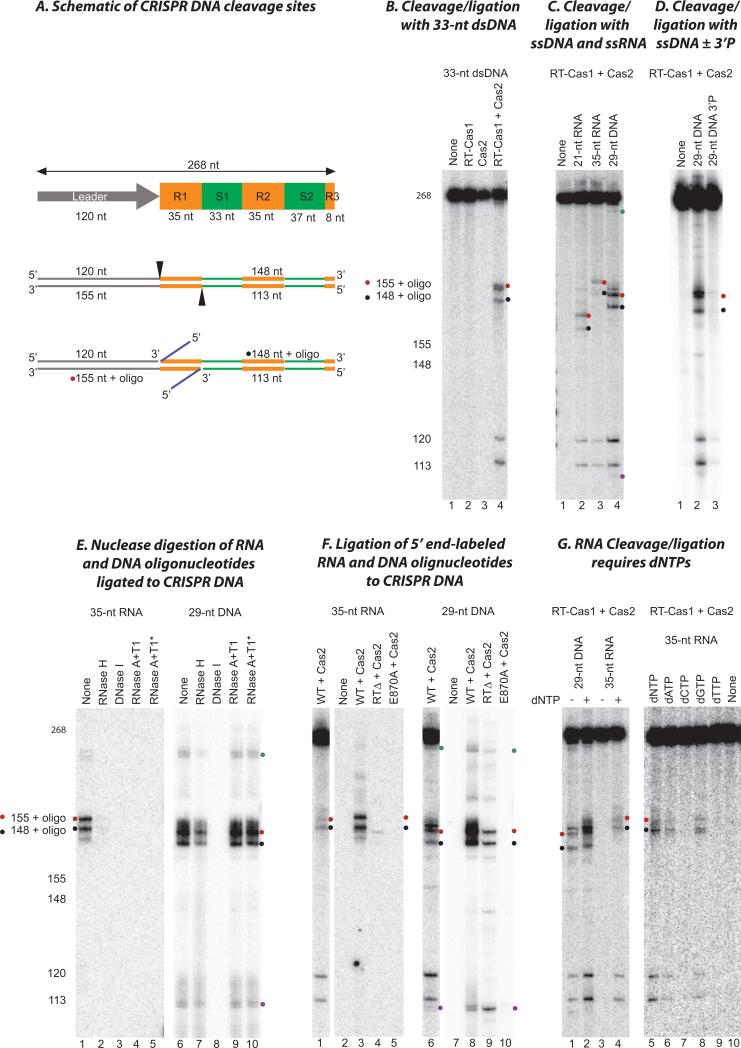
Figure 5. Site-specific CRISPR DNA cleavage/ligation by RT-Cas1/Cas2.
(A) Schematic of CRISPR DNA substrates and products of cleavage/ligation reactions. The substrate was a 268 bp DNA containing the leader (gray), the first two repeats (R1 and R2; orange) and spacers (S1 and S2; green), and part of the third repeat (orange) of the MMB-1 CRISPR03 array. Cleavages (arrowheads) occur at the boundaries of the first repeat with concomitant ligation of a DNA or RNA oligonucleotide (blue) to the 3’ fragment, yielding products of the sizes shown. (B) Internally labeled CRISPR DNA and a 33-nt dsDNA were incubated with no protein (None, lane 1), RT-Cas1 (lane 2), Cas2 (lane 3), or a 1:2 mixture of RT-Cas1 and Cas2 (lane 4). The sizes of products determined from sequencing ladders in parallel lanes are indicated (left). (C) Internally labeled CRISPR DNA was incubated with WT RT-Cas1 and Cas2 without (lane 1) or with a 21-nt RNA (lane 2), 35-nt RNA (lane 3), or 29-nt ssDNA (lane 4). (D) Internally labeled CRISPR DNA was incubated with WT RT-Cas1+Cas2 in the absence (none), or presence of a 29-nt ssDNA with either a 3’ OH (lane 2) or a 3’ phosphate (lane 3). (E) Nuclease digestion of 5’ end-labeled RNA and DNA oligonucleotides ligated to CRISPR DNA. Ligation reactions were done as in (C). After extraction with phenol-CIA and ethanol precipitation, the products were incubated with the indicated nucleases. An asterisk indicates that the sample was boiled to denature the DNA before adding the nuclease. (F) Ligation of 5’ end-labeled RNA and DNA oligonucleotides into CRISPR DNA by WT and mutant RT-Cas1 proteins. Lanes 1 and 6 show control reactions of internally labeled CRISPR with WT RT-Cas1+Cas2 and an unlabeled 35-nt ssRNA or 29-nt ssDNA oligonucleotide for comparison. Lanes 2-5 and 7-10 show reactions of unlabeled CRISPR DNA with 5’-end labeled 35-nt ssRNA and 29-nt ssDNA, respectively, and WT, E870A, and RTΔ RT-Cas1 plus Cas2. All reactions were done in the presence of dNTPs. (G) Effect of dNTPs. In the gel to the left, internally labeled CRISPR DNA was incubated with WT RT-Cas1 plus Cas2 in the presence of a 29-nt ssDNA (lanes 1 and 2) or 35-nt ssRNA (lanes 3 and 4) in the absence (lanes 1 and 3) or presence of 1 mM dNTPs (1 mM each of dATP, dCTP, dGTP, and dTTP; lanes 2 and 4). In the gel to the right, internally labeled CRISPR DNA was incubated with WT RT-Cas1+Cas2 in the presence of a 35-nt ssRNA oligonucleotide in the absence (none, lane 10) or presence of different dNTPs (1 mM) as indicated (lanes 5 to 9). Red and black dots indicate products resulting from cleavage and ligation of oligonucleotides at the junction of the leader and first repeat on the top strand and the junction of repeat 1 and spacer 1 on the bottom strand, respectively; cyan and purple dots indicate products of the size expected for cleavage and ligation of the oligonucleotide at the junctions of the second CRISPR repeat (see Fig. S10).
In initial assays using a dsDNA oligonucleotide, products derived from cleavage of the CRISPR substrate were readily detected in the presence of RT-Cas1+Cas2, but not with either protein alone (Fig. 5B). The sizes of these products were consistent with cleavage at the junctions between the leader and first repeat on the top strand and between the first repeat and spacer on the bottom strand, as expected for staggered cuts known to occur in Type I CRISPR systems (12). Structural features at the leader-repeat boundary might dictate cleavage at these sites (41). Bands of the sizes expected for free 3’ fragments (148 and 155 nt) were much weaker than those for the corresponding 5’ fragments (120 and 113 nt), reflecting their replacement with prominent bands of the sizes expected for ligation of the oligonucleotide to their 5’ ends (denoted 155+oligo and 148+oligo). Notably, similar products were also detected using ssDNA and RNA oligonucleotides of various sizes (ssDNA 19-59 nt; RNA 21-50 nt) (Figs. 5B,C, S9, S10), presumably reflecting that the more uniform spacer size of 34-36 bp in vivo is due to processing of the spacers prior to integration into the CRISPR array. Additionally, a 3’ phosphate modification of the ssDNA oligonucleotide almost completely abolished the cleavage/ligation reaction suggesting a crucial role of the 3’OH of the donor oligonucleotide in the integration reaction (Fig. 5D). The ligation of both DNA and RNA oligonucleotides into the CRISPR DNA was confirmed by their expected RNase and/or DNase sensitivity in reactions with 5’-end-labeled oligonucleotides and unlabeled CRISPR DNA (Figs. 5E). The ligated RNA oligonucleotide was sensitive to RNase H, indicating its presence in an RNA-DNA hybrid, as would be expected if it was used as a template for cDNA synthesis by RT-Cas1 (Fig. 5E).
While MMB-1 RT-Cas1/Cas2 functions similarly to E. coli Cas1/Cas2 to site-specifically integrate putative spacer precursors into CRISPR arrays, it differs in being able to utilize a linear CRISPR DNA substrate, and to insert not only dsDNA, but also ssDNA and RNA oligonucleotides. The ligation of RNA and DNA oligonucleotides into the CRISPR DNA substrate differs in two respects. First, while the E870A mutation at the Cas1 active site abolishes ligation of both RNA and DNA oligonucleotides, deletion of the RT domain (RTΔ) abolishes ligation of RNA but not DNA oligonucleotides (Fig. 5F). These findings mirror in vivo results showing that the E870 mutation abolishes acquisition of both RNA and DNA spacers, whereas the (RTΔ) mutation abolishes acquisition of RNA but not DNA spacers (Fig. 3B,E). Second, dNTPs are required for ligation of RNA but not DNA oligonucleotides, with dGTP or dATP alone sufficient to support RNA ligation (Fig. 5G). Together, these findings suggests that the RT-Cas1 protein is modular with the Cas1 domain catalyzing ligation of both RNA and DNA spacers into CRISPR repeats, but with ligation of RNA spacers requiring binding by the N-terminal and/or RT domains, possibly coupled to RT domain core closure and/or initiation of reverse transcription upon addition of dNTPs.
Integrated RNA oligonucleotides are reverse transcribed by the RT-Cas1/Cas2
We tested whether the RT-Cas1/Cas2 complex could reverse transcribe an integrated RNA oligonucleotide in vitro to generate the cDNA precursor of a fully integrated RNA spacer. The cleavage-ligation reactions on either side of repeat R1 generate products with 5’ overhangs that could potentially be substrates for target DNA-primed reverse transcription (TPRT) reactions in which the 3’ end of the opposite strand is extended to yield a DNA copy of the repeat plus the ligated RNA oligonucleotide (Fig. 6A). In order to detect synthesis of such cDNAs, we incubated the CRISPR DNA with RT-Cas1/Cas2 in the presence of a 21-nt RNA oligonucleotide and supplied radioactive dCTP and other unlabeled dNTPs during the incubation (Fig. 6A). cDNA synthesis during the reactions is evident by the labeled products of the same size as the two ligation products, as expected for a TPRT reaction extending through the R1 repeat and ligated RNA. The synthesis of these cDNAs depends on the presence of the RNA oligonucleotide, the CRISPR DNA, and RT-Cas1/Cas2 (Fig. 6B). The RTΔ mutant abolishes cDNA synthesis, while the E870A mutant, which retains RT activity (Fig. S8) but cannot integrate the RNA oligonucleotide or create the 3’OH required for priming cDNA synthesis (Fig. 5F), produces only a heterogeneous background of labeled products (Fig. 6B). The TPRT products detected in our assays may represent an intermediate in spacer acquisition, with additional steps potentially including digestion of the ligated RNA spacer strand by a host RNase H, synthesis of a fully double strand DNA containing the spacer sequence by RT-Cas1 or a host DNA polymerase, and ligation of the unattached ends of the dsDNA into the CRISPR array. Our in vivo and in vitro data suggest that this can occur in either orientation and may involve host enzymes that are present in MMB-1 but not in E. coli.
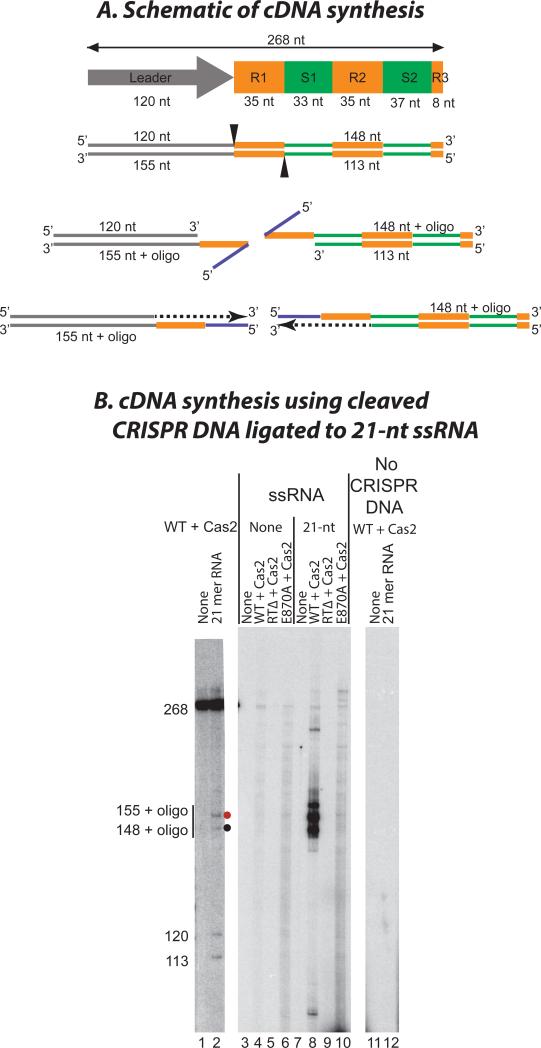
Figure 6. cDNA synthesis using RNA ligated to CRISPR DNA.
(A) Schematic shows the CRISPR DNA substrate (leader, gray; repeat, orange; spacer, green) and the expected products of cleavage/ligation (top) followed by TPRT of the ligated RNA oligonucleotide (blue). cDNAs are shown as black dashes with arrowheads indicating the direction of cDNA synthesis. (B) Wild-type (WT) or mutant RT-Cas1 proteins plus Cas2 were incubated with 268 bp CRISPR DNA in the presence of 21-nt RNA oligonucleotide, labeled dCTP and unlabeled dATP, dGTP, and dTTP. The wild-type RT-Cas1+Cas2 complex yields labeled bands of the sizes expected (148 and 155 nt+oligo) for target DNA-primed reverse transcription (TPRT) of the RNA oligonucleotide ligated site-specifically at opposite boundaries of the first CRISPR DNA repeat (R1, lane 8). The labeled products were not detected with the RT domain (RTΔ; lane 9) or Cas1 active site (E870A, lane 10) mutants, but a background of labeled products is seen in the E870A lane due to the RT activity of the protein in the absence of cleavage and ligation (see Fig. S8). Labeled products were not detected in the absence of the RNA oligonucleotide (lanes 3 to 6) or CRISPR DNA (lanes 11 and 12). Separate lanes from the same gel (lanes 1 and 2) show the positions of cleavage-ligation products for RT-Cas1+Cas2 with internally labeled CRISPR DNA substrate. “None” indicates no protein added.
Conclusion
We show that the MMB1 RT-Cas1 fusion protein can mediate the direct acquisition of spacers from donor RNA via direct ligation of an RNA protospacer into CRISPR DNA repeats using the Cas1 integrase activity. The 3’ end generated by cleavage of the opposite DNA strand is then poised for utilization as a primer for target DNA-primed reverse transcription (TPRT) (26). This mechanism shares features with group II intron retrohoming in which the intron RNA uses its ribozyme activity to insert itself directly into the host genome and is then converted to an intron cDNA by using the 3’ end generated by cleavage of the opposite DNA strand for TPRT (42). Since Type III CRISPR systems are known to target RNA for degradation, and RT-Cas1 genes are exclusively associated with such systems, RNA spacer acquisition makes these CRISPRs uniquely capable of generating immunity against parasitic RNA sequences, potentially including RNA phages and/or other selfish RNAs that maintain themselves through the action of host machinery (43-46). Acquisition of RNA spacers might also contribute to immune responses to highly transcribed regions of DNA phages and plasmids. This could then be coupled to an interference system that targets DNA, RNA, or both (15-21). It is possible that fusion between the RT and Cas1 domains may not be necessary to facilitate uptake of RNA spacers, as there are several examples of CRISPR loci in which genes encoding similar group II intron-like RTs are adjacent but not fused to Cas1 (29). Thus, the mechanisms described in this paper could potentially extend to species with separately encoded RT and Cas1 components. In addition, RNA spacer acquisition could be involved in gene regulation, providing a straightforward means for bacteria to down-regulate a set of target loci in response to activation of the CRISPR locus. To fully assess the prevalence and importance of CRISPR adaptation to RNA, a greater understanding of the impact of invasive RNAs in bacteria is necessary. However, our knowledge of the abundance and distribution of RNA phages and other RNA parasites is limited, with the vast majority restricted to the Escherichia and Pseudomonas genera. Future research on the distribution of spacers in RT associated CRISPR loci among natural populations of bacteria and their environment might help shed light on this topic.
Materials and Methods
RT-Cas1 genomic neighborhood analysis
The genomic neighborhoods (up to 20 kb) of RT-Cas1 genes were retrieved from 50 bacterial strains with a custom BioPython script that uses NCBI tblastn software. The HMMER 3.0 algorithm was then used to identify whether the RT-Cas1 genes were associated with Type I, II, or III CRISPR systems, using Cas3 (TIGR 01587, 01596, 02562, 02621, 03158), Cas9 (TIGR 01865, 3031), and Cas10 (TIGR 02577, 02578) HMMs as “signature” genes for each type, respectively (8). Each result was assessed manually by iterative BLAST and the CRISPRfinder online suite.
Monte Carlo simulation of expected spacer acquisition characteristics for random sampling of all genes
We used a Monte Carlo simulation to evaluate a null hypothesis based on random assortment of spacer acquisitions from genomic DNA, with no dependence on gene expression level. For each system, a series of samples of 500 spacers each were randomly chosen in silico from a list of all genes based on the sizes of the individual genes using the stochastic universal sampling algorithm. Sets of 1000 such trials were used to generate a range of null relationships between gene expression and spacer acquisition. The Monte Carlo bounds (black dotted lines on the respective figures) depict the envelope of such simulated random assortments. Traces above this envelope indicate preferential spacer acquisition from highly expressed genes, while traces below the envelope indicate spacer acquisition from poorly expressed genes more often than expected by random chance. E. coli K-12 RNAseq data were obtained from (49) (dataset without computational background subtraction). MMB-1 expression data were generated by RNAseq analysis of the transconjugants used in Fig. 3 in this study.
Construction of expression vectors
Plasmids for inducible over-expression of the MMB-1 Type III-B CRISPR operon in E. coli were built on the pBAD/Myc-His B backbone (Life Technologies). RT-Cas1 associated genes (Marme_0670, Marme_0669 (RT-Cas1), Marme_0668 (Cas2)) and GFP were driven by Para, and the CRISPR03 array was driven by Ptrc. The other 7 genes (Marme_0677–0672 (Cmr1–6), Marme_0671) and lacZα were driven by Plac. GFP and lacZα ORFs enabled verification of expression of the transcripts containing RT-Cas1 associated “adaptation” genes and Cmr “effector” genes, respectively. Point mutants of the Cas1 (E790A or E870A) and RT domains (YADD → YAAA at aa pos. 530-533) of the RT-Cas1 gene were tested with over-expression of the RT-Cas1 associated subset, with and without the remaining 7 genes. Deletion mutants of the RT domain of RT-Cas1 (Δ299-588), and Cas2 (Δ32-*) were tested with over-expression of the RT-Cas1 associated subset only.
Plasmids for the over-expression of the RT-Cas1 associated genes in MMB-1 cells were built on the pKT230 backbone (a gift from Prof. L. Banta, Williams College). The genes were driven by the 100 bp promoter-containing sequence (chr:306,879-306,978) upstream of an MMB-1 16S rRNA gene. Cas1 point mutants (E790A or E870A), as well as the RTΔ mutant were also tested. For experiments with td intron containing constructs, a copy of the CRISPR03 array with its leader sequence was also placed on the pKT230 vector to increase the concentration of CRISPR arrays per unit input DNA in the PCR amplification step, and thus increase the efficiency of our spacer detection assay.
Plasmids for protein expression and purification were built on the pMal-ct2 backbone (NEB) for RT-Cas1 (wild-type and mutants), and on the pET14b backbone (Novagene) for Cas2. Variants of RT-Cas1 were expressed with an N-terminal maltose-binding-protein tag attached via a non-cleavable rigid linker (50). Cas2 was expressed with an N-terminal 6xHis tag.
All plasmids were verified by sequencing. Plasmid structures are available upon request.
Strains and culture conditions
All bacterial strains used in this study were stored in 20% glycerol at −80°C. Two clones from each conjugation were maintained for each plasmid (referred to as independent “transconjugants”).
pBAD plasmids (AmpR) encoding MMB-1 Type III-B operon components were transformed into chemically competent TOP10F’ cells (Life Technologies). TOP10F’ derived strains were grown at 37°C on Luria-Bertani (LB) agar plates (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, 15 g/L agar) with 100 μg/mL ampicillin, 0.1% w/v arabinose, and 0.1 mM IPTG overnight.
pKT230 plasmids (KanR) encoding MMB-1 Type III-B operon components were mobilized into a spontaneous rifampicin-resistant mutant of M. mediterranea MMB-1 (ATCC 700492) from a donor E. coli strain carrying the pRL443 conjugal plasmid (a gift from Dr. M. Davison, Carnegie Institution) as described (51). All transformed MMB-1 strains were grown on 2216 marine agar (Difco) with 50 μg/mL kanamycin for 16 h at 25°C.
For experiments with MMB-1 transconjugants carrying td intron constructs, 150 mL cultures were subsequently prepared in 2216 broth (Difco) with 50 μg/mL kanamycin and shaken at 26-27°C in 1 L flasks for 20 h before midiprep.
E. coli DH5α (Life Technologies) was used for cloning and Rosetta2 and Rosetta2 (DE3) (Novagen) were used for protein expression. Bacteria were grown in LB medium with shaking at 200 rpm. Antibiotics were added at the following concentrations when needed: ampicillin, 100 mg/L; chloramphenicol, 25 mg/L.
Nucleic acid extraction
Plasmid DNA from E. coli strains was extracted using the QIAprep Spin Miniprep Kit (QIAGEN). Genomic DNA from MMB-1 strains was extracted using a modified SDS/Protease K method: briefly, cells were scraped from plates and resuspended in 1 mL lysis buffer (10 mM Tris, 10 mM EDTA, 400 μg/mL protease K, 0.5% SDS) and incubated at 55°C for 1 h. 50-100 μL of digest was subsequently purified using the gDNA Clean & Concentrator Kit (Zymo Research).
Total RNA was extracted from MMB-1 strains using a combined TRIZOL/RNeasy method: briefly, cells were scraped from plates and homogenized directly in 1 mL TRIZOL (Life Technologies) by vortexing and total RNA was extracted with 200 μL chloroform. 500 μL ethanol was added to an equal volume of the aqueous phase containing RNA, and the mixture was purified using the RNeasy Kit (QIAGEN) with On-Column DNase digestion according to the manufacturer's instructions. This protocol selects RNA >200-nt and thus depletes tRNAs.
Plasmid DNA was purified from large MMB-1 cultures using a custom midiprep method. Cells were harvested from 150-200 mL confluent cultures (3,000 × g, 30 min, 4°C) and homogenized in 12 mL alkaline lysis buffer (40 mM glucose, 10 mM Tris, 4 mM EDTA, 0.1 N NaOH, 0.5% SDS) at 37°C by pipetting until clear (10-15 min). 8 mL chilled neutralization buffer was added (3 M CH3COOK, 2 M CH3COOH) and lysates were immediately transferred to ice to prevent digestion of genomic DNA. Samples were mixed by inverting and the genomic DNA containing precipitate was removed by centrifugation (20,000 × g, 20 min, 4°C). Clarified lysates were extracted twice with a 1:1 mixture of Tris-saturated-phenol (Life Technologies) and ChCl3 (Fisher Scientific), and once with ChCl3 in Heavy Phaselock Gel tubes (5 Prime). 50 mL ethanol was added and DNA was pelleted by centrifugation (16,000 × g, 20 min, 4°C), washed twice in 80% ethanol, and resuspended in 500 μL elution buffer (10 mM Tris, pH 8.5). Samples were treated with 20 μg/mL RNase A (Life Technologies) at 37°C for 30 min, further digested with 150 μg/mL Protease K in 0.5% SDS at 50°C for 30 min, and purified by organic extraction. Plasmid DNA was resuspended in 0.5 mL elution buffer, desalted with Illustra NAP-5 G-25 Sephadex columns (GE Healthcare), and eluted with 1 mL water. 100 μL batches were linearized with PvuII-HF (NEB) to aid denaturation during PCR. Finally, each digest was purified using a Zymo gDNA Clean & Concentrator column.
DNA and RNA preparations were quantified using a Qubit 2.0 Fluorometer (Life Technologies).
Spacer Sequencing
Leader proximal spacers were amplified by PCR from 3-4 ng genomic DNA per μL PCR mix using forward primer AF-SS-119 (CGACGCTCTTCCGATCTNNNNNCTGAAATGATTGGAAAAAATAAGG) anchored in the leader sequence, and reverse primer AF-SS-121 (ACTGACGCTAGTGCATCACGTGGCGGAGATCTTTAA) in the first native spacer. 96 × 10 μL reactions were pooled for each sample. Sequencing adaptors were then attached in a second round of PCR with 0.01 volumes of the previous reaction as template, using AF-SS-44:55 (CAAGCAGAAGACGGCATACGAGAT NNNNNNNN GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCACTGACGCTAGTGCATCA) and AF-KLA-67:74 (AATGATACGGCGACCACCGAGATCTACAC NNNNNNNN ACACTCTTTCCCTACACGACGCTCTTCCGATCT) where the (N)8 barcodes correspond to Illumina TruSeq HT indexes D701-D712 (reverse complemented), and D501-D508 respectively. Template matching regions in primers are underlined. Phusion High-Fidelity PCR Master Mix with HF Buffer (Fisher Scientific) was used for all reactions. Cycling conditions were (98°C, 1 min), 2x (98°C, 10 s; 50°C, 20 s; 72°C, 30s), 24x (98°C, 15 s; 65°C, 15 s; 72°C, 30s), (72°C, 9 min) for Round 1, and (98°C, 1 min), 2x (98°C, 10 s; 54°C, 20 s; 72°C, 30s), 5x (98°C, 15 s; 70°C, 15 s; 72°C, 30s), (72°C, 9 min) for Round 2. The dominant amplicons containing the first native spacer from unmodified CRISPR templates after Rounds 1 and 2 were 123 bp and 241 bp, respectively. We prepared sequencing libraries by “blind” excision of gel slices at 300-320 bp (70 bp above the 241 bp band, consistent with the expected size of an amplicon from an expanded CRISPR array) following agarose electrophoresis (3%, 4.2 V/cm, 2 hrs) of Round 2 amplicons.
When amplifying spacers from plasmids, 1 ng DNA was used per μL PCR mix, synthesis time was shortened to 15 s, and 20 and 9 cycles were used in Rounds 1 and 2 instead of 24 and 5, respectively. Additionally, Round 1 amplicons were purified by “blind” excision of gel slices at 180-200 nt following denaturing PAGE (pre-run Novex TBE-Urea 10% gels, 180 V, 80 min in XCell SureLock Mini-Cells, Life Technologies), and agarose-gel-purified libraries were further PAGE purified by “blind” excision of gel slices at 300-320 nt (pre-run Novex TBE-Urea 6% gels, 180 V, 90 min as above). In this way, spacer detection efficiency was increased ~100-fold. Libraries were quantified by Qubit, and sequenced with Illumina MiSeq v3 kits (150 cycles, Read 1; 8 cycles, Index 1; 8 cycles, Index 2).
Spacers were trimmed from reads using a custom Python script, and considered identical if they differed only by 1 nucleotide. Protospacers were mapped using Bowtie 2.0 (--very-sensitive-local alignments). These methods preserve strand information.
Directional RNAseq profiling of MMB-1 strains
1 μg total RNA was incubated at 95°C in alkaline fragmentation buffer (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3, pH ~ 9.3) for 45 min, and PAGE purified (pre-run 15% TBE-Urea precast gels, 200 V, 45 min in Mini-PROTEAN electrophoresis cells, Bio-Rad) to select 30-80 nt fragments. RNA fragments were 3’-dephosphorylated with T4 polynucleotide kinase (PNK, NEB) at 37°C for 60 min in the supplied buffer, then desalted by ethanol precipitation. Desphosphorylated RNA was denatured again in adenylated ligation buffer (3.3 mM DTT, 10 mM MgCl2, 10 μg/mL acetylated BSA, 8.3% glycerol, 50 mM HEPES-KOH pH ~ 8.3) for 1 min at 98°C, and ligated to pre-adenylated adaptor AF-JA-34 (/5rApp/NNNNNNAGATCGGAAGAGCACACGTCT/3ddC/) at 22°C for 4 h using 10 U T4 RNA Ligase I (NEB). The (N)6 barcode for each RNA fragment allowed us to computationally collapse PCR bias. Excess adaptor was removed by treatment with 5’ deadenylase (NEB) followed by RecJf (NEB) treatment and organic extraction to purify ligation products. RNA was reverse transcribed using primer AF-JA-126 (/5Phos/AGATCGGAAGAGCGTCGTGT/iSp18/CACTCA/iSp18/GTGACTGGAGTTCAGACGTGTGCTCTTCC GATCT) with SuperScript II (Life Technologies) and subsequently hydrolyzed in 0.2 N NaOH at 70°C for 15 min. cDNA was PAGE purified (pre-run 10% TBE-urea gels, 200 V, 45 min in Mini-PROTEAN electrophoresis cells, Bio-Rad) to select 90-150 nt fragments, and circularized with 50U CircLigase I (Epicentre). Libraries were prepared by 6-14 cycles of PCR with universal adaptor AF-JA-158 (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT) and indexing primers AF-JA-118:125 (CAAGCAGAAGACGGCATACGAGAT NNNNNN GTGACTGGAGTTCAGACGTGTGCTCTTCCG) where the (N)6 barcodes correspond to Illumina TruSeq LT indexes AD001-AD008. 160-200 bp amplicons were gel-purified by agarose electrophoresis.
Construction and validation of td intron constructs
Constructs with the following features were ordered as gBlocks (IDT) and cloned downstream of the T7 promoter in pCR-Blunt II-TOPO (Life Technologies). Bases 208-216 (CTTAAGCGT) of the ribosomal protein S15 gene (Marme_0982), and bases 67-75 (CGTAAATCC) of the ssrA tmRNA gene (Marme_R0008) were replaced with the wild-type td intron splice junction (CTTGGGT|CT). The 393 bp intron sequence was inserted at the exon junction ‘|’. 128 bp of upstream sequence for Marme_0982, and 183 bp upstream sequence and 30 bp downstream sequence for Marme_R0008 were included. Transcripts were generated from linearized plasmids using the MEGAscript T7 Transcription kit (Life Technologies). Mostly unspliced RNA was obtained by arresting the transcription reaction after 5 min at 37°C, and subsequent extraction with acidified phenol:ChCl3 (Life Technologies). 1/3 of the reaction product was incubated in splicing buffer (40 mM Tris pH 7.5, 6 mM MgCl2, 100 mM KCl, 1 mM rGTP) at 37°C for 30 min, and desalted by ethanol precipitation. Spliced and unspliced transcripts were visualized by 1/4x TAE native agarose gel electrophoresis, with NEB 100 bp Quickload dsDNA ladder providing approximate sizing. Intron containing genes were then transferred to pKT230 derived MMB-1 over-expression vectors carrying RT-Cas1 associated genes and a copy of the CRISPR03 array. One clone each from two independent conjugations was isolated for each vector.
In vivo splicing efficiency was measured by high-throughput sequencing as follows. Total RNA was extracted and 1 μg reverse transcribed (SuperScript III, High GC content protocol, Life Technologies) with gene specific primers downstream of the splice junctions that would bind both spliced and unspliced transcripts – AF-SS-238 (CTTAGCGACGTAGACCTAGTTTTT) for Marme_0982, and AF-SS-241 (GGTTATTAAGCTGCTAAAGCGTAG) for Marme_R0008. cDNA was treated with RNase H and libraries were prepared by a two round PCR method adapted from the CRISPR spacer sequencing method above. Round 1 PCR was performed at annealing temperatures of 48°C and 65°C, for 2 and 19 cycles respectively with primers AF-SS-242 (CGACGCTCTTCCGATCTNNNNNGATTCGCATGGTAAAC) & AF-SS-243 (ACTGACGCTAGTGCATCAAACTAGTGTAACGTGCTG) for Marme_0982, and for 2 and 16 cycles respectively with primers AF-SS-247 (CGACGCTCTTCCGATCTNNNNNCACGAACCCTGAGGTG) and AFSS-248 (ACTGACGCTAGTGCATCACGTCGTTTGCGACTATATAATTGA) for Marme_R0008. This simultaneously generated amplicons of identical length for both spliced and unspliced transcripts, which were then attached to Illumina adaptors with a second round of PCR as before.
The presence of exon-junction sequences corresponding to the td intron constructs in DNA form outside the CRISPR arrays was also tested by high-throughput sequencing. Libraries consisting of the ~100 bp region containing the td intron-insertion sites in Marme_R0008 and Marme_0982 were prepared by a two round PCR method identical to the one described above for measuring splicing efficiency by RT-PCR, using 100 ng of genomic DNA (~2 × 107 copies) as a template instead of reverse-transcribed cDNA. Round 1 PCR was performed at annealing temperatures of 57°C and 68°C, for 2 and 16 cycles respectively with primers AF-SS-318 (CGACGCTCTTCCGATCTNNNNNCACATTCATGACCACCATTCTCG) & AF-SS-309 (ACTGACGCTAGTGCATCACTTCGGTCTTAGCGACGTAGAC) for Marme_0982, and primers AF-SS-310 (CGACGCTCTTCCGATCTNNNNNGGGGTGACATGGTTTCGACG) and AF-SS-311 (ACTGACGCTAGTGCATCAGCAGGTTATTAAGCTGCTAAAGCG) for Marme_R0008. The amplicons were then attached to Illumina adaptors with a second round of PCR as before. Each library was sequenced to a depth of ~5 million reads. To ensure that the PCR was not bottlenecked, we also included a spike-in (1 molecule per 1000 copies of the MMB-1 genome) of synthetic ssDNA templates – AF-SS-312 (TAAAAACATTGAAGGTCTACAAGGTCACTTTAAAGCTCACATTCATGACCACCATTCTCGTCGCNNNN NNNNNNNNATGGTAAACCAACGTCGTAAGTTGTTGGATTACCAGCTGCGTAAAGACGCAGCACGTTA CACTAGTTTGANNNNNNNNNNNNGTCTACGTCGCTAAGACCGAAG) for Marme_0982, and AF-SS-313 (GGGGTGACATGGTTTCGACGNNNNNNNNNNNNCCTGAGGTGCATGTCGAGAGTGATACGTGATCTCA GCTGTCCCCTCGTATCAATTATATAGTCGCAAANNNNNNNNNNNNCGCTTTAGCAGCTTAATAACCTG CTAGTGTGCTGCCCTCAGGTTGCTTGTAGCCCGAGATTCCGCAGT) for Marme_R0008 – that could be amplified concomitantly by the same primer sets to yield identically sized amplicons.
The spike-in-derived reads are easily identified by sequence, with the diversity of randomized (N)12 segments used to evaluate the degree to which distinct reads in the amplified pool represent independent molecules from the pre-amplification mixture. A large number of spike-in barcodes (ideally a different barcode for every spike-in read) indicate that a high fraction of reads from the amplified pool represent unique molecules in the initial sample, while repeated appearance of a small number of (N)12 barcodes in the amplified pool would be indicative of bottleneck formation during PCR (and hence a less-than-optimal relationship between read counts and molecules in the initial pool). For purposes of estimating the numbers of molecules sampled from an initial pool, we calculate a non-redundancy fraction, which is the ratio of spike-in-derived barcodes to total spike-in-derived reads. The non-redundancy fraction provides a multiplier that can be used to correct raw read counts from an amplified pool to obtain an estimate of the contributing number of molecules from the initial pool. This is particularly applicable for estimating a minimal incidence of a rare class (i.e. setting a detection limit for spliced copies of the td-intron containing DNA constructs in this work). Given non-redundancy fractions of >0.45 for all samples in these experiments, the observed totals of control (non-spliced, genomic) sequence reads (Fig. S6C) would have been sufficient to detect the presence of extended spliced td-intron-containing DNA molecules even at the low incidence of 10−6.
The same cultures of MMB-1 were used to assess both splicing efficiency and the presence of exon-junction sequences in DNA form.
PCR Fidelity
Analyzing sequence distributions through PCR and Sequencing entails certain “best practices” in terms of both experimental protocols and analysis. In particular, several precautions were observed in constructing sequencing libraries for spacer sequencing. PCR titrations were performed to ensure that the amplification kinetics were in the linear range of the reactions before any size selection step (e.g., band excision from native agarose gels); this avoids re-naturation artifacts in complex sequence pools. The overall error rate was empirically determined for every experiment by analyzing the distribution of mismatches in the sequences obtained from the first native spacer in the CRISPR03 array; this enabled the estimation of the error rate in the region of the sequencing reads that contained newly acquired spacers. PCR “bottlenecking” was also measured as the number of repeat occurrences of any given new spacer. All synthetic sequences that could lead to confounding contamination issues were avoided: no sequences from E. coli, MMB-1, or other sources have been synthesized as amplifiable substrates. As a benchmark for recovery of individual sequences, a non-bacterial sequence was synthesized as a spacer flanked by the appropriate CRISPR repeats. This repeat-flanked spacer sequence (CTGGGACATATAATATCGTCCCCGTAGATGCCTAT; a segment of the phage MS2) was indeed recovered effectively in experiments with an E. coli transformant carrying a plasmid with the indicated template. Appearance of MS2 sequences in other trials were limited to this single sequence, indicating a likely source due to a low level of cross-sample “bleeding”.
Protein purification
Expression plasmids were transformed into E. coli strains Rosetta2 (pMal derivatives) or Rosetta2(DE3) (pET derivatives), and single transformed colonies were grown in LB medium supplemented with appropriate antibiotics over night at 37°C with shaking. Six flasks each containing 1 L LB were inoculated with 1% of the overnight culture and grown at 37°C with shaking to log phase. After the culture reached an OD600 of ~0.8, IPTG was added to 1 mM final concentration and the cultures were incubated at 19°C for 20 to 24 h. Cells were harvested by centrifugation and the pellet was dissolved in A1 buffer (25 mM KPO4 pH 7; 500 mM NaCl; 10% glycerol; 10 mM ß-mercaptoethanol; 10 mL/g cell paste) on ice. Lysozyme was added to 1 mg/mL final concentration and incubated at 4°C for 0.5 h. Cells were then sonicated (Branson Sonifier 450; 3 bursts of 15 sec each with 15 sec between each burst). The lysate was cleared by centrifugation (29,400 × g, 25 min, 4°C), and polyethyleneimine (PEI) was added to the supernatant in six steps on ice with stirring to a final concentration of 0.4%. After 10 min, precipitated nucleic acids were removed by centrifugation (29,400 × g, 25 min, 4°C), and proteins were precipitated from the supernatant by adding ammonium sulfate to 60% saturation on ice and incubating for 30 min. Proteins were collected by centrifugation (29,400 × g, 25 min, 4°C), dissolved in 20 mL A1 buffer, and filtered through a 0.45 μm PES membrane (Whatman Puradisc).
Protein purification was done by using a BioRad Biologic FPLC system. RT-Cas1 was purified by loading the filtered crude protein onto an amylose column (30 mL; New England BioLabs Amylose High Flow resin), washing with 50 ml A1 buffer, followed by 30 mL A1 + 1.5 M NaCl and 30 ml A1 buffer. Bound proteins were eluted with 50 mL of 10 mM maltose in A1 buffer. Fractions containing RT-Cas1 were identified by SDS PAGE, pooled, and diluted to 250 mM NaCl. The protein was then loaded onto a 5 mL heparin-Sepharose column (HiTrap Heparin HP column; GE Healthcare) and eluted with a 0 to 1 M NaCl gradient. Peak fractions (~700 mM NaCl) were identified by SDS PAGE, pooled, and dialyzed into A1 buffer. The dialyzed protein was concentrated to >10 μM using Amicon Ultra Centrifugal Filter (Ultracel-50K). The protein was stable in A1 buffer on ice for about 3 months.
The initial steps in the Cas2 purification were similar, except that the cell paste was resuspended in N1 buffer (25 mM Tris-HCl, pH 7.5; 500 mM KCl; 10 mM imidazole; 10% glycerol; 10 mM DTT) and the ammonium sulfate precipitation step was omitted. Instead, the Cas2 PEI supernatant was loaded directly onto a 5 mL nickel column (HiTrap Nickel HP column; GE Healthcare) and eluted with an imidazole gradient (60 mL 10-500 mM in N1 buffer). Peak fractions containing Cas2 were identified by SDS PAGE and pooled. After adjusting the KCl concentration to 200 mM, the pooled fractions were loaded onto a 2 × 5 mL heparin-Sepharose column. The protein was eluted with a linear KCl gradient (50 mL, 100 mM to 1 M), and Cas2 peak fractions (~800 mM KCl) were identified by SDS PAGE and stored on ice in elution buffer. The protein was stable on ice for several months.
All protein concentrations were measured using the Qubit Protein assay kit (Life Technologies) according to the manufacturer's protocol. Proteins were >80% pure based on densitometry.
Formation of RTCas1+Cas2 complex
Purified RTCas1 (2,500 pMol) was mixed with a 2-fold excess of purified Cas2 in 250 mM KCl, 250 mM NaCl, 12.5 mM Tris-HCl, pH 7.5; 12.5 mM KPO4, pH7; 5 mM DTT;5 mM BME; 10% glycerol and incubated on ice for >16 h prior to reactions.
RT assay
RT assays with poly(rA)/oligo(dT)24 were done by pre-incubating poly(rA)/oligo(dT)24 (80 μM and 50 μM, respectively) in 200 mM KCl, 50 mM NaCl, 10 mM MgCl2, 20 mM Tris-HCl, pH 7.5, 1 mM unlabeled dTTP, and 5 μCi [α-32P]-dTTP (3,000 Ci/mmol; PerkinElmer) for 2 min at the desired temperature, and then initiating the reaction by adding the RT-Cas1 proteins (1-2 μM final concentration). The reactions (20 to 30 μL) were incubated for times up to 30 min. A 3 μL sample was withdrawn at each time point and added to 10 μL of stop solution (0.5% SDS, 25 mM EDTA). Reaction products were spotted onto Whatman DE81 paper (10 × 7.5-cm sheets; GE Healthcare Biosciences), which was then washed three times with 0.3 M NaCl and 0.03 M sodium citrate, dried, and scanned with a PhosphorImager (Typhoon Trio Variable Mode Imager; GE Healthcare Biosciences) to quantify the bound radioactivity.
CRISPR DNA cleavage/ligation assay
MMB-1 CRISPR DNA substrate was a PCR product amplified with primers MMB1crisp5b (CACTCGACCGGAATTATCGACGAA) and MMB1crisp3 (TCTGAAACTCTGAATACTAACGAAAAATAG) using Phusion High-fidelity DNA polymerase according to the manufacturer's protocol (New England Biolabs or Thermo Scientific). The resulting 268 bp PCR fragment contains 120 bp of the leader, 35 bp repeat 1, 33 bp spacer 1, 35 bp repeat 2, 37 bp spacer 2, and 8 bp of repeat 3. Internally labeled substrate was prepared by adding 25 μCi [α-32P]-dTTP or dCTP (Perkin Elmer) and 40 μM dTTP or dCTP, respectively, to the PCR reactions. Labeled DNA was purified by electrophoresis in a native 6% polyacrylamide gel, cutting out the labeled band, and electroeluting the DNA using midi D-Tube dialyzer cartridges (Novagen). The eluted DNA was extracted with phenol-CIA, ethanol precipitated, and quantitated using a Qubit dsDNA assay kit (Life Technologies).
CRISPR DNA cleavage-ligation assays contained RTCas1+Cas2 complex (500 nM final), MMB-1 CRISPR substrate (1 nM), 20 mM Tris pH 7.5, and 7.5 mM free MgCl2. DNA or RNA oligonucleotides and dNTPs/Mg2+ were added at 2.5 μM and 1 mM final concentrations as indicated for individual experiments. Reactions were incubated at 37°C for 1 h and stopped by adding phenol-CIA. The supernatant was mixed at a 2:1 ratio with loading dye (90% formamide, 20 mM EDTA, 0.25 mg/ml bromophenol blue and xyan cyanol), and nucleic acids were analyzed in a 6% polyacrylamide/7 M urea gel. Gels were dried and scanned with a PhosphorImager.
Labeled DNA or RNA oligonucleotide ligation assays were done as described above but using 22.5 μM unlabeled CRISPR PCR fragment and ~0.25 μM 5’ end labeled, gel-purified oligonucleotides. Control assays were done without adding CRISPR PCR fragment. For nuclease treatment of oligonucleotide ligation to CRISPR DNA, reactions were scaled up 4-fold, phenol-CIA treated, and ethanol precipitated. The precipitated nucleic acids were dissolved in 30 μl water. Equal amounts were then either untreated or treated with RNase H (2 units, Invitrogen), DNase I (RNase-free, 10 units, Roche), RNase A/T1 mix (0.5 μg RNase A (Sigma) and 500 units RNase T1 (Ambion)) in 40 mM Tris pH 7.9, 10 mM NaCl, 6 mM MgCl2, 1 mM CaCl2 for 20 min at 37°C. Samples were extracted with phenol-CIA to terminate the reaction and analyzed by electrophoresis in a denaturing polyacrylamide gel as described above.
Labeled cDNA extension reactions were done as above but using cold CRISPR DNA and oligonucleotides with 0.25 mM unlabeled dATP, dGTP, and dTTP and 5 μCi [α-32P]-dCTP (3000 Ci/mMol, PerkinElmer).
Oligonucleotides for cleavage/ligations assays were as follows: 29-nt DNA (TTTGGATCCTCATCTTTTAGGGCTCCAAG), 33-nt dsDNATop (GATGCTTATGGTTATTGCAGCTACCCTCGCCCT), 33-nt dsDNABot (AGGGCGAGGGTAGCTGCAATAACCATAAGCATC), 21-nt RNA (GCCGCUUCAGAGAGAAAUCGC), and 35-nt RNA (UUACGGUGCUUAAAACAAAACAAAACAAAACAAAA)
Supplementary Material
Acknowledgments
We thank J. Shor, S. Cohen, M. Bagdasarian, C. Pourcel, L. Mindich, C.P. Wolk, M. Poranen, and lab colleagues for help and advice, HHMI and Stanford for fellowship support (S.S.), and the NIH (R01-GM37706 [A.Z.F.], R01-GM37949 [A.M.L.], R01-GM37951 [A.M.L.]), and Welch Foundation (F-1607 [A.M.L.]) for grant support. Sequencing data are at SRA-SRP066108.
Footnotes
Author notes:
S.S., A.Z.F. conceived the project. S.S., G.M., A.M.L., A.Z.F. designed experiments, analyzed data, and wrote the paper with inputs from other authors. S.S. performed all genetics experiments. G.M., D.J.S., L.M.M. performed all biochemistry experiments. A.S-A., D.B. provided protocols and conceptual guidance.
References
- 1.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 4.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 5.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 6.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko KA, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 13.Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heler R, et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale CR, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamulaitis G, et al. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samai P, et al. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 23.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 24.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 25.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 26.Zimmerly S, Guo H, Perlman PS, Lambowitz AM. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 28.Kojima KK, Kanehisa M. Systematic survey for novel types of prokaryotic retroelements based on gene neighborhood and protein architecture. Mol Biol Evol. 2008;25:1395–1404. doi: 10.1093/molbev/msn081. [DOI] [PubMed] [Google Scholar]
- 29.Simon DM, Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36:7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toro N, Nisa-Martinez R. Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS One. 2014;9:e114083. doi: 10.1371/journal.pone.0114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 32.Blocker FJ, et al. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005;11:14–28. doi: 10.1261/rna.7181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohr G, Ghanem E, Lambowitz AM. Mechanisms used for genomic proliferation by thermophilic group II introns. PLoS Biol. 2010;8:e1000391. doi: 10.1371/journal.pbio.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solano F, Sanchez-Amat A. Studies on the phylogenetic relationships of melanogenic marine bacteria: proposal of Marinomonas mediterranea sp. nov. Int J Syst Bacteriol. 1999;49(Pt 3):1241–1246. doi: 10.1099/00207713-49-3-1241. [DOI] [PubMed] [Google Scholar]
- 35.Of the two RT-Cas1 associated Type III-B CRISPR arrays in this system, CRISPR03 was chosen for spacer acquisition assays since the other array (CRISPR02) has unusual truncated repeats at the leader-proximal end (1).
- 36.Grynberg M, Godzik A. NERD: a DNA processing-related domain present in the anthrax virulence plasmid, pXO1. Trends Biochem Sci. 2004;29:106–110. doi: 10.1016/j.tibs.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Kim TY, Shin M, Huynh Thi Yen L, Kim JS. Crystal structure of Cas1 from Archaeoglobus fulgidus and characterization of its nucleolytic activity. Biochem Biophys Res Commun. 2013;441:720–725. doi: 10.1016/j.bbrc.2013.10.122. [DOI] [PubMed] [Google Scholar]
- 38.One potential contributor to increased spacer acquisition frequency (in Fig. 2C) following RT deletion could be a higher growth rate observed for the cells expressing the RTΔ mutant.
- 39.Belfort M, Chandry PS, Pedersen-Lane J. Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb Symp Quant Biol. 1987;52:181–192. doi: 10.1101/sqb.1987.052.01.023. [DOI] [PubMed] [Google Scholar]
- 40.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 41.Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 43.Blumenthal T, Carmichael GG. RNA replication: function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 44.Biebricher CK, Orgel LE. An RNA that multiplies indefinitely with DNA-dependent RNA polymerase: selection from a random copolymer. Proc Natl Acad Sci U S A. 1973;70:934–938. doi: 10.1073/pnas.70.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konarska MM, Sharp PA. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989;57:423–431. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- 46.Flores R, Gago-Zachert S, Serra P, Sanjuan R, Elena SF. Viroids: survivors from the RNA world? Annu Rev Microbiol. 2014;68:395–414. doi: 10.1146/annurev-micro-091313-103416. [DOI] [PubMed] [Google Scholar]
- 47.Ludwig W, Klenk HP. In: Bergey's Manual of Systematic Bacteriology. Garrity GM, editor. Vol. 2. Springer; New York: 2001. pp. 49–65. [Google Scholar]
- 48.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics. 2012;13:734. doi: 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohr S, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solano F, Lucas-Elio P, Fernandez E, Sanchez-Amat A. Marinomonas mediterranea MMB-1 transposon mutagenesis: isolation of a multipotent polyphenol oxidase mutant. J Bacteriol. 2000;182:3754–3760. doi: 10.1128/jb.182.13.3754-3760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levy A, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.