Abstract
Temporal lobe epilepsy (TLE) is the most frequent drug-resistant epilepsy in adults and commonly associated with variable degrees of mesiotemporal atrophy on magnetic resonance imaging (MRI). Analyses of inter-regional connectivity have unveiled disruptions in large-scale cortico-cortical networks; little is known about the topological organization of the mesiotemporal lobe, the limbic subnetwork central to the disorder. We generated covariance networks based on high-resolution MRI surface-shape descriptors of the hippocampus, entorhinal cortex, and amygdala in 134 TLE patients and 45 age- and sex-matched controls. Graph-theoretical analysis revealed increased path length and clustering in patients, suggesting a shift toward a more regularized arrangement; findings were reproducible after split-half assessment and across 2 parcellation schemes. Analysis of inter-regional correlations and module participation showed increased within-structure covariance, but decreases between structures, particularly with regards to the hippocampus and amygdala. While higher clustering possibly reflects topological consequences of axonal sprouting, decreases in interstructure covariance may be a consequence of disconnection within limbic circuitry. Preoperative network parameters, specifically the segregation of the ipsilateral hippocampus, predicted long-term seizure freedom after surgery.
Keywords: connectivity, epilepsy, graph theory, MRI, temporal lobe
Introduction
Temporal lobe epilepsy (TLE) is the most common drug-resistant epilepsy in adults. Mesiotemporal lobe sclerosis, the histopathological hallmark of this disorder, is reflected on magnetic resonance imaging (MRI) as hippocampal, amygdalar, and entorhinal atrophy. The increasing accessibility of whole-brain MRI morphometry has unveiled that structural compromise in TLE is not limited to the mesiotemporal lobe, but present across multiple cortical and subcortical regions, as well as the underlying white matter ([Bernhardt, Hong et al. 2013] for review).
The concept of TLE as a system disorder has been reinforced by graph-theoretical analysis, which provides a formal framework to test topology-level hypotheses of interconnected networks (Bullmore and Sporns 2009; Sporns 2011). Graph-theoretical assessment of networks derived from structural, functional, and diffusion MRI have shown large-scale cortico-cortical network reorganization (Liao et al. 2010; Bernhardt et al. 2011; Bonilha et al. 2013; Wang et al. 2014). On the other hand, the organizational properties of the mesiotemporal structural network, known to host the primary epileptogenic substrate in this condition, have not been extensively assessed. So far, the most common approach has been regions-of-interest (ROI) based functional or diffusion connectivity analysis between individual mesiotemporal structures (largely limited to the hippocampus) and regions outside (Concha et al. 2005; Pittau et al. 2012; Kemmotsu et al. 2013; Kucukboyaci et al. 2013; Stretton et al. 2013; Holmes et al. 2014). A noteworthy exception is a study showing task-free functional connectivity increases within contralateral mesiotemporal networks, while ipsilateral connectivity decreased, suggestive of interstructure network breakdowns (Bettus et al. 2009). Constrained by the intrinsically low resolution of functional imaging, however, this work based on a small sample of patients did not assess intrastructure relationships.
MRI-based structural covariance provides an appropriate hypothesis-driven framework for population-based network analysis, given the availability of structural MRI scans essential to the clinical evaluation. Notably, compared with conventional diffusion and functional MRI acquisitions, structural scans offer higher anatomical resolution and only marginal geometric distortion confounds in antero-basal temporal areas. To probe mesiotemporal circuits at a subregional level, we generated high-resolution surface parcellations of the hippocampus, entorhinal cortex, and amygdala in a large cohort of patients with drug-resistant TLE. A battery of graph-theoretical covariance analyses compared network topology between patients and controls, complemented by split-half and multiscale reliability assessments. In a subset of patients who underwent surgery, we evaluated whether preoperative network topology has prognostic value to determine the postsurgical seizure outcome.
Materials and Methods
Subjects
We studied 134 patients referred to our hospital for the investigation of drug-resistant TLE (55 males; 36 ± 10 years; 16–63 years). Our sample included similar proportions of patients with unilateral left (left temporal lobe epilepsy ([LTLE]; n = 63, 29 males) and right unilateral TLE (right temporal lobe epilepsy [RTLE]; n = 71, 26 males). No patient presented with a mass lesion (malformations of cortical development, tumors, or vascular malformations), traumatic brain injury, or a history of encephalitis.
Demographic and clinical data were obtained through interviews with patients and their relatives. Diagnosis and lateralization of the seizure focus were determined by a comprehensive evaluation, including detailed seizure history, neurological examination, video-electroencephalography (EEG), neuroimaging, and neuropsychology.
Ninety-two patients underwent a selective amygdalohippocampectomy. We determined the surgical outcome according to Engel's modified classification (Engel et al. 1993), at the latest possible follow-up (mean ± standard deviation [SD] = 7 ± 2 years, range: 5–14 years).
Sixty-one patients (66%) became seizure-free (Engel I), 6 (7%) had rare disabling seizures (Engel II), 17 (18%) a worthwhile improvement (Engel III), and in 8 patients (9%) there was no change in seizure frequency (Engel IV). Among patients with Engel-I, 32% were completely seizure- and medication-free; in 68% antiepileptic medication therapy (often at a reduced dose) remained necessary to guarantee seizure freedom. Hippocampal specimens were available in 69 patients for histopathological analysis, which revealed hippocampal sclerosis in all (Blumcke et al. 2013). In the remaining 23, specimens were either incomplete or unsuitable for examination due to subpial aspiration.
The control group consisted of 45 healthy individuals (23 males; age mean ± SD = 32 ± 11 years; range = 20–66 years). Clinical and demographic data of patients and controls are presented in Table 1.
Table 1.
Demographic and clinical information
| Group | Male | Age | Duration | FC | Surgery | HS | Engel I |
|---|---|---|---|---|---|---|---|
| Controls (n = 45) |
23 | 33 ± 11 (20–66) |
– | – | – | – | – |
| LTLE (n = 63) |
29 | 36 ± 11 (16–57) |
19 ± 13 (0–42) |
19 (30%) |
42 (67%) |
34/34 | 25 (60%) |
| RTLE (n = 71) |
26 | 35 ± 11 (17–63) |
20 ± 11 (0–45) |
21 (30%) |
50 (70%) |
35/35 | 36 (72%) |
Age and duration of epilepsy are represented in mean ± SD (range) years; FC, history of febrile convulsions; HS, hippocampal sclerosis reported in available specimens; Engel I, seizure-free postsurgical outcome according to Engel's classification.
The Ethics Committee of the Montreal Neurological Institute and Hospital approved the study and written informed consent was obtained from all participants in accordance with the standards of the Declaration of Helsinki.
MRI Acquisition and Volumetry
MR images of all patients and controls were acquired on a 1.5-Tesla Gyroscan (Philips Medical Systems, Eindhoven, Netherlands) using a 3D T1-fast field echo sequence (time repetition = 18 ms; time echo = 10 ms; number-of-excitations = 1; flip angle = 30°; matrix size = 256 × 256; field-of-view = 256 × 256 mm2; slice thickness = 1 mm) that provides isotropic 1 × 1 × 1 mm3 voxels. Images underwent automated correction for intensity nonuniformity and intensity standardization (Sled et al. 1998) and were linearly registered to the MNI152 template (Collins et al. 1994). Manual segmentations of hippocampus (Watson et al. 1992), amygdala (Watson et al. 1992), and entorhinal cortex (Bernasconi et al. 1999) were carried out according to previously published protocols. The hippocampus was further subdivided into head, body, and tail (Fig. 1A) (Bernasconi et al. 2003). Segmentations were carried out by a single rater (N.B.), blinded to all clinical information.
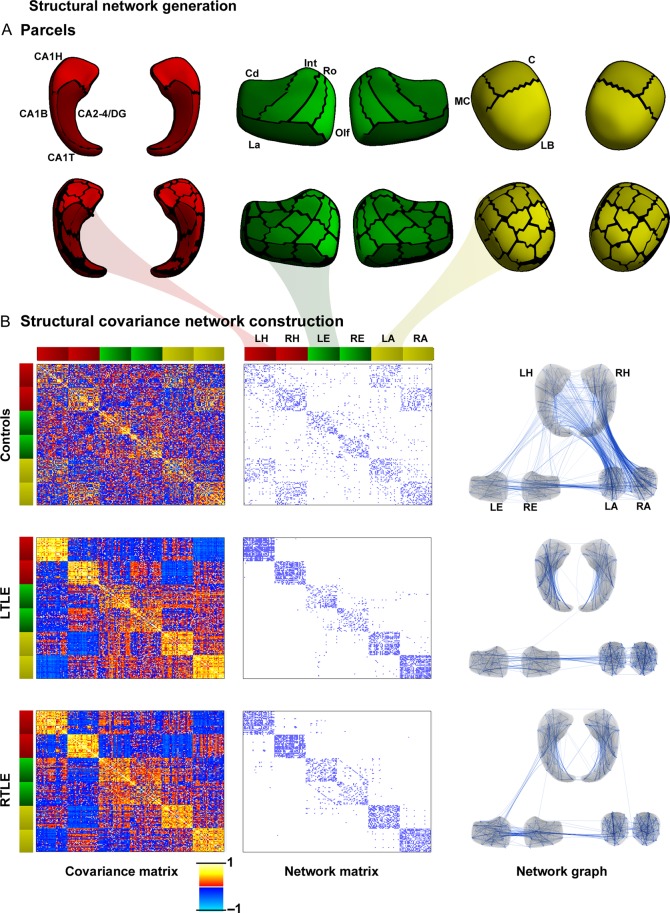
Figure 1.
Mesiotemporal circuit representation based on subregional volume covariance. (A) Low-resolution (upper panel) and high-resolution (lower panel) parcellation of the hippocampus (red), entorhinal cortex (green), and amygdala (yellow). (B) Covariance network in controls, and patients with left and right temporal lobe epilepsy (LTLE, RTLE). The left column displays high-resolution structural covariance matrices, the middle column displays the binary network matrices thresholded at a density of 8% (threshold ensuring fully connected networks in all 3 groups), the right column illustrates the corresponding network graphs. Color bars adjacent to the matrices signify the respective parcels as indicated in (A), with red colors representing the left/right hippocampus (LH/RH), green the left/right entorhinal cortex (LE/RE), and yellow the left and right amygdala (LA/RA). Hippocampus: CA, cornu ammonis; DG, dentate gyrus; H, hippocampal head; B, body; T, tail. Entorhinal cortex: Cd, caudal; Int, intermediate; La, lateral; Olf, olfactory; Ro, rostral. Amygdala nuclear groups: C, central; LB, laterobasal; MC, medial and cortical.
Surface-Based Mesiotemporal Subregional Morphometry
Notwithstanding recent progress in the MRI-based visualization of mesiotemporal lobe internal anatomy on postmortem brains (Adler et al. 2014), the in vivo identification of all subregional borders or subfields within a given structure lies beyond the resolution of clinically feasible exams. We circumvented this challenge by using heuristic boundaries drawn on MRI-derived surface templates instead of defining borders in each individual, a strategy opted in various studies (Hogan et al. 2004; Kim et al. 2008; Bernhardt, Kim et al. 2013). Note that, while this approach is not appropriate for brain structures with subdivisions hidden in the depth, it is conceptually sound to approximate boundaries for those that reach the surface; this is the case for all hippocampal subfields but CA4, all entorhinal subdivisions and all amygdalar nuclei aside from the magnocellular portion of the basal nucleus.
In brief, for each structure (hippocampus, amygdala, and entorhinal cortex), manual labels were converted to surface meshes and parameterized using spherical harmonics with a point distribution model (SPHARM-PDM) (Styner et al. 2006). For a given mesiotemporal structure, SPHARM-PDM surfaces of each individual were rigidly aligned to a template (constructed from the mean surface of controls and patients) with respect to the centroid and the longitudinal axis of the first-order ellipsoid (Gerig et al. 2001). To correct for differences in overall head size, each surface was inversely scaled with respect to intracranial volume (Styner et al. 2006). We calculated displacement vectors between each subject's SPHARM-PDM surface and the template across 1002 surface points (Styner et al. 2006). The signed surface-normal components of these vectors quantify inward/outward deformations. To compute pointwise volume changes, we applied a heat propagation approach to interpolate the pointwise displacement vectors within the volume enclosed by the SPHARM-PDM surface (Kim et al. 2008). The Jacobian determinant J of the resulting dense vector field was projected back onto the surface to quantify growth (J−1 > 0) or shrinkage (J−1 < 0) along the surface-normal direction.
Mesiotemporal Surface Parcellation
We schematically outlined hippocampal (Duvernoy 2005), entorhinal (Insausti et al. 1995), and amygdalar (Amunts et al. 2005) subdivisions (henceforth, ROIs) on the given surface templates according to histological atlases (see Fig. 1). The resulting 24 ROIs were further subdivided into parcels with comparable surface area using a k-means clustering approach (Cammoun et al. 2012), adapted to surface data. To achieve a good tradeoff between spatial resolution and computation time for graph-theoretical analysis, we chose a total of 198 parcels across all mesiotemporal structures in both hemispheres. While our main analysis findings are reported at the high-resolution 198-parcel scale, we also cross-validated the robustness of findings using the 24-ROI parcellation.
Subregional Network Construction
In each parcel (or ROI, in the low-resolution parcellation), we calculated the mean Jacobian determinant across all vertices and corrected for effects of age, gender, and overall mean Jacobian determinant of the 3 mesiotemporal structures (as a marker of global atrophy) through a statistical linear model, similar to our previous work on cortical thickness correlation networks (Bernhardt et al. 2011). Importantly, correction for global mesiotemporal volume permits topology-level assessment above and beyond effects of atrophy. Two inter-regional association matrices (R) were generated for each TLE group (i.e., LTLE and RTLE): R198 with 198 × 198 entries and R24 with 24 × 24 entries. In each matrix, an individual entry rij contained the pairwise Pearson product moment cross-correlation coefficients of the mean Jacobian determinant across subjects in regions i and j.
Assessment of Inter-regional Correlations
To assess differences in inter-regional correlation coefficients between LTLE and controls, entries rij of the R correlation matrices R were transformed using Fisher's R-to-Z transformation, where an individual entry was calculated as
The difference between 2 such correlation Z matrices (e.g., ZNC and ZLTLE) was
where nLTLE and nNC are the number of subjects in LTLE and NC, respectively. A similar approach assessed RTLE against controls.
Assessment of Network Topology
Network Thresholding
In each group (controls, LTLE, and RTLE), individual correlation matrices were thresholded at a fixed density prior to analysis. The matrix R198 (R24) in each group was thresholded to result in binarized connectivity matrices A198 (A24), where an entry aij equals 1 if rij exceeded a given threshold and 0 otherwise. We restricted our analysis to positive associations only. Such a binary matrix is equivalent to an undirected graph with 198 (24) nodes (i.e., regions) and K/2 edges (i.e., connections), where K is the total number of nonzero entries. Diagonal elements in A were set to 0. The density of a network with n nodes was defined as the percentage of the total number of actual connections K divided by the number of possible connections, that is, density = K/(n×(n−1)) × 100%. Density-based thresholding (only keeping the highest K edges in each group) ensured that networks in all groups had the same number of edges (Achard and Bullmore 2007) and that observed between-group differences reflect alterations in topological organization rather than differences in low-level correlations (He et al. 2008). For both matrices of R198 and R24, network properties for group comparison were computed over a wide range of density thresholds (5–40%) using previously described procedures (He et al. 2008; Bernhardt et al. 2011).
Clustering Coefficient and Characteristic Path Length
We computed the clustering coefficient C and characteristic path length L in controls, LTLE, and RTLE, using standard formulas (Watts and Strogatz 1998). These 2 quantities are the most widely used graph-theoretical parameters to describe the topology of complex networks. Clustering coefficient ci of a given node i, and mean clustering C were defined as:
where Ei is the number of existing connections among the neighbors of node i. As ki is the actual number of neighbors of node i (i.e., its degree), the denominator term ki(ki−1)/2 quantifies the number of all possible connections among the neighboring nodes. If a node i had only one edge or no edges, ci was set to 0. The mean clustering coefficient C of a network was defined as the average of ci over all nodes in the network N. C quantifies the cliquishness and is related to the local efficiency of a network (Latora and Marchiori 2001).
Average shortest path length li of a given node i, and characteristic path length L were defined as
In the above formula, min{lij} denotes the shortest absolute path length between 2 given nodes i and j. The characteristic path length L of a network was defined as the mean minimum number of edges that lie between any 2 nodes in the network. To overcome the problem of dramatically increased L values in networks with possibly disconnected components, L was calculated using a harmonic mean definition (Newman 2003; Bernhardt et al. 2011). The reciprocal of L is a measure of parallel information transfer or global efficiency of a network (Latora and Marchiori 2001).
Normalized Clustering Coefficient and Path Length
The normalized clustering coefficient γ was computed by dividing the clustering coefficient of the actual network C by the mean clustering coefficient Crand across 1000 randomly generated networks. These random networks had the same number of nodes, edges, and an identical degree distribution as the real network (Maslov and Sneppen 2002; Sporns and Zwi 2004). An analogous approach was used to compute the normalized path length λ. Compared with random networks, small-world networks have a similar characteristic path length, but higher clustering, that is γ = C/Crand > 1 while λ = L/Lrand ≈ 1 (Watts and Strogatz 1998).
Modularity Analysis
The modularity of a network refers to the degree of its decomposability into local communities, called “modules.” Advantages to modular organization include greater adaptability to changing environmental conditions. Networks with high modularity are assumed to have dense connections among nodes within each module and sparse connections between nodes across different modules. We quantified modularity Q in controls and patients using a previously developed algorithm (Blondel et al. 2008) that iteratively optimized local communities until global modularity was no longer improved. The modularity Q of a network was defined as
where aij corresponds to the connection between 2 anatomical regions, i and j. ki is the number of connections of node i to other nodes, ci is the community to which node i is assigned, the function δ(u, v) is 1 if u = v and 0 otherwise, and
Statistical Analysis
As in previous work (He et al. 2008; Bernhardt et al. 2011), analysis was performed separately for each TLE group (i.e., LTLE, RTLE) relative to controls using permutation tests with 1000 repetitions. Exact procedures are detailed below.
Inter-regional Correlation Coefficients
In each permutation, subregional volumes of a given subject were randomly reassigned to one of the two groups (i.e., LTLE or controls/RTLE or controls). Correlation matrices in each randomized group were converted to z-scores using Fisher R-to-Z transform. We calculated the differences between z-matrices of the random “TLE” and “control” groups (for formulas, see above). This generated a permutation distribution of between-group differences under the null hypothesis. The true between-group z-score difference was placed in this distribution to obtain the significance level.
Network Topology, Modularity, and Nodal Parameters
Differences in topological parameters C, L, γ, λ, σ, and modularity Q were assessed using an approach similar to a.
Following each random group assignment, we thresholded the correlation matrices and computed the above network parameters. For each parameter, the actual difference between a TLE group and controls was placed in its corresponding permutation distribution to obtain the significance level.
To localize alterations in network modularity across groups, we calculated inter-regional network stability matrices (which quantify the probability of any 2 nodes in the network to participate in the same module across different density thresholds, taken from 5% to 40% density) for each group. The actual difference in network stability for each inter-regional pair was placed into a permutation distribution of stability matrix differences (Bellec et al. 2010).
Correction for Multiple Comparisons
We corrected for multiple comparisons using the false discovery rate procedure (Benjamini and Hochberg 1995), controlling the proportion of false-positive findings to FDR <0.05.
Reproducibility Analysis
We split each group (i.e., controls, LTLE, RTLE) into 2 random subgroups. Repeating the main analysis between these smaller subgroups assessed reproducibility of our findings, even at markedly reduced sample sizes. To verify robustness of findings with regards to the spatial scale of the parcellation, group-differences in topological parameters and inter-regional correlation coefficients were assessed using the 24-ROI scheme. Note that the small number of ROIs precluded a meaningful modularity analysis at this scale.
Relation to the Surgical Outcome
We split the group of operated patients (n = 92) into those with the seizure-free (i.e., Engel-class I, n = 61) and nonseizure-free surgical outcome (i.e., Engel-class II–IV, n = 31). Permutation tests assessed the difference in inter-regional correlation coefficients, topological parameters, and modularity between these patient subgroups.
Results
Structural Covariance Analysis of the Mesiotemporal Circuitry
Findings are based on the high-resolution 198-parcellation unless otherwise stated.
Inter-regional Correlation Coefficients
Structural covariance matrices, binarized network matrices, and network graphs for controls and patients are shown in Figure 1. Networks in all groups were fully interconnected at a density of greater or equal to 8%. At this threshold, controls presented with dense patterns of structural covariance within and across all 3 mesiotemporal structures. Conversely, patient networks were largely characterized by a breakdown between the hippocampus and other structures, particularly the amygdala, while network links were maintained between the entorhinal cortex and amygdala.
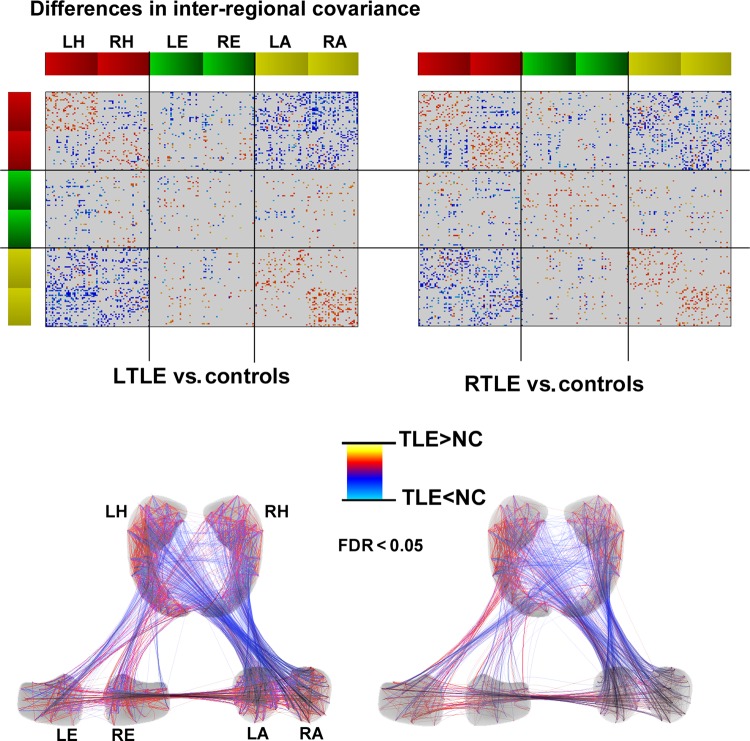
Statistical comparison of covariance patterns between patients and controls (Fig. 2) confirmed decreased correlations between most hippocampal and amygdalar subregions bilaterally in TLE (FDR <0.05). Patients also showed decreased correlations between the left and right hippocampus. On the other hand, intrahippocampal and intra-amygdalar covariance increased within the same hemisphere. Relative to amygdala and hippocampus showing marked anomalies in intra- and interstructure covariance, alterations between the entorhinal cortex and hippocampus were of similar nature but less marked. Results were virtually identical in LTLE and RTLE.
Figure 2.
Differences in inter-regional covariance between patients and controls. Increases/decreases in patients relative to controls are shown in red/blue, corrected for multiple comparisons at FDR <0.05. For further details, please see Figure 1 legend.
Global Network Topology Parameters
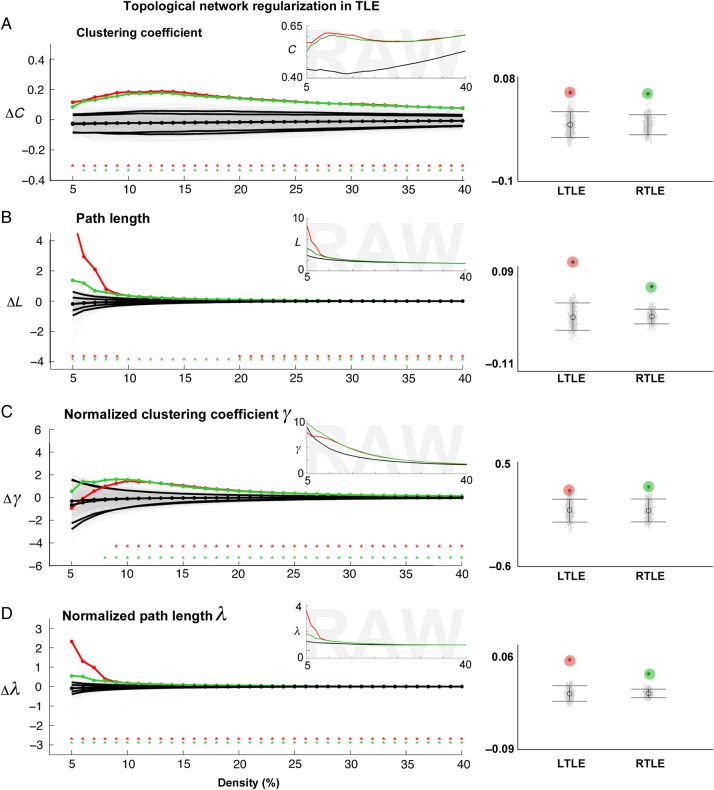
Relative to controls, we found increased clustering and path length almost over the entire density range in both TLE groups, resulting in a robust increase in these metrics when data were collapsed across all densities (FDR <0.05). Similarly, we observed increased normalized clustering coefficient γ (FDR <0.05) and normalized path length λ (FDR <0.05) in patients (Figure 3).
Figure 3.
Mesiotemporal network topology in TLE. (A) Left panel: Colored lines show differences between each TLE group (LTLE in red, RTLE in green) and controls for clustering coefficient (ΔC) as a function of network density. While ΔC = 0 indicates no difference, ΔC > 0/ΔC < 0 indicates increases/decreases in patients relative to controls. The black lines show the mean and 95% confidence interval of the null distribution of no between-group difference obtained from 1000 permutation tests at each density value, superimposed on individual permutations (gray). Inset displays raw parameter values in all 3 groups (LTLE in red, RTLE in green, controls in black). Right panel: Mean difference across the entire density range (5–40%), for LTLE (red) and RTLE (green), compared with permutation-based differences (i.e., each gray dot is representing the mean difference from one of the 1000 permutations). (B–D) Differences in path length, normalized clustering, and normalized path length between each TLE group and controls. Stars indicate a significant between-group difference, corrected at FDR <0.05.
We observed network regularization (i.e., significant increases in path length and clustering) in both LTLE and RTLE when separately analyzing the ipsilateral and contralateral subgraphs (i.e., ipsilateral-to-ipsilateral and contralateral-to-contralateral; P < 0.02).
Split-Half Reproducibility Assessment
After randomly splitting each of our groups (controls, LTLE, RTLE) into 2 equally sized groups, we could confirm increased clustering (P < 0.02), path length (P < 0.08) in each patient subgroup relative to their corresponding controls; furthermore, we observed similar alterations in inter-regional correlations, confirming a high reproducibility of findings even at markedly reduced sample sizes.
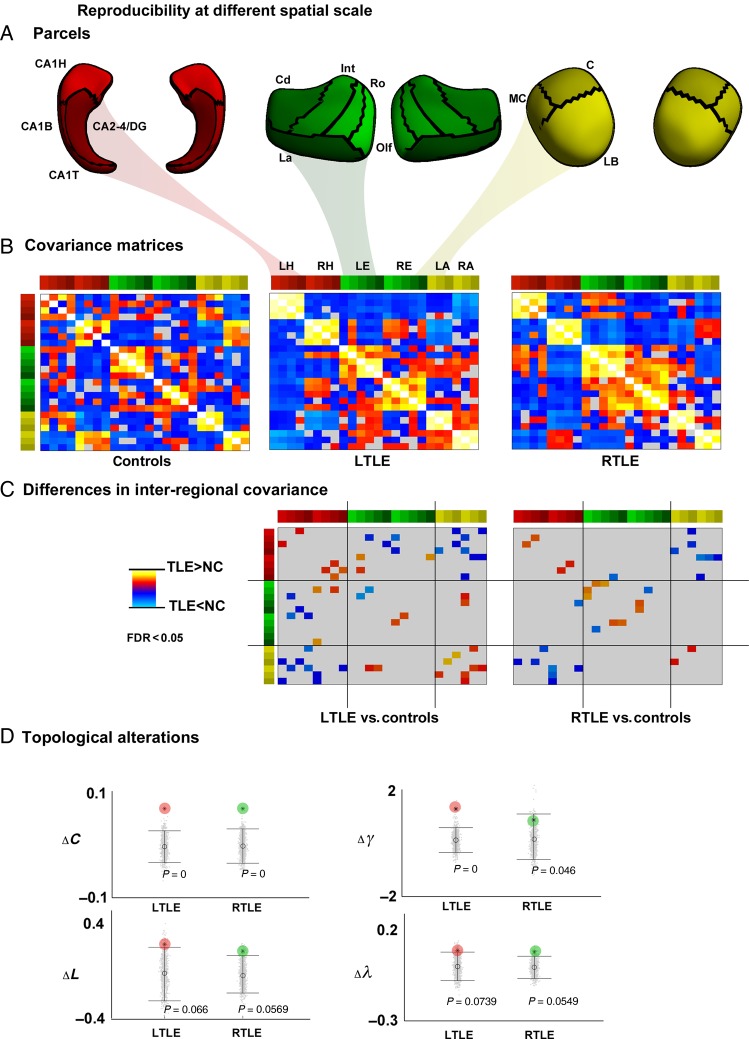
Robustness across Spatial Scales
Using a 24-ROI low-resolution parcellation, patterns of inter-regional covariance alterations between patients and controls were equivalent to those using the 198-parcel scale. Moreover, analyzing topological markers confirmed network regularization in patients, with increased clustering/normalized clustering (P < 0.05), as well as tendencies for increased path length/normalized path length in patients (P < 0.08) (Figure 4).
Figure 4.
Findings based on low-resolution (24 ROI) parcellation. (A) Parcellation. (B) Structural covariance matrices in controls, LTLE, and RTLE. (C) Inter-regional covariance differences between patients and controls. Increases/decreases in patients relative to controls are shown in red/blue, corrected at FDR <0.05. (D) Differences in clustering (ΔC), normalized clustering (Δγ), path length (ΔL), and normalized path length (Δλ) between patients and controls. For details on color bars, please see Figure 1 legend.
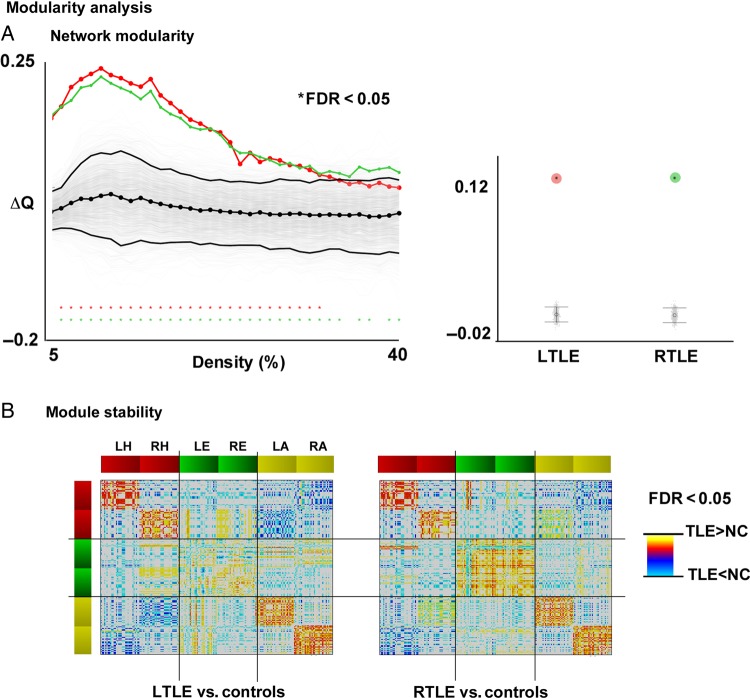
Modularity Assessment
Both TLE groups showed markedly increased modularity Q relative to controls across the entire range of network densities (FDR <0.05). We further visualized modules at a fixed density of 0.08, the minimal threshold at which all 3 groups had fully interconnected networks. Here, while modules in controls spanned different mesiotemporal structures across both hemispheres, in patients they were exclusively embedded in the same structure and hemisphere. Statistically comparing module stability validated the above observation across all densities, showing a divergence in “module participation” between patients and controls. In TLE, hippocampal subregions in one hemisphere were more likely to be aggregated within the same module, but were less frequently grouped with their counterparts of the other hemisphere (FDR <0.05). The same was observed for the amygdala. Furthermore, subregions in the hippocampus were less likely to be grouped into modules with amygdala subregions (FDR <0.05). Findings were generally consistent in LTLE and RTLE, aside for the entorhinal cortex, for which alterations in module participation were rather subtle and heterogeneous (Figure 5).
Figure 5.
Modularity in TLE. (A) Left panel: Difference in modularity (Q) between TLE and controls as a function of network density, compared with permutation distribution (see Fig. 3 for details on statistical procedures). While ΔQ = 0 indicates no difference, ΔC > 0/ΔC < 0 indicates increases/decreases in patients relative to controls. The black lines show the mean and 95% confidence interval of the null distribution of no between-group difference obtained from 1000 permutation tests at each density value, superimposed on individual permutations (gray). Right panel: Mean difference across entire density range (see Fig. 3 for further details), compared with permutation-based differences. (B) Group differences in module stability. Increases/decreases in patients relative to controls are shown in red/blue, corrected at FDR <0.05.
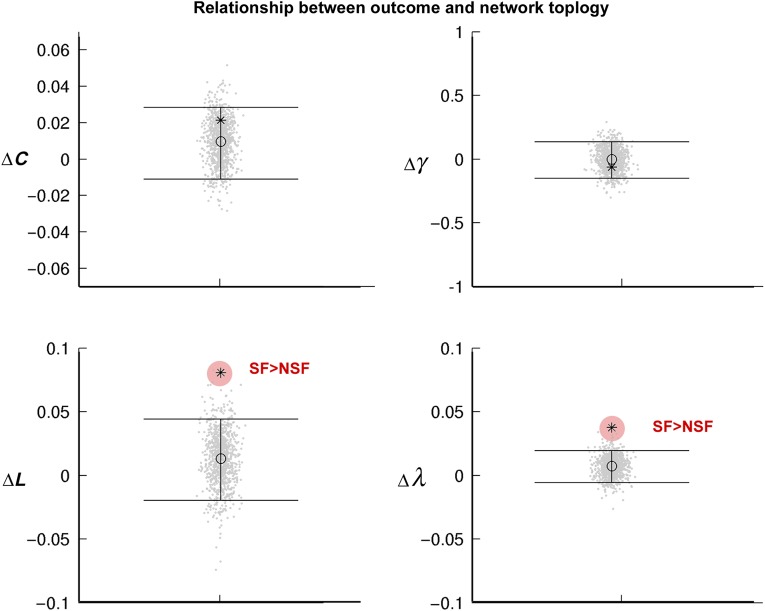
Network Markers of Seizure Outcome in Operated Patients
Compared with patients with residual seizures after surgery, seizure-free patients showed mainly a segregation of the ipsilateral hippocampus, with increased intrahippocampal covariance and largely decreased covariance to contralateral hippocampal and other mesiotemporal subregions. While there was no difference in network modularity Q between outcome groups, we observed increased path length and normalized path length of mesiotemporal network when considering left and right hemispheres together (FDR <0.05, Fig. 6) in seizure-free relative to nonseizure-free patients. Similar tendencies were observed when separately analyzing the ipsilateral and contralateral subgraphs.
Figure 6.
Relationship to postsurgical seizure outcome in 92 operated patients (at 7 ± 2 years of follow-up). Difference between seizure-free and nonseizure-free patients for clustering (ΔC), normalized clustering (Δγ), path length (ΔL), and normalized path length (Δλ). For further details, please see Figure 3.
Discussion
A small but rapidly increasing number of graph-theoretical analyses of neuroimaging data have addressed topological properties of structural and functional networks in TLE (Liao et al. 2010; Bernhardt et al. 2011; Bonilha et al. 2012; Wang et al. 2014). However, these studies have focused on large-scale neocortical networks and did not specifically target the mesiotemporal circuitry that hosts the core pathological substrate of this condition. Specifically, our results showed profound and highly reproducible shifts toward a more regularized, “lattice-like,” arrangement. We also found significant alterations of inter-regional correlations and increased network modularity, indicative of circuit “fragmentation,” i.e., decreased integration between different anatomical structures. Preoperative network parameters were associated with long-term seizure freedom after surgery.
Covariance analysis has been proposed as a noninvasive means to map structural brain networks (He et al. 2007; Alexander-Bloch, Giedd et al. 2013; Alexander-Bloch, Raznahan et al. 2013). According to its underlying assumptions, regions belonging to the same network have a high propensity to exchange trophic factors, participate in common molecular signaling pathways, and form functional assemblies. Indeed, cross-method studies have shown high correspondence between covariance patterns and maturational networks (Alexander-Bloch, Raznahan et al. 2013), and a moderate agreement to those derived from task-free functional MRI (Kelly et al. 2012; Alexander-Bloch, Raznahan et al. 2013; Hosseini and Kesler 2013) and diffusion MRI tractography (Gong et al. 2012). Applied to structural MRI that offers high anatomical precision and minimal geometric distortions, covariance analysis effectively probes networks in large clinical cohorts. Previous analyses in TLE evaluated low-level correlative relationships between single mesiotemporal structures and the neocortex (Bonilha et al. 2007; Bernhardt et al. 2008; Mueller et al. 2009). The current work, on the other hand, targeted the topology of networks within the assembly of regions forming the mesiotemporal circuitry. Notably, while our covariance analysis operated on surface-shape measures of local volume, a surrogate marker of limbic histopathology (Hogan et al. 2004, 2015; Kim et al. 2008; Bernhardt et al. 2012; Maccotta et al. 2015), these measures were corrected for confounds of global mean atrophy prior to network construction. Our findings, thus, likely reflect topology-level rearrangements in network organization above and beyond effects of global mesiotemporal atrophy.
The overall pattern of mesiotemporal network topology rearrangement parallels our previous findings in the neocortex (Bernhardt et al. 2011), yet with markedly higher effects. Confidence in our findings is high, as results were consistent in left and right TLE, when analyzing standard as well as normalized network parameters, and across different parcellation schemes. Moreover, split-half analysis, a conservative approach to assess reproducibility (He et al. 2009), confirmed robustness even at markedly reduced sample sizes. Separately assessing ipsi- and contralateral subgraphs demonstrated network alterations in patients in both hemispheres, a finding in accordance to data showing progressive atrophy in bilateral mesiotemporal regions in TLE (Bernhardt, Kim et al. 2013; Maccotta et al. 2015), and recent findings from a data-driven patient classification revealing bilateral anomalies in the majority of patients with a unilateral seizure focus (Bernhardt et al. 2015). In light of previous graph-theoretical analyses across independent modalities (Bonilha et al. 2012; Bartolomei et al. 2013; Wang et al. 2014), one may postulate that regularization is the prevailing network topology associated with drug-resistant TLE. Noteworthy, this property has also been shown at the time of seizure onset in intracerebral EEG signal analysis (Ponten et al. 2007; Kramer et al. 2008; Schindler et al. 2008).
In our study, analysis of inter-regional correlations and module participation showed increased within-structure covariance, but decreases between structures, particularly with regards to the hippocampus and amygdala. Similar results were found in a recent analysis of functional and diffusion MRI networks in a rodent model of focal epilepsy (Otte et al. 2012). Within-structure covariance increases likely drive networks toward a more clustered arrangement. A plausible explanation for these changes may be local excess in connectivity, as a consequence of axonal sprouting (Dyhrfjeld-Johnsen et al. 2007; Otte et al. 2012), a phenomenon commonly observed in both humans (de Lanerolle et al. 1989; Mathern et al. 1996; Blumcke et al. 1999; Sutula 2002) and models of limbic epilepsy (Cavazos et al. 2003; Kron et al. 2010). These pathologically interconnected networks may, in turn, generate hypersynchronous bursts of activity, which could ultimately spread throughout the limbic system (Bragin et al. 2000). On the other hand, decreases in interstructure covariance manifesting as increased path length may stem from the deafferentation of hippocampal connections, as previously shown in animal models (Knopp et al. 2005) and human diffusion MRI studies (Concha et al. 2005, 2010; Bonilha et al. 2010).
As for the hippocampus and amygdala, we observed mainly decreased covariance between ipsilateral hippocampal and entorhinal subregions that was, however, less extensive. The lower sensitivity in detecting group-differences in correlations between these 2 structures may have resulted from the lower magnitude of correlations in controls, as shown in Figure 1B.
Relating network parameters to the long-term postoperative outcome revealed increased path length in seizure-free patients relative to those with seizure relapse. While the inherent group-level character of covariance metrics precludes individualized assessment of sensitivity and specificity, our findings nevertheless suggest prognostic value of mesiotemporal circuit topology. Provided a sufficiently high anatomical resolution with only little signal distortions for the functional analysis of mesiotemporal subregions, this hypothesis could be further tested via task-free functional connectivity analysis that allows for single-subject prediction. Increased path length in seizure-free patients likely reflects structural disconnection, particularly between the hippocampus and the other structures. Considering that all patients underwent a selective resection, it is conceivable that an epileptogenic, yet topologically isolated hippocampus is easier to neutralize surgically. In a previous analysis of cortico-cortical networks, we observed more marked topological alterations in patients with seizure recurrence relative to those benefitting from surgery (Bernhardt et al. 2011). In the light of these findings, the current results suggest a regionally divergent impact of network-level alterations on the outcome; specifically, more marked topological rearrangements within mesiotemporal networks relate to more favorable outcome, while alterations in large-scale circuits beyond the mesiotemporal lobe correlate with seizure recurrence.
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR MOP-57840 and CIHR MOP-123520). B.C.B. was funded by CIHR and Jeanne Timmins Costello Fellowship of the Montreal Neurological Institute and Hospital.
Notes
We thank Hosung Kim for his help in image processing. Conflict of Interest: None declared.
References
- Achard S, Bullmore E. 2007. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, Yushkevich PA. 2014. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage. 84:505–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Giedd JN, Bullmore E. 2013. Imaging structural co-variance between human brain regions. Nat Neurosci. 14:322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. 2013. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci. 33:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah NJ, Habel U, Schneider F, Zilles K.. 2005. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat Embryol. 210:343–352. [DOI] [PubMed] [Google Scholar]
- Bartolomei F, Bettus G, Stam CJ, Guye M. 2013. Interictal network properties in mesial temporal lobe epilepsy: a graph theoretical study from intracerebral recordings. Clin Neurophysiol. 124:2345–2353. [DOI] [PubMed] [Google Scholar]
- Bellec P, Rosa-Neto P, Lyttelton OC, Benali H, Evans AC. 2010. Multi-level bootstrap analysis of stable clusters in resting-state fMRI. Neuroimage. 51:1126–1139. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 57:289–300. [Google Scholar]
- Bernasconi N, Bernasconi A, Andermann F, Dubeau F, Feindel W, Reutens DC. 1999. Entorhinal cortex in temporal lobe epilepsy: a quantitative MRI study. Neurology. 52:1870–1876. [DOI] [PubMed] [Google Scholar]
- Bernasconi N, Bernasconi A, Caramanos Z, Antel SB, Andermann F, Arnold DL. 2003. Mesial temporal damage in temporal lobe epilepsy: a volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain. 126:462–469. [DOI] [PubMed] [Google Scholar]
- Bernhardt BC, Bernasconi N, Kim H, Bernasconi A. 2012. Mapping thalamocortical network pathology in temporal lobe epilepsy. Neurology. 78:129–136. [DOI] [PubMed] [Google Scholar]
- Bernhardt BC, Chen Z, He Y, Evans AC, Bernasconi N. 2011. Graph-theoretical analysis reveals disrupted small-world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cereb Cortex. 21:2147–2157. [DOI] [PubMed] [Google Scholar]
- Bernhardt BC, Hong S, Bernasconi A, Bernasconi N. 2013. Imaging structural and functional brain networks in temporal lobe epilepsy. Front Hum Neurosci. 7:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt BC, Hong SJ, Bernasconi A, Bernasconi N. 2015. Magnetic resonance imaging pattern learning in temporal lobe epilepsy: classification and prognostics. Ann Neurol. 77(3):436–446. [DOI] [PubMed] [Google Scholar]
- Bernhardt BC, Kim H, Bernasconi N. 2013. Patterns of subregional mesiotemporal disease progression in temporal lobe epilepsy. Neurology. 81:1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt BC, Worsley KJ, Besson P, Concha L, Lerch JP, Evans AC, Bernasconi N. 2008. Mapping limbic network organization in temporal lobe epilepsy using morphometric correlations: insights on the relation between mesiotemporal connectivity and cortical atrophy. Neuroimage. 42:515–524. [DOI] [PubMed] [Google Scholar]
- Bettus G, Guedj E, Joyeux F, Confort-Gouny S, Soulier E, Laguitton V, Cozzone PJ, Chauvel P, Ranjeva JP, Bartolomei F et al. 2009. Decreased basal fMRI functional connectivity in epileptogenic networks and contralateral compensatory mechanisms. Hum Brain Mapp. 30:1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume JL, Lambiotte R, Lefebre E. 2008. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. P10008.
- Blumcke I, Beck H, Lie AA, Wiestler OD. 1999. Molecular neuropathology of human mesial temporal lobe epilepsy. Epilepsy Res. 36:205–223. [DOI] [PubMed] [Google Scholar]
- Blumcke I, Thom M, Aronica E, Armstrong DD, Bartolomei F, Bernasconi A, Bernasconi N, Bien CG, Cendes F, Coras R et al. 2013. International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a Task Force report from the ILAE Commission on Diagnostic Methods. Epilepsia. 54(7):1315–1329. [DOI] [PubMed] [Google Scholar]
- Bonilha L, Edwards JC, Kinsman SL, Morgan PS, Fridriksson J, Rorden C, Rumboldt Z, Roberts DR, Eckert MA, Halford JJ. 2010. Extrahippocampal gray matter loss and hippocampal deafferentation in patients with temporal lobe epilepsy. Epilepsia. 51:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Helpern JA, Sainju R, Nesland T, Edwards JC, Glazier SS, Tabesh A. 2013. Presurgical connectome and postsurgical seizure control in temporal lobe epilepsy. Neurology. 81:1704–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Nesland T, Martz GU, Joseph JE, Spampinato MV, Edwards JC, Tabesh A. 2012. Medial temporal lobe epilepsy is associated with neuronal fibre loss and paradoxical increase in structural connectivity of limbic structures. J Neurol Neurosurg Psychiatry. 83:903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Rorden C, Halford JJ, Eckert M, Appenzeller S, Cendes F, Li LM. 2007. Asymmetrical extra-hippocampal grey matter loss related to hippocampal atrophy in patients with medial temporal lobe epilepsy. J Neurol Neurosurg Psychiatry. 78:286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragin A, Wilson CL, Engel J Jr. 2000. Chronic epileptogenesis requires development of a network of pathologically interconnected neuron clusters: a hypothesis. Epilepsia. 41(Suppl 6):S144–S152. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Neurosci. 10:186–198. [DOI] [PubMed] [Google Scholar]
- Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O, Do KQ, Maeder P, Meuli R, Hagmann P. 2012. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods. 203:386–397. [DOI] [PubMed] [Google Scholar]
- Cavazos JE, Zhang P, Qazi R, Sutula TP. 2003. Ultrastructural features of sprouted mossy fiber synapses in kindled and kainic acid-treated rats. J Comp Neurol. 458:272–292. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. 1994. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 18:192–205. [PubMed] [Google Scholar]
- Concha L, Beaulieu C, Gross DW. 2005. Bilateral limbic diffusion abnormalities in unilateral temporal lobe epilepsy. Ann Neurol. 57:188–196. [DOI] [PubMed] [Google Scholar]
- Concha L, Livy DJ, Beaulieu C, Wheatley BM, Gross DW. 2010. In vivo diffusion tensor imaging and histopathology of the fimbria-fornix in temporal lobe epilepsy. J Neurosci. 30:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle NC, Kim JH, Robbins RJ, Spencer DD. 1989. Hippocampal interneuron loss and plasticity in human temporal lobe epilepsy. Brain Res. 495:387–395. [DOI] [PubMed] [Google Scholar]
- Duvernoy HM. 2005. The human hippocampus functional anatomy, vascularization and serial sections with MRI. 3rd ed Berlin, Heidelberg: Springer. [Google Scholar]
- Dyhrfjeld-Johnsen J, Santhakumar V, Morgan RJ, Huerta R, Tsimring L, Soltesz I. 2007. Topological determinants of epileptogenesis in large-scale structural and functional models of the dentate gyrus derived from experimental data. J Neurophysiol. 97:1566–1587. [DOI] [PubMed] [Google Scholar]
- Engel J Jr, Van Ness PC, Rasmussen T, Ojemann LM. 1993. Outcome with respect to epileptic seizures. In: Engel J., Jr Surgical treatment of the epilepsies. New York: Raven; 609–621. [Google Scholar]
- Gerig G, Styner M, Jones D, Weinberg DR, Lieberman J. 2001. Shape analysis of brain ventricles using SPHARM. MMBIA 171–178. doi:10.1109/MMBIA.2001.991731.
- Gong G, He Y, Chen ZJ, Evans AC. 2012. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. Neuroimage. 59:1239–1248. [DOI] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. 2007. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 17:2407–2419. [DOI] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. 2008. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci. 28:4756–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y et al. 2009. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS One. 4:e5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan RE, Moseley ED, Maccotta L. 2015. Hippocampal surface deformation accuracy in t1-weighted volumetric MRI sequences in subjects with epilepsy. J Neuroimaging. 25:452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan RE, Wang L, Bertrand ME, Willmore LJ, Bucholz RD, Nassif AS, Csernansky JG. 2004. MRI-based high-dimensional hippocampal mapping in mesial temporal lobe epilepsy. Brain. 127:1731–1740. [DOI] [PubMed] [Google Scholar]
- Holmes M, Folley BS, Sonmezturk HH, Gore JC, Kang H, Abou-Khalil B, Morgan VL. 2014. Resting state functional connectivity of the hippocampus associated with neurocognitive function in left temporal lobe epilepsy. Hum Brain Mapp. 35:735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Kesler SR. 2013. Comparing connectivity pattern and small-world organization between structural correlation and resting-state networks in healthy adults. Neuroimage. 78:402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insausti R, Tuñón T, Sobreviela T, Insausti AM, Gonzalo LM.. 1995. The human entorhinal cortex: a cytoarchitectonic analysis. J Comp Neurol. 355:171–198. [DOI] [PubMed] [Google Scholar]
- Kelly C, Toro R, Di Martino A, Cox CL, Bellec P, Castellanos FX, Milham MP. 2012. A convergent functional architecture of the insula emerges across imaging modalities. Neuroimage. 61:1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmotsu N, Kucukboyaci NE, Cheng CE, Girard HM, Tecoma ES, Iragui VJ, McDonald CR. 2013. Alterations in functional connectivity between the hippocampus and prefrontal cortex as a correlate of depressive symptoms in temporal lobe epilepsy. Epilepsy Behav. 29:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Besson P, Colliot O, Bernasconi A, Bernasconi N. 2008. Surface-based vector analysis using heat equation interpolation: a new approach to quantify local hippocampal volume changes. Med Image Comput Comput Assist Interv. 11:1008–1015. [DOI] [PubMed] [Google Scholar]
- Knopp A, Kivi A, Wozny C, Heinemann U, Behr J. 2005. Cellular and network properties of the subiculum in the pilocarpine model of temporal lobe epilepsy. J Comp Neurol. 483:476–488. [DOI] [PubMed] [Google Scholar]
- Kramer MA, Kolaczyk ED, Kirsch HE. 2008. Emergent network topology at seizure onset in humans. Epilepsy Res. 79:173–186. [DOI] [PubMed] [Google Scholar]
- Kucukboyaci NE, Kemmotsu N, Cheng CE, Girard HM, Tecoma ES, Iragui VJ, McDonald CR. 2013. Functional connectivity of the hippocampus in temporal lobe epilepsy: feasibility of a task-regressed seed-based approach. Brain Connect. 3:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V, Marchiori M. 2001. Efficient behavior of small-world networks. Phys Rev Lett. 87:198701. [DOI] [PubMed] [Google Scholar]
- Liao W, Zhang Z, Pan Z, Mantini D, Ding J, Duan X, Luo C, Lu G, Chen H. 2010. Altered functional connectivity and small-world in mesial temporal lobe epilepsy. PLoS One. 5:e8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccotta L, Moseley ED, Benzinger TL, Hogan RE. 2015. Beyond the CA1 subfield: local hippocampal shape changes in MRI-negative temporal lobe epilepsy. Epilepsia. 56(5):780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, Sneppen K. 2002. Specificity and stability in topology of protein networks. Science. 296:910–913. [DOI] [PubMed] [Google Scholar]
- Mathern GW, Babb TL, Leite JP, Pretorius K, Yeoman KM, Kuhlman PA. 1996. The pathogenic and progressive features of chronic human hippocampal epilepsy. Epilepsy Res. 26:151–161. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Laxer KD, Barakos J, Cheong I, Finlay D, Garcia P, Cardenas-Nicolson V, Weiner MW. 2009. Involvement of the thalamocortical network in TLE with and without mesiotemporal sclerosis. Epilepsia. 46(2):353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ME. 2003. The structure and function of complex networks. SIAM Rev Soc Ind Appl Math. 45:167–256. [Google Scholar]
- Otte WM, Dijkhuizen RM, van Meer MP, van der Hel WS, Verlinde SA, van Nieuwenhuizen O, Viergever MA, Stam CJ, Braun KP. 2012. Characterization of functional and structural integrity in experimental focal epilepsy: reduced network efficiency coincides with white matter changes. PLoS One. 7:e39078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittau F, Grova C, Moeller F, Dubeau F, Gotman J. 2012. Patterns of altered functional connectivity in mesial temporal lobe epilepsy. Epilepsia. 53:1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten SC, Bartolomei F, Stam CJ. 2007. Small-world networks and epilepsy: graph theoretical analysis of intracerebrally recorded mesial temporal lobe seizures. Clin Neurophysiol. 118:918–927. [DOI] [PubMed] [Google Scholar]
- Schindler KA, Bialonski S, Horstmann MT, Elger CE, Lehnertz K. 2008. Evolving functional network properties and synchronizability during human epileptic seizures. Chaos. 18:033119. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 17:87–97. [DOI] [PubMed] [Google Scholar]
- Sporns O. 2011. The human connectome: a complex network. Ann N Y Acad Sci. 1224:109–125. [DOI] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. 2004. The small world of the cerebral cortex. Neuroinformatics. 2:145–162. [DOI] [PubMed] [Google Scholar]
- Stretton J, Winston GP, Sidhu M, Bonelli S, Centeno M, Vollmar C, Cleary RA, Williams E, Symms MR, Koepp MJ et al. 2013. Disrupted segregation of working memory networks in temporal lobe epilepsy. Neuroimage Clin. 2:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Oguz I, Brechbuehler C, Pantazis D, Ger G. 2006. Statistical Shape Analysis of Brain Structures using SPHARM-PDM. In: MICCAI OpenSource Workshop. Copenhagen, Denmark. [PMC free article] [PubMed] [Google Scholar]
- Sutula T. 2002. Seizure-induced axonal sprouting: assessing connections between injury, local circuits, and epileptogenesis. Epilepsy Curr. 2:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qiu S, Xu Y, Liu Z, Wen X, Hu X, Zhang R, Li M, Wang W, Huang R. 2014. Graph theoretical analysis reveals disrupted topological properties of whole brain functional networks in temporal lobe epilepsy. Clin Neurophysiol. 125(9):1744–1756. [DOI] [PubMed] [Google Scholar]
- Watson C, Andermann F, Gloor P, Jones-Gotman M, Peters T, Evans A, Olivier A, Melanson D, Leroux G. 1992. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 42:1743–1750. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. 1998. Collective dynamics of ‘small-world’ networks. Nature. 393:440–442. [DOI] [PubMed] [Google Scholar]