Abstract
Brain regions with high connectivity have high metabolic cost and their disruption is associated with neuropsychiatric disorders. Prior neuroimaging studies have identified at the group-level local functional connectivity density (lFCD) hubs, network nodes with high degree of connectivity with neighboring regions, in occipito-parietal cortices. However, the individual patterns and the precision for the location of the hubs were limited by the restricted spatiotemporal resolution of the magnetic resonance imaging (MRI) measures collected at rest. In this work, we show that MRI datasets with higher spatiotemporal resolution (2-mm isotropic; 0.72 s), collected under the Human Connectome Project (HCP), provide a significantly higher precision for hub localization and for the first time reveal lFCD patterns with gray matter (GM) specificity >96% and sensitivity >75%. High temporal resolution allowed effective 0.01–0.08 Hz band-pass filtering, significantly reducing spurious lFCD effects in white matter. These high spatiotemporal resolution lFCD measures had high reliability [intraclass correlation, ICC(3,1) > 0.6] but lower reproducibility (>67%) than the low spatiotemporal resolution equivalents. GM sensitivity and specificity benchmarks showed the robustness of lFCD to changes in model parameter and preprocessing steps. Mapping individual's brain hubs with high sensitivity, specificity, and reproducibility supports the use of lFCD as a biomarker for clinical applications in neuropsychiatric disorders.
Keywords: aging, FCDM, global signal regression, motion, multiband, physiologic noise, resting state
Introduction
Functional connectivity density mapping (FCDM) is a powerful graph theory tool for exploring the topology of human brain function using magnetic resonance imaging (MRI) datasets collected at rest and task states (Tomasi and Volkow 2010; Tomasi, Wang, Wang, et al. 2014). FCDM quantifies local degree, the size of the local network cluster functionally connected to a brain network node, which measures the local centrality of every voxel in the human brain connectome. In contrast to seed-voxel correlation analysis (Biswal et al. 1995), data-driven FCDM is ideal for exploratory analyses because it does not require a priori hypotheses. Due to extremely high computational demand, it is almost impossible to perform voxelwise analysis using other graph theory metrics such as global degree, clustering, and path length, yet ultrafast FCDM quantifies the strength of the local functional connectivity hubs (network nodes with high connectivity to nearby brain regions) at 3-mm isotropic resolution in just a few minutes/subject (Tomasi and Volkow 2010). These characteristics make FCDM optimal for data mining of large image repositories of “resting-state” functional connectivity datasets such as the “1000 Functional Connectome Project” (Biswal et al. 2010) and the attention deficit hyperactivity disorder (ADHD)-200 (Tomasi and Volkow 2012a).
Using FCDM, we have demonstrated the distribution of local (lFCD) and global (gFCD) hubs with high functional connectivity density (i.e., degree; Tomasi and Volkow 2011a, 2011b) and the functional asymmetry of the human brain (Tomasi and Volkow 2011d), as well as the effects of task performance (Tomasi, Wang, Wang, et al. 2014), gender (Tomasi and Volkow 2011c), and aging (Tomasi and Volkow 2012b) on lFCD and gFCD, and the metabolic cost of functional connectivity (Tomasi, Wang, et al. 2014). Recent studies using FCDM have demonstrated modulatory effects of catechol-O-methyltransferase and dopamine D2 receptors (Tian et al. 2013) and the influence of the oxytocin receptor gene (Wang, Qin, et al. 2013) on the strength of the functional connectivity hubs. Independent laboratories (including ours) have shown disrupted lFCD and gFCD in ADHD (Tomasi and Volkow 2012a), traumatic axonal injury (Caeyenberghs et al. 2014), non-epileptic seizures (Ding et al. 2014), schizophrenia (Tomasi and Volkow 2014; Zhuo et al. 2014; Liu et al. 2015), congenital blindness (Qin et al. 2015), and cocaine addiction (Konova et al. 2015). lFCD and gFCD hubs have also shown to be sensitive to stimulants (Konova et al. 2015), fluid reasoning capacity (Lang et al. 2015), and brain development (Tomasi and Volkow 2014).
Whereas functional connectivity metrics promise to have a major impact on neuroscience research, several limitations still prevent their translation as clinical biomarkers in neurology and psychiatry. For instance, at 3-mm isotropic resolution (standard native spatial resolution in fMRI studies), the functional connectivity metrics include significant contamination from blood vessels and have suboptimal tissue specificity (Menon 2002; Turner 2002). More importantly, functional connectivity metrics from data collected with low temporal resolution (1.5–3.0 s) include significant aliasing artifacts of physiological origin (Birn et al. 2006). Thus, similar to other functional connectivity metrics, FCDM would benefit significantly from greater accuracy in spatial localization and faster image acquisition.
The HCP (https://db.humanconnectome.org/; Van Essen et al. 2012) recently released a large fMRI database including “resting-state” functional connectivity datasets with unprecedented spatiotemporal resolution (2-mm isotropic; 0.72 s) that allow mapping the distribution of functional connectivity hubs at the individual level (Cohen et al. 2008) with high spatial specificity and reduced physiologic noise contamination. We hypothesized that spontaneous fluctuations in the resting state are driven by neuronal populations (Logothetis et al. 2001; Logothetis 2002; Shmuel and Leopold 2008; Lei et al. 2014; Li et al. 2014; Siero et al. 2014), and we predicted that the better spatiotemporal resolution of the data would enable us to observe these fluctuations only within the gray matter (GM). Therefore, the higher spatial resolution and faster sampling rate of the HCP datasets would lead to sharper lFCD patterns with greater precision in spatial localization at the individual and group levels and with reduced physiologic noise artifacts.
Here, we assess for the first time the distribution of lFCD hubs with high spatiotemporal resolution at the individual and group levels in 40 healthy adults using the HCP-Q1 dataset. Aiming to identify the brain location of functional hubs with unprecedented spatial resolution, we assess GM sensitivity and specificity, reproducibility and test–retest reliability, as well as the effects of MRI acquisition (i.e., LR vs. RL phase encodings), and specific lFCD parameters (i.e., correlation threshold) and preprocessing steps (i.e., motion covariates, 0.08 Hz low-pass filtering, and global signal normalization) on the strength of the lFCD patterns. We hypothesized that higher spatiotemporal resolution would enable precise identification of hub locations within occipito-parietal and primary sensory cortices, which are the regions that house the most prominent hubs in the human brain (Tomasi and Volkow 2010).
Materials and Methods
Subjects
Data were drawn from the publicly available repository of the WU-Minn HCP (http://www.humanconnectome.org/). The scanning protocol was approved by Washington University in the St. Louis's Human Research Protection Office (HRPO), IRB# 201204036. No experimental activity with any involvement of human subjects took place at the author's institutions.
The 40 participants (age: 31 ± 3 years; 31 females) of the WU-Minn HCP-Q1 data release included in this study provided written informed consent and were scanned on a 32-channel 3.0-T Siemens Skyra unit according to procedures approved by the IRB at Washington University. Resting-state functional images were acquired while the participant relaxed with eyes open using a gradient-echo-planar imaging (EPI) sequence with multiband factor 8, time repetition (TR) 720 ms, time echo 33.1 ms, flip angle 52°, 104 × 90 matrix size, 72 slices, 2 mm isotropic voxels, and 1200 time points (Smith et al. 2013; Uğurbil et al. 2013). Scans were repeated twice using different phase-encoding directions (RL and RL) in each of the 2 sessions (REST1 and REST2). To capitalize on the high-quality data offered by the HCP, we used the “minimal preprocessing” datasets (hp2000_clean.nii files), which include gradient distortion correction, rigid body realignment, field map processing, spatial normalization to the stereotactic space of the Montreal Neurological Institute (MNI), high-pass filtering (1/2000 Hz frequency cutoff; Glasser et al. 2013), independent component analysis (ICA)-based denoising (Salimi-Khorshidi et al. 2014), and brain masking. Thus, 160 “resting-state” image time series with 902 629 2-mm isotropic voxels and 1200 time points (4.3 Gb memory space) collected over 864 s were used in this study. In addition, we used the HCP's gray and white matter parcellations (guided by “wmparc” and “ROIs” files) of each subject's brain structural scans, to create a GM template. This template was used to assess the GM specificity of the lFCD.
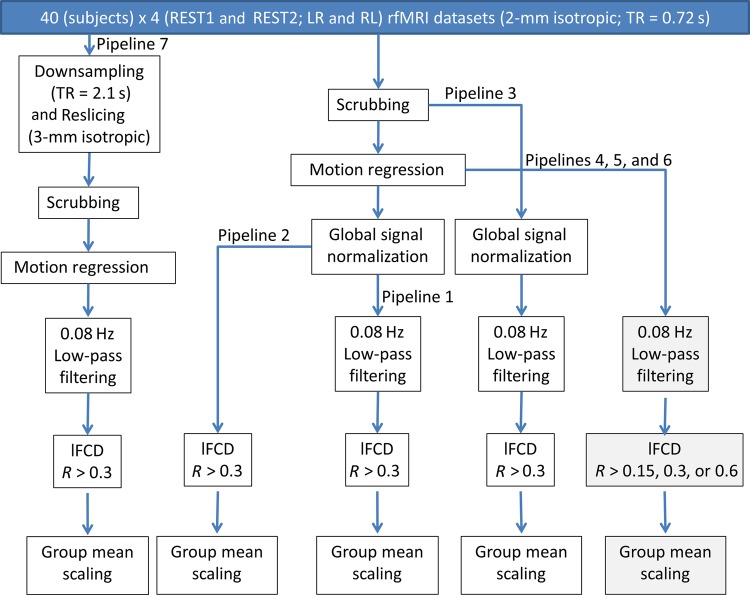
lFCD Pipelines
The interactive data language (IDL, ITT Visual Information Solutions, Boulder, CO, USA) and a workstation with 2 Intel® Xeon® X5680 processors and 48 Gb random access memory were used in subsequent FCDM-processing steps (Fig. 1). Seven lFCD pipelines were implemented. Pipeline 1 included “scrubbing” to remove image time points that could be severely contaminated with motion. Specifically, framewise displacements, FDs, were computed for every time point from head translations and rotations, using a radius of r = 50 mm to convert angle rotations to displacements. The mean FD was not different for the REST1 (0.17 ± 0.05 mm; mean ± standard deviation) and REST2 (0.18 ± 0.05 mm) sessions or for the LR (0.17 ± 0.05 mm) and RL (0.18 ± 0.05 mm) phase-encoding directions (P > 0.24; paired t-test). Scrubbing was implemented in IDL to remove image time points with temporal derivative of the root mean square variance of the signal over voxels, DVARS >0.5% and FD >0.5 mm (Power et al. 2012). The number of time points removed per time series by scrubbing was not significantly different for REST1 (0.5 ± 1.1; mean ± SD) and REST2 (0.8 ± 1.1) or for LR (0.5 ± 1.2) and RL (0.8 ± 1.1; P > 0.12, t-test).
Figure 1.
lFCD processing pipelines. Seven lFCD maps (see text) were computed for each subject, session, and phase-encoding direction. A total of 960 lFCD maps covering the whole brain (white matter and cerebrospinal fluid regions were not masked out to assess the strength of the lFCD in these regions) with 2-mm isotropic resolution and 91 × 109 × 91 voxels were computed using 160 HCP datasets with “minimal preprocessing” (Glasser et al. 2013) from the Q1 release. Smoothing was not used to preserve the high spatial resolution of the resting-state functional dataset.
Pipeline 1 also included multilinear regression to minimize motion-related fluctuations in the MRI signals (Tomasi and Volkow 2010), 3D global signal intensity normalization across time points to minimize global fluctuations, and standard 0.08 Hz low-pass filtering to remove magnetic field drifts and minimize physiologic noise of high frequency components. Note that these standard lFCD preprocessing steps partially overlap with those used by the HCP (ICA-based denoising) to remove unwanted noise sources (Salimi-Khorshidi et al. 2014).
In addition, pipeline 1 assessed the lFCD at every voxel in the brain, computed as the number of elements in the local functional connectivity cluster, using a “growing” algorithm written in IDL (Tomasi and Volkow 2010). The Pearson correlation was used to assess the strength of the functional connectivity, Rij, between voxels i and j in the brain, and a correlation threshold, Rij > 0.3, was selected to ensure significant correlations between time-varying signal fluctuations are corrected at PFWE < 0.05. A voxel (xj) was added to the list of voxels functionally connected with x0 only if it was adjacent to a voxel that was linked to x0 by a continuous path of functionally connected voxels and R0j > 0.3. This calculation was repeated for all brain voxels that were adjacent to those that belonged to the list of voxel functionally connected to x0 in an iterative manner until no new voxels could be added to the list. Spatial smoothing was not used to preserve the high spatial resolution of the native datasets. Note that lFCD was evaluated in the whole brain without any masking procedure (227 372 ± 2461 voxels; mean ± SD).
Three alternative pipelines avoiding either (pipeline 2) low-pass filtering; or (pipeline 3) regressing out motion parameters; or (pipeline 4) avoiding global signal normalization were implemented to assess the effects of additional preprocessing steps on the ICA-based denoising approach; and 2 alternative pipelines with (pipeline 5) R0j > 0.6; or (pipeline 6) R0j > 0.15 and with no global signal normalization were also implemented to assess the effects of model parameters on lFCD patterns at high spatiotemporal resolution. Furthermore, time series with low spatiotemporal resolution (3-mm isotropic, spatial resolution of typical fMRI studies; 2.16 s = 3TR, temporal resolution of typical fMRI studies) were created from high-resolution scans (REST1 and REST2) using reslicing and downsampling (pipeline 7) in order to assess potential gains in sensitivity, specificity, and reproducibility for lFCD maps (R0j > 0.3; no global signal normalization) with high versus low spatiotemporal resolution (pipeline 4 vs. pipeline 7). Group mean normalization using a single scaling factor reflecting the average lFCD across subjects was performed independently as the last step of the 7 pipelines to allow comparison of results.
Reproducibility, Sensitivity, and Specificity Indices
The “reproducibility” of the lFCD hubs in the brain across resting-state sessions was quantified across voxels for each subject or across subjects for each voxel as:
| (1) |
The gray and white matter parcellations provided with the Q1 release of the HCP dataset were used to compute 2 additional indices assessing tissue-specific differences in lFCD across image voxels for each subject or across subjects for each image voxel:
| (2) |
gauges the proportion of lFCD within the tissue of interests (i.e., cortical and subcortical GM and white matter), normalized by tissue volume to that of GM. Note that brain counts,, must be larger than tissue counts,, in order to assess the true positive rate of lFCD in the tissue. This criterion is naturally satisfied across voxels for each individual (gray matter volume < brain volume) but is not warranted across subjects for each image voxel because normalized brain anatomy in the MNI space is highly similar across subjects. Thus for subcortical structures and cerebellum, and major sulci and gyri sensitivity could not be determined.
| (3) |
a true negative rate test, gauges the proportion of white matter measures with lower lFCD than the whole-brain average. Specificity could not be determined across subjects for voxels that were either classified as GM or had lower lFCD than the whole-brain average in all subjects.
Reliability
The test–retest reliability of the lFCD patterns was evaluated for each imaging voxel using two-way mixed single-measures intraclass correlation (Shrout and Fleiss 1979)
| (4) |
Specifically, ICC(3,1) was mapped in the brain in terms of between-subjects (BMS) and residuals mean-square (EMS) values computed for each voxel using the IPN matlab toolbox (http://www.mathworks.com/matlabcentral/fileexchange/22122-ipn-tools-for-test-retest-reliability-analysis) and the lFCD maps corresponding to REST1 and REST2 sessions (k = 2). Note that ICC(3, 1) coefficients range from 0 (no reliability) to 1 (perfect reliability).
Physiologic Noise
The HCP datasets included physiologic data (cardiac and respiratory) corresponding to the resting-state functional scans, which were collected with 2.5 ms temporal resolution. The fast Fourier transform was used to assess the amplitude of the different frequencies of the cardiac and respiratory signals in the 0- to 200-Hz bandwidth of the frequency domain representation. In addition, the physiologic data were downsampled to the 0.72-s temporal resolution of the multiband acquisition using the trigger signal embedded in the physiologic data file, which render frequency domain representations of the cardiac and respiratory signals in the 0.0- to 0.7-Hz bandwidth. The power amplitudes in the low-frequency bandwidth, relative to the full power of the Fourier spectra, were computed for each scan.
Statistical Methods
A full factorial design was used to compare lFCD between sessions (REST1 vs. REST2) and phase encoding directions (LR vs. RL) and to compare lFCD differences associated with specific lFCD preprocessing steps (i.e., multilinear motion regression, low-pass filtering, and global signal normalization). The statistical parametric mapping package (SPM8) was used for this purpose. Statistical significance was set by PFWE <0.05, corrected for multiple comparisons at the cluster level with the random field theory and a family-wise error correction with a cluster-forming threshold of P < 0.001 and a minimum cluster size of 625 voxels (5 mL).
Results
lFCD Patterns
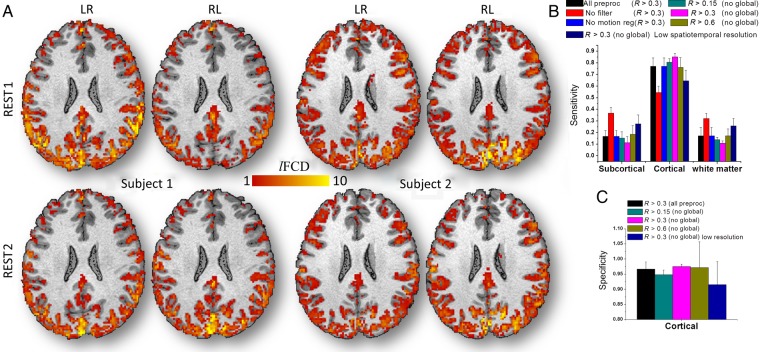
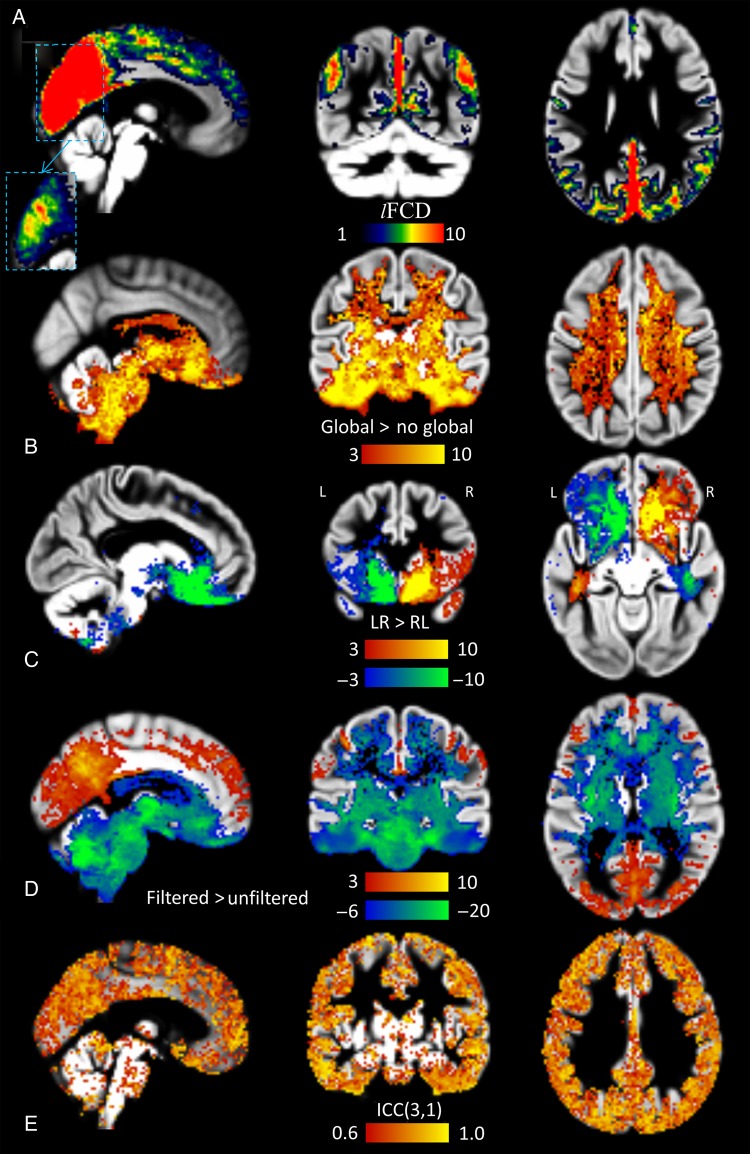
For all resting-state scans, the individual's lFCD patterns predominantly followed the shape of cortical GM, and had minimal overlap with white matter and cerebrospinal fluid (Fig. 2A). These individual's cortical lFCD patterns were highly reproducible across sessions and phase-encoding conditions. Figure 2 demonstrates the reproducibility of the lFCD patterns from pipeline 1 (see Materials and Methods) across sessions and phase-encoding conditions for 2 randomly selected subjects. In most cortical regions, the average lFCD across subjects was stronger than the whole-brain mean lFCD. Inferior ventral, orbitofrontal, and insular cortices and subcortical regions showed attenuated lFCD, perhaps reflecting the lower sensitivity of the 32-channels coil used to collect the data in these regions (Anteraper et al. 2013). The remarkable GM specificity of the lFCD in cortical regions across all subjects was used as one of the benchmark criteria to assess the effect of image preprocessing steps on lFCD.
Figure 2.
GM sensitivity and specificity across subjects. (A) Four exemplary lFCD maps from test–retest sessions (REST1 and REST2) with different multiband acquisition (phase-encoding directions: LR and RL) for 2 randomly selected subjects, superimposed on the subjects' T1-weighted MRI structures. Average sensitivity for subcortical and cortical GM and white matter (B) and specificity (C) of lFCD across subjects for each of the processing pipelines in Figure 1, for REST1 and LR phase encoding. Error bars are standard deviations.
Reproducibility
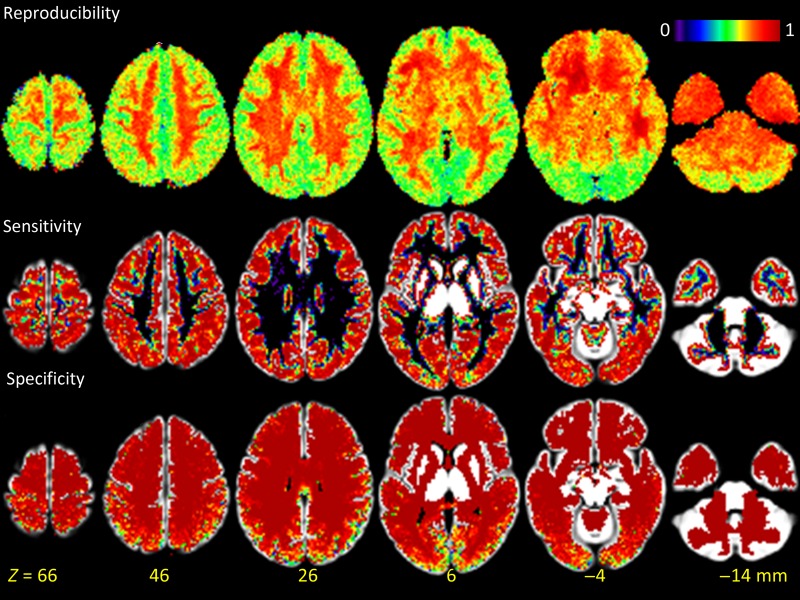
The lFCD patterns computed with global signal normalization and correlation threshold R > 0.3 had reproducibility of 70 ± 4% across sessions. The lack of global signal normalization improved the sensitivity and specificity (Fig. 2B,C) indices but reduced lFCD reproducibility to 67 ± 5% (P < 0.002; paired t-test, df = 79). lFCD patterns (without global normalization) computed with R > 0.6 had higher reproducibility (88 ± 4%) and those with R > 0.15 had lower reproducibility (59 ± 6%) than at R > 0.3. The reproducibility of the lFCD patterns did not differ between LR and RL runs. The reproducibility index was maximal in white matter and minimal in the visual cortex (Fig. 3). Voxelwise analysis contrasting REST1 and REST2 did not show statistically significant lFCD differences between sessions in any brain region. To assess the impact of higher spatiotemporal resolution, we compared the reproducibility of lFCD maps for high versus low spatiotemporal resolution (downsampled) datasets, which were computed with R > 0.3 and without global signal normalization (pipeline 4 vs. 7). The reproducibility index was significantly lower for datasets with high spatiotemporal resolution (67 ± 5%; mean ± SD) than for those with low spatiotemporal resolution (82 ± 3%; P < 10−26, paired t-test).
Figure 3.
Reproducibility and GM sensitivity and specificity of lFCD patterns computed across subjects superimposed on axial view of the GM template. Sensitivity and specificity could not be computed for major sulci and gyri, thalamus, caudate putamen, amygdala, hippocampus, midbrain, brainstem, and cerebellar GM, due to the absence of white matter in these regions in all subjects.
Sensitivity
Across subjects, the “sensitivity” index (i.e., the proportion of lFCD within a tissue of interest; see Materials and Methods) was higher for cortical gray matter than for white matter and subcortical GM (including cerebellum; Fig. 2B). This differential pattern was highly significant across subjects and reproducible across sessions and phase-encoding conditions (P < 10−9; paired t-test, df = 39; Supplementary Fig. 1). For cortical GM, the sensitivity index reached a maximal value (85 ± 3%; mean ± SD) for lFCD maps computed without global signal normalization and using a correlation threshold R > 0.3 (Fig. 2B). Decreasing or increasing the correlation threshold from R > 0.3 to >0.15 or >0.6 reduced the sensitivity index to 80 ± 3% or 76 ± 8% (P < 10−10; paired t-test, df = 39) for cortical GM. The lack of 0.08 Hz low-pass filtering significantly reduced the sensitivity index in cortical regions to 55 ± 5% (P < 10−17). Multilinear motion regression did not have a significant effect on the sensitivity index (P > 0.4), suggesting that ICA-based denoising properly corrects for motion artifacts in lFCD.
The cortical GM sensitivity index was significantly higher for datasets with high spatiotemporal resolution (85 ± 3%; mean ± SD) than for those with low spatiotemporal resolution (65 ± 9%; P < 10−18, paired t-test). Figure 3 shows that the sensitivity index was maximal in cortical regions, minimal in white matter, and null in the ventricles. Note that sensitivity could not be assessed for major sulci and gyri, thalamus, caudate putamen, amygdala, hippocampus, midbrain, and cerebellar GM, due to the absence of white matter in these regions in all subjects.
Specificity
The specificity of the lFCD patterns for cortical GM was higher than 93.6% across threshold and global normalization conditions, sessions, and phase-encoding directions (“specificity index” = 96.3 ± 1.7; mean ± SD; Fig. 2C). Global normalization improved the specificity index (P < 0.04, paired t-test, df = 39). The correlation threshold used in the computation of lFCD did not have a significant impact on the specificity index, in different sessions or phase-encoding directions. The specificity index was significantly higher for datasets with high spatiotemporal resolution (97 ± 1%; mean ± SD) than for those with low spatiotemporal resolution (92 ± 7%; P = 0.0001, paired t-test; pipeline 4 vs. pipeline 7). The specificity index was maximal in white matter regions, minimal in cortical regions, and null in the ventricles (Fig. 3). Similar to sensitivity, specificity could not be assessed for major sulci and gyri, thalamus, caudate putamen, amygdala, hippocampus, midbrain, brainstem, and cerebellar GM, due to the absence of white matter in these regions in all subjects.
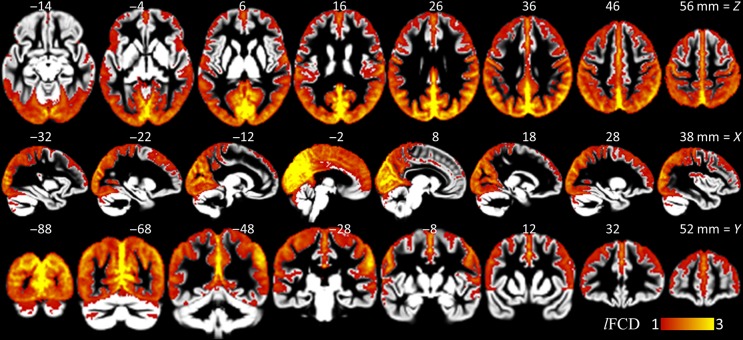
Strength
Figure 4 highlights the distribution of the lFCD in the human brain, which is consistent with our previous reports at lower spatiotemporal resolution (Tomasi and Volkow 2010, 2011b). Specifically, in cuneus, precuneus, and primary visual cortices, the lFCD was 3 times or higher than the whole brain average. Within this large region, the cuneus and the precuneus, encompassing posterior ventral regions of the parieto-occipital fissure, exhibited maximal lFCD. The angular gyrus, posterior cingulum, and the posterior regions of the default-mode network also demonstrated prominent lFCD with 6 times or higher strength than the whole brain average. Other regions with high lFCD included precentral gyrus, anterior cingulum, as well as primary auditory, somatosensory, and motor cortices.
Figure 4.
lFCD patterns. Average lFCD strength across subjects showing brain areas with higher lFCD than the whole brain average (pipeline 1), superimposed on axial (top row), sagittal (middle row), and coronal (bottom row) views of cortical and subcortical GM template developed using the HCP structural scans.
Hubs
The local maxima of the average lFCD highlight the precise location of the hubs (i.e., nodes with high local connectivity) in the human brain (Fig. 5). The strongest hubs (lFCD > 5.0) were densely located in a posterior occipito-parietal network, which included cuneus, middle and superior occipital, supramarginal and angular gyri, and inferior parietal cortex (BAs 2, 7, 18, 19, 39, and 40; Supplementary Table 1). The probability of lFCD local maxima within a 1-mL cubic volume, P, was used to assess the reproducibility of hub locations across subjects. The locations and intensity of the hubs varied across subjects (see Supplementary Fig. 2), but were highly reproducible within the 1-mL cubic search volume (P > 0.8, Supplementary Table 1). Similarly, the locations and intensity of the lFCD hubs were highly reproducible across sessions and phase-encoding directions (Fig. 6). The linear associations between GM sensitivity and specificity, reproducibility, and strength of the lFCD hubs were not statistically significant (P > 0.07, Pearson correlation).
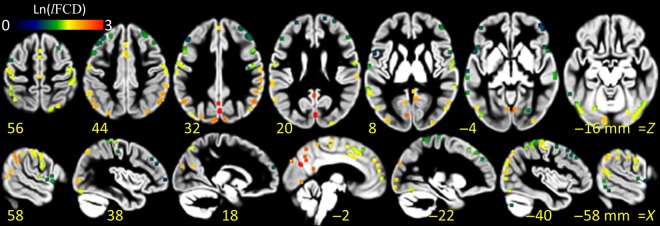
Figure 5.
lFCD hubs. Local maxima of the average lFCD detected with probability P > 0.5 across subjects, sessions, and phase-encoding directions within a 1-mL cubic volume, superimposed on axial (top row) and sagittal (bottom row) views of a GM template of the human brain. The color pattern indicates the average strength of the lFCD hubs in a logarithmic scale.
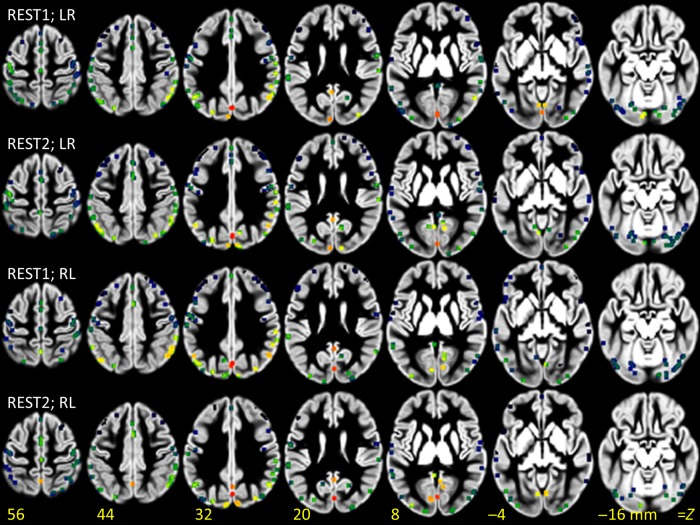
Figure 6.
lFCD hubs: Reproducibility. Local maxima of the average lFCD across subjects, detected with probability P > 0.5 within a 1-mL cubic volume, for each session (REST1 and REST2) and phase-encoding direction (LR and RL), superimposed on axial (top row) views of a GM template of the human brain. The colored pattern indicates the strength of the lFCD hubs in a logarithmic scale.
Global Signal Normalization
Global signal normalization did not alter the lFCD in most cortical regions, but caused significant increases in lFCD in white matter, ventral temporal cortex, basal ganglia, thalamus, and brainstem and in the lateral ventricles (Fig. 7B).
Figure 7.
lFCD: Effects of global signal, phase encoding and low-pass filtering, and test–retest reliability. (A) Brain regions with high lFCD superimposed on 3 orthogonal views of the GM template (pipeline 1). The dashed rectangle insert depicts the lFCD distribution in ventral occipital and posterior parietal regions using a higher upper threshold (color map: 6 < lFCD < 21). (B) A t-score map demonstrating the significant lFCD increases in white matter and subcortical regions elicited by global signal normalization (pipeline 1 vs. pipeline 4). (C) t-score maps highlighting brain regions with significant phase-encoding-related artifacts in the lFCD. (D) t-score maps highlighting regions with significant lFCD increases (red-yellow) and decreases (blue-green) associated with 0.8 Hz low-pass filtering (pipeline 1 vs. pipeline 2). Two-way mixed single-measures intraclass correlation ICC(3,1) maps at 2-mm isotropic resolution depicting regional variability in reliability for lFCD measures for processing pipeline 1 with LR phase-encoding data.
Phase-Encoding Direction
The lFCD was sensitive to the phase-encoding direction of the multiband EPI acquisition, particularly in brain regions affected by magnetic field gradients induced by magnetic susceptibility differences at air-tissue interfaces such as the sinus cavity. In contrast to LR phase encoding, RL phase encoding increased lFCD in right orbitofrontal gray and white matter and left ventral temporal white matter and decreased it in the contralateral regions (Fig. 7C).
Physiologic Noise
When sampled at TR = 0.72 s, physiological (cardiac and respiratory) signals are aliased into the 0- to 0.7-Hz bandwidth and lack of proper low-pass filtering could spuriously increase MRI signal fluctuations (see Supplementary Fig. 3). The lFCD maps corresponding to datasets processed without low-pass filtering had lower strength in cortical regions and higher strength in white matter, ventral temporal cortex, basal ganglia, thalamus, brainstem, and the lateral ventricles than those corresponding to datasets processed with low-pass filtering (Fig. 7D).
Reliability
Intraclass correlation analyses of test–retest datasets demonstrated the high reliability (ICC(3,1) > 0.6) of the lFCD in cortical regions (Fig. 7E). lFCD measures with global signal normalization showed increased reliability in cortical regions, compared with those without global signal normalization (see Supplementary Fig. 4). Test–retest reliability was higher for R > 0.3 than for R > 0.15. The high correlation threshold (R > 0.6) had poor reliability.
Discussion
Here, we show for the first time patterns of lFCD with high spatiotemporal resolution that can reliably identify the precise location of functional connectivity hubs in each individual. Specifically, we quantified the sensitivity and specificity of lFCD for GM as well as the reproducibility and reliability of the lFCD patterns while assessing effects of image preprocessing steps and lFCD model parameters. Based on these analyses, optimal sensitivity and specificity were obtained for the threshold R > 0.3, 0.01–0.08 Hz frequency bandwidth, and without global normalization.
The lFCD is a graph theory metric that measures the degree of the local cluster functionally connected to a given network node. Using MRI data (Achard et al. 2006; van den Heuvel et al. 2008; Beu et al. 2009; Buckner et al. 2009; Tomasi and Volkow 2010, 2011b), magnetoencephalography and electroencephalography (Stam 2004), and optical imaging (Bonifazi et al. 2009), others and we have demonstrated that brain network architecture is based on few hubs (regions with high degree) and numerous nodes with low degree (Bullmore and Sporns 2009). This distribution of the nodes is also in agreement with the structure of the small-world (Watts and Strogatz 1998) and scale-free (Barabasi and Albert 1999) networks. The hubs of the human brain are energy demanding (Tomasi, Wang, et al. 2013) and prone to malfunction in neuropsychiatric disorders of metabolic origin (Bassett et al. 2008; He et al. 2008; Buckner et al. 2009; Supekar et al. 2009; Crossley et al. 2014). Even in normal subjects, the strength of the functional connectivity hubs is influenced by aging (Tomasi and Volkow 2012b) and genetic background (Kelly et al. 2007; Biswal et al. 2010; Van Dijk et al. 2010; Wang, Qin, et al. 2013). This work demonstrates for the first time the precise location of the functional connectivity hubs in the brain, as defined by lFCD local maxima. Using high spatiotemporal resolution FCDM, we demonstrate that cortical hubs are densely located in occipito-parietal cortices, which are known to be disrupted in patients with schizophrenia (van den Heuvel et al. 2013), autism spectrum disorders (ASDs) (Blanken et al. 2015), and Alzheimer's disease (Sheline and Raichle 2013). At lower spatiotemporal resolution (∼3-mm isotropic; ∼2 s), we have quantified the variability across subjects in the position of the 2 more prominent hubs as 4.4 ± 2.7 and 6.7 ± 4.1 mm, respectively, which are located in posterior and inferior parietal cortices (Tomasi and Volkow 2010). However, in our previous study, the variability in the spatial location of other hubs was larger (>11 mm). In the present work, we identify the precise locations of 120 cortical hubs that can be detected across subjects with probability higher than 80% within a 1-mL cubic volume (i.e., within 5 mm from MNI coordinates in Supplementary Table 1). The locations of these cortical hubs were highly reproducible across subjects and sessions.
This is also the first study to quantify sensitivity, specificity, and reproducibility of functional connectivity measures in GM tissue, which is where neuronal activity gives rise to oscillations in the blood oxygenation level-dependent MRI signals (Logothetis et al. 2001; Lu et al. 2016; Carandini et al. 2015). The lFCD patterns were highly reproducible within and across subjects and exhibited high GM specificity such that they accurately delineated the shape of cortical gyri and sulci for each individual subject. Reproducibility ranged from 59 ± 6%, for the lowest correlation threshold, to 88 ± 4%, for the highest correlation threshold, and the fraction of lFCD (sensitivity index) in GM reached a maximum of 85 ± 3% for the medium correlation threshold. In this study, the reliability of the lFCD was high in cortical regions (ICC(3,1) = 0.65 ± 0.14; mean ± SD) and moderate in basal ganglia and cerebellum (ICC(3,1) < 0.40 ± 0.20). Previous studies have reported low to moderate reliability (0.15 < ICC(3,1) < 0.6) of functional connectivity measures (Zuo et al. 2010; Braun et al. 2012; Guo et al. 2012; Yang et al. 2012; Birn et al. 2013; Wang, Jiao, et al. 2013), in part due to intrinsic limitations in the spatiotemporal resolution of the MRI data. The present work shows that MRI acquisition protocols with high spatial and temporal resolutions, as those used under the HCP (Van Essen et al. 2012; Uğurbil et al. 2013), can provide a quantum leap in sensitivity, specificity, reproducibility, and reliability of the cortical lFCD hubs at the individual- and group-level analyses. Precise identification of hubs at the individual level is needed in order to develop biomarkers that assess variability of these hubs through an individual's brain developmental trajectory and in disease, and to assess the effects of therapeutic interventions.
The predominance of lFCD in occipito-parietal cortices at the group level is consistent with findings from our previous studies at lower spatiotemporal resolution (Tomasi and Volkow 2010, 2011b). The strength of the lFCD hubs in cuneus and primary visual cortex was 10 times or higher than the whole brain average. The cuneus and visual cortex have higher neuronal density than other cortical regions (Changeux 1997) and show high glucose metabolic rates (Raichle and Gusnard 2002) that are associated with their functional connectivity (Tomasi, Wang, et al. 2013).
Many sources of fMRI signal variability confound the detection of spontaneous fluctuations in resting-state functional connectivity studies. The data used in this work were cleaned by the HCP using FIX (“FMRIB's ICA-based X-noiseifier”), an ICA-based automatic noise detection algorithm that can minimize the effect of various types of noise sources (Salimi-Khorshidi et al. 2014). Here, we assessed residual effects of global signal normalization, low-pass filtering, and motion regression, preprocessing steps that overlap with those implemented in FIX. We show that the GM localization of the lFCD patterns was significantly improved by 0.08 Hz low-pass filtering. These improvements are consistent with the prediction that proper low-pass filtering attenuates the spurious effect of physiologic noise (cardiac and respiration fluctuations) on resting-state functional connectivity patterns (Birn et al. 2006, 2008). Whereas previous resting-state fMRI studies at lower temporal resolutions (TR > 1 s) were unable to effectively filter out physiologic noise, the higher temporal resolution (TR = 0.72 s) enabled by the HCP's multiband EPI acquisition provided the necessary bandwidth to filter out spurious signal fluctuations of physiologic origin. However, the lFCD in orbitofrontal and ventral temporal cortices was sensitive to the multiband phase-encoding scheme, which might be a concern for studies targeting these brain region such as in ADHD, obsessive compulsive and addiction disorders, and for those on impulsivity and reward processing.
We also studied the effect of global signal normalization on the strength of lFCD in GM, as well as the specificity, reproducibility, and reliability of the lFCD patterns. Global signal normalization increased the sensitivity, reproducibility, and reliability of the lFCD in cortical GM, but spuriously increased lFCD in white matter and subcortical regions, which might confound results in these regions (Wong et al. 2012). Techniques such as global signal regression/normalization are proposed to control for scanner instabilities in resting-state functional connectivity. However, global signal fluctuations could reflect true electrophysiological activity (Schölvinck et al. 2010) and global normalization may lead to side effects such as creating spurious anticorrelations (Murphy et al. 2009; Chai et al. 2012; Saad et al. 2012).
Overall, this work demonstrates the robustness of FCDM at high spatiotemporal resolution. The high sensitivity, specificity, and reliability make high-resolution lFCD ideal for the development of imaging biomarkers for neurological and psychiatric disorders and for studying normal variability in human brain development and aging. Finally, the established reproducibility and robustness of the high spatiotemporal resolution lFCD metric offer promise as a potential biomarker for the evaluation of therapeutic interventions at the individual level.
Supplementary material
Supplementary material can be found at http://www.cercor.oxfordjournals.org/.
Funding
Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research and by the McDonnell Center for Systems Neuroscience at Washington University. This work was accomplished with support from the National Institutes of Alcohol Abuse and Alcoholism (2RO1AA09481).
Supplementary Material
Notes
Conflict of Interest: None declared.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. 2006. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci. 26:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anteraper S, Whitfield-Gabrieli S, Keil B, Shannon S, Gabrieli J, Triantafyllou C. 2013. Exploring functional connectivity networks with multichannel brain array coils. Brain Connect. 3:302–315. [DOI] [PubMed] [Google Scholar]
- Barabasi A, Albert R. 1999. Emergence of scaling in random networks. Science. 286:509–512. [DOI] [PubMed] [Google Scholar]
- Bassett D, Bullmore E, Verchinski B, Mattay V, Weinberger D, Meyer-Lindenberg A. 2008. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 28:9239–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beu M, Baudrexel S, Hautzel H, Antke C, Mueller H-W. 2009. Neural traffic as voxel-based measure of cerebral functional connectivity in fMRI. J Neurosci Methods. 176:263–269. [DOI] [PubMed] [Google Scholar]
- Birn R, Diamond J, Smith M, Bandettini P. 2006. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 31:1536–1548. [DOI] [PubMed] [Google Scholar]
- Birn R, Molloy E, Patriat R, Parker T, Meier T, Kirk G, Nair V, Meyerand M, Prabhakaran V. 2013. The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage. 83:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn R, Murphy K, Bandettini P. 2008. The effect of respiration variations on independent component analysis results of resting state functional connectivity. Hum Brain Mapp. 29:740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Mennes M, Zuo X, Gohel S, Kelly C, Smith S, Beckmann C, Adelstein J, Buckner R, Colcombe S et al. . 2010. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin F, Haughton V, Hyde J. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 34:537–541. [DOI] [PubMed] [Google Scholar]
- Blanken L, Mous S, Ghassabian A, Muetzel R, Schoemaker N, El Marroun H, van der Lugt A, Jaddoe V, Hofman A, Verhulst F et al. . 2015. Cortical morphology in 6- to 10-year old children with autistic traits: a population-based neuroimaging study. Am J Phychiatry. doi:10.1176/appi.ajp.2014.14040482. [DOI] [PubMed]
- Bonifazi P, Goldin M, Picardo M, Jorquera I, Cattani A, Bianconi G, Represa A, Ben-Ari Y, Cossart R. 2009. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science. 326:1419–1424. [DOI] [PubMed] [Google Scholar]
- Braun U, Plichta M, Esslinger C, Sauer C, Haddad L, Grimm O, Mier D, Mohnke S, Heinz A, Erk S et al. . 2012. Test-retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage. 59:1404–1412. [DOI] [PubMed] [Google Scholar]
- Buckner R, Sepulcre J, Talukdar T, Krienen F, Liu H, Hedden T, Andrews-Hanna J, Sperling R, Johnson K. 2009. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 29:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 10:186–198. [DOI] [PubMed] [Google Scholar]
- Caeyenberghs K, Siugzdaite R, Drijkoningen D, Marinazzo D, Swinnen S. 2014. Functional connectivity density and balance in young patients with traumatic axonal injury. Brain Connect. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Carandini M, Shimaoka D, Rossi L, Sato T, Benucci A, Knöpfel T. 2015. Imaging the awake visual cortex with a genetically encoded voltage indicator. J Neurosci. 35:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai X, Castañón A, Ongür D, Whitfield-Gabrieli S. 2012. Anticorrelations in resting state networks without global signal regression. Neuroimage. 59:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux J. 1997. Neuronal man. New Jersey: Princeton University Press. [Google Scholar]
- Cohen A, Fair D, Dosenbach N, Miezin F, Dierker D, Van Essen D, Schlaggar B, Petersen S. 2008. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 41:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N, Mechelli A, Scott J, Carletti F, Fox P, McGuire P, Bullmore E. 2014. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 137:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, An D, Liao W, Wu G, Xu Q, Zhou D, Chen H. 2014. Abnormal functional connectivity density in psychogenic non-epileptic seizures. Epilepsy Res. 108:1184–1194. [DOI] [PubMed] [Google Scholar]
- Glasser M, Sotiropoulos S, Wilson J, Coalson T, Fischl B, Andersson J, Xu J, Jbabdi S, Webster M, Polimeni J et al. . 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Kurth F, Zhou J, Mayer E, Eickhoff S, Kramer J, Seeley W. 2012. One-year test-retest reliability of intrinsic connectivity network fMRI in older adults. Neuroimage. 61:1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. 2008. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci. 28:4756–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Margulies D, Castellanos F. 2007. Recent advances in structural and functional brain imaging studies of attention-deficit/hyperactivity disorder. Curr Psychiatry Rep. 9:401–407. [DOI] [PubMed] [Google Scholar]
- Konova A, Moeller S, Tomasi D, Goldstein R. 2015. Effects of chronic and acute stimulants on brain functional connectivity hubs. Brain Res. doi:10.1016/j.brainres.2015.1002.1002. [DOI] [PMC free article] [PubMed]
- Lang X, Liu H, Qin W, Zhang Y, Xuan Y, Yu C. 2015. Brain functional connectivity density and individual fluid reasoning capacity in healthy young adults. Neuroreport. 26:17–21. [DOI] [PubMed] [Google Scholar]
- Lei X, Wang Y, Yuan H, Mantini D. 2014. Neuronal oscillations and functional interactions between resting state networks. Hum Brain Mapp. 35:3517–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, van Zijl P, Thakor N, Pelled G. 2014. Study of the spatial correlation between neuronal activity and BOLD fMRI responses evoked by sensory and channelrhodopsin-2 stimulation in the rat somatosensory cortex. J Mol Neurosci. 53:553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fan L, Cui Y, Zhang X, Hou B, Li Y, Qin W, Wang D, Yu C, Jiang T. 2015. DISC1 Ser704Cys impacts thalamic-prefrontal connectivity. Brain Struct Funct. 220:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis N. 2002. The neural basis of the blood-oxygen-level-dependent functional magnetic resonance imaging signal. Philos Trans R Soc Lond B Biol Sci. 357:1003–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis N, Pauls J, Augath M, Trinath T, Oeltermann A. 2001. Neurophysiological investigation of the basis of the fMRI signal. Nature. 412:150–157. [DOI] [PubMed] [Google Scholar]
- Lu H, Wang L, Rea W, Brynildsen J, Jaime S, Zuo Y, Stein E, Yang Y. 2016. Low- but not high-frequency LFP correlates with spontaneous BOLD fluctuations in rat whisker barrel cortex. Cereb Cortex. 26:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon R. 2002. Postacquisition suppression of large-vessel BOLD signals in high-resolution fMRI. Magn Reson Med. 47:1–9. [DOI] [PubMed] [Google Scholar]
- Murphy K, Birn R, Handwerker D, Jones T, Bandettini P. 2009. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 44:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J, Barnes K, Snyder A, Schlaggar B, Petersen S. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Xuan Y, Liu Y, Jiang T, Yu C. 2015. Functional connectivity density in congenitally and late blind subjects. Cereb Cortex. 25:2507–2516. [DOI] [PubMed] [Google Scholar]
- Raichle M, Gusnard D. 2002. Appraising the brain's energy budget. Proc Natl Acad Sci USA. 99:10237–10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad Z, Gotts S, Murphy K, Chen G, Jo H, Martin A, Cox R. 2012. Trouble at rest: how correlation patterns and group differences become distorted after global signal regression. Brain Connect. 2:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi-Khorshidi G, Douaud G, Beckmann C, Glasser M, Griffanti L, Smith S. 2014. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 90:449–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölvinck M, Maier A, Ye F, Duyn J, Leopold D. 2010. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci USA. 107:10238–10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline Y, Raichle M. 2013. Resting state functional connectivity in preclinical Alzheimer's disease. Biol Psychiatry. 74:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A, Leopold D. 2008. Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: implications for functional connectivity at rest. Hum Brain Mapp. 29:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout P, Fleiss J. 1979. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 86:420–428. [DOI] [PubMed] [Google Scholar]
- Siero J, Hermes D, Hoogduin H, Luijten P, Ramsey N, Petridou N. 2014. BOLD matches neuronal activity at the mm scale: a combined 7T fMRI and ECoG study in human sensorimotor cortex. Neuroimage. 101:177–184. [DOI] [PubMed] [Google Scholar]
- Smith S, Beckmann C, Andersson J, Auerbach E, Bijsterbosch J, Douaud G, Duff E, Feinberg D, Griffanti L, Harms M et al. . 2013. Resting-state fMRI in the Human Connectome Project. Neuroimage. 80:144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam C. 2004. Functional connectivity patterns of human magnetoencephalographic recordings: a “small-world” network? Neurosci Lett. 355:25–28. [DOI] [PubMed] [Google Scholar]
- Supekar K, Musen M, Menon V. 2009. Development of large-scale functional brain networks in children. PLoS Biol. 7:e1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Qin W, Liu B, Jiang T, Yu C. 2013. Functional connectivity in healthy subjects is nonlinearly modulated by the COMT and DRD2 polymorphisms in a functional system-dependent manner. J Neurosci. 33:17519–17526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2010. Functional connectivity density mapping. Proc Natl Acad Sci USA. 107:9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2011a. Association between functional connectivity hubs and brain networks. Cereb Cortex. 21:2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2011b. Functional connectivity hubs in the human brain. Neuroimage. 57:908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2011c. Gender differences in brain functional connectivity density. Hum Brain Mapp. 33:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2011d. Laterality patterns of brain functional connectivity: gender effects. Cereb Cortex. 22:1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2012a. Abnormal functional connectivity in children with attention-deficit/hyperactivity disorder. Biol Psychiatry. 71:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2012b. Aging and functional brain networks. Mol Psychiatry. 17:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2014. Mapping small-world properties through development in the human brain: disruption in schizophrenia. PLoS ONE. 9:e96176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Wang G, Volkow N. 2013. Energetic cost of brain functional connectivity. Proc Natl Acad Sci USA. 110:13642–13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Wang R, Wang G, Volkow N. 2014. Functional connectivity and brain activation: a synergistic approach. Cereb Cortex. 24:2619–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. 2002. How much cortex can a vein drain? Downstream dilution of activation-related cerebral blood oxygenation changes. Neuroimage. 16:1062–1067. [DOI] [PubMed] [Google Scholar]
- Uğurbil K, Xu J, Auerbach E, Moeller S, Vu A, Duarte-Carvajalino J, Lenglet C, Wu X, Schmitter S, Van de Moortele P et al. . 2013. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage. 80:80–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Sporns O, Collin G, Scheewe T, Mandl R, Cahn W, Goñi J, Hulshoff Pol H, Kahn R. 2013. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry. 70:783–792. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Stam C, Boersma M, Hulshoff Pol H. 2008. Small-world and scale-free organization of voxel-based resting-state functional connectivity in the human brain. Neuroimage. 43:528–539. [DOI] [PubMed] [Google Scholar]
- Van Dijk K, Hedden T, Venkataraman A, Evans K, Lazar S, Buckner R. 2010. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 103:297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D, Ugurbil K, Auerbach E, Barch D, Behrens T, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss S et al. . 2012. The Human Connectome Project: a data acquisition perspective. Neuroimage. 62:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qin W, Liu B, Wang D, Zhang Y, Jiang T, Yu C. 2013. Variant in OXTR gene and functional connectivity of the hypothalamus in normal subjects. Neuroimage. 81:199–204. [DOI] [PubMed] [Google Scholar]
- Wang X, Jiao Y, Tang T, Wang H, Lu Z. 2013. Investigating univariate temporal patterns for intrinsic connectivity networks based on complexity and low-frequency oscillation: a test-retest reliability study. Neuroscience. 254:404–426. [DOI] [PubMed] [Google Scholar]
- Watts D, Strogatz S. 1998. Collective dynamics of “small-world” networks. Nature. 393:440–442. [DOI] [PubMed] [Google Scholar]
- Wong C, Olafsson V, Tal O, Liu T. 2012. Anti-correlated networks, global signal regression, and the effects of caffeine in resting-state functional MRI. Neuroimage. 63:356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zuo X, Wang P, Li Z, LaConte S, Bandettini P, Hu X. 2012. Generalized RAICAR: discover homogeneous subject (sub)groups by reproducibility of their intrinsic connectivity networks. Neuroimage. 63:403–414. [DOI] [PubMed] [Google Scholar]
- Zhuo C, Zhu J, Qin W, Qu H, Ma X, Tian H, Xu Q, Yu C. 2014. Functional connectivity density alterations in schizophrenia. Front Behav Neurosci. 8:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Kelly C, Adelstein J, Klein D, Castellanos F, Milham M. 2010. Reliable intrinsic connectivity networks: test-retest evaluation using ICA and dual regression approach. Neuroimage. 49:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.