Abstract
The dynamics of spontaneous fluctuations in neural activity are shaped by underlying patterns of anatomical connectivity. While numerous studies have demonstrated edge-wise correspondence between structural and functional connections, much less is known about how large-scale coherent functional network patterns emerge from the topology of structural networks. In the present study, we deploy a multivariate statistical technique, partial least squares, to investigate the association between spatially extended structural networks and functional networks. We find multiple statistically robust patterns, reflecting reliable combinations of structural and functional subnetworks that are optimally associated with one another. Importantly, these patterns generally do not show a one-to-one correspondence between structural and functional edges, but are instead distributed and heterogeneous, with many functional relationships arising from nonoverlapping sets of anatomical connections. We also find that structural connections between high-degree hubs are disproportionately represented, suggesting that these connections are particularly important in establishing coherent functional networks. Altogether, these results demonstrate that the network organization of the cerebral cortex supports the emergence of diverse functional network configurations that often diverge from the underlying anatomical substrate.
Keywords: connectome, multivariate, network, partial least squares
Introduction
Cognitive functions and complex behaviors are thought to emerge from the links and interactions among brain areas (McIntosh 2000). Recent advances in imaging and tracing of neuronal connections have resulted in the creation of comprehensive network maps (connectomes) of neural elements and their white matter connections (Hagmann et al. 2008; Iturria-Medina et al. 2008). The evolution of neural dynamics on top of this anatomical substrate gives rise to coherent electrophysiological, hemodynamic, and metabolic activity. A concurrent literature emphasizing the temporal correlations between remote neural elements has revealed highly organized global coupling patterns, termed functional connectivity (FC) (Friston 1994; Fox et al. 2005; Power et al. 2011).
Although functional interactions are shaped and constrained by the underlying anatomy, the precise nature of the relationship between structure and function remains a fundamental question (Honey et al. 2010; Park and Friston 2013). Several studies have demonstrated that the presence and/or strength of a structural connection between two brain areas predicts the strength of the functional connection between those areas (Hagmann et al. 2008; Skudlarski et al. 2008; Honey et al. 2009; Shen et al. 2012; Hermundstad et al. 2014), and vice versa (Hermundstad et al. 2014; Deco et al. 2014). Similarly, the structural and functional connectivity profiles of brain areas are often correlated, such that central, well-connected structural hubs (Hagmann et al. 2008; Gong et al. 2009) are also likely to play central roles in shaping functional connectivity (Buckner et al. 2009; Tomasi and Volkow 2010; Shen et al. 2012).
Despite the link between structural and functional connectivity at the level of individual connections or nodes, it remains unknown whether network-level interactions among neural elements may give rise to global functional patterns. In other words, can a network of structural connections predict the strength of a functional network? A consistent finding in computational modeling is that biophysical dynamical systems (e.g., neural masses) coupled using anatomically realistic connectivity patterns can simulate functional connectivity patterns that are more similar to empirically observed functional connectivity than static structural connectivity (SC) (Ghosh et al. 2008; Deco et al. 2009; Adachi et al. 2012; Haimovici et al. 2013; Goni et al. 2014; Messé et al. 2014; Hansen et al. 2015; Mišić et al. 2015; Ponce-Alvarez et al. 2015; Stam et al. 2015). This theoretical work suggests that coordinated coupling between multiple brain areas may lead to indirect, network-level effects, whereby a particular structural configuration collectively supports a different, nonoverlapping functional configuration, in addition to the “one-to-one” mapping between individual structural and functional connections.
Here, we use a multivariate statistical technique (partial least squares; PLS; McIntosh et al. 1996; McIntosh and Lobaugh 2004) to investigate the relationship between structural and functional networks, derived from diffusion-weighted imaging (DWI) and functional magnetic resonance imaging (fMRI). We simultaneously search for structural and functional patterns that optimally covary with each other and empirically test the hypothesis that the integrated structural connectome gives rise to network-wide associations, whereby structural networks support divergent, nonoverlapping functional network configurations. In addition, we investigate how network-level structure-function relationships determine the interaction among resting-state networks (RSNs; Supplementary Fig. 1) (Damoiseaux et al. 2006; Smith et al. 2009; Yeo et al. 2011; Power et al. 2011), and how the emergence of these relationships depends on the topological properties of individual nodes and connections.
Materials and Methods
Data Acquisition
Data were collected as part of the Human Connectome Project, a Washington University—Minnesota Consortium (Van Essen et al. 2013). Healthy participants were recruited from the Washington University (St. Louis, MO) area. All participants provided informed consent. Data used were from third quarter (Q3) release, comprising a total of N = 156 participants.
Functional Connectivity (FC)
Whole-brain echo-planar imaging was acquired using a 32-channel head coil on a modified 3T Siemens Skyra scanner, with the following parameters: time to repetition = 720 ms, time to echo = 33.1 ms, flip angle = 52o, bandwidth = 2290 Hz/pixel, in-plane field of view = 208 × 180 mm, 72 slices, 2 mm isotropic voxels, and multiband acceleration factor = 8 (Ugurbil et al. 2013; Smith et al. 2013). FMRI data were collected for 28 min during rest, with eyes open. The first 13 volumes (corresponding to 10 s) were removed to allow signal stabilization. Preprocessing included motion correction, regression of white matter, cerebrospinal fluid and movement signals, linear detrending, motion scrubbing, and low-pass filtering (Power et al. 2012).
Structural Connectivity (SC)
The cortex was parcellated into 114 distinct regions using a subdivision of FreeSurfer's Desikan–Killiany atlas (Cammoun et al. 2012). White matter edges were reconstructed from each participant's diffusion-weighted MR images using deterministic streamline tractography. All streamlines were initiated in white matter voxels. If a streamline intersected more than two parcels, it was interpreted as comprising multiple connections. Crossing fibers were modeled using generalized q-sampling imaging (GQI; Yeh et al. 2010). The resulting matrices were not thresholded, thus preserving their original density (mean across subjects = 15.24%, standard deviation = 0.86%). For more details regarding the tractography procedure, we refer the reader to van den Heuvel, Scholtens, Feldman Barrett, et al. (2015); van den Heuvel, Scholtens, de Reus, et al. (2015).
PLS Analysis
PLS is a multivariate statistical technique used to relate two sets of variables to each other (Wold 1966; McIntosh and Lobaugh 2004; Abdi 2010; Krishnan et al. 2011; McIntosh and Mišić 2013). The goal of the analysis is to simultaneously find linear combinations of variables in each block that maximally covary with each other. In the present analysis, this corresponds to a weighted combination of structural connections (interpreted as a structural network) and a weighted combination of functional connections (interpreted as a functional network).
Singular Value Decomposition
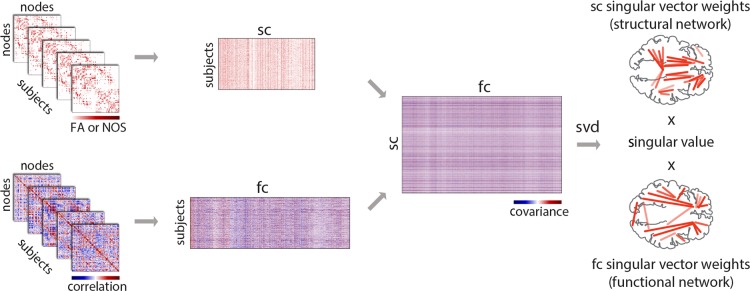
Group structural and functional data were organized in two separate data matrices, X and Y, respectively (Fig. 1). Unique connections (corresponding to the elements of the upper triangle of the adjacency matrix) were extracted from each individual participant's connectivity matrix and stacked on top of each other. Given n = 114 nodes, this corresponds to k = 114 × (114−1)/2 = 6441 unique connections. Thus, X and Y had N = 156 rows and k = 6441 columns. Due to the sparse nature of structural connectivity, several connections are not observed in tractography-reconstructed data for any individual, resulting in several all-zero columns in matrix X. These columns were removed from further analysis, such that X had N = 156 rows and k = 3896 columns. To ensure that the differences in structural and functional connection magnitudes do not dominate the statistical model, the structural and functional data matrices were z-scored columnwise, by subtracting the mean from each column and dividing by the standard deviation of that column. Thus, in the present analyses the covariance matrix was effectively converted to a correlation matrix prior to decomposition.
Figure 1.
PLS analysis. SC and FC data matrices were organized by stacking the upper triangle elements from individual participants' matrices to form two new matrices. The rows of these matrices correspond to participants, and columns correspond to either structural or functional connections. Structural connections that were zero-valued for all participants were removed, and the remaining data were z-scored columnwise. The covariance between structural and functional connections was computed across participants, resulting in a rectangular SC–FC covariance matrix. The SC–FC covariance matrix was then subjected to singular value decomposition (SVD), resulting in a vector of weights for all structural connections (interpreted as a structural network), a vector of weights for all functional connections (interpreted as a functional network) that optimally covary with each other. Each SC–FC pattern was associated with a scalar singular value, which indicates the proportion of SC–FC covariance accounted for by the extracted structural and functional networks.
The structure-function covariance matrix is then computed, representing the covariation of all structural and functional connections across participants. The resulting covariance matrix, with ksc = 3956 rows (due to the removal of zero-valued connections) and kfc = 6441 columns, is then subjected to singular value decomposition (SVD; Eckart and Young 1936):
such that
The outcome of the analysis is a set of mutually orthogonal latent variables (LVs), where U and V are matrices of left and right singular vectors, and Δ is a diagonal matrix with singular values along the diagonal. The ith LV is a triplet of the ith left and right singular vectors and the ith singular value. The number of LVs is equal to the rank of the covariance matrix , which is the smaller of its dimensions or the dimensions of its constituent matrices. In the present study, the number of participants is smaller than the number of structural or functional connections (N << k), so the rank of can at most be equal to N. Accordingly, the dimensions of U are ksc × N, V is kfc × N and Δ is N × N.
Singular vectors weigh the contribution of individual variables (i.e., structural and functional connections) to the overall multivariate pattern. Thus, the column vectors of U and V weigh the structural and functional connections such that they maximally covary with each other, and can be interpreted as an optimal combination of structural and functional networks. Each such structural–functional network combination is associated with a scalar singular value from the diagonal matrix Δ, which reflects the covariance between the original structural and functional connectivity that is captured by the LV. The effect size (proportion of cross-block covariance) of a LV can be estimated as the ratio of the squared singular value to the sum of squared singular values from the decomposition.
Significance of SC–FC Patterns
We assessed the statistical significance of each LV using permutation tests. Permutation tests were performed by randomly reordering the rows of the original structural data matrix X, generating a set of permuted data matrices where the original ordering of individual participants had been destroyed. Structure-function covariance matrices were then computed for the permuted structural and nonpermuted functional data matrices, and subjected to SVD as described above. This procedure generated a distribution of singular values under the null hypothesis that there is no relation between structure and function. Given that singular values are proportional to the magnitude of a statistical relationship captured by a LV, a P-value for a LV was estimated as the proportion of times the permuted singular values exceeded the original singular value.
Contribution and Reliability of Individual Connections
The contribution and reliability of individual connections was estimated by bootstrap resampling, which allows patterns derived from the analysis to be cross-validated. Bootstrapping was performed by randomly resampling participants with replacement (i.e., the rows of data matrices X and Y). The resampled data matrices were subjected to SVD as described above, to generate a sampling distribution for each of the weights in the singular vectors. The bootstrap distribution was used to estimate the standard error for each weight, which reflects the stability of the weight. Finally, a bootstrap ratio was calculated for each structural and functional connection by dividing the weight from the singular vector by its bootstrap-estimated standard error.
Thus, a bootstrap ratio with a large magnitude indicates that the connection with which it is associated has both a large singular vector weight (i.e., contributes to the LV) and a small standard error (i.e., is stable across participants). Thus, bootstrapping serves as a form of cross-validation, allowing us to select connections that are stable across participants. When the bootstrap distribution is approximately normal, the bootstrap ratio is equivalent to a z-score (Efron and Tibshirani 1986). Bootstrap ratio maps were thresholded at values corresponding to the 95% confidence interval. Connections were included only if their bootstrap-estimated 95% confidence interval did not include zero. Finally, it is important to note that the bootstrap procedure was not used for hypothesis testing, eschewing the need to correct for multiple comparisons. Rather, hypothesis testing was done using the permutation procedure described above, at the level of the entire multivariate pattern.
Matching Decompositions From Original and Resampled Matrices
The decompositions of both permuted and bootstrap-resampled data matrices are not guaranteed to produce a set of LVs comparable with the ones derived from the original data matrix, because both types of resampling could induce arbitrary axis rotation (a change in the order of LVs) or axis reflection (a sign change for the weights). As a result, Procrustes rotations are used to match the resampled LVs to the original LVs (McIntosh and Lobaugh 2004).
Graph Theoretic Measures
All graph theoretic metrics and analyses were performed using the Brain Connectivity Toolbox (https://sites.google.com/site/bctnet/), including degree/strength, modularity, and the rich club coefficient (Rubinov and Sporns 2010).
Community Detection
Functional network communities (RSNs) were derived using a modularity maximization procedure (Newman and Girvan 2004; Rubinov and Sporns 2011; Sporns and Betzel 2016). The goal of the procedure was to find a partition that maximizes the quality function
where is the connectivity matrix containing only the positive correlations, while contains only correlations less than zero. The term denotes the expected density of connections between nodes i and j given a particular null model, where and . The variable represents the community assignment of node i, and is the Kronecker function and is equal to 1 when and 0 otherwise. The resolution parameter scales the relative importance of the null model, potentiating the discovery of either larger or smaller communities.
The procedure was performed across a range of resolution parameters, from γ = 0.5 to 2.5 in increments of 0.1. At each resolution parameter (corresponding to a particular scale), we ran the Louvain algorithm 250 times (Blondel et al. 2008) to identify partitions that produced large Q values. For each resolution parameter, we quantified the similarity of all pairs of community assignments as the z-score of the Rand index, and selected the parameter (γ = 1.8) with the greatest average similarity (Traud et al. 2011). Rather than treat any of the 250 individual partitions as representative, we derived a consensus partition using the method presented in Bassett et al. (2013). The final consensus partition had a modularity score of Q(γ) = 0.48 and contained Nc = 6 communities which was visually compared and matched with the topographical profiles of previously reported RSNs (Power et al. 2011; Yeo et al. 2011), including the ventral attention (VA), frontoparietal network (FPN), default mode network (DMN), salience (SAL), somato-motor (SM), and visual (VIS) networks (Supplementary Fig. 1).
Rich Club Detection
A rich club is defined as a subgraph of high-degree nodes that are densely interconnected among each other, above and beyond what would be expected on the basis of their degrees alone (Colizza et al. 2006). Typically, rich club detection is performed across a range of degrees k, selecting the subset of nodes >k and comparing the density of the subgraph with a null model. In the present study, we sought to detect the presence of a rich club for a population of n = 156 participants. Due to inter-participant variance in density and the presence of individual connections, we did not perform the analysis for a group-averaged connectivity matrix. Rather, we used a method for rich club detection that operates at the group level, pooling together subgraphs for the entire population to create an ensemble subgraph.
Rich club detection was performed across a range of degrees. Each individual matrix was first binarized, and for each degree k, all nodes with degree ≤k were removed from the network. The number of edges and nodes in the remaining subgraph was recorded, and the procedure was repeated for all individual participant matrices. For each k, we calculated a pooled rich club coefficient φ(k) as the ratio of all remaining connections to all possible connections, representing the density of the ensemble subgraph. This procedure was simultaneously performed for a null distribution of 1000 randomized networks with preserved degree sequences (Maslov and Sneppen 2002). The resulting null distribution of rich club coefficients was used to normalize the empirical rich club coefficient φnorm(k) = φ(k)/φrandom(k).
This procedure was repeated for a range of k. A φnorm(k) that is consistently >1 over a range of k suggests the presence of rich club organization. In the present study, we observed consistent, statistically significant φnorm(k) > 1 for k ≥ 27, resulting in a rich club with 14 nodes, including the following bilateral regions: medial prefrontal cortex, superior frontal gyrus, superior parietal cortex, insula, superior temporal gyrus, and precuneus. Once nodes are labeled as belonging to the rich club or not, edges can be labeled in a similar way. Using the nomenclature described by van den Heuvel et al. (2012), we labeled edges connecting non-rich club nodes to non-rich club nodes as “local”, edges connecting non-rich club nodes to rich club nodes as “feeder” and edges connecting rich club nodes to rich club nodes as “rich club”.
Results
We investigated multivariate relationships between structural and functional connectivity using PLS analysis (McIntosh et al. 1996; McIntosh and Lobaugh 2004; Abdi 2010; Krishnan et al. 2011; McIntosh and Mišić 2013), a multivariate statistical technique that relates one set of variables (in this case, structural connections) to another (functional connections) by decomposing the covariance between them and assigning weights to each connection (Fig. 1). Structural and functional data were derived from the Human Connectome Project (Van Essen et al. 2013), with n = 156 healthy participants. SC was estimated in terms of FA, while FC was estimated from 28 min-long resting-state multiband fMRI scans in the same participants. The analyses were subsequently repeated for SC estimated in terms of the number of streamlines (NOS; see Sensitivity to diffusion imaging, below).
The analysis revealed a set of 156 components or LVs(Fig. 2), each of which represents a particular weighted pattern of structural and functional connections that optimally covary with each other. These weighted combinations are naturally interpreted as structural and functional networks, respectively. The number of LVs (156) corresponds to the rank of the SC–FC covariance matrix (in the present case, the number of participants; see Materials and Methods). We selected latent variables for further analysis based on 3 criteria. First, latent variables must be statistically significant by permutation test (see Materials and Methods; Fig. 2, orange). Second, latent variables must account for at least 1/156th of cross-block covariance, corresponding to covariance accounted for by the average single latent variable (also known as the Kaiser criterion; Fig. 2, blue). Third, each latent variable must account for a substantially larger proportion of covariance than the subsequent one, also known as Cattell's scree test (Cattell 1966). The scree test is a heuristic that identifies the most important LVs based on the shape of the singular value curve. If there is a change in the slope of the curve, from a steep slope associated with the larger LVs to a shallow slope associated with the smaller LVs, the criterion suggests to retain only the LVs before the change in slope. In the present analysis, the change in effect size between successive LVs was most pronounced at the 5th LV (change between 1st–2nd, 2nd–3rd, 3rd–4th, 4th–5th, 5th–6th = 2.67, 2.06, 0.42, 0.39, 0.48; change between 6th–7th, 7th–8th, 8th–9th, 9th–10th, 10th–11th = 0.12, 0.09, 0.11, 0.15, 0.07).
Figure 2.
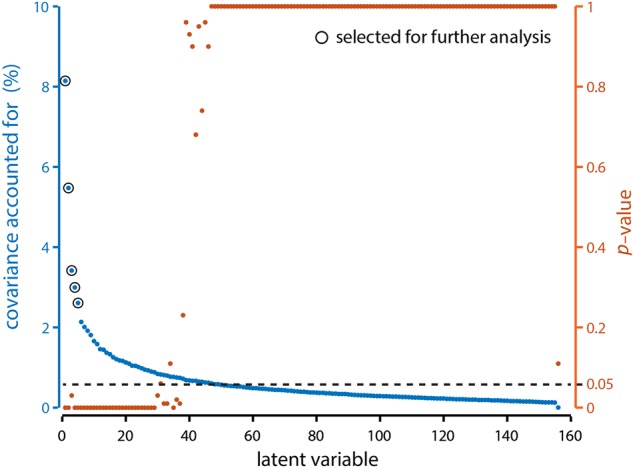
Latent variable magnitudes. The PLS analysis produced k = 156 LVs, each of which represents a particular SC–FC relationship, and is associated with a singular value. The squared magnitude of each singular value is proportional to the strength of the SC–FC relationship. (Blue) percent covariance (i.e., effect size) accounted for by each LV. (Orange) P-value associated with each LV, as determined by permutation tests.
Divergent SC–FC Patterns
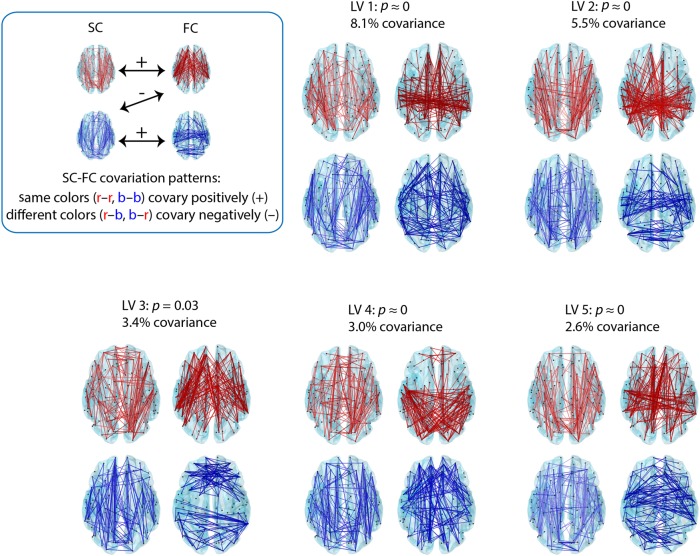
We first explore the topographic arrangement of the 5 LVs (LVs 1–5) that were selected for further analysis based on the criteria described above (Fig. 3). The 5 LVs were statistically significant by permutation test (P = 0.03 for LV3 and P < 10−5 for all others) and accounted for 8.1, 5.5, 3.4, 3.0, and 2.6% of covariance between structure (DWI) and function (fMRI). Each connection represents a bootstrap ratio: the weight of a particular connection, divided by its bootstrap-estimated standard error. The most salient feature of these patterns is that they generally do not show a simple one-to-one correspondence between structural and functional edges. Rather, the patterns are rich and heterogeneous, rarely conforming or perfectly matching, suggesting that structural connections have the capacity to support and influence remote functional connections (Fig. 3). To quantify this lack of correspondence, we correlated the structural and functional weights for each LV, showing generally poor and insignificant correlations between the 2 (r = 0.01, 0.008, −0.025, 0.021, 0.0091 and P = 0.41, 0.53, 0.04, 0.08, 0.48 for LVs 1–5). Supplementary Figures 2–6 provide more detail regarding the specific structural and functional connections involved in these networks by showing the weighing of these connections between all pairs of ROIs.
Figure 3.
Spatial configuration of optimally covarying SC–FC networks. PLS analysis revealed 5 statistically significant SC–FC patterns. Statistical significance is assessed using permutation tests, and effect size is estimated from the magnitude of the singular value associated with each pattern (see Materials and Methods). Each LV is comprised of a weighted pattern of structural (left) and functional (right) connections. Bootstrap resampling is used to construct a 95% confidence interval for each connection weight; the top 5% most reliable connections are shown. Connections with positive weights are shown in red and connections with negative weights are shown in blue. SC–FC patterns with the same color covary positively with each other, while SC–FC with different colors covary negatively.
At the same time, the patterns recapitulate several previously reported SC–FC relationships. For instance, the first LV (LV1) prominently captures functional connectivity within the DMN, including connectivity between medial prefrontal cortex and posterior cingulate, as well as between lateral parietal cortices (see also Fig. 4). The corresponding structural pattern shows that this functional configuration strongly covaries with the anatomical projections between medial prefrontal cortex and the posterior cingulate, but does not find any covarying projections between posterior cingulate and lateral parietal cortices. This is consistent with previous reports, which have shown that DMN FC is supported by projections between medial frontal cortex and medial and lateral parietal cortices, yet have repeatedly failed to find evidence of anatomical projections between medial parietal cortex (precuneus and posterior cingulate) and lateral parietal cortices cortices (Greicius et al. 2009; Honey et al. 2009; van den Heuvel et al. 2009).
Figure 4.
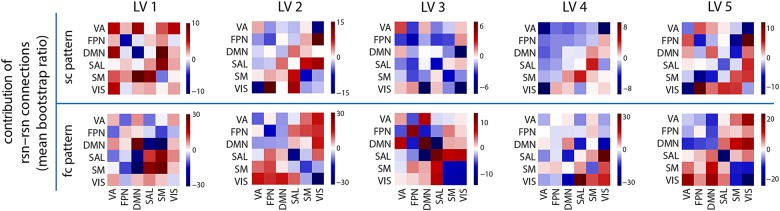
RSN relationships. The 5 SC–FC patterns revealed by PLS are stratified by their RSN membership. The RSNs are abbreviated as follows: ventral attention (VA), frontoparietal network (FPN), default mode network (DMN), salience (SAL), somato-motor (SM), and visual (VIS). The mean contribution (bootstrap ratio) is computed for all within- and between-RSN connections.
Anatomical Basis of RSN Connectivity
We next investigate patterns of connectivity within and between functional communities (RSNs; Supplementary Fig. 1), and relate these to anatomical connectivity patterns. RSNs are communities of brain regions that, at rest, display greater functional connectivity with each other than with other regions, yet are often anatomically separated. They have a distinct functional profile and are thought to support specific cognitive functions (Damoiseaux et al. 2006; Smith et al. 2009; Yeo et al. 2011; Power et al. 2011).
Figure 4 shows, for each LV, the mean connectivity within and between RSNs (top of each panel), as well as the most reliable individual connections (bottom of each panel). Stratifying connections by their RSN membership again displays the considerable divergence in multivariate SC–FC patterns. In many cases, FC within an RSN is not exclusively supported by SC within the same RSN. Rather, functional connections within and between RSNs are often associated with structural connections within and between other RSNs.
LV2 shows that structural connectivity between the visual and FPN is associated with a considerably broader set of functional interactions, encompassing not just visual-frontoparietal interactions, but also visual-default mode, visual-salience, and visual–VA interactions. This suggests that the anatomical projections between the visual and FPN act as a conduit to the rest of the brain, facilitating communication between the relatively peripheral visual network and other networks, including the default mode, salience and VA networks. LV 4 shows a related effect, whereby the anatomical connections between the visual and VA networks support functional interactions between the visual and DMNs (Fig. 4, blue connections). This statistical effect again highlights the integrative role of polysensory association networks, in this case facilitating communication between the visual system and the rest of the brain.
LV3 captures the emergence of bilateral frontal and lateral occipito-parietal functional clusters, encompassing lateral and superior temporal cortices and anterior cingulate, as well as superior and inferior parietal cortices, cuneus and precuneus, pericalcarine, and inferior temporal cortices. The analysis indicates that these functional patterns are supported by frontoparietal, fronto-temporal, and fronto-occipital structural connections, but interestingly, despite common covariation in the frontal and parietal functional connections (Fig. 3, blue connections), there are no statistically reliable functional connections between these frontal and posterior clusters. This demonstrates that the presence of a structural connection does not always predict the corresponding functional connection. In addition, LV3 shows that much of the within-module functional connectivity for large, polysensory association networks such as default mode, salience and FPN occurs in the absence of direct anatomical projections (Figs 3 and 4, red connections).
Finally, LV5 shows that occipito-temporal and temporo-parietal anatomical connections are associated with long-distance, bilateral temporal functional interactions, particularly in association cortex, including bilateral transverse temporal and superior temporal gyri. This LV suggests that temporal homotopic functional connectivity and synchrony may at least in part be mediated by indirect projections (for example, the superior longitudinal fasciculus), in addition to direct rostral and midsaggital callosal projections that interconnect the superior temporal lobes (Cipolloni and Pandya 1985). We also note that, since the present parcellation and subsequent fiber tracking do not include subcortical structures and subcortical projection fibers, such as those from the medial geniculate to the transverse temporal gyrus, their influence on interhemispheric communication cannot be assessed.
High-degree Hubs Shape SC–FC Relationships
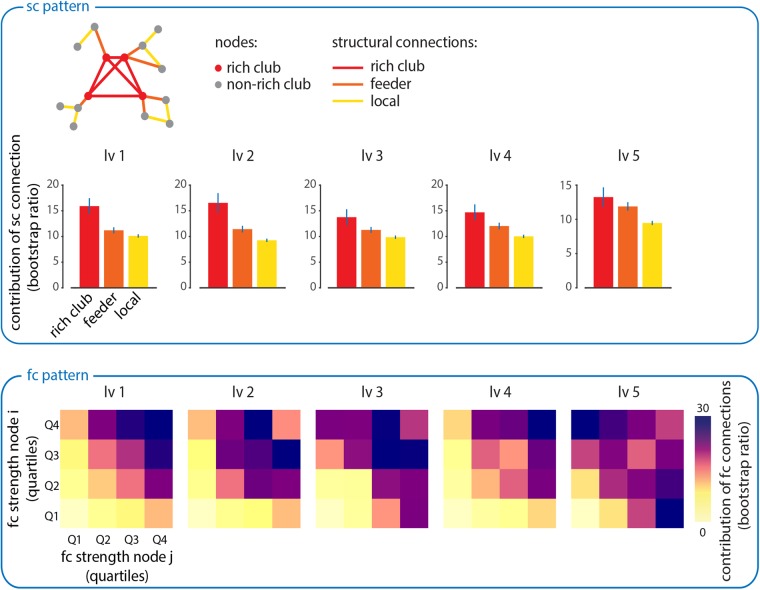
Finally, we investigate the contribution of high-degree nodes (hubs) to the establishment of structure-function relationships. Due to their dense connectivity, hubs are hypothesized to act as bridges for signal traffic, facilitating the spread and integration of information in brain networks (Sporns et al. 2007; Zamora-Lopez et al. 2010; van den Heuvel and Sporns 2013). This suggests that hubs should be disproportionately important in mediating the emergence of large-scale functional networks. To investigate the role of high-degree hubs on structural networks extracted using PLS, we first perform a rich club analysis (see Materials and Methods; van den Heuvel and Sporns 2011; van den Heuvel et al. 2012). The rich club is a central collective of densely interconnected high-degree hubs. Once nodes have been classified as belonging to the rich club or not, the edges between them can also be stratified into distinct classes (Fig. 5): connections between two rich club nodes (“rich club”), connections between rich club nodes and non-rich club nodes (“feeder”) and connections between two non-rich club nodes (“local”).
Figure 5.
Hubs shape structure-function relationships. Top: mean reliability for each type of structural connection. Structural connections are stratified according to whether they connect rich club nodes to each other (“rich club”), rich club nodes to non-rich club nodes (“feeder”) or non-rich club nodes to other non-rich club nodes (“local”). This classification system is illustrated in the accompanying schematic. Bottom: all functional connections are plotted as a function of the strength of the participating brain regions (strength is calculated as the mean absolute functional connectivity of an individual region). The strengths of participating nodes are stratified into quartiles and the contribution of all connections in those quartiles is averaged. Each quartile is colored according to how reliably it contributes to the FC pattern derived from the PLS analysis.
For each PLS-derived structural pattern, we calculate the mean contribution of each class of structural connections (rich club, feeder, local). Figure 5 shows that structural connections are significantly more involved in network-level SC–FC associations if they connect nodes that are members of the rich club of hubs (“rich club” connections, Student's t-test, P < 10−5), compared with connections that involve only one rich club node (“feeder” connections) or those that involve no rich club nodes (“local” connections; van den Heuvel et al. 2012).
A similar effect is observed for the corresponding functional patterns (Fig. 5, bottom). For each functional connection, we compare the strength (mean absolute functional connectivity) of the participating nodes with the contribution that the connection makes to the functional network extracted by PLS. The nodes participating in each functional connection were stratified into quartiles to facilitate interpretation. We find that connections between high-strength functional hubs make a greater-than-expected contribution to the functional networks captured by each of the 5 LVs (Fig. 5, bottom). For instance, functional connections between nodes with strengths in the top quartile (top right-hand corner in each plot) make the greatest average contribution to each of the 5 functional patterns. These results suggest that highly connected brain regions are disproportionately involved in establishing network-wide communication, acting as the focal points in large-scale structural and functional networks.
Sensitivity to Diffusion Imaging
The estimation of SC from diffusion imaging entails several important methodological choices and limitations. To determine the extent to which the present results depend on how SC is estimated, we performed 2 additional control analyses. First, to test how sensitive the results are to whether SC is estimated in terms of FA (reported thus far) versus NOS, we compared SC–FC patterns from both types of data. Without applying any post hoc rotation, we correlated the weighted patterns derived from the PLS analyses (Supplementary Fig. 7). All correlations were statistically significant, for both SC patterns (r = 0.68, 0.64, 0.69, 0.68 and 0.61 for LV1–5, P < 10−5, Bonferroni corrected) and FC patterns (r = 0.99, 0.95, 0.97, 0.96 and 0.86 for LV1-5, P < 10−5, Bonferroni corrected). This suggests that the global SC–FC patterns derived from PLS are stable for both FA- and NOS-defined SC.
Second, we sought to determine the extent to which the present results depend on the ability of diffusion imaging to capture interhemispheric connections. We repeated the PLS analysis relating SC and FC, but only focused on the connections within a single hemisphere (right hemisphere). We then correlated the patterns derived from the whole-brain and single-hemisphere analyses (Supplementary Fig. 8). All correlations were statistically significant, for both SC patterns (r = 0.95, 0.89, 0.77, 0.57 and 0.42 for LV1–5, P < 10−5, Bonferroni corrected) and FC patterns (r = 0.94, 0.90, 0.80, 0.58 and 0.37 for LV1-5, P < 10−5, Bonferroni corrected), though there was a large variation in the magnitude of the correlation. This indicates that PLS is sensitive to interhemispheric projections, but that their inclusion does not dominate the resulting patterns.
Discussion
In the present report, we provide evidence for network-level associations between brain structure and function. Using a versatile multivariate framework, we demonstrate systematic, statistically significant relationships between structural and functional network patterns. Importantly, we find that FC patterns generally do not conform to SC patterns. Rather, functional networks often diverge from the topology of structural networks, likely as a result of complex network-wide interactions.
These results speak to a core feature of brain networks: while the presence of an individual structural connection partially predicts the magnitude of the corresponding functional connection, the reverse is not necessarily true (Koch et al. 2002; Skudlarski et al. 2008; Honey et al. 2009; Shen et al. 2012; Hermundstad et al. 2014, but see also Hermundstad et al. 2013). We find that that a structural network can predict a functional network, but that the two do not necessarily overlap. This suggests that, in the context of a network, structural connections may influence downstream functional connections, consistent with the notion that coordinated functional interactions may produce statistical associations (functional connectivity) that diverge from the anatomical substrate. For instance, computational studies have investigated the emergence of functional networks from structural networks by modeling how the biophysical dynamics of individual brain regions (using neural masses, for instance) interact when coupled by anatomically realistic connectivity patterns. These studies suggest that through indirect, network-level interactions, the stable anatomical network can support a wide range of functional network configurations (Honey et al. 2007; Ghosh et al. 2008; Deco et al. 2009; Adachi et al. 2012; Goni et al. 2014; Messé et al. 2015; Hansen et al. 2015; Mišić et al. 2015; Ponce-Alvarez et al. 2015; Stam et al. 2015). As a result, functional configurations need not necessarily match the underlying structural network. The present study provides empirical support for the predictions generated by dynamic models and further highlights the utility of such models for understanding the emergence of large-scale functional network configurations.
A salient example of diverging SC–FC patterns are the RSNs or intrinsic connectivity networks. These networks of brain areas display coherent functional connectivity, yet are often found to be anatomically distributed (Damoiseaux et al. 2006; Smith et al. 2009; Yeo et al. 2011; Power et al. 2011). Although many of the RSNs are known to have an anatomical basis (van den Heuvel et al. 2009), including several subcomponents of the DMN (Greicius et al. 2009), the anatomical basis for the prominent interactions among RSNs displayed over time is unknown (de Pasquale et al. 2012). Network analyses have so far failed to demonstrate a clear one-to-one correspondence between network communities in FC (corresponding to RSNs) and communities in the underlying SC. Indeed, several studies using theoretical models have raised the possibility that FC within an RSN may be supported by SC outside that RSN (Haimovici et al. 2013; Hansen et al. 2015; Ponce-Alvarez et al. 2015). The present report provides a comprehensive analysis of connectivity within and between RSNs, demonstrating that these interactions are often influenced by remote anatomical projections.
Despite the topographic divergence between structural and functional network configurations, our results suggest that high-degree hub regions are disproportionately involved in mediating the emergence of large-scale interactions. In particular, high-degree structural and functional hubs appear to be central in the PLS-derived network patterns. This result provides further evidence that, by virtue of their high connectivity, hub regions integrate information and facilitate communication among multiple brain regions (Zamora-Lopez et al. 2010; van den Heuvel and Sporns 2011; van den Heuvel et al. 2012; Mišić et al. 2014). As a result, the formation of global network patterns naturally revolves around hub regions, which serve to promote synchronization among distributed areas.
The idea that a structural connection between two brain areas has the capacity to influence the expression of a functional connection between two different areas is closely related to the concept of effective connectivity (Friston 1994; McIntosh and Gonzalez-Lima 1994; McIntosh 2012). Namely, the apparent statistical dependencies between two brain areas may be subject to indirect, intervening influences from other areas due to their network embedding. Although PLS cannot be used to infer causal influences mediated by individual connections, what the present method does show is the collective effect of these causal influences: a structural network may support a functional network with a different configuration.
We also note that the structural and functional patterns reported in the present study encompass multiple systems. These multivariate patterns represent an optimal mapping between individual variation in structure and function: individuals who have a particular structural pattern also tend to express a particular functional pattern. Each LV is a statistical model, showing how increased integrity or thickness in a single or multiple structural edges tends to result in greater functional connectivity in a particular network of areas. In other words, all connections may mutually influence one another, and importantly, there are multiple pathways or patterns through which these influences can be exerted. Thus, these patterns represent particular modes that embody both the physical constraints imposed by the brain's anatomy, as well as the tendency for functional coactivation among distributed areas, and therefore naturally involve multiple cognitive systems.
Altogether, the present multivariate approach is part of an emerging effort to construct models that simultaneously take into account multiple modalities (Crossley et al. 2016; Mišić and Sporns, 2016). Correlative or relational techniques, such as PLS, have been used to study the relationship between white matter integrity and task-related BOLD signal (Burzynska et al. 2013), as well as the relationship between structural covariance and functional connectivity in the face processing network (Shaw et al. 2016). A mathematically related technique, canonical correlation analysis, also seeks to relate two sets of variables to one another, but in contrast to PLS, corrects for within-set correlations prior to the decomposition (McIntosh and Mišić 2013). For instance, a recent study used canonical correlation in the same data set to relate functional connectivity patterns to cognitive-behavioral profiles (Smith et al. 2015). The increasing depth and availability of imaging and phenotypic data will further drive the development and application of multivariate techniques capable of modeling complex relationships between multiple modalities.
Methodological Considerations
The present results are subject to several important limitations regarding the estimation of structural and functional connectivity. At present, computational tractography based on diffusion imaging is the leading technique for reconstructing human anatomical connectivity patterns in vivo. However, diffusion-based tractography has also been shown to be susceptible to both false positives and false negatives (Jones et al. 2013; Thomas et al. 2014). Thus, the structural patterns captured by the PLS analysis should be interpreted with caution. The limitations of computational tractography highlight the need for more.
It is important to note that the networks derived from PLS may not have the same statistical and topological properties as networks defined using other methods, notably correlations among time series. Functional connections identified by PLS collectively covary, and in this sense they embody the statistical association that is the principal hallmark of functional connectivity. However, PLS-derived patterns are fully connected, weighted networks, where the weight of each connection is determined by its contribution to a linear combination across all functional connectivity strengths. Unlike correlation-based functional brain networks, PLS networks are not guaranteed to be positive semi-definite and are unlikely to show properties that are inherent to networks mapped with pairwise correlations, such as increased transitivity (Zalesky et al. 2012), closed triangle motifs (Sporns & Kotter 2004), and local clustering (Sporns & Zwi 2004). As such, networks derived from PLS weights should not be confused with correlation-based networks. This is an important consideration if subsequent analyses are performed on the PLS networks, particularly with respect to the choice of null model.
We also note that FC was estimated over the entire recording epoch, which precludes the study of temporal dynamics. There is an emerging literature emphasizing the dynamic properties of functional connections on faster time scales (Allen et al. 2012; Tagliazucchi et al. 2012; Hutchison et al. 2013; Gonzalez-Castillo et al. 2014; 2015; Shen et al. 2015), and the dependence of these fluctuations on anatomical connectivity is an important future question. The present analysis was not configured to capture such fluctuations in FC, but in principle, this PLS-based framework could be readily adapted to include multiple functional configurations (e.g., by treating each one as a separate variable and concatenating them together in a single FC matrix prior to the analysis; Fig. 1).
Conclusion
As the focus of neuroscience shifts towards anatomical and functional relations between brain regions, there is a growing recognition that cognitive operations and complex behaviors emerge from the network interactions. Using a novel application of multivariate PLS analysis, we demonstrate that the structure-function relationship in the cerebral cortex transcends one-to-one correspondences and is present at the network level. More generally, the present results showcase the utility and flexibility of a multivariate statistical framework for addressing questions related to large-scale, multimodal datasets. By examining the effect of experimental manipulations on all variables (e.g., connections) simultaneously, multivariate models naturally embrace the notion of coherent networks and can be readily extended to other data types and empirical questions, including brain–behavior relationships, multimodal relationships (e.g., electrophysiological and hemodynamic neural activity), and brain–gene relationships.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
B.M. was supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (NSERC PDF). O.S. was supported by the J.S. McDonnell Foundation (#220020387), the National Science Foundation (#1212778), and the National Institutes of Health (NIH R01 AT009036-01). R.F.B. was supported by the National Science Foundation/Integrative Graduate Education and Research Training Program in the Dynamics of Brain-Body-Environment Systems at Indiana University. M.G.B. was supported by the TKF Foundation. Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. Funding to pay the Open Access publication charges for this article was provided by a National Institutes of Health grant to OS (NIH R01 AT009036-01).
Supplementary Material
Notes
Conflict of Interest: None declared.
References
- Abdi H. 2010. Partial least squares regression and projection on latent structure regression (PLS regression). Wiley Interdiscip Rev Comput Stat. 2(1):97–106. [Google Scholar]
- Adachi Y, Osada T, Sporns O, Watanbe T, Matsui T, Miyamoto K, Miyashita Y. 2012. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb Cortex. 22(7):1586–1592. [DOI] [PubMed] [Google Scholar]
- Allen E, Damaraju E, Plis S, Erhardt E, Eichele T, Calhoun V. 2012. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 24(3):663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D, Porter M, Wymbs N, Grafton S, Carlson J, Mucha P. 2013. Robust detection of dynamic community structure in networks. Chaos. 23(1):013142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel V, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech Theory E. 2008(10):P10008. [Google Scholar]
- Buckner R, Sepulcre J, Talukdar T, Krienen F, Liu H, Hedden T, Andrews- Hanna J, Sperling RA, Johnson K. 2009. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 29(6):1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzynska AZ, Garrett DD, Preuschhof C, Nagel IE, Li S-C, Backman L, Heekeren HR, Lindenberger U. 2013. A scaffold for efficiency in the human brain. J Neurosci. 33(43):17150–17159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammoun L, Gigandet X, Meskaldji D, Thiran J, Sporns O, Do K, Maeder P, Meuli R, Hagmann P. 2012. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods. 203(2):386–397. [DOI] [PubMed] [Google Scholar]
- Cattell R. 1966. The scree test for the number of factors. Multivar Behav Res. 1(2):245–276. [DOI] [PubMed] [Google Scholar]
- Cipolloni PB, Pandya DN. 1985. Topography and trajectories of commissural fibers of the superior temporal region in the rhesus monkey. Exp Brain Res. 57(2):381–389. [DOI] [PubMed] [Google Scholar]
- Colizza V, Flammini A, Serrano M, Vespignani A. 2006. Detecting rich-club ordering in complex networks. Nat Phys. 2(2):110–115. [Google Scholar]
- Crossley NA, Fox PT, Bullmore ET. 2016. Meta-connectomics: human brain network and connectivity meta-analyses. Psychol Med. 10.1017/S0033291715002895. [DOI] [PMC free article] [PubMed]
- Damoiseaux J, Rombouts S, Barkhof F, Scheltens P, Stam C, Smith SM, Beckmann C. 2006. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 103(37):13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa V, McIntosh A, Sporns O, Kotter R. 2009. Key role of coupling, delay, and noise in resting brain fluctuations. Proc Natl Acad Sci USA. 106(25):10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, McIntosh AR, Shen K, Hutchison RM, Menon RS, Everling S, Hagmann P, Jirsa VK. 2014. Identification of optimal structural connectivity using functional connectivity and neural modeling. J Neurosci. 34(23):7910–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder A, Marzetti L, Pizzella V, Romani G, Corbetta M. 2012. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron. 74(4):753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart C, Young G. 1936. The approximation of one matrix by another of lower rank. Psychometrika. 1(3):211–218. [Google Scholar]
- Efron B, Tibshirani R. 1986. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat Sci. 1(1):54–75. [Google Scholar]
- Fox M, Snyder A, Vincent J, Corbetta M, Van Essen D, Raichle M. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA. 102(27):9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K. 1994. Functional and effective connectivity in neuroimaging: a synthesis. Hum Brain Mapp. 2(1–2):56–78. [Google Scholar]
- Ghosh A, Rho Y, McIntosh A, Kotter R, Jirsa V. 2008. Noise during rest enables the exploration of the brains dynamic repertoire. PLoS Comput Biol. 4(10):e1000196–e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross D, Evans A, Beaulieu C. 2009. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex. 19(3):524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi J, van den Heuvel MP, Avena-Koenigsberger A, Velez de Mendizabal N, Betzel RF, Griffa A, Hagmann P, Corominas-Murtra B, Thiran J-P, Sporns O et al. 2014. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci USA. 111(2):833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Handwerker DA, Robinson ME, Hoy CW, Buchanan LC, Saad ZS, Bandettini PA. 2014. The spatial structure of resting state connectivity stability on the scale of minutes. Front Neurosci. 8, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Hoy C, Handwerker D, Robinson M, Buchanan L, Saad Z, Bandettini P. 2015. Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proc Natl Acad Sci USA. 112(28):8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius M, Supekar K, Menon V, Dougherty R. 2009. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 19(1):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey C, Wedeen V, Sporns O. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6(7):e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovici A, Tagliazucchi E, Balenzuela P, Chialvo DR. 2013. Brain organization into resting state networks emerges at criticality on a model of the human connectome. Phys Rev Lett. 110:178101. [DOI] [PubMed] [Google Scholar]
- Hansen ECA, Battaglia D, Spiegler A, Deco G, Jirsa VK. 2015. Functional connectivity dynamics: modeling the switching behavior of the resting state. NeuroImage. 105:525–535. [DOI] [PubMed] [Google Scholar]
- Hermundstad A, Bassett D, Brown K, Aminoff E, Clewett D, Freeman S, Frithsen A, Johnson A, Tipper C, Miller M et al. 2013. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc Natl Acad Sci USA. 10(15):6169–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermundstad A, Brown K, Bassett D, Aminoff E, Frithsen A, Johnson A, Tip- per C, Miller M, Grafton S, Carlson J. 2014. Structurally-constrained relationships between cognitive states in the human brain. PLoS Comput Biol. 10(5):e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey C, Kotter R, Breakspear M, Sporns O. 2007. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci USA. 104(24):10240–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey C, Sporns O, Cammoun L, Gigandet X, Thiran J-P, Meuli R, Hagmann P. 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 106(6):2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey C, Thivierge J-P, Sporns O. 2010. Can structure predict function in the human brain? NeuroImage. 52(3):766–776. [DOI] [PubMed] [Google Scholar]
- Hutchison R, Womelsdorf T, Allen E, Bandettini P, Calhoun V, Corbetta M, Della Penna S, Duyn J, Glover G, Gonzalez-Castillo J et al. 2013. Dynamic functional connectivity: promise, issues, and interpretations. NeuroImage. 80:360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero R, Canales-Rodŕıguez E, Aleman-Gomez Y, Melie- Garcıa L. 2008. Studying the human brain anatomical network via diffusion- weighted MRI and graph theory. NeuroImage. 40(3):1064–1076. [DOI] [PubMed] [Google Scholar]
- Jones D, Knosche T, Turner R. 2013. White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI. NeuroImage. 73:239–254. [DOI] [PubMed] [Google Scholar]
- Koch M, Norris D, Hund-Georgiadis M. 2002. An investigation of functional and anatomical connectivity using magnetic resonance imaging. NeuroImage. 16(1):241–250. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Williams L, McIntosh A, Abdi H. 2011. Partial least squares (PLS) methods for neuroimaging: a tutorial and review. NeuroImage. 56(2):455–475. [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K. 2002. Specificity and stability in topology of protein networks. Science. 296(5569):910–913. [DOI] [PubMed] [Google Scholar]
- McIntosh A. 2000. Towards a network theory of cognition. Neural Networks. 13(8):861–870. [DOI] [PubMed] [Google Scholar]
- McIntosh A. 2012. Tracing the route to path analysis in neuroimaging. NeuroImage. 62(2):887–890. [DOI] [PubMed] [Google Scholar]
- McIntosh A, Bookstein F, Haxby J, Grady C. 1996. Spatial pattern analysis of functional brain images using partial least squares. Neuroimage. 3(3):143–157. [DOI] [PubMed] [Google Scholar]
- McIntosh A, Gonzalez-Lima F. 1994. Structural equation modeling and its application to network analysis in functional brain imaging. Hum Brain Mapp. 2(1–2):2–22. [Google Scholar]
- McIntosh A, Mišić B. 2013. Multivariate statistical analyses for neuroimaging data. Annu Rev Psychol. 64:499–525. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Lobaugh NJ. 2004. Partial least squares analysis of neuroimaging data: applications and advances. NeuroImage. 23:S250–S263. [DOI] [PubMed] [Google Scholar]
- Messé A, Hutt M-T, Konig P, Hilgetag CC. 2015. A closer look at the apparent correlation of structural and functional connectivity in excitable neural networks. Sci Rep. 5:7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messé A, Rudrauf D, Benali H, Marrelec G. 2014. Relating structure and function in the human brain: relative contributions of anatomy, stationary dynamics, and non-stationarities. PLoS Comput Biol. 10(3):e1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mišić B, Betzel R, Nematzadeh A, Goni J, Griffa A, Hagmann P, Flammini A, Ahn Y-Y, Sporns O. 2015. Cooperative and competitive spreading dynamics on the human connectome. Neuron. 86(6):1518–1529. [DOI] [PubMed] [Google Scholar]
- Mišić B, Sporns O. (forthcoming, 2016) From regions to connections and networks: new bridges between brain and behavior. Curr Opin Neurobiol. In press. [DOI] [PMC free article] [PubMed]
- Mišić B, Sporns O, McIntosh A. 2014. Communication efficiency and congestion of signal traffic in large-scale brain networks. PLoS Comput Biol. 10(1):e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M, Girvan M. 2004. Finding and evaluating community structure in networks. Phys Rev E. 69(2):026113. [DOI] [PubMed] [Google Scholar]
- Park H-J, Friston K. 2013. Structural and functional brain networks: from connections to cognition. Science. 342(6158):1238411. [DOI] [PubMed] [Google Scholar]
- Ponce-Alvarez A, Deco G, Hagmann P, Romani GL, Mantini D, Corbetta M. 2015. Resting-state temporal synchronization networks emerge from connectivity topology and heterogeneity. PLoS Comput Biol. 11(2):e1004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 59(3):2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J, Cohen A, Nelson S, Wig G, Barnes K, Church J, Vogel A, Laumann T, Miezin F, Schlaggar B et al. 2011. Functional network organization of the human brain. Neuron. 72(4):665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 52(3):1059–1069. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2011. Weight-conserving characterization of complex functional brain networks. NeuroImage. 56(4):2068–2079. [DOI] [PubMed] [Google Scholar]
- Shaw DJ, Marecek R, Grosbra M-H, Leonard G, Pike GB, Paus T. 2016. Co-ordinated structural and functional covariance in the adolescent brain underlies face processing performance. Soc Cogn Affect Neurosci. 10.1093/scan/nsv138. [DOI] [PMC free article] [PubMed]
- Shen K, Bezgin G, Hutchison R, Gati J, Menon R, Everling S, McIntosh A. 2012. Information processing architecture of functionally defined clusters in the macaque cortex. J Neurosci. 32(48):17465–17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Mišić B, Cipollini BN, Bezgin G, Buschkuehl M, Hutchison RM, Jaeggi SM, Kross E, Peltier SJ, Everling S et al. 2015. Stable long-range interhemispheric coordination is supported by direct anatomical projections. Proc Natl Acad Sci USA. 112(20):6473–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skudlarski P, Jagannathan K, Calhoun V, Hampson M, Skudlarska B, Pearlson G. 2008. Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. NeuroImage. 43(3):554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Fox P, Miller K, Glahn D, Fox P, Mackay C, Filippini N, Watkins K, Toro R, Laird A et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 106(31):13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Vidaurre D, Beckmann C, Glasser M, Jenkinson M, Miller K, Nichols T, Robinson E, Salimi-Khorshidi G, Woolrich M et al. 2013. Functional connectomics from resting-state fMRI. Trends Cogn Sci. 17(12):666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Nichols TE, Vidaurre D, Winkler AM, Behrens TEJ, Glasser MF, Ugurbil K, Barch DM, Van Essen DC, Miller KL. 2015. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat Neurosci. 18(11):1565–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Betzel RF. 2016. Modular brain networks. Annu Rev Psychol 67:613–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey C, Kotter R. 2007. Identification and classification of hubs in brain networks. PLoS ONE. 2(10):e1049–e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Kotter R. 2004. Motifs in brain networks. PLoS Biology. 2(11):e369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. 2004. The small world of the cerebral cortex. Neuroinformatics. 2(2):145–162. [DOI] [PubMed] [Google Scholar]
- Stam C, van Straaten E, Van Dellen E, Tewarie P, Gong G, Hillebrand A, Meier J, Van Mieghem P. 2015. The relation between structural and functional connectivity patterns in complex brain networks. Int J Psychophysiol. 10.1016/j.ijpsycho.2015.02.011. [DOI] [PubMed]
- Tagliazucchi E, von Wegner F, Morzelewski A, Brodbeck V, Laufs H. 2012. Dynamics BOLD functional connectivity in humans and its electrophysiological correlates. Front Hum Neurosci. 6, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Frank Q, Irfanoglu M, Modi P, Saleem K, Leopold D, Pierpaoli C. 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci USA. 111(46):16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow N. 2010. Functional connectivity density mapping. Proc Natl Acad Sci USA. 107(21):9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traud A, Kelsic E, Mucha P, Porter M. 2011. Comparing community structure to characteristics in online collegiate social networks. SIAM Rev. 53(3):526–543. [Google Scholar]
- Ugurbil K, Xu J, Auerbach E, Moeller S, Vu A, Duarte-Carvajalino J, Lenglet C, Wu X, Schmitter S, Van de Moortele P et al. 2013. Pushing spatial and temporal resolution for functional and diffusion MRI in the human connectome project. NeuroImage. 80:80–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Kahn R, Goni J, Sporns O. 2012. High-cost, high-capacity backbone for global brain communication. Proc Natl Acad Sci USA. 109(28):11372–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Kahn R, Pol H, Hilleke E. 2009. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp. 30(10):3127–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Sporns O. 2013. Network hubs in the human brain. Trends Cogn Sci. 17(12):683–696. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Sporns O. 2011. Rich-club organization of the human connectome. J Neurosci. 31(44):15775–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, Feldman Barrett L, Hilgetag CC, de Reus MA. 2015a. Bridging cytoarchitectonics and connectomics in human cerebral cortex. J Neurosci. 35(41):13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, de Reus MA, Kahn RS. 2015b. Associated microscale spine density and macroscale connectivity disruptions in schizophrenia. Biol Psychiatry. 10.1016/j.biopsych.2015.10.005. [DOI] [PubMed]
- Van Essen D, Smith S, Barch D, Behrens T, Yacoub E, Ugurbil K. 2013. The Wu-Minn human connectome project: an overview. NeuroImage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold H. 1966. Estimation of principal components and related models by iterative least squares. In Krishnaiah P, editor. Multivariate analysis, Vol. 1 New York: Academic Press, p. 391–420. [Google Scholar]
- Yeh F, Wedeen V, Tseng W. 2010. Generalized-sampling imaging. IEEE T Med Imaging. 29(9):1626–1635. [DOI] [PubMed] [Google Scholar]
- Yeo B, Krienen F, Sepulcre J, Sabuncu M, Lashkari D, Hollinshead M, Roff- man J, Smoller J, Zollei L, Polimeni J et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106(3):1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET. 2012. On the use of correlation as a measure of network connectivity. NeuroImage. 60(4):2096–2106. [DOI] [PubMed] [Google Scholar]
- Zamora-Lopez G, Zhou C, Kurths J. 2010. Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front Neuroinformatics. 4: Article number 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.