Summary
The Phosphoinositide 3-Kinase (PI3K) pathway regulates multiple steps in glucose metabolism but also cytoskeletal functions, such as cell movement and attachment. Here we show that PI3K directly coordinates glycolysis with cytoskeletal dynamics in an AKT-independent manner. Growth factors or insulin stimulate the PI3K-dependent activation of Rac, leading to disruption of the actin cytoskeleton, release of filamentous actin-bound aldolase A and an increase in aldolase activity. Consistently, PI3K-, but not AKT-, SGK- or mTOR-inhibitors, cause a significant decrease in glycolysis at the step catalyzed by aldolase, while activating PIK3CA mutations have the opposite effect. These results point towards a master regulatory function of PI3K that integrates an epithelial cell’s metabolism and its form, shape and function, coordinating glycolysis with the energy-intensive dynamics of actin remodeling.
Introduction
Glucose avidity and cytoskeletal plasticity are hallmarks of epithelial cancers, including breast cancers. The phosphoinositide 3-kinase (PI3K)-pathway regulates cytoskeletal functions such as cell movement and intracellular compartmentalization, reviewed in (Cantley, 2002), and also modulates multiple steps in glucose uptake and metabolism (Rathmell et al., 2003). Binding of insulin and other growth factors to their specific cell membrane receptors activates PI3K, resulting in production of phosphatidylinositol-3,4,5-trisphosphate (PIP3) and recruitment of PIP3-binding proteins to the cytosolic side of the plasma membrane, thereby initiating signaling events that control glucose metabolism, cell growth and movement. While there is extensive evidence that glucose uptake and phosphorylation are mediated by PIP3-dependent activation of the protein Ser/Thr kinase AKT, actin remodeling is mediated by PIP3-dependent activation of guanine nucleotide exchange factors (GEFs), namely the Rho/Rac/CDC42 family members (Hanna and El-Sibai, 2013). Here we show that full activation of glycolysis by PI3K requires both AKT activation and Rac-dependent actin remodeling. We show that in quiescent epithelial cells aldolase is trapped in the actin cytoskeleton in a low activity state and that activation of PI3K releases aldolase A, resulting in enhanced flux through glycolysis. We propose that coordination of actin remodeling with glycolysis may facilitate macromolecular biosynthesis needed for cell growth and cell division.
Results
PI3K inhibition blocks the aldolase step of glycolysis in an AKT-independent manner
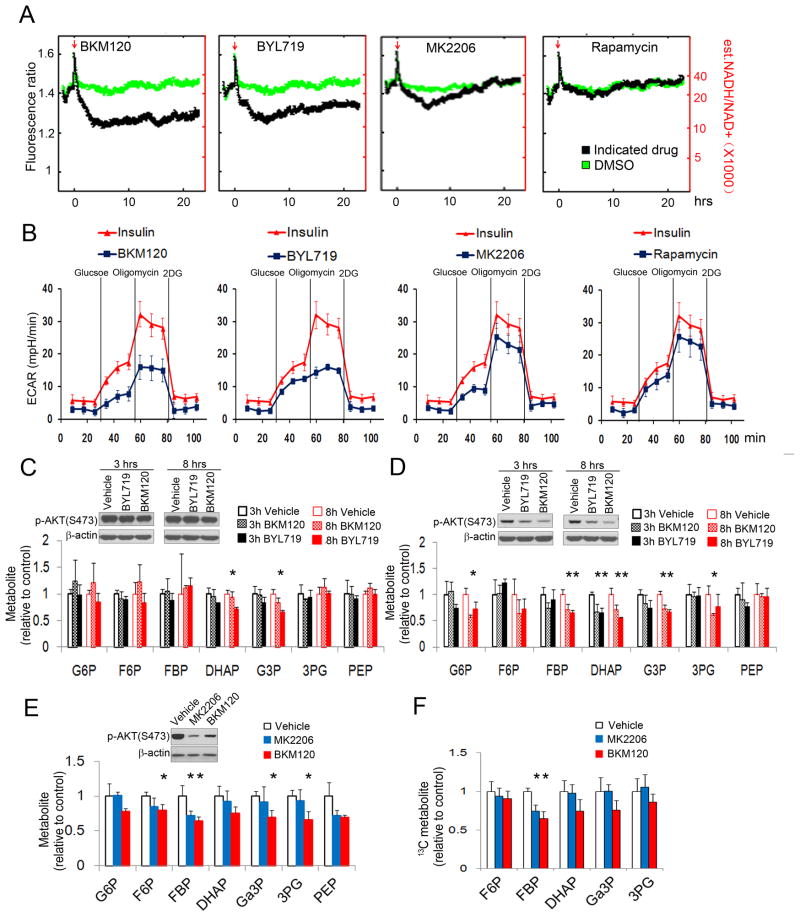
In order to dissect the contributions of PI3K pathway components to the regulation of glycolysis, we examined the effects of specific enzyme inhibitors on the reduction of NAD(+) (Nicotinamide adenine dinucleotide) to NADH, occurring at the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) step (Fig. 1 A) as well as on extracellular acidification rate (ECAR, Fig. 1 B), as read-outs for glycolysis in mammary epithelial cells (MCF10A). The pan-PI3K inhibitor BKM120, Buparlisib (Maira et al., 2012) and the PI3Kα specific inhibitor BYL719, Alpelisib (Furet et al., 2013) led to a decrease in the NADH/NAD(+) ratio in MCF10A cells starting within minutes and reaching a minimum plateau at 4 hours (Fig. 1 A, first two panels), while inhibition of AKT with MK2206 or mTOR with rapamycin caused only a transient decline in the NADH/NAD(+) ratio (Fig. 1A, 3d and 4th panel). Both, BKM120 and BYL719 reduced the initial ECAR increase in response to insulin stimulation and a glucose challenge, and drastically reduced the cells’ ability to mobilize the glycolytic reserve, i.e. to respond with increased glycolysis after addition of Oligomycin to the medium (Fig. 1 B, first two panels). MK2206 and rapamycin also decreased the ECAR after addition of glucose, as expected (Rathmell et al., 2003), but different from the PI3K-inhibitors, the AKT- and the mTOR-inhibitor did not block mobilization of the glycolytic reserve (Fig. 1 B, right two panels). When we examined the ECAR in MCF10A cells expressing constitutively active, myristoylated AKT, mAKT, (Barthel et al., 1997), the PI3K-inhibitors BYL719 and BKM120 prevented mobilization of the glycolytic reserve (Fig. S1A), suggestive of a specific role for PI3K for the maximum achievable glycolytic rate that can not be compensated for by constitutive activation of AKT. The PI3Kβ inhibitor TGX221 and GSK650394, an inhibitor of serum and glucocorticoid-induced protein kinase (SGK), had little effect on the NADH/NAD(+) ratio (Fig. S1 B) or the ECAR (Fig. S1C). The concentration of drugs used achieved target inhibition (Fig. S1 D). All the inhibitors caused a variable degree of glucose uptake inhibition (Fig. S1 E, F) while the prolonged effects on the NADH/NAD+ ratio (Fig. 1A) and on mobilization of the glycolytic reserve (Fig. 1B) were specific to pan-PI3K and PI3Kα-inhibition. These data suggest that PI3K exerts a regulatory role on the maximal glycolytic capacity that cells can mount and that this regulatory role is independent of AKT, SGK or mTOR.
Figure 1.
Inhibition of AKT does not phenocopy the effects of PI3K inhibition on glycolysis. A, B. PI3K-, but not AKT- or mTOR-inhibitors decrease the cytosolic NADH/NAD+ ratio and glycolysis in MCF10A cells. The NADH/NAD+ ratio (A) was determined in MCF10A cells expressing the fluorescent biosensor Peredox (Hung et al., 2011) treated with inhibitors of pan-PI3K (BKM120, 2.5 μM), PI3Kα (BYL719, 2.5 μM), AKT (MK2206, 200 nM), or mTOR (Rapamycin, 100 nM; see Tab S1 for a list of the inhibitors). The ratio of NADH/NAD+ (right scale, red numbers) was estimated based on in vitro calibration experiments shown in (Hung et al., 2011). The red arrow indicates start of treatment. Each curve represents the median biosensor response for a population of ~300–700 cells, acquired in two independent assays. B. Glycolysis, including glycolytic reserve (mobilized and extinguished with Oligomycin or 2-deoxy-glucose (2DG) respectively) were measured using a Seahorse instrument after pre-incubation with drugs or insulin. Shown are ECAR means ± SD of experimental triplicates. C, D. Effect of the pan-PI3K inhibitor BKM120 and the PI3Kα inhibitor BYL719 on glycolytic intermediates in MCF10A cells expressing constitutively active mAKT1 (C) or WT AKT1 (D). Cells were treated with BKM120 (1 μM) or BYL719 (1 μM) for 3 or 8 hours, and metabolite abundance determined by mass spectrometry. E, F. Effects of BKM120 or MK2206 on glycolytic intermediates in HCC1937 cells, E. Steady-state after 16 hours of treatment; F. cells were treated with inhibitors as indicated for 3 hours, glucose was replaced with [U-13C]-glucose for 0.5 min before extraction flux analysis. All metabolite levels (Tab. S2) were normalized to vehicle control, each bar represents the mean ± SD of experimental triplicates.
To distinguish PI3K versus AKT effects on glycolytic intermediates, we examined responses to BKM120 or BYL719 in mAKT MCF10A cells (Fig. 1C). Quantification of steady-state metabolite levels by LC-MS/MS revealed that BKM120 or BYL719 caused a decrease in glyceraldehyde 3-phosphate (Ga3P) and dihydroxyacetone-phosphate (DHAP), the products of the aldolase reaction. While a change in the steady-state levels of DHAP and Ga3P could be caused by either a decrease in aldolase activity or an increase in conversion to downstream metabolites, the decrease in glycolytic rate implied by the reduction in the NADH/NAD(+) ratio (Fig. 1A) and ECAR (Fig. 1B) supports a selective decrease in aldolase activity. In WT-AKT MCF10A, both BKM120 and BYL719 inhibited AKT phosphorylation and caused a general decrease in glycolytic intermediates (Fig. 1 D), consistent with the well-known role of AKT in glucose uptake and phosphorylation (Rathmell et al., 2003). Similar effects of BKM120 on glycolysis were seen in HCC1937 breast cancer cells, while the AKT inhibitor MK2206 caused a decrease in F1,6BP, but not the decline in Ga3P or DHAP observed with the PI3K-inhibitor (Fig. 1E). Measuring the breakdown of U-13C6-glucose, we observed that both MK2206 and BKM120 decreased flux into the aldolase substrate F1,6BP but only BKM120 also decreased the aldolase products DHAP and Ga3P (Fig. 1F). These results are consistent with a model in which PI3K, but not AKT, regulates net flux through lower glycolysis through the selective regulation of aldolase.
PI3K pathway activation increases the cytosolic abundance and activity of aldolase
To examine the regulation of aldolase activity by the PI3K pathway, we measured its enzymatic activity directly. Following insulin stimulation, cells were lysed with digitonin and aldolase activity measured in the culture plates with the hydrazine assay (Jagannathan et al., 1956). Total aldolase activity increased up to 4-fold (Fig. 2A), suggestive of post-transcriptional regulation. Previous studies had shown that binding of aldolase to actin filaments (F-Actin) inhibits its activity (Arnold and Pette, 1970; Harris and Winzor, 1987; Wang et al., 1996). Activation of PI3K signaling regulates actin dynamics, independent from signaling through AKT (Cantley, 2002; Cantley et al., 1991), raising the possibility that PI3K-initiated actin dynamics and release of aldolase from the cytoskeleton are coordinated and regulate glycolysis.
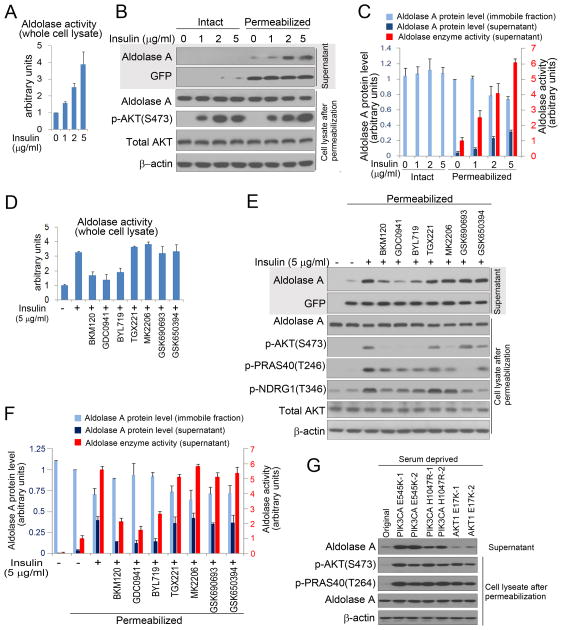
Figure 2.
PI3K pathway activation promotes mobilization of aldolase and increases its catalytic activity. A. Aldolase activity in response to insulin stimulation. GFP-expressing MCF10A were serum-starved overnight, insulin-stimlated for 3 hours, lysed with Digitonin (100 μg/ml), followed by aldolase enzyme activity determination. B. Cells were treated as in A. Where indicated, cells were permeabilized with digitonin (30 μg/ml) for 5 min. Supernatant (upper two panels) and cell lysate (lower 4 panels) for each assay were subjected to immunoblotting as indicated. C. Quantification of aldolase A in the immobile (cell lysate) and the diffusible fraction (supernatant) by immunoblotting (for scan see Fig. S2A), and of aldolase activity in the immobile fraction (supernatant). Bar graphs represent means ± SD of three independent experiments. D. Serum- and growth factor deprived MCF10A cells were prepared as in A, pre-treated with drugs for 15 min, followed by addtion of insulin for 3 hours, lysed with Digitonin (100 μg/ml) and aldolase activity was determined. Bar graphs represent means ± SD of three independent experiments. E. MCF10 cells were treated as in D, and permeabilized with Digitonin (30 μg/ml). Supernatant and cell lysate were collected separately for immunoblotting. F. Quantification of aldolase A protein in the immobile fraction (cell lysate) and in the diffusible fraction (supernatant), and determination of aldolase activity in the supernatant (for scan see Fig. S2B and C). Bar graphs represent means ± SD of experimental triplicates. G. Determination of free aldolase in MCF10A cells with knock-in mutations of activating mutations in PIK3CA (E545K, H1047 R) or AKT1 (E17K). Digitonin permeabilization as in B; quantification of aldolase release in three independent experiments is provided in Fig. S3F. See Tab. 1 for details on drugs used.
To test this hypothesis, we examined the cytoskeletal binding and enzymatic activity of aldolase A in response to experimental manipulation of PI3K (Fig. 2, S2, S3). Acute PI3K pathway activation was achieved with insulin for 3 hours unless indicated otherwise (Fig. 2A–F, S2, S3 A,B) or IGF (Fig. S3C). The PI3K pathway was blocked with pharmaceutical inhibitors (Fig. 2 D–F, S3 A–C) or siRNA ablation of PI3K p110 subunits (Fig. S3 D, E). Constitutive PI3K pathway activation was achieved with knock-in of the clinically relevant PIK3CA mutants H1047R and E545K (Fig. 2 G, S3F). To estimate the fraction of aldolase A in the soluble versus immobilized state we permeabilized cells with Digitonin to allow for efflux of diffusible aldolase A, followed by separate collection of supernatant and cell lysates for aldolase analyses (Fig. 2 B–G, Fig. S2, Fig. S3 A–F). In GFP-expressing MCF10A cells, insulin treatment resulted in the release of aldolase A in a time- (Fig. S3A) and concentration-dependent (Fig. 2B, C and S3B) manner, that paralleled the increase in AKT phosphorylation (Fig. 2B, S3A, S3B), while GFP was released independent of insulin addition. Insulin-dependent mobilization of aldolase A occurred within 20 minutes with a maximum effect by 3 hours (Fig. S3A). We estimate that ~30% of the aldolase A is mobilized by treatment with insulin (Fig. 2B, C and Fig. S2 A). Proportionate to the increasing amounts of aldolase A protein in the supernatant, increased enzymatic activity was detected (Fig. 2 C),
When we examined the effect of insulin (Fig. 2D–F, S3A,B) or IGF1 (Fig. S3C) pathway inhibition, the pan-PI3K inhibitors BKM120 and GDC0941, and to some extent the PI3Kα inhibitor BYL719, blocked release of aldolase A, but inhibitors of PI3Kβ (TGX221), AKT ((MK2206 (allosteric inhibitor) or GSK690693 (ATP-competitive), SGK1/2 (GSK650394) or mTOR (rapamycin) did not (Fig. 2 E-F, Fig. S3 C). Consistently, BKM120 and BYL719 also blocked the insulin-induced increase in aldolase activity in the total cell lysate (Fig. 2 D) and supernatant (Fig. 2 F). Consistently, siRNA ablation of the p110 α but not β subunit prevented the insulin-induced release of aldolase (Fig. S3D, E). Activating mutations of PI3K occur frequently in cancer, especially in breast cancer, mostly at two ‘hotspots’ within the kinase (H1047R) and helical domains (E542K or E545K) of p110α (Samuels et al., 2004). In knock-in cell lines (Gustin et al., 2009) both activating PIK3CA mutations, but not the activating AKT mutation (E17K) led to an increase of aldolase A release (Fig. 2G, S3F).
In summary, these data indicate that the effects of insulin on aldolase activity and on overall control of glycolysis are dependent on the activation of PI3K, independent of AKT, SGK or mTOR, and correlate with the release of aldolase A from the cytoskeleton.
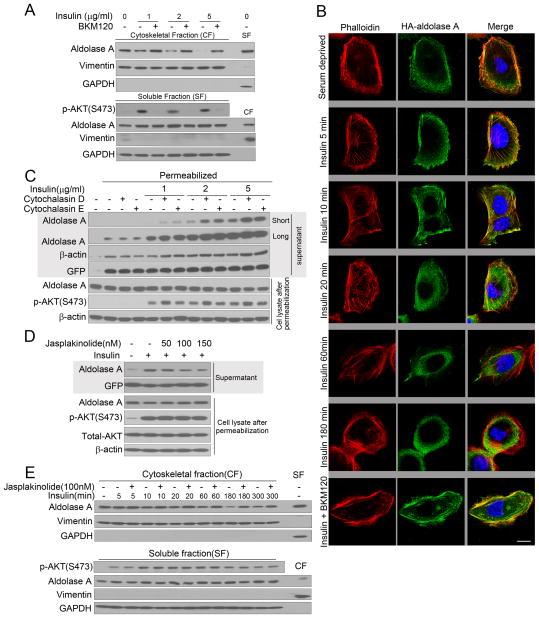
Cell fractionation confirmed that, insulin caused a shift of aldolase A from the cytoskeletal to the cytosolic fraction, preventable by BKM120 (Fig. 3A, S3G). To visualize the association of aldolase A with F-actin, we expressed HA-tagged aldolase A in immortalized human mammary epithelial cells (HMECs), which have a flat morphology that allows visualization of actin structures. In serum-deprived HMECs much of the HA-aldolase co-localized with F-actin, (Fig. 3B, upper panels). Stimulation with insulin led to actin remodeling (Fig. 3B, panels 2–5); less HA-aldolase A co-localized with F-actin at the cell periphery and instead concentrated centrally in the cytoplasm (Fig. 3B, panels 2–5). The dissociation of HA-aldolase A from F-actin was prevented by BKM120 (Fig. 3B, bottom panel). Inhibition of actin polymerization with membrane-permeable mycotoxins (cytochalasins D and E) that by themselves do not affect cellular glucose uptake (Jijakli et al., 2002), enhanced the release of aldolase A (Fig. 3C, S3H). Conversely, the actin-stabilizing agent Jasplakinolide prevented the mobilization of aldolase (Fig. 3 D, S3I) and led to retention of aldolase in the cytoskeletal fraction (Fig. 3 E, S3J).
Figure 3.
PI3K activation mobilizes aldolase from F-actin. A. MCF10A cells were prepared as in Fig. 2 D, lysed and fractionated. Vimentin is used as a marker for the cytoskeletal (CF) and GAPDH for the soluble fraction (SF). Fractions from untreated cells are controls for the fractionation procedure (far-right lane) (also see Fig. S3G). B. Co-localization of aldolase with F-actin is disrupted by PI3K activation. HA-ALDOA (visualized with Alexa-green) was transfected into HMEC cells. Cells were serum-starved, pretreated with BKM120 or vehicle control for 15 min, followed by addition of insulin, fixation, immunostaining (green) and phalloidin staining (red). Scale bar represents 10 μm. C. The actin polymerization status affects insulin-induced aldolase mobilization from the cytoskeleton. MCF-10A were treated as in Fig. 2 D, except that cytochalasin D or E were used, permeabilized and supernatant and lysates were collected (also see Fig. S3H). D. MCF10A were treated as in Fig. 3C, except that Jasplakinolide was used to stabilize F-actin (also see Fig. S3I). E. Time course of aldolase mobilization from the cytoskeleton and its reversal by actin stabilization (also see Fig. S3J).
The dynamics of the aldolase-actin interaction are regulated by PI3K and determine glycolytic flux
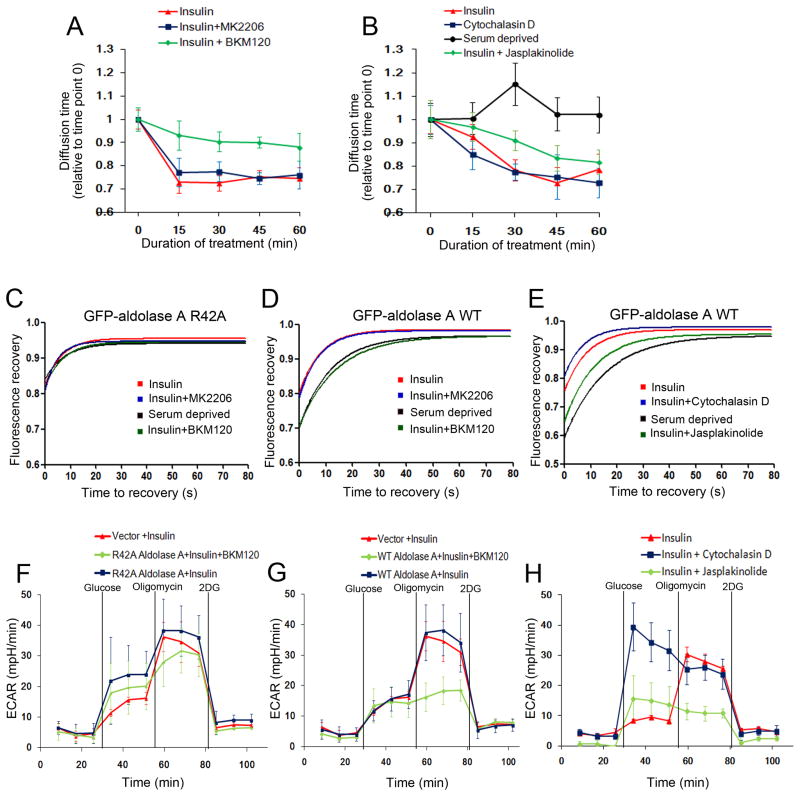
To determine the interdependence of cytoskeletal dynamics, aldolase binding and PI3K signaling, we examined the mobility of GFP-aldolase A and glycolytic flux in response to experimental manipulation of the PI3K pathway and actin polymerization (Fig. 4). Assuming that binding of aldolase to F-actin should slow its rate of diffusion, we used 2-photon fluorescence correlation spectroscopy (FCS) in live cells to measure the diffusion time of GFP-aldolase A and its R42A mutant which is unable to bind F-actin (Wang et al., 1996, Fig. S4 A). GFP-aldolase A paralleled endogenous aldolase in the digitonin release and F-actin precipitation assays while the R42 MT was not affected by either insulin or PI3K-inhibition (Fig. S4 A, B). PI3K pathway activation decreased the diffusion time within 20 min (Fig. 4 A, B, Fig. S4 C,D), consistent with increased aldolase mobility. Decreased diffusion time was sustained for at least 2 hours, consistent with the timing of the release of aldolase (Fig. S3A, S4) and blocked by BKM120 but not MK2206 (Fig. 4A). Consistent with sequestration of aldolase A by F-actin, cytochalasin D led to a decrease in diffusion time while the actin-stabilizing Jasplakinolide partially prevented the acceleration of diffusion (Fig. 4 B). Consistent results were obtained when we determined the rate of fluorescence recovery after photobleaching (FRAP, Fig. 4 C–E, S5, S6) of GFP-aldolase A. The aldolase mutant unable to bind actin but with full catalytic activity (R42A) had rapid FRAP kinetics unaffected by PI3K pathway manipulation (Fig. 4 C, S5A). In serum-deprived cells FRAP of wild-type (WT) GFP-aldolase A was slower than FRAP of R42A GFP-aldolase A (Fig. 4 D, S5B). With insulin stimulation, FRAP of WT GFP-aldolase A accelerated (Fig. 4 D), and acceleration was prevented by BKM120 but not MK2206 (Fig. 4D, S5B), indicative of a PI3K-mediated release of aldolase from the cytoskeleton. Treatment with Jasplakinolide partially reversed FRAP acceleration, while the actin-destabilizing Cytochalasin D accelerated FRAP further (Fig. 4 E, S6).
Figure 4.
Regulation of aldolase dynamics and glycolytic flux by PI3K and F-actin remodeling. Fluorescence correlation spectroscopy (FCS) measures the mobility of GFP-aldolase molecules as they cross the path of a laser beam focused on a given point in the cytoplasm (also see Fig. S4 C, D). Fluorescence recovery after photobleaching (FRAP) measures the influx of fluorescent GFP-aldolase into a cytoplasmic area after photobleaching (Fig. S5 and S6). A, B. FCS of GFP-aldolase transfected into MCF10A cells. Serum or growth factor-deprived cells were scanned and treated with insulin in the absence or presence of drugs as indicated. Every 15 min, the diffusion time was obtained for at least 8 different cells. Displayed are means +/− SD relative to control, i.e. time point 0 prior to insulin and/or drug treatment. C, D, E. FRAP analysis of GFP-aldolase. HMEC cells expressing the aldolase mutant GFP-R42A ALDOA (C) or GFP-WT ALDOA (D, E) were serum-deprived and treated with BKM120, MK2206 (C, D) and the actin de-stabilizing Cytochalasin D or actin-stabilizing Jasplakinolide (E). Insulin was added 15 min after drugs for a total of 3 hours. Time point zero is set to the start of the recording of the recovery signal following completion of the laser pulsing (7 seconds) and recovery was recorded every 4 seconds. For each condition, at least 14 different cells were analyzed over time. Note that diffusion of the GFP-proteins continues during the bleach, leading to a y-intercept of 0.8 for GFP-R42A aldolase A (C) and 0.7 for GFP-WT aldolase A under serum starvation (D and E). F–H. The actin-aldolase interaction regulates glycolysis. MCF10A cells were transfected with vector control, HA-R42A-ALDOA (F) or HA-WT-ALDOA (G) (also see Fig. S4E) and treated with BKM120 (F, G) or Cytochalasin D or Jasplakinolide (H) and insulin and subjected to ECAR determination as described in Fig. 1 B. See Tab. S1 for details on drugs used.
Glycolytic flux, as determined by ECAR, tightly correlated with the mobility of aldolase A (Fig. 4 F–H). In R42A GFP-aldolase A expressing cells, glycolytic flux was similar to that of control cells stimulated with insulin, and not significantly affected by BKM120 (Fig. 4 F, S4E). In WT GFP-aldolase A expressing cells, BKM120 prevented the mobilization of the glycolytic reserve (Fig. 4 G). As expected from FRAP and FCS, the actin-stabilizing Jasplakinolide prevented mobilization of the glycolytic reserve while the actin-destabilizing Cytochalasin D led to a maximum increase in glycolytic flux (Fig. 4 H).
Aldolase mobilization is mediated by PI3K-signaling through activation of Rac
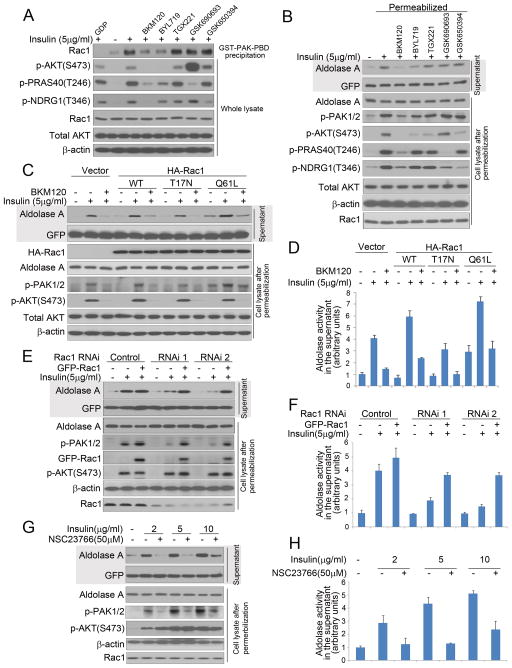
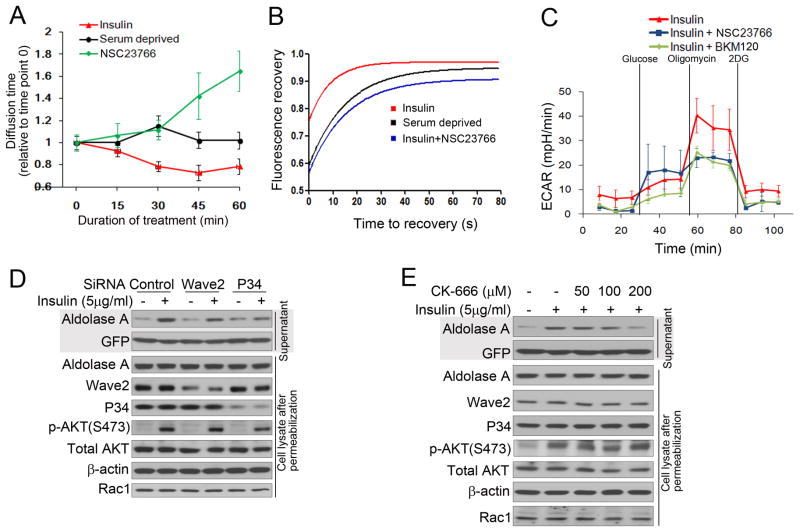
Activation of the PI3K pathway including activation of Rac leads to cytoskeletal reorganization (Brachmann et al., 2005) (Cantley, 2002). Binding of GTP-Rac to the Rac/Cdc42 (p21) binding domain (PBD) of PAK promotes actin remodeling (Benard et al., 1999). PBD domain-dependent pull-down of endogenous Rac (Fig. 5A, S7A) or GFP-Rac (Fig. S7B) was enhanced by insulin and blocked by BKM120 and BYL719 but not by inhibitors of AKT or SGK. Downstream targets of Rac include the p21-activated protein kinase (PAK) (Joneson et al., 1996) as well as the WAVE/p34 complex (Miki et al., 1998). Insulin-induced phosphorylation of PAK was blocked by BKM120 and BYL719, but not by inhibitors of PI3Kβ, AKT or SGK (Fig. 5 B) and paralleled aldolase release (Fig. 5 B). PAK activation, aldolase release and activity were decreased after transfection of a dominant negative form of Rac1 (Rac1T17N) while over-expression of WT or constitutively active Rac1 (Rac1Q61L) augmented PAK phosphorylation, aldolase release and activity (Fig. 5C,D and S7 C,D). Similarly, depletion of Rac1, restored through transfection of a siRNA-resistant GFP-Rac1, decreased the response to insulin stimulation with regard to aldolase mobilization, aldolase activity or PAK1/2 phosphorylation, but had little effect on AKT S473 phosphorylation (Fig. 5 E, F, S7E). Consistently, an inhibitor of Rac1, NSC 23766, abolished insulin-induced PAK phosphorylation and aldolase A mobilization and aldolase activity, but not AKT phosphorylation (Fig 5G, H, S7F). As expected, the RAC1-inhibitor also decreased aldolase A mobility (Fig. 6 A,B) as determined by FCS or FRAP, and the glycolytic reserve (Fig. 6 C).
Figure 5.
Aldolase mobilization from the cytoskeleton is regulated by PI3K through the Rac pathway. A. Binding of endogenous, activated Rac1 to the PAK1-PBD is prevented by PI3K-inhibition. MCF10A cells were prepared as in Fig 2D and lysed. In lane 1, GDP (1 mM final concentration) was added prior to precipitation as a negative control. Immunoblotting of GST-PAK-PBD precipitates (first row) or total lysates (rows 2–7) as indicated (also see Fig. S7A). B Inhibition of total PI3K and PI3Kα but not inhibition of AKT, SGK or PI3Kβ, prevents insulin-induced PAK1/2 phosphorylation and aldolase mobilization (see Tab. S1 for details on drugs used). Treatment of cells and collection of supernatant and immobile fractions were as in Fig. 2E. C. Effects of Rac1 mutants on insulin-induced aldolase mobilization. MCF10A cells were transfected with control vector, HA-Rac1, dominant negative HA-Rac1T17N, or constitutively active HA-Rac1Q61L, treated with BKM120 and insulin, permeabilized and lysed as in Fig. 2E (also see Fig. S7D). D. Quantification of aldolase activity in the supernatant of cells treated in C as described in 2F. Bar graphs represent means ± SD of experimental triplicates. E. Depletion of Rac1 decreases insulin-induced aldolase mobilization from F-actin. MCF10A cells were transfected with Rac1 siRNAs or controls and treated as in Fig. 2B. SiRNA resistant GFP-Rac1 expression constructs were used to rescue the siRNA-mediated effects (also see Fig. S7E). F. Quantification of aldolase activity in the supernatant of cells treated in E, as described in Fig. 2C. Bar graphs represent means ± SD of three independent experiments. G. Rac inhibition prevents insulin induced mobilization of aldolase A. MCF10A cells were treated as in Fig. 2E except that a Rac inhibitor (NSC23766) was used (also see Fig. S7F). H. Quantification of aldolase activity in the supernatant of permeabilized cells treated in G. Bar graphs represent means ± SD of experimental triplicates.
Figure 6.
Rac1 inhibition reduces insulin-induced aldolase mobilization from F-actin and glycolytic flux A. Two-photon fluorescence correlation spectroscopy of GFP-aldolase transfected into MCF10A cells. Serum or growth factor-deprived cells were scanned and then treated with insulin in the absence or presence of the Rac inhibitor NSC23766. FCS was performed as described in Fig. 4 A. B. FRAP analysis of GFP-aldolase. Cells expressing GFP-WT ALDOA were serum-deprived overnight, stimulated for 3 hours with insulin in the absence or presence of the Rac inhibitor and FRAP was determined as in Fig. 4C (also see Fig. S6).
C. Rac-inhibtion decreases the glycolytic reserve similar to PI3K-inhibition. MCF10A cells were treated with PI3K- or Rac-inhibitor and insulin and subjected to ECAR determination as described in Fig. 1 B. D. Depletion of Wave2 or P34 decreases insulin-induced aldolase mobilization from F-actin. MCF10A cells were transfected with pooled siRNAs or controls and then treated as in Fig. 2B (also see Fig. S7I). E. Arp2/3 complex inhibition prevents insulin induced mobilization of aldolase A. MCF10A cells were treated as in Fig. 2E, except that a Arp2/3 complex inhibitor (CK-666) was used (also see Fig. S7J).
Consistent with Rac activation, WAVE2 and p34, components of the WAVE and ARF2/3 complexes, co-localized with F-actin in response to insulin (Fig. S7 G,H). SiRNA ablation of p34 or WAVE2 (Fig. 6 D, S8 I) or the Arp2/3 inhibitor CK-666 (Fig. 6 E, S7J) decreased the ability of cells to mobilize aldolase A from F-actin. These data show that mobilization of aldolase A in response to PI3K pathway activation is mediated by Rac and its downstream effectors.
PI3K-inhibitors block the aldolase step in breast cancers in vivo
Since alterations in PI3K signaling are highly prevalent in epithelial cancers, we tested if PI3K-inhibition had a similarly profound effect on mid- and lower glycolysis in cancer in vivo. We used a mouse model of BRCA1-related breast cancer, where we had previously observed a synergistic effect of PI3K- and PARP-inhibition (Juvekar et al., 2012). Cohorts of tumor-bearing mice were created through syngeneic transplantation (Rottenberg et al., 2007).
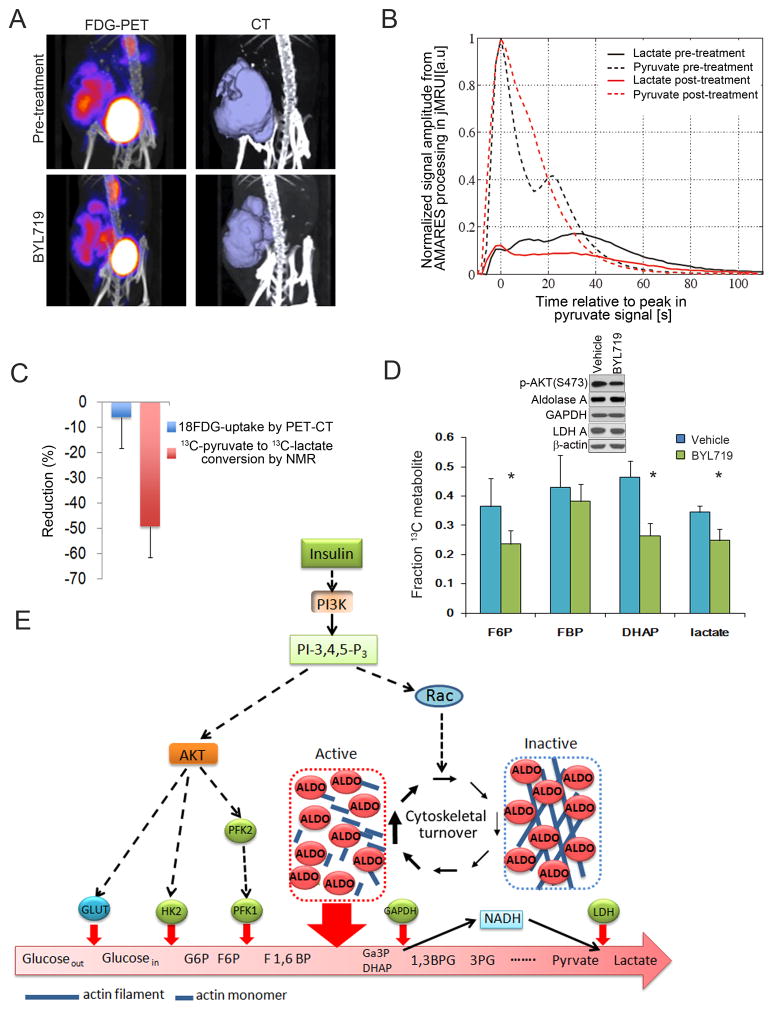
Upper glycolysis was probed with positron emission tomography of 18FDG (18fluoro-deoxyglucose) uptake (Fig. 7 A, C). FDG-PET measures glucose uptake and phosphorylation, and previous studies have shown that PI3K inhibitors can suppress FDG-PET uptake in breast tumors (Mayer et al., 2014). We used NMR detection to determine the interconversion of hyperpolarized 13C-pyruvate to 13C-lactate (Fig. 7 B, C), which is influenced by glucose uptake, LDH activity, NADH availability and endogenous lactate concentration (Witney et al., 2011). For each tumor-bearing mouse (n=4 for PET-CT and NMR), baseline imaging was performed. Then, two doses of BYL719 were given to the mice, 12–16 hours and 2–4 hours before the second scan. All measurements are expressed as change relative to the pre-treatment baseline (Fig. 7C). Tumors proved highly glucose-avid at baseline (Fig. 7A) and, similarly, the inter-conversion of pyruvate to lactate was readily detected (Fig. 7B). BYL719 caused only a minor, insignificant reduction in FDG-uptake during this period of time similar to what was seen with 2-DG in MCF10A (Fig. S1E) or HCC1937 cells (Fig. S1F). However, BYL719 caused a 40 to 50% decrease in the rate of conversion of 13C-pyruvate to 13C-lactate (Fig. 7B, quantified in Fig. 7C), i.e. there was a disproportionate reduction of the 13C-pyruvate to 13C-lactate conversion relative to the minor reduction in FDG-uptake, while levels of total aldolase A, GAPDH and LDH in tumors did not change (Fig. 7D). The in vitro studies (Fig. 1) showed that BYL719 suppresses production of the substrate for GAPDH, Ga3P and the product of GAPDH, NADH. Although there may be multiple explanations for the ability of BYL719 to inhibit the conversion of 13C-pyruvate to 13C-lactate in the tumor, a reduction in NADH required to convert pyruvate to lactate via lactate dehydrogenase (LDH) would explain this finding and is consistent with the regulation of mid- or lower glycolysis by PI3K independent from glucose uptake and phosphorylation.
Figure 7.
The PI3K-inhibitor BYL719 blocks glycolysis in breast cancers in vivo. Cohorts of mice with breast cancer were created through syngeneic transplantation of K14-Cre BRCA1f/f p53f/f tumors and randomized to control or BYL719. Mice were treated twice with BYL719, 12 hours and again 2 hours prior to the MRI or PET-CT scans. A. Effect of BYL719 on tumoral 18FDG uptake. Mice were scanned and tumor uptake determined before (upper panel) and after (lower panel) treatment with BYL719. FDG-PET (left panels) and computer tomography (right panels) were obtained simultaneously. B. Time dependence of the 1-13C-pyruvate and 1-13C-lactate signals in a representative tumor following injection of hyperpolarized 1-13C-pyruvate solution. Tumor-bearing mice (n=4) were scanned at baseline and after BYL719. For each scan a ~10 second bolus of 13C-pyruvate was given via tail vein injection and slice-selective 13C spectra were acquired every 2 seconds starting at the time of the injection. C. Summary of the reduction in 18FDG-uptake and pyruvate to lactate conversion upon in vivo treatment with BYL719. The bar graph represents the mean reduction observed between pre- and post-treatment scans (done within a 2–4 day window) for each of the two modalities. D. Effect of BYL719 on the metabolism of [U-13C6] glucose in breast tumors in vivo. Mice were injected intraperitoneally with a [U-13C6]-glucose solution and metabolites extracted from the tumors. Data are presented as the fractional labeling of the pool (Tab. S3). Error bars represent the means ± SD of four tumors per treatment group. Immunoblot insert shows target inhibition (pAKT) and metabolic enzyme levels in tumors. E. PI3K activation promotes glycolysis through mobilization of aldolase from the cytoskeleton. PI3K-initiated AKT activation leads to increased glucose import and positively regulates the hexokinase and phosphofructokinase reactions, providing increased substrate for the aldolase reaction. In parallel, PI3K-activation accelerates actin dynamics via Rac increasing levels of free, cytoplasmic aldolase thereby coordinates the generation of ATP and biomass with the energy-intensive process of cytoskeletal remodeling.
In vivo isotope tracing analyses with ([U]-13C)-glucose pinpointed the block to the aldolase step. A bolus of [U]-13C-glucose was administered intraperitoneally 90 min prior to euthanasia, followed by tumor harvest and mass spectrometry (Fig. 7D). The sharpest drop in 13C-glucose metabolites was again seen in the aldolase product DHAP (Fig. 7D). Immunoblotting of tumor tissue lysates showed that enzyme levels did not decrease in response to BYL719, and hence the differences in glycolytic flux were not explained by transcriptional or post-transcriptional regulation of aldolase A (Fig. 7D, insert), but by PI3K-induced mobilization of aldolase.
Discussion
Regulation of glycolysis through mobilization of aldolase
Systemically elevated aldolase activity was first observed in cancer, in vivo, 70 years ago (Warburg, 1945). The tumor tissue was subsequently found to be the source of high aldolase activity (Sibley, 1958) and specifically high aldolase A levels (Schapira, 1966). Our data provide a mechanism for this finding and may explain its biological significance: Signal transduction via PI3K allows for the physical dissociation of aldolase from F-actin into the cytoplasm where it is active (Fig. 7E). This simple, biophysical mechanism of activating aldolase through recruitment from the cytoskeleton is a rapid and efficient way for cells to increase metabolic flux. Redistribution of aldolase in response to PI3K signaling achieves coordination of cytoskeletal dynamics and glycolysis, while avoiding the time- and energy-consuming path of transcriptional activation and biosynthesis of new enzyme molecules.
While a number of glycolytic enzymes associate with the cytoskeleton, and will presumably be released when actin dynamics increase, the change of glycolytic flux in response to insulin signaling appears to be primarily mediated by the mobilization and activation of aldolase. Aldolase catalyzes a reverse aldol condensation leading to break-up of F-1,6-BP into DHAP and Ga3P in an energetically unfavorable multi-step reaction that is slow relative to the preceding phosphorylation and subsequent oxidation steps of glycolysis. With its unique mechanism, aldolase A has a low turnover rate (Kcat) relative to the upstream Hexokinase or Phosphofructokinase (Albe et al., 1990; Morris et al., 1996). Hence when insulin stimulation leads to a rapid increase in cytosolic F1,6 BP, the cytoplasmic abundance of active aldolase A may become rate-limiting to pace mid- and lower glycolysis. Conversely, PI3K-inhibition can limit glycolytic flux by tethering aldolase to the actin cytoskeleton, even under conditions where AKT remains active and supports glucose uptake and phosphorylation (Fig. 1C).
The differential response to PI3K- versus AKT-inhibition is most clearly seen after 3–4 hours (Fig. 1A) when the PI3K-inhibitor effect persists, while the NADH/NAD ratio in cells treated with the AKT-inhibitor recovers (Fig 1A). Consistent with recent reports showing that disruption of mitochondrial metabolism can lead to feedback activation of the PI3K pathway (Pelicano et al., 2006), this recovery may be due to feed-back activation of enzymes upstream from aldolase, such as phosphofructokinase (PFK1). In the case of AKT-inhibition, such feed-back activation would compensate for increasing ATP depletion and rescue the NADH/NAD ratio (Fig. 1A, 3d and 4th panel). However, with PI3K-inhibition blocking glycolysis at the aldolase step downstream from PFK1, such feedback activation of PFK1 could not result in a recovery of the NADH/NAD ratio (Fig. 1A, first two panels). The acute disruption of mitochondrial electron flux with oligomycin resulting in the mobilization of the glycolytic reserve may be the equivalent of ATP depletion due to prolonged AKT-inhibition (Fig. 1B).
Metabolic implications of a selective block at the aldolase step
Recent studies from our and other laboratories have shown that in some tumors, the non-oxidative pentose phosphate pathway, which requires the product of aldolase A, Ga3P, is a major source of the ribose needed for nucleotide synthesis and 3PG conversion to Ser and Gly can play a critical role in purine synthesis (Locasale et al., 2011) and glycolysis supplies ATP needed for deoxynucleotide phosphorylation. Thus, regulation of glycolysis at the step of aldolase can potentially have a profound effect on deoxynucleotide synthesis, perhaps explaining the dramatic effect of PI3K inhibitors in combination with a PARP-inhibitor on rapidly growing BRCA1-related tumors (Juvekar et al., 2012) that need high rates of nucleotide synthesis both for cell cycle progression and DNA repair.
The combined analysis of In vivo imaging with 18FDG-PET scanning and MRI of hyperpolarized 13C-pyruvate to lactate conversion suggests that PI3K α regulates glycolysis in mid- and lower glycolysis, independent from and in addition to its role in regulating initial steps in glycolysis. While the data could be explained by a decrease of the cytosolic NADH/NAD+- ratio resulting from effects of PI3K inhibition outside of lower glycolysis, our in vitro observations support the concept of regulation of the aldolase reaction by PI3K, mediated by Rac and independent from AKT. They are consistent with and may explain the recent observation that AKT-independent PI3Kα regulation of Rac1 is required for KRAS-induced pancreatic tumorigenesis in mice (Wu et al., 2014). Recent clinical studies demonstrated that the ability of PI3K-inhibitor treatments to cause a decrease in 18FDG-PET uptake in tumors may be predictive of response to these agents (Mayer et al., 2014) It is possible that metabolic imaging of 13C-pyruvate to 13C-lactate conversion (Fig. 7B), which has recently been shown to be safe and feasible in humans (Nelson et al., 2013), could potentially be helpful in guiding the use of PI3K- versus AKT- or mTOR-inhibitors for cancer treatment, although such approach would require prospective clinical evaluation.
Coordination of glycolysis and cytoskeletal remodeling by PI3K signaling
Functional epithelial cells are polar and respond to localized growth factor stimulation with PI3K activation leading to cytoskeletal remodeling. The structure and dynamics of F-actin are determined by the Arp2/3 complex, the initiation site for the polymerization of new, branched actin filament strands (Rotty et al., 2013). The Arp2/3 complex itself is regulated by signaling events, specifically the small GTPase Rac and phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which jointly activate the SCAR–WAVE complex which in turn further activates the Arp2/3 complex (Lebensohn and Kirschner, 2009). Recent studies have shown the spatial and temporal integration of F-actin polymerization, Arp2/3 nucleation and PI3K signaling on the subcellular level in fibroblasts (Johnson et al., 2015) and suggest that PI3K signaling can both promote and respond to Arp2/3-mediated actin polymerization in subcellular areas where architecture rapidly changes, such as membrane protrusions. Our results add to this model as they suggest that PI3K regulates epithelial function through the temporary and spatially controlled release of aldolase from the actin cytoskeleton, resulting in the coordination of glycolysis and cytoskeletal dynamics. In functional epithelial cells it may ensure the proximity of energy production and cytoskeletal rearrangement. Conversely, in epithelial cancer cells, constitutive activation of the PI3K pathway, for example as a result of an activating PIK3CA mutation, may lead to uncoordinated, increased cytoskeletal turnover resulting in loss of epithelial cell function and excess of free, cytoplasmic aldolase that drives tumoral glycolysis, effectively leading to the de-differentiation characteristic of cancer cells.
Experimental Procedures
For additional information see Supplemental Experimental Procedures Drugs and treatments
BKM120 (Buparlisib) and BYL719 (Alpelisib) were obtained via a MTA from Novartis pharmaceuticals. See Table S1 for sources and concentrations of drugs used.
Cell culture and transfection
Immortalized mammary epithelial cells (HMEChtert, in short HMEC, a gift from Dr. Robert Weinberg) and MCF-10A and HCC1937 (both from ATCC) were cultured in standard conditions. MCF-10A cells expressing inducible myristoylated HA-AKT1 or WT HA-AKT1 (gift from Dr. Alex Toker) were treated with doxycycline at 0.5 μg/ml 16 hours prior to lysis to induce AKT1 expression. MCF10-A cells with knock-in of PIK3CA or AKT mutations were generated as described (Lauring et al., 2010). Unless noted otherwise, cells were serum- and growth factor deprived for 18 hours overnight, and then stimulated with insulin for 3 hours prior to permeabilization and/or lysis. For drug treatments, drugs were added to the cell cultures 15 min prior to insulin.
Plasmids and siRNA experimentation
For a list of plasmids and transfected cells see table S4. Two siRNA duplexes that target P110α were used., 5,-GCTTAGAGTTGGAGTTTGA-3′, and 5,-GCGAAATTCTCACACTATT-3′. siRNA duplexes that target Rac1 were 5′-AGACGGAGCTGTAGGTAAA-3′ and 5′-CCTTTGTACGCTTTGCTCA-3′). For controls and transfection procedures see Supplemental Experimental Procedures.
Syngeneic tumor implants
Animal experiments were conducted in accordance with Institutional Animal Care and Use Committee (IACUC) approved protocols at Beth Israel Deaconess Medical Center. Tumors generated in K14-Cre Brca1 f/f, Tp53 f/f mice were syngeneically transplanted as described (Rottenberg et al., 2007) into the mammary pad of Cre-negative females to generate cohorts of mice.
NADH/NAD+ determination with Wide-Field Time-Lapse Microscopy
MCF10A cells stably expressing nuclear-targeted NADH biosensor Peredox-mCitrine were maintained at ~37°C and ~5% CO2. T-Sapphire and YFP images were acquired on a Nikon Eclipse Ti microscope at intervals of 5–6 minutes. After baseline imaging, cells were treated with vehicle control or inhibitors and imaged for additional ~20 hours. We generated T-Sapphire-to-YFP ratios for a population of ~300–700 cells over time in two independent assays for each condition.
Immunofluorescence
MCF10A cells were examined with a Zeiss Axiovert 200M fluorescence microscope or a Zeiss LSM 510 Inverted Live-Cell Confocal System.
Fluorescence Recovery After Photobleaching
FRAP was performed on a Zeiss LSM 510 confocal microscope at 37°C, using a 63× objective. Bleaching was done using the 488 nm argon ion laser and the 543 nm Helium-Neon laser, set to 100% output. Fluorescence recovery curves represent the median of the fluorescence recovery of at least 14 cells. Curves were modeled using GraphPad Prism software and the one phase exponential decay algorithm.
Cell permeabilization, fractionation and determination of aldolase A levels
2 x 105 MCF10A cells/well were seeded into 6-well plates and treated with as indicated. For permeabilization, cells were washed with 3× PBS and then incubated in 30 μg/ml digitonin/PBS for 5 min at 4°C. After incubation, the supernatant was collected and cells lysed with 200 μl of RIPA buffer for each well. The supernatant was centrifuged at 2000 rpm to remove cellular components. 40 μl of supernatant (8% of total supernatant) or 20 μl (10% of total lysate) of cell lysate were run on the same SDS-page, transferred to PVDF membrane for immunoblotting and scanned using a LI-COR system. Another 40 μl of supernatant was used for an enzymatic aldolase assay. The abundance of aldolase protein was calculated based on the intensity of the aldolase A signal determined with Image J and corrected for the dilution.
Aldolase enzymatic assay
The aldolase enzymatic assay was performed based on Boyer’s modification of the hydrazine assay (Jagannathan et al., 1956) in which 3-phosphoglyceraldehyde reacts with hydrazine to form a hydrazone which absorbs at 240 nm. Assays were done on either cell lysates (Digitoning 100 μg/ml) or supernatants of permeabilized cells (Digitonin 30 μg/ml).
Metabolism analysis
For steady state studies metabolite fractions were resuspended in HPLC-grade water and analyzed by targeted liquid chromatography - tandem mass spectrometry (LC-MS/MS) using a 5500 QTRAP mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) with Amide HILIC chromatography (Waters). Data were acquired in selected reaction monitoring (SRM) mode using positive/negative ion polarity switching for steady-state polar profiling. SRM transitions were also created for 13C labeled precursor and fragment ions for monitoring 13C incorporation. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/SCIEX).
FDG-PET Scanning
Mice were examined on a NanoPET/CT (Bioscan/Mediso) scanner under isoflurane anesthesia as described previously (Juvekar et al., 2012).
Hyperpolarized NMR Studies
100mM hyperpolarized pyruvate solution was prepared by dynamic nuclear polarization as described previously (Varma et al., 2015). A 28 mm transmit/receive 13C surface coil was placed over the tumor of an anaesthetized mouse. T2-weighted proton images were acquired. 250μl of hyperpolarized pyruvate solution was administered intravenously as a 10s bolus, and slice-selective 13C spectra were acquired from the tumors. Acquisition parameters and quantification of relative lactate signal are described in the supplementary methods.
Two-photon fluorescence correlation spectroscopy
MCF10A cells expressing eGFP-labeled aldolase were grown on a cover glass and mounted onto a custom-made heating stage with temperature controlled by thermocouple. eGFP fluorophores were excited by two-photon excitation: 3 mW at 850 nm, generated by a mode-locked Ti-sapphire laser. FCS curves were acquired over a 2-hour time period at 15 min intervals. The FCS curves obtained during the experiments were fit by a model function for freely diffusing particles and a 3-dimensional Gaussian confocal volume.
Glucose uptake
Cells were treated with 1 mM 13C labeled 2DG for 30 seconds and metabolites collected with 70% methanol extraction for LC-MS/MS. Alternatively, glucose uptake was measured with the a kit from abcam (ab136955).
Seahorse assay
An XF24 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA) was used to determine the effects of the inhibitors on MCF10A or mAKT-MCF10A cells which were seeded at 20,000 cells/well Measurements of extracellular acidification rate (ECAR) were performed according to the manufacturer’s instructions.
Statistical analysis
Unless otherwise noted, data are presented as the means ± SD; a two-tailed t test was used to determine significance.
Supplementary Material
Acknowledgments
LCC and GMW are supported by a Stand Up to Cancer Dream Team Translational Research Grant, (SU2C-AACR-DT0209), by the Breast Cancer Research Foundation (BCRF), the Mary Kay Ash Foundation, the Men’s Initiative of DFHCC and the Breast Cancer Alliance; LCC by NIH grant GM041890; JMA in part by NIH NCI grants 5P01CA120964-05 and 5P30CA006516-46; AKG, PS, G. in part by NIH R21 EB014471 and R01 CA169470.; PS in part by R01CA152330-0. DJN is supported in part by NSF DBI-0959721, DMR-0820484, and United States – Israel Binational Science Foundation, BSF 2009271; CAL by a Dale F. Frey award from the Damon Runyon Cancer Research Foundation.
Footnotes
Conflicts of interest: LCC has consulted for Novartis, which is developing BKM120 and BYL719 for cancer treatment; he is a member of the board of directors and a consultant/advisory board member for Agios, for which he also has ownership interests (including patents).
Author Contributions: Conceptualization GMW, HH, LCC; Methodology GMW, HH, LCC, DO, DJN, JMA, PS, MRL, AKG, JL; Validation HH, CAL, ECL, GMW; Formal Analysis HH, CAL, ECL, JGA, DO, YPH, SU, AKG, MRL, JMA, GMW; Investigation HH, AJ, ECL, JGA, DO, GV, SU, PS, MRL, AKG, LMA, GMW; Resources AJ, ECL, JGA, GV, AKG, DRT, LCC; Data Curation HH, AJ, CAL, ECL, JGA, DO, PS; Writing HH, LCC, GMW; Visualization HH, DO, AKG, GMW; Supervision LCC, DRT, DJN, GMW; Project Administration HH, GMW; Funding Acquisition DJN, JMA, LCC, GMW
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J Theor Biol. 1990;143:163–195. doi: 10.1016/s0022-5193(05)80266-8. [DOI] [PubMed] [Google Scholar]
- Arnold H, Pette D. Binding of aldolase and triosephosphate dehydrogenase to F-actin and modification of catalytic properties of aldolase. European journal of biochemistry/FEBS. 1970;15:360–366. doi: 10.1111/j.1432-1033.1970.tb01016.x. [DOI] [PubMed] [Google Scholar]
- Barthel A, Kohn AD, Luo Y, Roth RA. A constitutively active version of the Ser/Thr kinase Akt induces production of the ob gene product, leptin, in 3T3-L1 adipocytes. Endocrinology. 1997;138:3559–3562. doi: 10.1210/endo.138.8.5263. [DOI] [PubMed] [Google Scholar]
- Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. The Journal of biological chemistry. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Yballe CM, Innocenti M, Deane JA, Fruman DA, Thomas SM, Cantley LC. Role of phosphoinositide 3-kinase regulatory isoforms in development and actin rearrangement. Mol Cell Biol. 2005;25:2593–2606. doi: 10.1128/MCB.25.7.2593-2606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Auger KR, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- Furet P, Guagnano V, Fairhurst RA, Imbach-Weese P, Bruce I, Knapp M, Fritsch C, Blasco F, Blanz J, Aichholz R, et al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett. 2013;23:3741–3748. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, Cosgrove D, Tamaki A, Konishi H, Konishi Y, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna S, El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cellular signalling. 2013;25:1955–1961. doi: 10.1016/j.cellsig.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Harris SJ, Winzor DJ. Enzyme kinetic evidence of active-site involvement in the interaction between aldolase and muscle myofibrils. Biochim Biophys Acta. 1987;911:121–126. doi: 10.1016/0167-4838(87)90279-2. [DOI] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan V, Singh K, Damodaran M. Carbohydrate metabolism in citric acid fermentation. 4. Purification and properties of aldolase from Aspergillus niger. Biochem J. 1956;63:94–105. doi: 10.1042/bj0630094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijakli H, Zhang HX, Dura E, Ramirez R, Sener A, Malaisse WJ. Effects of cytochalasin B and D upon insulin release and pancreatic islet cell metabolism. Int J Mol Med. 2002;9:165–172. [PubMed] [Google Scholar]
- Johnson HE, King SJ, Asokan SB, Rotty JD, Bear JE, Haugh JM. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J Cell Biol. 2015;208:443–455. doi: 10.1083/jcb.201406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, Rajendran A, Papa A, Spencer K, Lyssiotis CA, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2:1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring J, Cosgrove DP, Fontana S, Gustin JP, Konishi H, Abukhdeir AM, Garay JP, Mohseni M, Wang GM, Higgins MJ, et al. Knock in of the AKT1 E17K mutation in human breast epithelial cells does not recapitulate oncogenic PIK3CA mutations. Oncogene. 2010;29:2337–2345. doi: 10.1038/onc.2009.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira SM, Pecchi S, Huang A, Burger M, Knapp M, Sterker D, Schnell C, Guthy D, Nagel T, Wiesmann M, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther. 2012;11:317–328. doi: 10.1158/1535-7163.MCT-11-0474. [DOI] [PubMed] [Google Scholar]
- Mayer IA, Abramson VG, Isakoff SJ, Forero A, Balko JM, Kuba MG, Sanders ME, Yap JT, Van den Abbeele AD, Li Y, et al. Stand up to cancer phase Ib study of pan-phosphoinositide-3-kinase inhibitor buparlisib with letrozole in estrogen receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol. 2014;32:1202–1209. doi: 10.1200/JCO.2013.54.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AJ, Davenport RC, Tolan DR. A lysine to arginine substitution at position 146 of rabbit aldolase A changes the rate-determining step to Schiff base formation. Protein Eng. 1996;9:61–67. doi: 10.1093/protein/9.1.61. [DOI] [PubMed] [Google Scholar]
- Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PE, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Science translational medicine. 2013;5:198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol. 2003;23:7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S, Nygren AO, Pajic M, van Leeuwen FW, van der Heijden I, van de Wetering K, Liu X, de Visser KE, Gilhuijs KG, van Tellingen O, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schapira F. Aldolase isozymes in cancer. Eur J Cancer. 1966;2:131–134. doi: 10.1016/0014-2964(66)90005-3. [DOI] [PubMed] [Google Scholar]
- Sibley JA. Significance of serum aldolase levels. Annals of the New York Academy of Sciences. 1958;75:339–348. doi: 10.1111/j.1749-6632.1958.tb36881.x. [DOI] [PubMed] [Google Scholar]
- Varma G, Wang X, Vinogradov E, Bhatt RS, Sukhatme VP, Seth P, Lenkinski RE, Alsop DC, Grant AK. Selective spectroscopic imaging of hyperpolarized pyruvate and its metabolites using a single-echo variable phase advance method in balanced SSFP. Magn Reson Med. 2015 doi: 10.1002/mrm.26004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Morris AJ, Tolan DR, Pagliaro L. The molecular nature of the F-actin binding activity of aldolase revealed with site-directed mutants. J Biol Chem. 1996;271:6861–6865. [PubMed] [Google Scholar]
- Warburg OaCW. Gaerungsfermente im Blutserum von Tumor-ratten. Biochemische Zeitschrift. 1945;314:399–408. [Google Scholar]
- Witney TH, Kettunen MI, Brindle KM. Kinetic modeling of hyperpolarized 13C label exchange between pyruvate and lactate in tumor cells. J Biol Chem. 2011;286:24572–24580. doi: 10.1074/jbc.M111.237727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Carpenter ES, Takeuchi KK, Halbrook CJ, Peverley LV, Bien H, Hall JC, DelGiorno KE, Pal D, Song Y, et al. PI3K regulation of RAC1 is required for KRAS-induced pancreatic tumorigenesis in mice. Gastroenterology. 2014;147:1405–1416. e1407. doi: 10.1053/j.gastro.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.