Abstract
PIWI proteins and PIWI-interacting RNAs (piRNAs) are part of a cellular pathway that has evolved to protect genomes against the proliferation of transposable elements (TEs). PIWIs and piRNAs assemble into complexes that are involved in epigenetic and post-transcriptional repression of TEs. Most of our understanding of the mechanisms of piRNA-mediated TE silencing comes from fruit fly and mouse models. However, even in these well-studied animals it is unclear how piRNA responses relate to variable TE expression and whether the strength of the piRNA response affects TE content over time. Here, we assessed the evolutionary interactions between TE and piRNAs in a statistical framework using three nonmodel laurasiatherian mammals as a study system: dog, horse, and a vesper bat. These three species diverged ∼80 million years ago and have distinct genomic TE contents. By comparing species with distinct TE landscapes, we aimed to identify clear relationships among TE content, expression, and piRNAs. We found that the TE subfamilies that are the most transcribed appear to elicit the strongest “ping-pong” response. This was most evident among long interspersed elements, but the relationships between expression and ping-pong pilRNA (piRNA-like) expression were more complex among SINEs. SINE transcripts were equally abundant in the dog and horse yet new SINE insertions were relatively rare in the horse genome, where we identified a stronger piRNA response. Our analyses suggest that the piRNA response can have a strong impact on the TE composition of a genome. However, our results also suggest that the presence of a robust piRNA response is apparently not sufficient to stop TE mobilization and accumulation.
Keywords: small RNAs, comparative genomics, evolution, ping-pong cycle, PIWI proteins
Introduction
Transposable elements (TEs) are selfish DNA sequences that have the ability to invade and propagate in host genomes and are classified as either DNA transposons or retrotransposons based on their mechanism of mobilization and cycle of replication. Retrotransposons (Class I TEs) mobilize exclusively through “copy-and-paste” mechanisms, by transcribing an RNA intermediate that is reverse transcribed and inserted into a new genomic location. In mammalian genomes, the most common retrotransposons are Long INterspersed Elements (LINEs), Short INterspersed Elements (SINEs), and Long Terminal Repeat elements (LTRs). DNA transposons (Class II TEs) do not use an RNA intermediate and may mobilize either through a “cut-and-paste” mechanism (Terminal Inverted Repeat elements), by excising themselves from one locus and reinserting into a novel one (Kapitonov and Jurka 2007), or by “copy-and-paste” mechanisms (e.g., Helitrons and Mavericks).
TEs are major components of vertebrate genomes, and in the case of mammals, TEs can account for up to 70% of the genomic content (De Koning et al. 2011), most of which is derived from retrotransposon insertions (Yohn et al. 2005). Not surprisingly, TEs are an important source of variation within and among species. In addition to increasing genome size, TE insertions can disrupt gene-reading frames or alter gene expression by inserting within or close to a gene, promote genomic deletions and reorganize genome structure via TE-mediated nonhomologous recombination (Gilbert et al. 2002; Liu et al. 2003; Callinan et al. 2005; Han et al. 2005; Sen et al. 2006). Because of these potential impacts, TE mobilization is generally considered deleterious and their unrestricted proliferation can have profound biological effects. As a result, the question of how organisms control TE mobilization has attracted great interest.
Data from multiple metazoans indicate that proteins in the PIWI and Argonaute gene families, referred to as PIWI proteins from here onwards, and PIWI-interacting RNAs (piRNAs), a class of small noncoding RNAs predominantly expressed in the germline, play a major role reducing TE expression and mobilization (Aravin, Hannon, et al. 2007; Aravin, Sachidanandam, et al. 2007; Brennecke et al. 2007; O’Donnell and Boeke 2007; Saito and Siomi 2010). piRNAs are the most abundant class of small RNAs expressed in testis and range in size from ∼24 to 32 nucleotides (Aravin et al. 2006, 2008; Girard et al. 2006; ; Höck and Meister 2008). piRNA and PIWI proteins associate in complexes that are involved in epigenetic and post-transcriptional repression of TEs (Siomi et al. 2011). Post-transcriptional silencing of TEs occurs via a feed-forward amplification loop known as the “ping-pong” cycle. In brief, primary piRNAs direct PIWIs to complementary TE-derived transcripts. These transcripts are cleaved by the PIWIs to generate secondary piRNAs. The secondary piRNA is then loaded onto a new PIWI protein and the cycle is repeated, amplifying the pool of both primary and secondary piRNAs while reducing the threat of TE transcripts.
Two distinct populations of piRNAs have been described in mammals, pre-pachytene and pachytene, which differ in their expression, biogenesis, and genomic origins (Aravin, Sachidanandam, et al. 2007; Li et al. 2013). Expression of pre-pachytene piRNAs begins in pre-meiotic and early prophase 1 spermatogonia whereas expression of pachytene piRNAs starts in the pachytene stage of prophase 1 through sperm maturation (Aravin, Sachidanandam, et al. 2007; Aravin et al. 2008; Reuter et al. 2011). Both classes of piRNAs are present in mature testes; however, pachytene piRNAs greatly outnumber pre-pachytene piRNAs (Li et al. 2013). Pachytene piRNAs are largely derived from unannotated regions of the genome and appear to regulate and eliminate gene transcripts from the cytoplasm in a manner similar to the miRNA pathway (Guo et al. 2014). In contrast, the pre-pachytene population of piRNAs appears to be heavily involved with post-transcriptional silencing of TEs via the ping-pong cycle. In regards to the mammalian ping-pong cycle, Aravin et al. (2008) proposed that a primary piRNA is derived from a TE transcript. This contrasts with the Drosophila ping-pong cycle where anti-sense piRNAs are derived from transcribed piRNA clusters and subsequently bind sense TE transcripts (Brennecke et al. 2007).
The evolutionary relationships between TE families and piRNAs have not been extensively examined. Lukic and Chen (2011) and Mourier (2011) found a correlation between the age of TE families and piRNA density in humans and mice, respectively. However, very few other studies of piRNAs have been attempted outside of mice (Liu et al. 2012; Rozenkranz et al. 2015). In other words, there has not been a thorough investigation into which TE parameters elicit the strongest piRNA response and an understanding of the general relationship between piRNAs and TE families in mammals is therefore lacking.
The goal of this research is to better understand the relationships between TE abundance at the genome and transcriptome level and piRNA abundance among mammals. To do so, we compared genome-wide TE composition, TE expression, and the strength of the piRNA response elicited by TEs in three laurasiatherian mammals with very distinct TE landscapes. The three species diverged from one another within a relatively short period, ∼80 million years ago (Meredith et al. 2011), and the combination of distinct TE loads and similar evolutionary divergences allowed us to explore the piRNA response to TE-related variables within the context of the ping-pong model. In brief, our analyses indicate that TE expression was a strong predictor of the level of piRNA response, in agreement with predictions of the ping-pong model, and suggest that the level of piRNA response may modulate the relative contribution of the different TEs to the genome.
Materials and Methods
Sample Collection and Library Prep
We collected discarded testicular tissue from one adolescent dog and one adolescent horse after the animals were sedated and neutered by licensed veterinarians from the College of Veterinary Medicine at Mississippi State University. We killed a wild caught adult big brown bat, Eptesicus fuscus, in accord with IACUC standards to collect testis tissue. In each case, a cross section of testis was snap frozen in liquid nitrogen immediately following castration and stored at −70 °C prior to RNA isolation. We isolated total RNA using Trizol (Invitrogen, USA) according to the manufacturer’s specifications. Small RNA libraries were prepped using the Illumina TruSeq small RNA kit and 1×50 bp reads were sequenced on the Illumina HiSeq2000 platform. Directional RNASeq libraries were prepped using the Illumina TruSeq v2 kit and 2×100 bp reads were sequenced on a single lane of a HiSeq2000.
TE Composition and Expression
We masked the dog (CanFam3.1) and horse (EquCab2) genomes using RepeatMasker 4.0.5 using the “-species dog” and “-species horse” parameters, respectively. The big brown bat genome was obtained from NCBI (EptFus1.0, GenBank accession ALEH00000000, 1.806 gigabases). Contigs were first masked with RepeatMasker with the “-species Chiroptera” option then secondarily masked with a de novo repeat library constructed from the Eptesicus genome draft (supplementary data S1, Supplementary Material online) (Platt et al. 2014). To estimate genetic distances, we used the calcDivergenceFromAlign.pl script included with RepeatMasker to calculate Kimura two-parameter (Kimura 1980) distances between each insertion and its respective consensus sequence. The option –noCpG was invoked to exclude highly mutable CpG sites from distance calculations. We calculated the total number of insertions, total number of bases (expressed as a proportion of the genome), average insertion length, and the median genetic distance among insertions for each TE family from the RepeatMasker output. Novel TE insertions, especially among retrotransposons, are expected to be identical to the source element, and the consensus sequence of a given subfamily is inferred to be the best estimate of the sequence of the source element for that subfamily. Within the framework of the master element model proposed by Brookfield and Johnson (2006), the distance between an element of a given subfamily and the corresponding consensus provides an estimate of the age of that insertion, and the median distance among insertions of a given subfamily provide an estimate of the peak of accumulation in that subfamily. Thus, insertions with high similarity to the corresponding consensus, that is, low pairwise genetic distances, are assumed to have occurred in the recent past, whereas insertions with low similarity (high genetic distance) are thought to be older.
We estimated the relative expression of TE families by mapping RNASeq reads to the corresponding TE consensus sequences representing families found in each genome. For each species, we mapped ∼30 million RNASeq reads to the consensus elements using the default parameters of RSEM (Li and Dewey 2011), which used Bowtie to initially map reads. The default parameters allow two mismatches in a seed region of the first 25 bases of an alignment, then unlimited mismatches in the remainder of the sequence alignment. Expression estimates were measured in transcripts per million.
piRNA Processing and Cluster Annotation
Prior to small RNA mapping, we clipped barcodes, removed reads that had bases with Phred quality score <25, and removed identical reads using modules in the fastx toolkit. We also removed low complexity small RNA sequences using a custom python script. We mapped piRNA-like (pilRNA) sequences 24–32 bases long to the complete genomes using Bowtie (Langmead et al. 2009) allowing one mismatch in the alignment. pilRNAs that mapped to only one locus were reported. A cluster was defined as a group of at least 50 pilRNAs where contiguous pilRNAs were separated by <1,500 bases (Beyret et al. 2012). Only clusters >10 kb that had a normalized small RNA count of ten pilRNAs/cluster length (in thousands)/number of mapped sequences (in millions) were analyzed for TE insertions. We calculated the same TE parameters within clusters as we did for the whole genome (see above).
Ping-Pong piRNA Expression
pilRNA sequences were mapped to a library of consensus sequences representing the TE families annotated in each genome. We mapped pilRNAs to the consensus elements, allowing three mismatches, and allowed pilRNAs to map to all possible loci. Because primary and secondary piRNAs 5′ ends are cleaved ten nucleotides downstream of the complementary piRNA, the 5′ ends of piRNAs in ping-pong pairs should be complementary for these first ten nucleotides. Accordingly, we identified the ping-pong signature by partitioning mapped reads into putative primary or secondary pilRNAs. pilRNAs that had a U in the first position and did not have an A in the 10th position were considered “primary” pilRNAs, whereas those pilRNAs that had an A in the 10th position and did not have a U in the first position were classified as “secondary” pilRNAs. Pairs of primary and secondary pilRNAs that overlapped at the first ten nucleotides were assumed to have resulted from the ping-pong cycle. Ping-pong pilRNA expression was estimated for each TE family by summing the number of ping-pong pilRNAs for each element and dividing the pilRNA counts by the length of the consensus sequence (in thousands of bp) and the number of ping-pong pilRNA that mapped to the entire consensus libraries for each species (in millions). We refer to this metric as ping-pong pilRNA expression (PPE) throughout and consider it as a proxy for the strength of the piRNA response against a given TE because the abundance of ping-pong pairs would indicate where PIWI proteins are most concentrated.
Statistical Analyses
Because they have different mechanisms of transposition, the major types of elements (LINEs, SINEs, LTRs, and DNA transposons) were analyzed independently within each species. We log +1 transformed all variables (both dependent and independent) associated with TE families and first performed simple linear regression between PPE and all independent variables. We then used bi-directional stepwise regression analyses using Akaike’s Information Criteria to choose the best sub-model from a full model that included all independent variables to explain the most PPE variation. To explore the relevance of each independent variable in the chosen sub-model, we used the lmg method available in the R package relaimpo which averages sequential sums of squares over the ordering of regressors. The final data used for these analyses are available in supplementary data S2, Supplementary Material online.
Results
We sought to better understand the interplay between TEs and piRNAs among mammals by comparing genomic TE landscapes, TE expression, and piRNA repertoires in dog, horse, and the big brown bat. Patterns of TE activity are often inferred based on the relative abundance of TEs in a given genome. However, for the purpose of this study, it was critical to distinguish between genome-wide patterns of TE accumulation and levels of TE expression, two different facets of TE activity. The first reflects historical patterns of TE deposition and retention, whereas the second reflects the population of TEs currently challenging a given genome. Both of these factors could impact piRNA production, as the abundance of a given TE in the genome could directly relate to its potential as a source of primary piRNAs, and TEs that are actively transcribed are expected to contribute more to the pool of piRNAs in the ping-pong cycle.
SINE, LINE, LTR, and DNA transposon insertions are grouped into discrete families based on overall similarity and the families are often represented by single consensus sequences. These consensus sequences are considered the best approximation of the mobilizing elements for any particular family (Brookfield and Johnson 2006). There were 745, 787, and 976 distinct TE families annotated by RepeatMasker in the dog, horse, and bat genomes, respectively, corresponding to 150–159 LINE, 20–26 SINE, 283–430 LTR, and 280–376 DNA transposon families (supplementary table S1, Supplementary Material online). For each separate TE family, we calculated 1—the number of insertions, 2—the relative age of the family, 3—the average length of insertions (estimated for 3.1 all insertions in the genome, and for 3.2 all insertions within piRNA clusters), and 4—the abundance of transcripts. We found clear differences in patterns of TE accumulation and expression among the three genomes that may allow us to tease apart what drives the production of TE-related ping-pong piRNAs when the types of TE families, insertion numbers, expression, and genomic proportion vary.
Genomic TE Composition and Properties
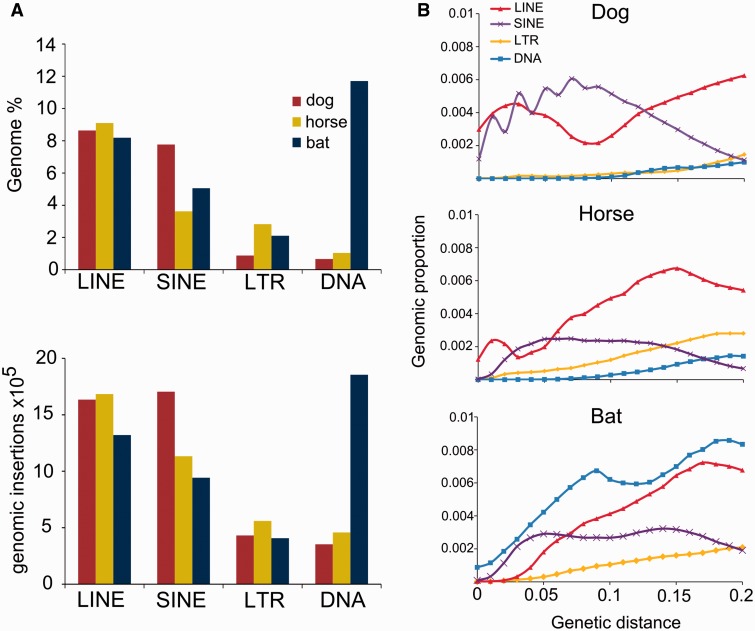
Among retrotransposons, LINEs occupied the most genomic space in all three species, accounting for ∼10% of the genome, followed by SINEs, which ranged from ∼3% to 8%, and LTR retrotransposons, which ranged from ∼1% to 3% (fig. 1A). Based on number of insertions, LINEs were the most abundant TEs in the horse genome, but SINEs and DNA transposons were the most abundant in the dog and the bat, respectively, with over 1.65×106 insertions in all cases (fig. 1A). The bat genome stands out in this regard, as it has experienced a resurgence of DNA transposons when compared with most mammals, a characteristic shared with the closely related little brown bat, Myotis lucifugus (Pritham and Feschotte 2007; Ray et al. 2007, 2008; Mitra et al. 2013; Platt et al. 2014). We estimated that ∼11% of the bat genome derives from DNA transposon insertions, in contrast to ∼1% in dog or horse (fig. 1A).
Fig. 1.—
Relative contribution of the different TEs to the dog, horse and bat genome. (A) The percentage of each genome contributed by the major TE types (top) and the number of insertions for each type (bottom). Only insertions that were <0.2 divergent from the consensus were considered. (B) The temporal contribution of major TE types in each genome. Insertions with lower genetic distances were deposited more recently.
Historical patterns of TE accumulation also vary among these three species (fig. 1B). In the recent past, the dog genome has accumulated LINE and SINE insertions at a higher rate than either the horse or bat. Some LINEs have been deposited relatively recently in the horse genome, but young LINE insertions are almost undetectable in the bat genome. Similarly, recent SINE insertions are very uncommon in the bat and horse genomes while SINEs have accumulated at a relatively high rate in the dog. Recent DNA transposon insertions are uncommon in all three species. However, the bat differs from dog and horse in that there was a high rate of DNA transposon deposition in the recent past (fig. 1B). This has also been seen in M. lucifugus, which diverged from the big brown bat lineage ∼25 million years ago (Miller-Butterworth et al. 2007; Ray et al. 2007; Pagán et al. 2012; Platt et al. 2014). Despite the clear slowdown in DNA transposon accumulation in the genome of the big brown bat, these elements have remained the dominant TE type.
pilRNAs Formed Clusters, Which Were Not Enriched for TEs
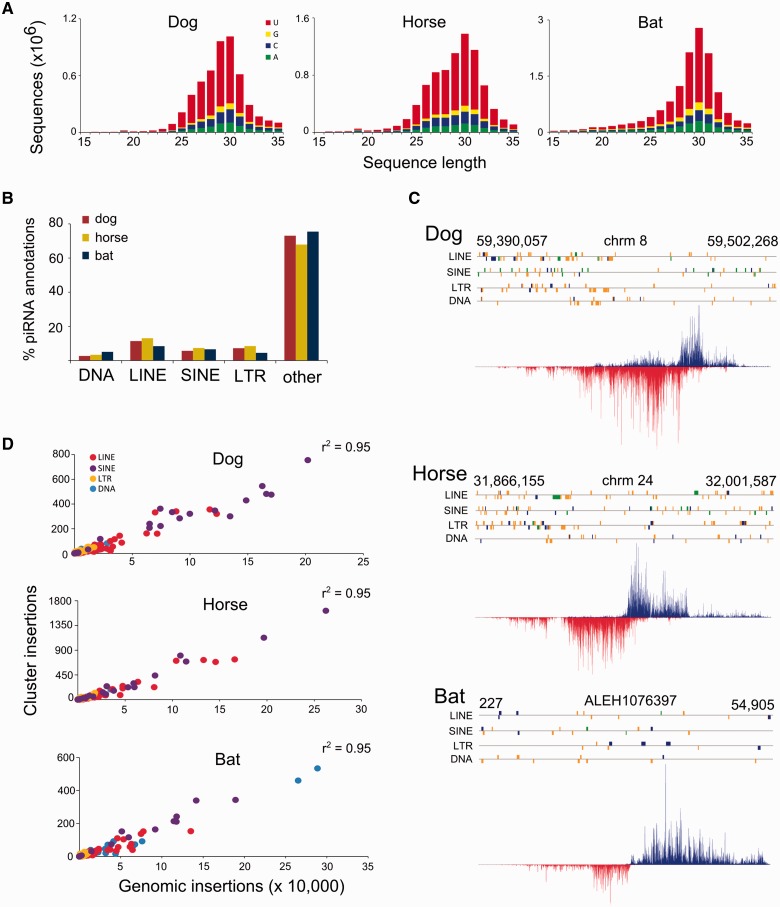
We then moved on to characterize pilRNA diversity in these three species. The sequenced small RNAs were similar to previously characterized piRNAs extracted from other mammalian testes (Lau et al. 2006; Yan et al. 2011; Liu et al. 2012). Specifically, >75% of the sequenced small RNAs were between 24 and 32 nucleotides long and there was a strong uridine bias in the first base position (fig. 2A), consistent with previously described pilRNAs. Allowing one mismatched base between the pilRNA and genome alignment, between 51% and 72% of the unique pilRNA sequences mapped to each genome (supplementary table S2, Supplementary Material online), and the majority of these pilRNAs mapped to non-TE-related genomic space (fig. 2B), which is characteristic of the pachytene piRNAs. Interestingly, the proportions of LINE, SINE, LTR, and DNA transposon derived pilRNAs were similar among the three species.
Fig. 2.—
Characteristics of the pilRNA sequences extracted from the sequenced small RNAs. (A) The length and distribution of unique small RNA sequences presented with the frequency of the first nucleotide illustrating the 5′ U bias. (B) Proportion of singly mapping pilRNAs that mapped to TE and non-TE space. (C) The TE content of one homologous cluster found in the dog, horse, and bat. TE insertions with genetic distances <0.1 from the family consensus are colored green, between 0.1 and 0.2 divergent are blue, and >0.2 are orange. pilRNAs that mapped anti-sense relative to the contig are red, and sense pilRNA are blue. (D) The raw number of genomic insertions plotted against the total number of cluster insertions per TE family. r2 values from simple linear regressions between the two variables are reported, P < 0.001 in all cases.
We then compared TE content within pilRNA clusters against genome wide patterns of TE accumulation. We restricted our analyses to clusters >10 kb because these are more likely to contain full length TE insertions. Our annotated clusters generally occupied unannotated space and generated ∼50% of the unique pilRNAs. We annotated 290, 376, and 221 clusters in the dog, horse, and bat genomes, respectively. By comparison, groups have annotated ∼100 clusters in the mouse (Girard et al. 2006; Beyret et al. 2012; Li et al. 2013). Although we annotated many more clusters in these genomes, cluster variation among species is typical. For example, Chirn et al. (2015) found that most piRNA clusters were species specific, few were conserved among species, and the number of piRNA clusters varied drastically.
TE content for these clusters varied among the species as well. For example, in a large cluster shared between the three species there were between 53 and 191 TE insertions, most of which were >20% diverged from the consensus sequence (fig. 2C). More than 98% of the pilRNA clusters included at least one TE insertion. However, we did not find that clusters were enriched for insertions relative to the genome. Rather, the number of genomic insertions from each family was tightly correlated with the total number of insertions among all clusters (fig. 2D), as observed by Hirano et al. (2014).
Ping-Pong Response
The next step was to explore relationships between different TE family characteristics and PPE using bivariate and multivariate regression analyses. To estimate the ping-pong pilRNA response, we mapped all pilRNA sequences to the TE consensus sequences and restricted our estimates of expression to pilRNAs that exhibited the signature of the ping-pong cycle, that is, 10 bp overlap between pairs of pilRNAs where a uridine is in the first position of the primary pilRNA, and an adenine is in the 10th position of the secondary pilRNA. A small percentage of pilRNAs mapped to the consensus sequences (∼3–6%), half of which were found as ping-pong pairs (supplementary table S2, Supplementary Material online). This was expected because pachytene piRNAs are the most abundant in mature testes and are generated independently of the ping-pong cycle (Beyret et al. 2012).
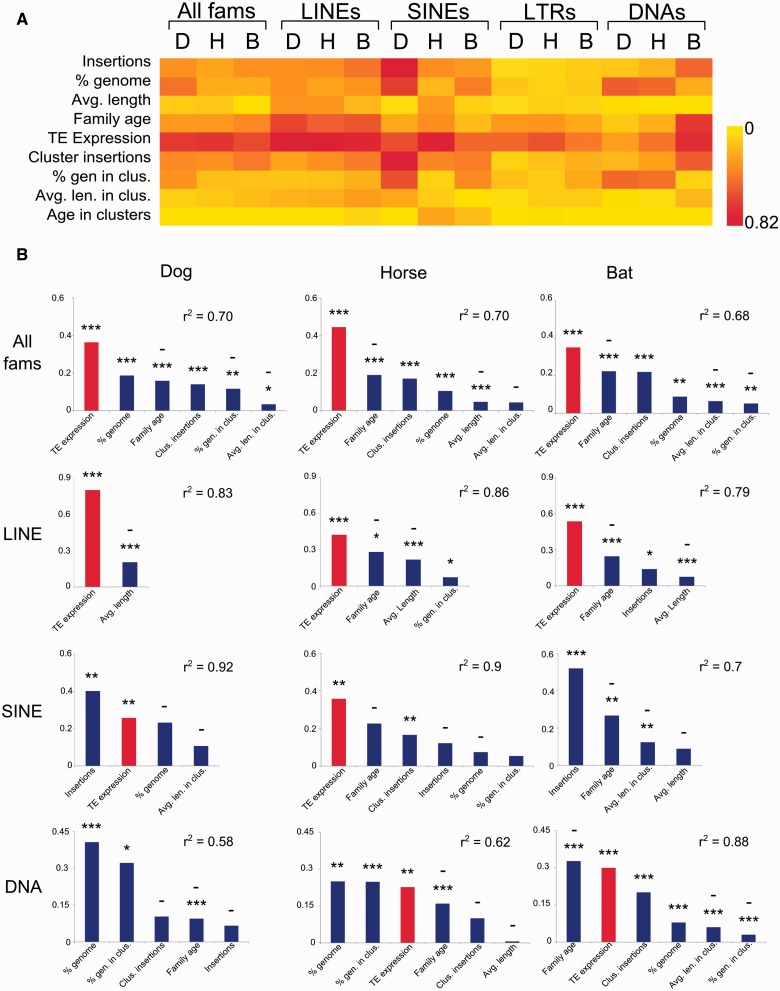
To estimate the level of pilRNA response, we initially discriminated between sense, antisense, and total PPE. However, because of the high correlation (r2 > 0.95) observed among the three measurements, we only measured the impact of TE parameters on total PPE. In each species, analyses were performed for all TE families combined and for LINEs, SINEs, LTRs, and DNA transposons. When we examined parameters individually, the largest r2 values were generally associated with estimates of TE family expression, especially in LINEs, SINEs, and DNA transposons in the bat (r2 = 0.47–0.81, P < 0.001; fig. 3A), that is, the most expressed families also generate the most ping-pong pilRNAs. In addition, the estimated age of the TE family was also a strong predictor of ping-pong pairs among LINEs in all three species (r2 = 0.51–0.60, P < 0.001; fig. 3A). We tested whether RSEM’s mapping parameters potentially biased RNASeq reads to younger elements by increasing the number of allowed mismatches in the seed region and found that increasing mismatches did not change the overall pattern that younger elements had higher expression and only made inferences from the default parameters. This aligns with previous predictions of the ping-pong model, given that younger TE families are often the most expressed (Lukic and Chen 2011; Mourier 2011). In contrast, abundance of TEs in the genome and piRNA clusters, measured as insertion number, total bases, and average length, typically had much lower r2 values suggesting that they are not as important with regards to the ping-pong response, with the exception of SINEs in the dog, which we discuss below in more detail.
Fig. 3.—
Results from univariate and multivariate statistical analyses relating different aspects of TE abundance to the pilRNA response, measured as PPE. (A) Heat map representing r2 values for independent linear regressions between PPE and each independent variable for dog (D), horse (H), and bat (B). (B) The independent variables selected for each step-wise regression analysis and relative importance of each variable in the model. TE expression is colored red. Negative interactions are indicated by a “–” above the variable. r2 values are reported for each model. Abbreviations: avg: average, gen: genome, clus: cluster, len: length. *P < 0.05, **P < 0.001, ***P < 0.0001.
We next explored relationships between TE metrics and PPE in a multivariate framework. We combined all variables into a single model and used stepwise regression to find an optimal sub-model, based on r2 scores. Stepwise regression also selects optimal variables when one or more variables correlate, such as genome insertions and cluster insertions (fig. 2D). Only estimates of subfamily expression had any meaningful and significant relationship with PPE for LTRs. However, since LTR families appear to be largely inactive, with negligible transcription or accumulation, they were excluded from further analyses. For SINEs, LINEs, and DNA transposons, the parameters selected and their relative contribution to PPE varied by type and species. Between two and six independent variables were selected for each multivariate regression model (fig. 3B). With the exception of DNA transposons in dog and horse, which are restricted to relatively old families that are no longer accumulating, the models yielded high r2 values (between 0.7 and 0.92), and in most cases included TE expression as the most important variable. A second common parameter selected among species and TE types was TE family age, which when selected, always had a negative relationship with PPE, that is, younger families had higher PPE. When all TE families were combined, the number of cluster insertions was selected in all three species. However, when TEs were separated by type, cluster insertions were only selected as part of the horse SINE and bat DNA transposon models (fig. 3B). The remaining pilRNA cluster parameters, if selected, were typically not among the most influential, and had negative relationships with PPE.
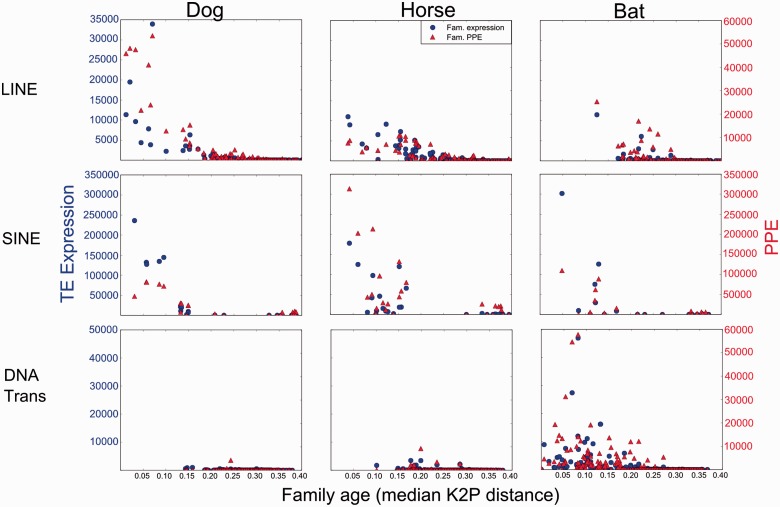
In the ping-pong model, TE expression is predicted to be a major determinant of ping-pong piRNA abundance. Our bivariate and multivariate regression analyses generally conform to this prediction, with variation among species and TE types. In general, TE families that are the youngest and most transcribed appear to elicit the strongest ping-pong response (fig. 4), particularly in LINEs. The relationships between expression and PPE appear to be more complex in SINEs, which contributed the largest fraction of TE transcripts in all species, ranging from ∼50% in bat to 80% in dog. However, the piRNA response to SINE expression varies greatly among the three taxa. In horse, SINEs are the most highly expressed TEs and also elicit the strongest pilRNA response (fig. 4). In contrast, expression of ping-pong pilRNAs in the dog correlates more with the total number of SINE family insertions (fig. 3A) than with SINE expression, and the dog SINE PPE is generally much lower than SINE transcription levels (fig. 4). In the bat, the Ves SINE family is the only significantly expressed family and elicits a weak ping-pong response (fig. 4). Finally, we found a correlation between DNA transposon expression and PPE in the bat (fig. 3A). However, because recent DNA transposon activity is unique to the bat, a meaningful comparison with dog and horse is not possible.
Fig. 4.—
Separate dot plots illustrating the relationship between TE expression, TE family age, and PPE. For each TE family, we plotted the expression values (blue) and PPE (red) plotted against family age (median K2P) separately for LINEs, SINEs, and DNA transposons in each species.
Discussion
The relationships among TE expression, piRNAs, and TE accumulation are not entirely clear, and it is challenging to summarize the outcome of these interactions. In an attempt to identify general patterns, we examined three laurasiatherian mammals with markedly different TE landscapes, patterns of TE expression and pilRNA repertoires to better understand the complex relationship between host defenses and TE accumulation over time. We explored the contribution of different measures of TE activity to the piRNA response, and in line with predictions from the mammalian ping-pong model, univariate and multivariate analyses identified the abundance of TE transcripts as a strong predictor of the abundance of ping-pong pilRNAs.
Ping-Pong piRNAs Target the Most Highly Transcribed Families
The ping-pong model suggests that the TE families that are most transpositionally active, probably the most deleterious, would be the most important contributors to piRNA repertoires. We generally found that this prediction was satisfied (see fig. 4). When all TE families were considered, the abundance of family transcripts was the best predictor of ping-pong piRNA abundance in bivariate and multivariate analyses. When comparing within the different TE types, families that were the most transcriptionally active, usually elicited a stronger pilRNA response, in agreement with results from mouse, reported by Mourier (2011).
DNA transposons are the most recently active elements in the big brown bat genome. Thomas et al. (2014) found evidence of low-level ongoing Helitron accumulation and Mitra et al. (2013) and Ray et al. (2008) suggested that piggyBac elements were still accumulating in the closely related little brown bat, M. lucifugus, raising the possibility that these elements are still actively inserting in this genome at low rates. There was an abundance of DNA transposon transcripts and a statistically significant pilRNA response to these elements in the testis transcriptome of the big brown bat (figs. 3A, 3B and 4). This was unexpected for several reasons. DNA transposons do not require an RNA intermediate for transposition; therefore, it was unexpected to detect RNA associated with these elements, especially elements that are annotated as nonautonomous, which do not encode the proteins needed to mobilize. There are at least two plausible explanations as to why we observed expressed DNA transposons and a subsequent piRNA-response. First, there are large numbers of DNA transposon insertions in the bat genome. The sheer density of these insertions suggests that at least a subset will exist in close proximity to a promoter, leading to spurious transcription and incorporation into the ping-pong cycle. Second, several families of transposons harbor promoters that act to encourage transcription of their transposase and those transcripts could be targeted by piRNAs. Regardless of the mechanism, our results indicate a statistically significant relationship between DNA transposons and ping-pong pilRNAs that may suggest a defensive response. The strong response to DNA transposons may suggest an adaptable defense to both Class I and Class II TEs. Because vesper bats are the only known vertebrate to harbor actively mobilizing DNA transposons, this relationship could be worthy of additional investigation.
piRNA Cluster Likely Do Not Regulate TEs in Mammals
In contrast to Drosophila, where piRNA clusters are thought to give rise to primary piRNAs (Kelleher and Barbash 2013), TE transcripts are proposed as the substrates for primary piRNA processing in mammals (Girard et al. 2006; Aravin et al. 2008). However, dissenting views exist. For example, Ha et al. (2014) and Hirano et al. (2014) both suggested pachytene piRNA clusters could be a source of antisense piRNAs used in the ping-pong cycle. Our results suggest that there is no enrichment for mammalian piRNAs derived from these clusters in the TE silencing pathway.
We tested the role of piRNA clusters as determinants of the ping-pong response in a statistical framework and found that the abundance of insertions within clusters strongly correlated with overall genome insertions in all three species, but was not the most important factor with regard to PPE. Furthermore, we found total bases in clusters and the median age of cluster insertions did not correlate well with PPE. Interestingly, when all TE families were taken into consideration under the multivariate framework, cluster insertions were included in the final models, but TE expression was always the most important contributor to PPE. These results suggest that piRNA clusters may play some role in TE silencing but the extent of that role is yet to be determined.
Because of the many TE insertions that exist in mammalian genomes, it is difficult to determine the ultimate source of any single TE-derived piRNA, much less whether it arose from a mobilizing TE transcript or from an insertion that lies within a piRNA cluster. However, the families with the most cluster insertions were generally older, had few mapped ping-pong pilRNAs, and there was ultimately little relationship between the number of insertions in clusters and the abundance of ping-pong pilRNAs. Perhaps piRNAs that are processed from cluster insertions are incorporated into the ping-pong cycle, but unlike in Drosophila where clusters seem to act as TE “traps” (Malone and Hannon 2009), pachytene piRNA clusters do not appear to function this way in mammals.
Complex Relationship between TE Accumulation and Genome Defense
Our results have implications for understanding the relationship between TE transcription and accumulation. We generally assume a dearth of recent deposition for a given TE is related to decreases in TE transcription in the recent past. This assumption holds for LINEs. There is an abundance of young LINE transcripts in the dog and a corresponding abundance of young LINE insertions. Furthermore, there is little expression of full-length LINEs in the bat and very few recent insertions. The horse is intermediate between the two.
SINEs presented a different case. Based on their genomic abundance, we expected that SINEs would only be highly expressed in the dog. However, after taking their sequence length into account, young SINE families were the most transcribed elements in all three species, but recent SINE accumulation is only seen in the dog genome. Figure 4 suggests that the abundance of SINE insertions in the dog could be the result of a reduced pilRNA response. Although young SINE families have the highest abundance of ping-pong pilRNAs in the dog, the ping-pong response appears weak in relation to the level of SINE expression. The opposite is true in the horse genome, where SINEs are being expressed at comparable levels but the ping-pong piRNAs appear to offer a formidable response, potentially preventing the eventual reverse transcription and insertion. In bats, yet a third scenario appears to have played out, revealing the complexity of this system. There, the Ves SINE family is highly transcribed, the piRNA response appears limited, but there are very few recent Ves insertions. Unlike LINEs, which are completely autonomous, SINEs depend on the reverse transcriptase and endonuclease genes encoded by LINE elements to mobilize and this might account for the lack of insertions.
In summary, our data indicate that the level of piRNA response against a given TE subfamily is most strongly associated to the abundance of the corresponding transcripts, with other factors, such as the age of the subfamily playing a more modest role. Our analyses suggest that piRNA responses are able to provide protection against TE invasion in mammalian genomes, but that TEs are still able to propagate even in the presence of a putatively robust piRNA response. Furthermore, it appears that the interplay between TEs and piRNAs is distinct among species and TE types. Expanding comparative studies of piRNAs and TEs to a broader array of mammals could help uncover a general model to account for the relationship between TE abundance at the genome and the piRNA response.
Supplementary Material
Supplementary tables S1 and S2 and supplementary data S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We acknowledge the support from the National Science Foundation (EPS-0903787, DBI-1004842, DBI-1262901, DEB-1354147, MCB-0841821, and DEB-1020865). Additional support was provided by the College of Agriculture and Life Sciences at Mississippi State University and the College of Arts and Sciences at Texas Tech University. Tissues were provided by the College of Veterinary Medicine at Mississippi State University, Jeremiah Dumas, and the Natural Sciences Research Laboratory at the Museum of Texas Tech University. The Broad Institute Genomics Platform and Genome Sequencing and Analysis Program, Federica Di Palma, and Kerstin Lindblad-Toh made the data for E. fuscus available. Raw small RNA and transcriptome sequences are available under the BioProject ID PRJNA290346.
Literature Cited
- Aravin AA, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207. [DOI] [PubMed] [Google Scholar]
- Aravin AA, et al. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 31:785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747. [DOI] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H. 2012. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 22:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103. [DOI] [PubMed] [Google Scholar]
- Brookfield JFY, Johnson LJ. 2006. The evolution of mobile DNAs: when will transposons create phylogenies that look as if there is a master gene? Genetics 173:1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, et al. 2005. Alu retrotransposition-mediated deletion. J Mol Biol. 348:791–800. [DOI] [PubMed] [Google Scholar]
- Chirn G-W, et al. 2015. Conserved piRNA expression from a distinct set of piRNA cluster loci in eutherian mammals. PLoS Genet. 11:e1005652.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315–325. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202. [DOI] [PubMed] [Google Scholar]
- Gou L-T, et al. 2014. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24:680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H, et al. 2014. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 15:545.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, et al. 2005. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 33:4040–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, et al. 2014. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 20:1223–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Meister G. 2008. The Argonaute protein family. Genome Biol. 9:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2007. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 23:521–529. [DOI] [PubMed] [Google Scholar]
- Kelleher ES, Barbash DA. 2013. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 30:1816–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Slazberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, et al. 2006. Characterization of the piRNA complex from rat testes. Science 313:363–367. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 50:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, et al. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13:358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, et al. 2012. Discovery of potential piRNAs from next generation sequences of the sexually mature porcine testes. PLoS One 7:e34770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukic S, Chen K. 2011. Human piRNAs are under selection in Africans and repress transposable elements. Mol Biol Evol. 28:3061–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. 2009. Small RNAs as guardians of the genome. Cell 136:656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334:521–524. [DOI] [PubMed] [Google Scholar]
- Miller-Butterworth CM, et al. 2007. A family matter: conclusive resolution of the taxonomic position of the long-fingered bats, Miniopterus. Mol Biol Evol. 24:1553–1561. [DOI] [PubMed] [Google Scholar]
- Mitra R, et al. 2013. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc Natl Acad Sci U S A. 110:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier T. 2011. Retrotransposon-centered analysis of piRNA targeting shows a shift from active to passive retrotransposon transcription in developing mouse testes. BMC Genomics 12:440.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Boeke JD. 2007. Mighty Piwis defend the germline against genome intruders. Cell 129:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán HJT, et al. 2012. Survey sequencing reveals elevated DNA transposon activity, novel elements, and variation in repetitive landscapes among vesper bats. Genome Biol Evol. 4:575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, et al. 2014. Large numbers of novel miRNAs originate from DNA transposons and are coincident with a large species radiation in bats. Mol Biol Evol. 31:1536–1545. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 104:1895–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, et al. 2008. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 18:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Pagan HJT, Thompson ML, Stevens RD. 2007. Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Mol Biol Evol. 24:632–639. [DOI] [PubMed] [Google Scholar]
- Reuter M, et al. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480:264–267. [DOI] [PubMed] [Google Scholar]
- Rozenkranz D, Rudloff S, Bastuck K, Ketting RF, Zischler H. 2015. Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals. RNA 21:911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Siomi MC. 2010. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell. 19:687–697. [DOI] [PubMed] [Google Scholar]
- Sen SK, et al. 2006. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 79:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defense. Nat Rev Mol Cell Biol. 12:246–258. [DOI] [PubMed] [Google Scholar]
- Thomas J, Phillips CD, Baker RJ, Pritham EJ. 2014. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol. 6:2595–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, et al. 2011. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39:6596–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CT, et al. 2005. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 3:e110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.