Abstract
Understanding the evolution of animal societies, considered to be a major transition in evolution, is a key topic in evolutionary biology. Recently, new gateways for understanding social evolution have opened up due to advances in genomics, allowing for unprecedented opportunities in studying social behavior on a molecular level. In particular, highly eusocial insect species (caste-containing societies with nonreproductives that care for siblings) have taken center stage in studies of the molecular evolution of sociality. Despite advances in genomic studies of both solitary and eusocial insects, we still lack genomic resources for early insect societies. To study the genetic basis of social traits requires comparison of genomes from a diversity of organisms ranging from solitary to complex social forms. Here we present the genome of a subsocial bee, Ceratina calcarata. This study begins to address the types of genomic changes associated with the earliest origins of simple sociality using the small carpenter bee. Genes associated with lipid transport and DNA recombination have undergone positive selection in C. calcarata relative to other bee lineages. Furthermore, we provide the first methylome of a noneusocial bee. Ceratina calcarata contains the complete enzymatic toolkit for DNA methylation. As in the honey bee and many other holometabolous insects, DNA methylation is targeted to exons. The addition of this genome allows for new lines of research into the genetic and epigenetic precursors to complex social behaviors.
Keywords: small carpenter bee, epigenetics, maternal care, social evolution, insect genome
Introduction
The goal of sociogenomics is to explore the molecular genetic basis of social life (Robinson et al. 2005). This field was spearheaded by sequencing the first genome of an eusocial species, the honey bee, Apis mellifera (Honeybee Genome Sequencing Consortium 2006). Following the publication of the honey bee genome, researchers sequenced the genomes of many other eusocial insect species and their solitary relatives, including seven ants (Bonasio et al. 2010; Nygaard et al. 2011; Smith, Smith, et al. 2011; Smith, Zimin, et al. 2011; Suen et al. 2011; Wurm et al. 2011) and nine additional bees (Kocher et al. 2013; Kapheim et al. 2015; Sadd et al. 2015). These genomes revealed much about the importance of gene family expansions, chemical communication, and epigenetics to caste determination (Gadau et al. 2012; Simola et al. 2013; Yan et al. 2014). Although we are beginning to elucidate the genetic basis of highly eusocial behaviors, hallmarked by overlapping generations, reproductive division of labor, and cooperative brood care, we still understand very little about the genetic architecture of incipient sociality. Indeed, identification of the genetic precursors to advanced eusociality necessitates a comparison of highly social species with those species possibly transitioning to highly social behaviors.
A prominent theory in the transition to sociality is that incipient groups consisted of a mother and her offspring (Alexander et al. 1991). Maternal care is thus a key step in the evolution of social behavior (Michener 1974; Anderson 1984; Gadagkar 1990). Subsocial species often display behaviors thought to be necessary precursors to highly social behavior, such as maternal care, nest loyalty, cohabitation, and adult longevity (Sakagami and Maeta 1977; Tallamy and Wood 1986; Crespi 1994; Costa 2006; Wilson 2008). To date, there are published genomes of ten bee species (Kocher et al. 2013; Kapheim et al. 2015; Sadd et al. 2015), but there are no sequenced genomes of a subsocial bee species.
Because the evolutionary path from solitary and eusocial behavior likely involves a subsocial intermediate, comparisons between solitary and subsocial species closely related to eusocial taxa may provide evidence of the genetic scaffolding for more complex social behaviors (West-Eberhard 1967; Rehan and Toth 2015). Ceratina small carpenter bees (Hymenoptera: Apidae) are a diverse and cosmopolitan genus (Michener 2007). Most species of this genus are considered solitary, but sociality occurs naturally in some species (Sakagami and Laroca 1971; Sakagami and Maeta 1984; Rehan et al. 2009; Rehan et al. 2014) and can be induced artificially in others (Chandler 1975; Sakagami and Maeta 1987, 1989, 1995). This natural variation provides a unique opportunity to explore the transition from solitary to social life (Michener 1985). Ceratina are members of the subfamily Xylocopinae in the bee family Apidae and provide a phylogenetic contrast and independent origin of eusociality to honey bees and bumble bees, members of the subfamily Apinae (Cardinal and Danforth 2011; Rehan et al. 2012).
Ceratina calcarata (Hymenoptera: Apidae) is a small carpenter bee native to eastern North America (Rehan and Sheffield 2011; Shell and Rehan 2016). Unlike solitary species, such as the leaf cutter bee Megachile rotundata, C. calcarata displays prolonged maternal care, mother–adult offspring interactions, and mutual tolerance (Rehan et al. 2014). Ceratina females make their nests in the pith of dead, broken twigs (Daly 1973). The mother lays eggs within the stem and provides each offspring with its own nutrition-rich pollen ball (McFrederick and Rehan 2016). As the offspring grow, the mother will protect and groom her offspring (Michener 1985; Rehan and Richards 2010a). Ceratina calcarata females are long-lived and nest loyal, and provide care to their offspring throughout development and even into adulthood (Rehan and Richards 2010b). In spring, overwintered females emerge from their hibernacula, dispersing to construct new nests in twigs and stems. In summer, mothers remain in the nest to guard against predators and parasites. In autumn, offspring emerge and are primarily fed by the mother or female siblings prior to overwintering in the natal nest (Rehan et al. 2014). Male and female offspring overwinter together as adults and all individuals disperse and mate in the spring.
Ceratina calcarata is of special interest to the study of social evolution because in addition to prolonged mother–offspring interactions and overlapping generations this species also exhibits reproductive division of labor (Rehan et al. 2014). Mothers have complete control over the size and sex of their offspring and produce a special class of dwarf eldest daughters as well as regular daughters (Rehan and Richards 2010b). Regular daughters emerge in the natal nest, do not forage, and wait until spring to disperse and establish their own nests solitarily. Conversely, dwarf eldest daughters are first to emerge in autumn, forage and feed siblings prior to overwintering, and do not survive the winter (Rehan and Richards 2010a). The sibling care without reproduction exhibited in C. calcarata is comparable in some ways with worker behavior exhibited in eusocial species such as the honey bee. However, C. calcarata exhibits alloparental care but not cooperative brood care in the strict sense (Michener 1974). The distinction here is that dwarf eldest daughters feed adult siblings but do not contribute to the next generation by provisioning immature offspring. Thus, C. calcarata exhibits two of the three hallmarks of eusociality: Reproductive division of labor and overlapping generations, yet this species does not exhibit cooperative brood care and therefore remains a bee on the brink of eusociality (Rehan et al. 2014). This system provides an excellent model for studying the genetic and epigenetic factors associated with maternal care, sibling care, and transitional stages of social behavior.
Here we present the genome of the small carpenter bee, C. calcarata. We also compare these data with the previously published bee genomes to identify genomic regions under selection at the brink of sociality. Finally, we provide the DNA methylome of C. calcarata. The hypothesis that DNA methylation in bee genomes may be correlated with social complexity has garnered recent support from an indirect measure of DNA methylation (Kapheim et al. 2015). However, empirical assessment of DNA methylation has remained limited among bees to A. mellifera. Our study provides the first direct comparison of DNA methylation in A. mellifera with a bee with markedly less complex social interactions.
Methods and Materials
Sample Preparation and Sequencing
DNA was isolated from the whole body of two haploid male C. calcarata collected in Durham, NH, in the winter of 2013. DNA extractions were performed using a Qiagen Genomic-tip 20/G kit (Valencia, CA) and standard protocol. Genomic DNA was used to generate nonamplified 150-bp DNA libraries for Illumina sequencing. In order to improve genome assembly, genomic DNA was extracted from the whole body of ten female C. calcarata as above, pooled, and used to construct 3–4 kb and 5–8 kb mate pair libraries. Four paired-end libraries with insert sizes ranging from 150 bp to 8 kb were constructed and sequenced on Illumina HiSeq 2500. Before filtering this produced 45 Gb of raw sequence data (150.5 million read pairs, 2 × 150 bp each). Low quality reads, reads with a high proportion of Ns, or poly-A structures were filtered and paired-end reads whose mates overlapped were merged prior to assembly. PRINSEQ (Schmeider and Edwards 2011) was used to filter out reads containing a high number of Ns or poly-A 5′ or 3′ runs (≥15 poly-A nts, ≥50% Ns). FLASH (Magoč and Salzberg 2011) was used to merge overlapping reads of the same pair (due to fragment size < read length × 2 on many reads). All genomic data have been submitted to public repositories and can be found using the NCBI BioProject number PRJNA299559.
Genome Assembly
Reads were checked with FastQC (Patel and Jain 2012) for quality metrics (both before and after correction/adapter removal). Trimmomatic (Bolger et al. 2014) was then used to remove adapter contamination using Illumina nextera-related adapter sequences. FastUniq was then used to remove duplicates from each library (Xu et al. 2012). SOAPdenovo’s error correction module (Luo et al. 2012) was then used to correct overt errors in the reads using a kmer size of 23. Postfiltering, 32 Gb of sequence remained (107,384,903 read pairs; supplementary table S1, Supplementary Material online).
Contigs and initial scaffolds were constructed using SOAPdenovo (Luo et al. 2012) with a kmer size of 57. We then used REAPR to break putatively misassembled scaffolds at weak points (Hunt, Kikuchi, et al. 2013). SSpace was then used to rescaffold broken scaffolds/contigs (Boetzer et al. 2011). GapFiller (Boetzer and Pirovano 2012) was then used to fill assembly gaps left after scaffolding. Finally, L_RNA_Scaffolder (Xue et al. 2013) was used along with independently assembled transcripts (Trinity; Haas et al. 2013) to further scaffold contigs and scaffolds that were broken in the middle of gene annotations, followed by a final round of gap filling with GapFiller (Boetzer and Pirovano 2012). VecScreen (NCBI) was used to scan the assembly for contamination from sequence adapters and vectors, and BLASTN was used to search the nucleotide database for foreign DNA (Wolbachia, Escherichia coli, bacterial). Scaffolds showing contamination were then removed or trimmed accordingly.
Genome Annotation
Gene annotation was performed with the MAKER2 pipeline (Holt and Yandell, 2011). Briefly, the gene prediction algorithms of SNAP (Korf 2004), Augustus (Stanke and Waack 2003), and GeneMark (Lukashin and Borodovsky 1998) were iteratively trained by C. calcarata RNA-seq data and homologous proteins in other bee taxa (see Methods in supplementary material S1, Supplementary Material online). Annotations with a MAKER AED score above 0.75 or no significant InterProScan domains were flagged as weakly supported. Our filtered gene set was produced requiring a MAKER2 AED of less than 0.75. AED corresponds to the level of external support (transcriptome coverage or homology to other bee gene sets) a given MAKER2 gene prediction received, with our cutoff (< 0.75) identified as reasonable in a previous publication (Holt and Yandell 2011).
RepeatMasker (Smit et al. 2015) was used with default parameters to identify repetitive elements based on the Repbase transposable element library (“species = all”) and a custom species-specific repeat library generated by RepeatModeler (Jurka et al. 2005). Functional annotation of genes was performed with Blast2GO (Conesa et al. 2005), using default settings for GO assignment based on querying of the NR database (May 1, 2015) using BLASTP (E-value < 10−5), along with all significant InterProScan hits. KEGG orthology was assigned online using the KAAS server (Moriya et al. 2007). Genome assembly and annotation completeness were assessed with Benchmarking Universal Single-Copy Orthologs (BUSCO; Simão et al. 2015). Specifically, the OrthoDB BUSCO pipeline (v.1.2; October 2013) was employed to test for the presence and completeness of orthologs to A. mellifera sequences from arthropod-level BUSCOs in C. calcarata.
Gene Family Expansion and Contraction
To identify orthologous groups for analysis of gene family expansion and contraction, we used OrthoMCL (Li et al. 2003) to assign proteins from our study species, C. calcarata, and ten other bee species: Apis florae, A. mellifera, Bombus impatiens, Bombus terrestris, Dufourea novaeangliae, Eufriesea mexicana, Habropoda laboriosa, Lasioglossum albipes, Melipona quadrifasciata, and M. rotundata into gene families. Briefly, the longest transcript isoform only for each gene was used and we only considered proteins larger than 50 amino acids. An all-versus-all sequence search was performed using BLASTP with default parameters and cut-off E-values of 10−7. The output was uploaded to the OrthoMCL website and run using the default parameters (cutoff of 10−5 and 50% match).
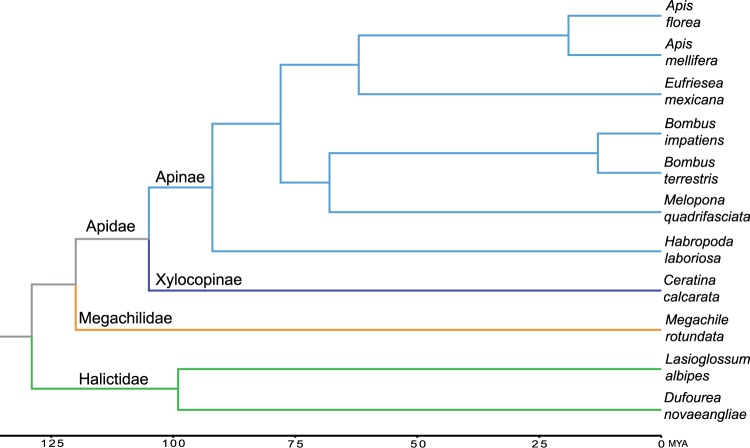
To identify gene families that have undergone significant size changes, we used the program COUNT (Csűros 2010). COUNT uses phylogenetic birth-and-death models in probabilistic inference to calculate the probability of gene family evolution across the phylogeny. As an input phylogenetic tree (fig. 1), we used the following: (Hlab:91,(Ccal:65,((Mqua:68,(Bimp:13,Bter:13):55):10,(Emex:62,(Amel:19,Aflo:19):43):16):14):13)):15,Mrot:106):9,(Dnov:85,Lalb:85):30) based on Kapheim et al. (2015).
Fig. 1.—
Phylogenetic placement of Ceratina calcarata. Divergence dates taken from Kapheim et al. (2015) and Rehan and Schwarz (2015). Ceratina and Apis diverged approximately 105 Ma. Tree topology was confirmed by an analysis of orthologous 4-fold degenerate sites (supplementary material, Supplementary Material online).
The rate optimization was computed by COUNT using the default parameters and posterior probabilities for each gene family were given. These gene families were then described using Pfam (Finn et al. 2014) domains and keywords and subsequently sorted into gene ontology (GO) terms (Ashburner et al. 2000). By comparing the GO/IRP/KEGG enrichment results for C. calcarata with the previously published bee genomes, we identified genes specific to C. calcarata. To identify genes lost or absent from the C. calcarata genome, we identified genes found in the other ten bee species, but not in C. calcarata (see Methods in supplementary material S1, Supplementary Material online).
Molecular Evolution
For alignments used in PAML analyses, we identified sequences with homology to putative 1-to-1 ortholog groups previously established among ten bee genomes by OrthoDB (Kriventseva et al. 2015). We first performed reciprocal BLASTP searches between the C. calcarata OGS and published gene sets of each of the ten bee species. Ceratina calcarata genes were then assigned to a given ortholog group if the C. calcarata gene was a reciprocal best hit to the respective ortholog in at least 60% of the ten bees.
For multiple sequence alignment, PRANK was used to align amino acid sequences then back-translate to coding sequences (CDS) (with the option “+F”) from within the Guidance wrapper (v.2.0; Löytynoja and Goldman 2005; Sela et al. 2015). Initial gene trees produced by guidance were then improved using RAxML (Stamatakis 2014) with the GTRGAMMA model. Gblocks was then used to extract higher confidence alignment blocks, while maintaining reading frame, for downstream analysis (Talavera and Castresana 2007).
PAML (v.4.7) was then used to estimate synonymous and nonsynonymous substitution rates (dS and dN) and ω (dN/dS) with free ratios for each branch (Yang 2007). Values corresponding to the C. calcarata branch were used in analyses. To test for further evidence of positive selection, we used the branch-site test of positive selection (branch-site model A, test 2; Zhang et al. 2005), which uses likelihood ratio tests (LRTs) to detect positive selection on a small number of sites along a specific lineage. Ceratina calcarata was set as the foreground branch, with two dN/dS ratios for branches (model = 2). For the alternative model, ω2 was estimated from the data (fix_omega = 0, omega = 1, NSsites = 2), while the null model fixes ω2 at 1 (fix_omega = 1, omega = 1, NSsites = 2) as described in PAML documentation and sample files (Yang 2007). The log-likelihoods for the null and alternative models were used to calculate an LRT statistic, which was then compared against the χ2df = 1 distribution. P values were then corrected for multiple testing using the method of Benjamini and Hochberg (1995), with a corrected P value threshold of 0.05 being used to assign significance.
Species Phylogeny
Four-fold degenerate sites were extracted from codon sequence alignments after treating with Gblocks. The msa_view utility (from the PhastCons program suite; Siepel and Haussler 2004) was used to extract and concatenate 4-fold degenerate sites from the alignments. PhyloFit (REV model) was then used to fit tree models to subwindows (l = 20,000; step = 10,000) of the concatenated 4-fold degenerate sites, which were then averaged using phyloBoot (supplementary material S1, Supplementary Material online).
Gene Expression
RNA-seq reads (Rehan et al. 2014; Durant et al. 2016) were aligned to the reference genome using Tophat2 with default options (bar limiting maximum intron length to -I 25000). Cuffnorm was used to produce normalized (geometric) FPKM values for genes (Trapnell et al. 2012). Raw data are available on the NCBI Sequence Read Archive with accession number SRX541305. Six classes of females were assayed and are represented in these studies: Spring mothers, summer mothers, autumn mothers, autumn workers, autumn gynes, and overwintering gynes. Each of the six RNA-seq libraries comprised whole brain tissue samples from 20-pooled females. Reads were filtered for quality (threshold greater than or equal to 20) with a length threshold of 50 bases. After adapter removal, approximately 20% of the reads were removed from the libraries after quality filtering (98 million raw Illumina reads; 79 million reads after preprocessing).
DNA Methylation
In order to assess DNA methylation at a single-nucleotide resolution, genomic DNA was extracted from ten whole body females using a standard phenol–chloroform protocol and then pooled into one sample for bisulfite conversion and sequencing. Unmethylated enterobacteria phage lambda DNA (GenBank accession: J02459.1) was added to C. calcarata genomic DNA as a control for bisulfite conversion efficiency. Bisulfite conversion and sequencing on the Illumina HiSeq platform was performed by Beijing Genomics Institute (Shenzhen, China). We obtained 37.9× mean coverage of genomic CpG sites. We used the program Bismark (Krueger and Andrews 2011) to align raw sequencing reads to the C. calcarata reference genome. Apis mellifera bisulfite-sequencing data from a worker whole body (Zemach et al. 2010) and from queen and worker brains (Lyko et al. 2010) were previously published, but raw reads were mapped and processed in the same manner as C. calcarata. For comparison with C. calcarata DNA methylation, we first examined data from an A. mellifera worker whole body library (Zemach et al. 2010) to achieve comparability in tissues assayed. Second, we examined data produced by merging worker whole body, worker brain, and queen brain libraries (Lyko et al. 2010; Zemach et al. 2010) to achieve comparability in read depth between species. Results from each comparison were qualitatively similar.
Significantly methylated CpG sites were assessed using a binomial test, which incorporated deamination rate (from our unmethylated control) as the probability of success, and assigned a significance value to each CpG site related to the number of unconverted reads (putatively methylated Cs) as they compare with the expected number from control (Lyko et al. 2010). Resulting P values were then adjusted for multiple testing using the method of Benjamini and Hochberg (1995). Only sites with false discovery rate (FDR) corrected binomial P values <0.05 and >3 reads sequence coverage were considered “methylated.”
Fractional methylation values were calculated for each CpG site as mCG/CG, where mCG is the number of reads with a methylated cytosine at a CpG site and CG is the total number of reads mapped to the site. Fractional methylation was calculated for specific genomic features (e.g., exons and introns) as the mean of all CpG fractional methylation values within that feature.
Results and Discussion
Genome and Annotation
The C. calcarata genome size is estimated at 364 Mb after using kmer-based error correction in QUAKE (Kelley et al. 2010). The final assembly has an N50 of 74 kb and a total length of 261.9 Mb (table 1). Completeness of the assembly was first assessed using the CEGMA pipeline (Parra et al. 2007). Of the 248 core eukaryotic genes (CEGs), 247 CEGs were completely assembled in the C. calcarata genome. A combination of RNA-sequencing, de novo, and homology-based predictions generated the unfiltered gene set including 20,693 predicted genes. A more conservative “filtered” official gene set was produced including 11,310 predicted genes. The closest relative to C. calcarata with a genome sequence is the honey bee, A. mellifera, and our results of genome size and gene count are comparable with that of the other ten sequenced bee species (supplementary table S2, Supplementary Material online).
Table 1.
Genome Assembly Statistics
| Contigs (n) | 46,222 |
| Largest contig (bp) | 52,361 |
| Scaffolds > 1 kb (n) | 3,577 |
| N50 scaffolds (bp) | 73,643 |
| Scaffolds > N50 (n) | 83 |
| Largest scaffold (bp) | 3,675,446 |
| Predicted genes | Filtered: 11,310; unfiltered: 20,693 |
| Ultraconserved core eukaryotic genes (complete/partial, %) | 99.6/100 |
| Arthropod BUSCOs in genome assembly | 99.3% (97.0% single copy, 2.3% multicopy) |
| BUSCOs in predicted genes | 96.6% (94.2% single copy, 2.4% multicopy; same results for filtered and unfiltered gene predictions) |
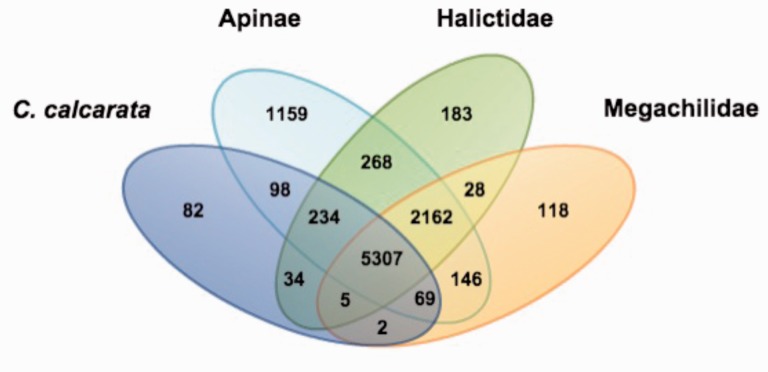
The 11,310 predicted genes comprise 7,888 gene families (supplementary table S3, Supplementary Material online). A total of 5,307 gene families are shared among all bee species and there are 82 predicted orthogroups unique to C. calcarata (fig. 2). The orthogroup and Pfam protein family results for C. calcarata-specific genes are listed in supplementary table S4, Supplementary Material online. Among these 82 unique orthogroups many are associated with zinc finger protein binding and have overrepresentation of protein domain-specific binding (OG5_158738).
Fig. 2.—
Gene family overlap among four bee lineages. The 11 species were sorted into 4 groups based on taxonomic classification: (1) Ceratina calcarata (subfamily Xylocopinae) versus (2) subfamily Apinae (Apis florae, Apis mellifera, Bombus impatiens, Bombus terrestris, Eufriesea mexicana, Habropoda laboriosa, and Melipona quadrifasciata) versus (3) family Halictidae (Lasioglossum albipes and Dufourea novaeangliae) versus (4) family Megachilidae (Megachile rotundata). Numbers indicate the gene families in each comparison. A total of 5,307 gene families are shared among all 11 bee species.
Gene Family Expansion
A total of 306 gene families had been significantly expanded in C. calcarata (supplementary table S5, Supplementary Material online). Two notable gene families appear to be expanded in the C. calcarata lineage: Zinc finger and odorant receptors proteins. Zinc finger proteins have been implicated in gene regulation in female reproduction and are reportedly expanded in the eusocial termite (Zootermopsis nevadensis; Terrapon et al. 2014). We found an expansion of zinc finger proteins in C. calcarata (OG5_126627) with 50 copies in their genome compared with 16 in L. albipes and only 2 copies in A. mellifera. Zinc finger proteins are known transcription factors and recent analyses of ten bee genomes revealed increasing number of zinc finger binding sites in eusocial versus solitary species (Kapheim et al. 2015). Detailed comparative analyses within and between lineages of bees will be required to reveal the role and regulation of transcription factors across the social spectrum from solitary to eusocial (Rehan and Toth 2015).
Odorant receptors are a large family of transmembrane chemosensory proteins that detect odor molecules and initiate a signal transduction cascade to neurons in the brain (Sanchez-Gracia et al. 2009). The C. calcarata genome contains 16 members of this gene family, in contrast to 4 in M. rotundata and 2 in L. albipes (OG5_168510). Recent analysis of ten bee genomes (Kapheim et al. 2015) found odorant receptors to be expanded in complex eusocial lineages (Apinae) compared with solitary (Megachilidae and Halictidae) species. However, we found expansion of odorant receptors in a subsocial apid bee, consistent with gene family expansion common to the Apidae and likely preceding the evolution of eusociality.
Gene Family Contraction
A total of 281 gene families have been significantly contracted in C. calcarata (supplementary table S6, Supplementary Material online). One underrepresented protein domain of interest is the Perilipin gene family. Perilipin is also known as lipid droplet-associated protein or PLIN, a protein that, in Drosophila, is encoded by the PLIN gene. The perilipins are a family of proteins that associate with the surface of lipid droplets. Phosphorylation of perilipin is essential for the mobilization of fats in adipose tissue (Beller et al. 2010). Past studies have implicated lipid storage and metabolism as key to overwintering physiology in this and other Hymenopteran species (Strassmann et al. 1984; Toth et al. 2009; Wasielewski et al. 2014; Durant et al. 2016). Moreover, previous studies have linked honey bee immune status to lipid store gene expression profiling (Mao et al. 2013; Salmela et al. 2015). Future studies on lipid metabolism and the role of fat bodies on bee health are an important line of research and will benefit from additional genome-wide comparisons. Ceratina calcarata is an emerging model species to understand the lipid metabolism and overwintering diapause and nutritional regulation of caste determination (Rehan et al. 2014; Durant et al. 2016).
DNA Methylation
Epigenetic marks can be influenced by environmental variation, and are capable of influencing postembryonic development (Jaenisch and Bird 2003). Such a role for DNA methylation has been highlighted in the honey bee A. mellifera, where larval knockdown of DNA methyltransferase 3 (DNMT3), an enzyme essential to de novo DNA methylation (Klose and Bird 2006), resulted in a developmental shift from the phenotype of workers to the phenotype of queens (Kucharski et al. 2008). Reversible changes in DNA methylation are also associated with changes in behavior linked to the division of labor among workers in A. mellifera (Herb et al. 2012). These findings, along with the analysis of mutational signatures of DNA methylation in ten bee genomes, have given rise to the hypothesis that increased gene regulatory capacity may contribute to the evolution of social complexity in bees (Kapheim et al. 2015). One potential mechanism implicating DNA methylation in such regulatory complexity is its role in the regulation of alternative mRNA splicing (Shukla et al. 2011; Li-Byarlay et al. 2013).
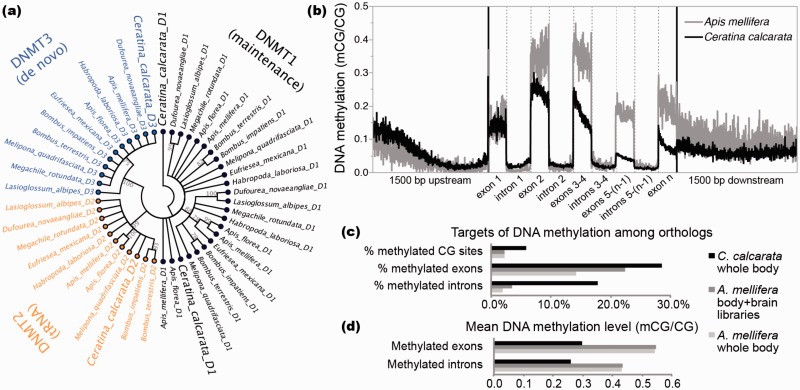
Here, we present results of whole genome bisulfite sequencing (WGBS) for C. calcarata (supplementary table S7, Supplementary Material online). Like A. mellifera (Wang et al. 2006), the genome of C. calcarata contains a complete enzymatic toolkit for DNA methylation (fig. 3a; Klose and Bird 2006), comprised in C. calcarata of two copies of DNMT1 (Ccalc.v2_000349 and Ccalc.v2_004812) and one copy of DNMT3 (Ccalc.v2_012237); DNMT2 is not known to methylate DNA. Moreover, genes targeted by DNA methylation in C. calcarata are enriched for GO biological processes associated with core cellular functions, including translation, and have detectable expression in our RNA-seq data (fractional CDS DNA methylation vs. gene expression level [FPKM], Spearman's rho = 0.58, P < 0.0001; same correlation when removing genes with mCG/CG < 0.01 and FPKM < 1, Spearman’s rho = 0.14, P < 0.0001). These findings are consistent with those from diverse invertebrate taxa, including A. mellifera (Sarda et al. 2012; supplementary table S8, Supplementary Material online). As in other investigated holometabolous insects (Hunt, Glastad, et al. 2013), DNA methylation is preferentially targeted to exons positioned proximal to the translation start site (TSS) in C. calcarata (fig. 3b; Wilcoxon rank-sum test between exons 1–3 and 4–n, P < 0.0001). We previously putforth the hypothesis that TSS-proximal DNA methylation plays a role in differentiating states of transcriptional initiation and elongation (Glastad et al. 2015).
Fig. 3.—
DNA methylation in Ceratina calcarata. (a) Unrooted neighbor-joining cladogram showing relationships among the DNA methyltransferase proteins DNMT1 (D1), DNMT2 (D2), and DNMT3 (D3) from C. calcarata and 10 other bee species, demonstrating the presence of a fully functional DNA methylation toolkit in C. calcarata. Polytomies are plotted for nodes with less than 75% support among 1,000 bootstrap replicates. (b) Spatial profiles of mean DNA methylation levels in genes with significant methylation from C. calcarata and the honey bee Apis mellifera. Exons are divided into 150 proportional bins and introns are divided into 200 proportional bins. DNA methylation is preferentially targeted to exons 1–3, and levels decay toward the 3′ end of genes in each species (Wilcoxon rank-sum tests between exons 1–3 and 4–n, P < 0.0001). (c) Percent of CG dinucleotides, exons, and introns targeted by DNA methylation in 5,698 shared orthologs suggests more widespread targeting of DNA methylation in C. calcarata versus A. mellifera, whereas (d) mean methylation levels suggest that C. calcarata exhibits lower levels of DNA methylation, where targeted, compared with A. mellifera.
To gain insight into the potential relationship between social complexity and DNA methylation, we compared C. calcarata DNA methylation with that observed in orthologs of the advanced eusocial honey bee A. mellifera using WGBS data generated from female adults of each species (Lyko et al. 2010; Zemach et al. 2010; supplementary table S9, Supplementary Material online). We observed that a greater proportion of CpG dinucleotides were methylated in C. calcarata, which is generally considered subsocial, compared with A. mellifera, which exhibits complex eusociality (fig. 3c). Moreover, among 3,841 ortholog pairs with significant DNA methylation detected in CDS of at least one species, 3,147 (81.9%) were targeted by DNA methylation in both species, 456 (11.9%) were targeted by DNA methylation in C. calcarata only, and 238 (6.2%) were targeted by DNA methylation in A. mellifera only. Thus, our results stand in contrast with the prediction that DNA methylation targets are more widespread in species with complex social organization in bees (Kapheim et al. 2015). However, we did observe that methylated cytosines in A. mellifera exhibited higher DNA methylation levels (i.e., were markedly more frequently methylated across sequenced cells) than in C. calcarata (fig. 3b and d). Overall, our results highlight the multidimensionality of DNA methylation—the number of targets and the frequency with which cells are methylated each stand to influence gene regulation. Thus, detailed investigation of DNA methylation patterns in specific tissues from numerous taxa will be required to resolve whether generalities exist in the relationship between DNA methylation and the evolution of sociality.
Molecular Evolution
Ceratina calcarata is the first bee of the subfamily Xylocopinae and the first subsocial bee species with genome characterization. Thus, this species represents an important point of comparison for the molecular evolution of sociality (Rehan and Toth 2015). To identify genes showing signatures of positive selection in the C. calcarata lineage, we conducted branch-site tests in PAML. We included all currently published bee genomes to perform these analyses. These taxa include ten species (A. mellifera, A. florea, E. mexicana, B. terrestris, B. impatiens, Mel. quadrifasciata, H. laboriosa, M. rotundata, L. albipes, D. novaeangliae). We used the “branch-site A” model in PAML to search for sites showing signatures of positive selection in the C. calcarata lineage using a LRT to calculate significance. Following correction for multiple testing, 877 genes showed signatures of positive selection in the C. calcarata lineage (FDR P < 0.05; supplementary table S10, Supplementary Material online). Among the noteworthy genes with signatures of positive selection were two genes associated with regulation of TOR functioning (Ccalc.v2_007251 and Ccalc.v2_008102; supplementary table S11, Supplementary Material online). TOR genes are highly conserved among bees (Kapheim et al. 2015) and have been shown to play a functionally important role in honey bee caste development (Patel et al. 2007; Mutti et al. 2011).
In general, genes associated with lipid transport, protein binding, and ribonuclease activity appear to have undergone positive selection in C. calcarata relative to the other species examined (table 2). Genes associated with DNA recombination are also evolving more quickly in C. calcarata (Ccalc.v2_012690, Ccalc.v2_001846, Ccalc.v2_013055, Ccalc.v2_004327; supplementary table S11, Supplementary Material online). High rates of DNA recombination are known in advanced eusocial insects (Wilfert et al. 2007; Sirviö et al. 2011; Kapheim et al. 2015) and could be linked to accelerated evolution of caste related traits (Kent et al. 2012). It is possible that the accelerated evolution of DNA recombination genes may reflect this group’s transitional form of sociality including the presence of an incipient worker caste (Rehan et al. 2014).
Table 2.
GO Enrichment for Genes Subject to Positive Selection according to a Branch-Site Test (FDR P < 0.05 vs. FDR P > 0.05). Gene ontology categories include molecular function (F) and biological processes (P)
| Term | GO-ID | Category | No. of Genes | Fold Enrichment | P Value | FDR |
|---|---|---|---|---|---|---|
| Nuclease activity | GO:0004518 | F | 19 | 2.24 | 3.49E−03 | 1.00E+00 |
| Lipid metabolic process | GO:0006629 | P | 35 | 1.45 | 3.32E−02 | 1.00E+00 |
| DNA metabolic process | GO:0006259 | P | 34 | 1.43 | 4.11E−02 | 1.00E+00 |
| Secondary metabolic process | GO:0019748 | P | 3 | 5.66 | 4.73E−02 | 1.00E+00 |
Conclusions
The maternal care and sibling care expressed in an incipiently social bee make C. calcarata ideal for developmental and sociodemographic studies to characterize the genomic effects of social experience on behavioral plasticity. Our results suggest that mechanisms associated with DNA methylation and nutrition are candidate targets of evolutionary change during the origin of more complex social forms. The addition of C. calcarata to the published bee genomes provides the first subsocial bee species to ask questions on the evolutionary origins of social behavior. Moreover, the characterization of a fully functional C. calcarata methylome allows for new lines of research into the epigenetic regulation of maternal care, sibling care, and worker behavior.
Supplementary Material
Supplementary tables S1–S11 and supplementary material S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Wyatt Shell for assistance with field collections and DNA extractions as well as the staff at the Hubbard Center for Genome Studies at the University of New Hampshire for DNA library preparation and sequencing. This work was supported by funding from the University of Georgia to B.G.H., the University of New Hampshire and a National Science Foundation grant IOS-1456296 to S.M.R. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution Number 2667. This work was also supported by the USDA National Institute of Food and Agriculture Hatch Project 1004515.
Literature Cited
- Alexander RD, Noonan KM, Crespi BJ. 1991. The evolution of eusociality In: Sherman PW, Jarvis JUM, Alexander RD, editors. The biology of the naked mole rat. Princeton (NJ: ): Princeton University Press; p. 3–44. [Google Scholar]
- Anderson M. 1984. The evolution of eusociality. Annu Rev Ecol Syst. 15:165–189. [Google Scholar]
- Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, et al. 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12:521–532. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13:R56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, et al. 2010. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329:1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal S, Danforth BN. 2011. The antiquity and evolutionary history of social behavior in bees. PLoS One 6:e21086.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler L. 1975. Eusociality in Ceratina calcarata Robertson. Proc Indiana Acad Sci. 84:283–284. [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Costa JT. 2006. The other insect societies. Cambridge (MA: ): Belknap Press of Harvard University Press. [Google Scholar]
- Crespi BJ. 1994. Three conditions for the evolution of eusociality: are they sufficient? Insectes Soc. 41:395–400. [Google Scholar]
- Csűros M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. [DOI] [PubMed] [Google Scholar]
- Daly HV. 1973. Bees of the genus Ceratina in America north of Mexico (Hymenoptera: Apoidea). Los Angeles (CA: ): University of California Press. [Google Scholar]
- Durant DR, Berens AJ, Toth AL, Rehan SM. 2016. Transcriptional profiling of overwintering gene expression in the small carpenter bee, Ceratina calcarata. Apidologie Advance Access published November 20, 2015; doi:10.1007/s13592-015-0402-x. [Google Scholar]
- Finn RD, et al. 2014. The Pfam protein families database. Nucleic Acids Res. 42:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar R. 1990. Evolution of eusociality: the advantage of assured fitness returns. Phil Trans R Soc Lond B. 329:17–25. [Google Scholar]
- Gadau J, et al. 2012. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet. 28:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glastad K, Hunt BG, Goodisman MAD. 2015. DNA methylation and chromatin organization in insects: insights from the ant Camponotus floridanus. Genome Biol Evol. 7:931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq: reference generation and analysis with Trinity. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb BR, et al. 2012. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nature Neurosci. 15:1371–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Glastad K, Yi SV, Goodisman MAD. 2013. The function of intragenic DNA methylation: insights from insect epigenomes. Integr Comp Biol. 53:319–328. [DOI] [PubMed] [Google Scholar]
- Hunt M, Kikuchi T, et al. 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14:R47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 33:245–254. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Kapheim, et al. 2015. Genomic signatures of evolutionary transitions from solitary to group living. Science 348:1139.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Schatz MC, Salzberg SL. 2010. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 11:R116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CF, Minaei S, Harpur BA, Zayed A. 2012. Recombination is associated with the evolution of genome structure and worker behavior in honey bees. Proc Natl Acad Sci U S A. 109:18012–18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 31:89–97. [DOI] [PubMed] [Google Scholar]
- Kocher SD, et al. 2013. The draft genome of a socially polymorphic halictid bee, Lasioglossum albipes. Genome Biol. 14:R142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, et al. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 43:D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319:1827–1830. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Byarlay H, et al. 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc Natl Acad Sci U S A. 110:12750–12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102:10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, et al. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8:e1000506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W, Schuler MA, Berenbaum MR. 2013. Honey constituents up-regulate detoxification and immunity genes in the western honey bee Apis mellifera. Proc Natl Acad Sci U S A. 110:8842–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFrederick QS, Rehan SM. 2016. A novel approach for simultaneous investigation of pollen and bacterial communities in brood provisions of a small carpenter bee Mol Ecol. Advance Access publication April 6, 2016; doi:10.1111/mec.13608. [DOI] [PubMed] [Google Scholar]
- Michener CD. 1974. The social behavior of the bees. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- Michener CD. 1985. From solitary to eusocial: need there be a series of intervening species? Fortschr Zool. 31:293–305. [Google Scholar]
- Michener CD. 2007. The bees of the world. 2nd ed Baltimore (MD: ): The John Hopkins University Press. [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti NS, et al. 2011. IRS and TOR nutrient-signalling pathways act via juvenile hormone to influence honey bee caste fate. J Exp Biol. 214:3977–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard S, et al. 2011. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 21:1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Patel A, et al. 2007. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS One 6:e509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan SM, Berens AJ, Toth AL. 2014. At the brink of eusociality: transcriptomic correlates of worker behavior in a small carpenter bee. BMC Evol Biol. 14:260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan SM, Leys R, Schwarz MP. 2012. A mid-cretaceous origin of sociality in xylocopine bees with only two origins of true worker castes. PLoS One 7:e34690.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan SM, Richards MH. 2010a. Nesting biology and subsociality of Ceratina calcarata (Hymenoptera: Apidae). Can Entomol. 142:65–74. [Google Scholar]
- Rehan SM, Richards MH. 2010b. The influence of maternal quality on brood sex allocation in the small carpenter bee, Ceratina calcarata. Ethology 116:876–887. [Google Scholar]
- Rehan SM, Richards MH, Schwarz MP. 2009. Evidence of social nesting in the Ceratina of Borneo. J Kansas Entomol Soc 82:194–209. [Google Scholar]
- Rehan SM, Schwarz MP. 2015. A few steps forward and no steps back: long-distance dispersal patterns in small carpenter bees suggest major barriers to back-dispersal. J Biogeogr. 42:485–494. [Google Scholar]
- Rehan SM, Sheffield CS. 2011. Morphological and molecular delineation of a new species in the Ceratina dupla species-group (Hymenoptera: Apidae: Xylocopinae) of eastern North America. Zootaxa 2873:35–50. [Google Scholar]
- Rehan SM, Toth AL. 2015. Climbing the social ladder: molecular evolution of sociality. Trends Ecol Evol. 30:426–433. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Grozinger CM, Whitfield CW. 2005. Sociogenomics: social life in molecular terms. Nature Rev Genet. 6:257–270. [DOI] [PubMed] [Google Scholar]
- Sadd BM, et al. 2015. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16:76.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami SF, Laroca S. 1971. Observations of the bionomics of some neotropical xylocopine bees with comparative and biofaunistic notes (Hymenoptera, Anthophoridae). J Fac Sci Hokkaido Univ. 18:57–127. [Google Scholar]
- Sakagami SF, Maeta Y. 1977. Some presumably presocial habits of Japanese Ceratina bees with notes on various social types in Hymenoptera. Insectes Soc. 24:319–343. [Google Scholar]
- Sakagami SF, Maeta Y. 1984. Multifemale nests and rudimentary castes in the normally solitary bee Ceratina japonica (Hymenoptera: Xylocopinae). J Kansas Entomol Soc. 57:639–656. [Google Scholar]
- Sakagami SF, Maeta Y. 1987. Multifemale nests and rudimentary castes of an ‘almost’ solitary bee Ceratina flavipes, with additional observations on multifemale nests of Ceratina japonica (Hymenoptera, Apoidea). Kontyu 55:391–409. [Google Scholar]
- Sakagami SF, Maeta Y. 1989. Compatibility and incompatibility of solitary life with eusociality in two normally solitary bees Ceratina japonica and Ceratina okinawana (Hymenoptera, Apoidea), with notes on the incipient phase of eusociality. Jpn J Entomol. 57:417–739. [Google Scholar]
- Salmela H, Amdam GV, Freitak D. 2015. Transfer of immunity from mother to offspring is mediated via egg-yolk protein vitellogenin. PLoS Pathog. 1:e1005015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gracia A, Vieira FG, Rozas J. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103:208–216. [DOI] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV. 2012. The evolution of invertebrate gene body methylation. Mol Biol Evol. 29:1907–1916. [DOI] [PubMed] [Google Scholar]
- Schmeider R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43:W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell WA, Rehan SM. 2016. Recent and rapid diversification of the small carpenter bees in eastern North America. Biol J Linn Soc. 117:633–645. [Google Scholar]
- Shukla S, et al. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. 2004. Combining phylogenetic and hidden Markov models in biosequence analysis. J Comput Biol. 11:413–428. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simola DF, et al. 2013. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 23:1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirviö A, et al. 2011. A high recombination rate in eusocial Hymenoptera: evidence from the common wasp Vespula vulgaris. BMC Genet. 12:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2015. RepeatMasker Open-4.0. 2013–2015. Available from: http://www.repeatmasker.org.
- Smith CD, Zimin A, et al. 2011. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci U S A. 108:5673–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Smith CD, Robertson HM, et al. 2011. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci U S A. 108:5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Waack S. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19:215–225. [DOI] [PubMed] [Google Scholar]
- Strassmann JE, Lee RR, Rojas RR, Baust JG. 1984. Caste and sex-differences in cold-hardiness in the social wasps, Polistes annularis and Polistes exclamans (Hymenoptera, Vespidae). Insectes Soc. 31:291–301. [Google Scholar]
- Suen G, et al. 2011. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 7:e1002007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Tallamy DW, Wood TK. 1986. Convergence patterns in subsocial insects. Annu Rev Entomol. 31:369–390. [Google Scholar]
- Terrapon, et al. 2014. Molecular traces of alternative social organization in a termite genome. Nat Commun. 5:3636.. [DOI] [PubMed] [Google Scholar]
- Toth AL, Bilof KBJ, Henshaw MT, Hunt JH, Robinson GE. 2009. Lipid stores, ovary development, and brain gene expression in Polistes metricus females. Insectes Soc. 56:77–84. [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. 2006. Functional CpG methylation system in a social insect. Science 314:645–647. [DOI] [PubMed] [Google Scholar]
- Wasielewski O, et al. 2014. The potential role of adiponectin- and resistin-like peptides in the regulation of lipid levels in the hemolymph of over-wintering adult females of Osmia bicornis. Apidologie 45:491–503. [Google Scholar]
- West-Eberhard MJ. 1967. Foundress associations in polistine wasps: dominance hierarchies and the evolution of social behaviour. Science 157:1584–1585. [DOI] [PubMed] [Google Scholar]
- Wilfert L, et al. 2007. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 98:189–197. [DOI] [PubMed] [Google Scholar]
- Wilson EO. 2008. One giant leap: how insects achieved altruism and colonial life. Bioscience 58:17–25. [Google Scholar]
- Wurm Y, et al. 2011. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci U S A. 108:5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, et al. 2012. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One 7:e52249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, et al. 2013. L_RNA_scaffolder: scaffolding genomes with transcripts. BMC Genomics 14:604.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, et al. 2014. Eusocial insects as emerging models for behavioural epigenetics. Nat Rev. 15:677–688. [DOI] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.