Abstract
The lone star tick, Amblyomma americanum, is an important disease vector and the most frequent tick found attached to humans in the eastern United States. The lone star tick has recently experienced a rapid range expansion into the Northeast and Midwest, but despite this emerging infectious threat to wildlife, livestock, and human health, little is known about the genetic causes and consequences of the geographic expansion. In the first population genomic analysis of any tick species, we characterize the genetic diversity and population structure of A. americanum across its current geographic range, which has recently expanded. Using a high-throughput genotyping-by-sequencing approach, we discovered more than 8,000 single nucleotide polymorphisms in 90 ticks from five locations. Surprisingly, newly established populations in New York (NY) and Oklahoma (OK) are as diverse as historic range populations in North and South Carolina. However, substantial population structure occurs among regions, such that new populations in NY and OK are genetically distinct from historic range populations and from one another. Ticks from a laboratory colony are genetically distinct from wild populations, underscoring the need to account for natural variation when conducting transmission or immunological studies, many of which utilize laboratory-reared ticks. An FST-outlier analysis comparing a recently established population to a long-standing population detected numerous outlier sites, compatible with positive and balancing selection, highlighting the potential for adaptation during the range expansion. This study provides a framework for applying high-throughput DNA sequencing technologies for future investigations of ticks, which are common vectors of diseases.
Keywords: lone star tick, genotyping-by-sequencing, range expansion, nonmodel organism, adaptation
Introduction
Population genomic studies of disease vectors are valuable because they provide insights into basic biological properties of species, such as genetic diversity and adaptation potential, the structure and spatial extent of populations, and mechanisms of dispersal and gene flow. For example, genetic variation of a disease vector may affect the patterns of transmission of its pathogenic microorganisms at large geographic scales (Gooding 1996). Thus, knowledge about the genetics and natural history of arthropod vectors is essential for understanding and predicting disease dynamics and developing molecular methods to control disease transmission (Gooding 1996; McCoy 2008).
The lone star tick (Amblyomma americanum) is a major vector of several viral, bacterial, and protozoan pathogens affecting humans and other animals in the United States (Childs and Paddock 2003; Goddard and Varela-Stokes 2009). For example, A. americanum transmits the pathogens that cause human ehrlichiosis, southern tick-associated rash illness, and tularemia; additionally, A. americanum may be a competent vector of other emerging bacterial and viral agents (Mixson, Campbell, et al. 2006; Goddard and Varela-Stokes 2009; Tokarz, Sameroff, et al. 2014; Tokarz, Williams, et al. 2014). Amblyomma americanum is also the most frequently reported tick attached to humans in the Southeast and Atlantic states (Merten and Durden 2000). An increase in the incidence of lone star tick-borne diseases is projected in the coming decades owing to its aggressive and nondiscriminatory biting habits at all active life stages (Childs and Paddock 2003), its competence in transmitting a wide array of pathogenic infectious agents (Goddard and Varela-Stokes 2009), its high local population densities and expanding distribution (Springer et al. 2014; Dahlgren et al. 2016), and other ecological and human sociological factors (reviewed in Childs and Paddock 2003). The public health significance of A. americanum thus merits careful investigation and monitoring. Yet, although A. americanum was the first North American tick to be scientifically described (Linnaeus 1758), its relevance to public and veterinary health has been overshadowed by the American dog tick (Dermacentor variabilis) and the blacklegged tick (Ixodes scapularis), the primary vectors of Rocky Mountain spotted fever and Lyme disease, respectively (Childs and Paddock 2003). This study aims to address the paucity of knowledge about genetic variation in A. americanum, which is likely to affect the ecology, physiology, and competence of this important disease vector.
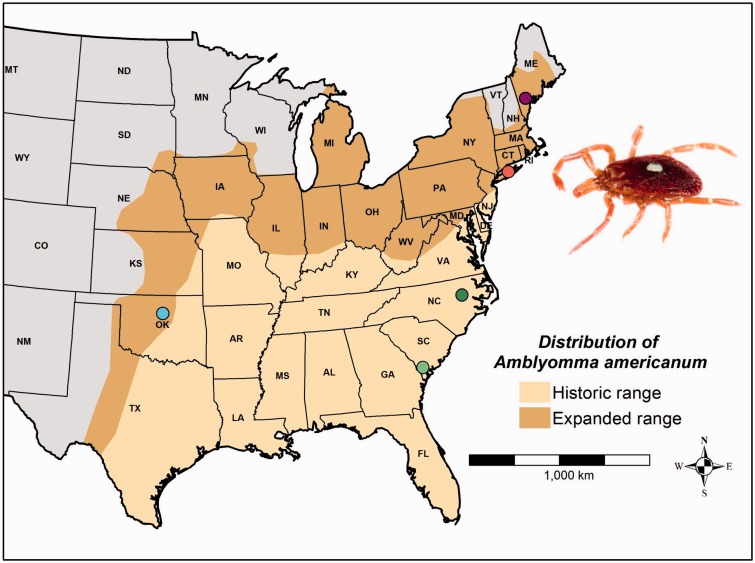
Historically a southern pest, the lone star tick has recently experienced a remarkably rapid expansion in geographic distribution and population densities in the Northeast and Midwest. Prior to this expansion in the late 20th century, the northern range limit was southern New Jersey (fig. 1; Bishopp and Trembley 1945; Schulze et al. 1984). Amblyomma americanum was first reported in New York (NY) in 1969, established small populations in far eastern Long Island in the early 1970s, and by 1990 was widely distributed throughout the southern coast of Long Island (Ginsberg et al. 1991; Means and White 1997). The first report of A. americanum in NY outside of Long Island is from 1987, when two specimens were identified in Westchester County. By 1996, the distribution of A. americanum extended to all boundaries of NY, with reports from 46 of NY’s 62 counties (Means and White 1997). The geographic expansion also progressed rapidly throughout the New England states, with A. americanum becoming established in Maine (ME) by the late 1990s (Keirans and Lacombe 1998). In the Midwest, the range of lone star ticks recently expanded in Central Oklahoma (OK) (Barrett et al. 2015), Missouri (Brown et al. 2011), and Nebraska (Cortinas and Spomer 2013). The most updated species distribution map depicts “established” and “reported” populations of lone star tick on a county level in the United States since 1898 (Springer et al. 2014). This map shows disjunct populations in South Dakota, Minnesota, and Michigan (fig. 1; Springer et al. 2014). It is important to note that some of these recent expansions may be recolonizations of areas inhabited by A. americanum long ago. Hooker et al. (1912) provided a map of “the probable range of the species” which included Michigan, NY, and the New England states. However, Bishopp and Trembley (1945) redrew the distribution map and excluded these northern areas, stating that only one specimen had been collected in NY in the 1830s, and that the few specimens collected in the northern states are probably accidental.
Fig. 1.—
Historic and current expanded distribution of Amblyomma americanum, showing sampling locations in ME, NY, OK, NC, and SC. Historic range from Bishopp and Trembley (1945). Expanded range from Barrett et al. (2015), Cortinas and Spomer (2013), and Springer et al. (2014). Photo credit: J.P. Lawrence.
To date, only four molecular studies have investigated the population genetics of A. americanum, but these studies have been limited in geographic scope and genomic sampling. Hilburn and Sattler (1986) surveyed nine populations of A. americanum (from Texas, Oklahoma, Kentucky, Florida, South Carolina, Virginia, and New Jersey) for electrophoretic variations in 21 enzymes, three of which were not polymorphic. They found a high degree of similarity among the nine populations and concluded that the species is genetically homogeneous throughout its range. Two studies examined the sequence of the mitochondrial 16S rRNA gene: Mixson, Lydy, et al. (2006) sampled lone star ticks from three distinct ecoregions in Georgia, and Trout et al. (2010) sampled lone star ticks from six distinct ecoregions in Arkansas. Both studies failed to detect appreciable levels of genetic differentiation and concluded that lone star ticks are actively intermixing among ecoregions. The only molecular investigation yet to support genetic divergence of A. americanum populations analyzed the sequence variation in the nuclear rRNA ITS-2 region in two spatially distinct populations, both from OK (Reichard et al. 2005). Studies examining genetic variation using many loci from across the genome and a large number of A. americanum (or any tick species) across its geographic distribution are thus notably lacking. Modern genomic sequencing approaches, such as the genotyping-by-sequencing (GBS) method employed here, allow for the assessment of variation across the entire genome, rather than limited to a single gene or chromosomal region. These methods thus provide a more extensive and accurate quantification of genomic variation and population structure across geographic regions.
In this study, the first population genomics investigation of any tick species, we characterize the genetic diversity and population structure of A. americanum across its current geographic distribution, including its newly expanded range, using next-generation sequencing technology. We also investigate the potential role of adaptation in the recent range expansion of lone star ticks; specifically, we test the hypothesis that local adaptive evolution at a geographic margin accompanied range expansion. Knowledge of the spatial distribution of genetic variation and the role of natural selection in population expansion of the lone star tick is critical for furthering our understanding of the transmission, geographic spread, epidemiology, and control of tick-borne diseases (Tabachnick and Black 1995; Gooding 1996; Tibayrenc 1998; McCoy 2008).
Materials and Methods
Sample Collection and DNA Extraction
Adult lone star ticks were sampled from five locations across three regions of the geographic range (table 1, fig. 1, and supplementary table S1, Supplementary Material online). The NY, ME, and OK populations are recent (Ginsberg et al. 1991; Means and White 1997; Keirans and Lacombe 1998; Barrett et al. 2015) and represent the expanded portion of the species range. In contrast, the populations from South Carolina (SC) and North Carolina (NC) represent the historic portion of the species range, where A. americanum has been plentiful in the South Atlantic coast since at least the early 1900s (Cooley and Kohls 1944; Bishopp and Trembley 1945). NY, NC, and SC ticks were sampled from the local vegetation using standard flagging methods (Ginsberg and Ewing 1989). ME ticks were obtained from the collection of Maine Medical Center Research Institute’s passive surveillance tick submission program. Human hosts contributing these ticks did not have a travel history, providing confidence that the ticks were acquired in ME. OK ticks were randomly sampled from the A. americanum colony in the Tick Rearing Facility at Oklahoma State University. This colony was started with engorged females collected from the natural population in OK, and is annually or biannually supplemented with females from the natural population in Payne County, OK, which was recently colonized by A. americanum. Many tick-borne disease transmission studies examine colony-reared ticks, so this sample provides a means to assess how such a laboratory situation affects genomic patterns of diversity.
Table 1.
Number of Lone Star Ticks (N) Sampled from Suffolk County, NY; Various Counties in ME; Payne County, OK; Martin County, NC; and Jasper County, SC
| Location | N | Region | Range |
|---|---|---|---|
| NY | 26 | Northeast | Expanded |
| ME | 5 | Northeast | Expanded |
| OK | 20 | Southwest | Expanded |
| NC | 22 | Southeast | Historic |
| SC | 17 | Southeast | Historic |
| Total | 90 |
Live ticks were washed in 70% ethanol for 5 min and rinsed in sterile distilled water for 5 min to remove environmental contaminants from the external surface (Carpi et al. 2011). Genomic DNA was extracted from each tick using the DNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s recommended protocol (Purification of Total DNA from Insects). Genomic DNA was treated with RNase and then further cleaned and concentrated using a modified QIAamp DNA Micro kit (Qiagen) protocol with SpinSmart PCR purification columns (Denville Scientific, South Plainfield, NJ, USA) (Monzón et al. 2014). DNA quality was assessed by running all 90 samples on a 1% agarose gel and by conducting a trial digestion of ten samples with restriction enzyme HaeIII (New England Biolabs, Ipswich, MA, USA).
Genotyping-by-Sequencing
Extracted DNA was sent to the Cornell University Biotechnology Resource Center (BRC), where GBS and a corresponding bioinformatics pipeline for single nucleotide polymorphism (SNP) discovery were first developed (Elshire et al. 2011; Lu et al. 2013). GBS is a technique for constructing reduced representation libraries and is conceptually similar to RAD-seq (Davey et al. 2011). BRC scientists optimized the GBS protocol for the lone star tick. The restriction enzyme PstI was used to reduce genome complexity, and unique barcodes were ligated to the digested DNA to construct a sequencing library. The resulting library was single-end sequenced in 100-bp reads on the Illumina HiSeq 2000. The final quality-filtered and demultiplexed dataset contained 235,335,191 reads from two lanes of sequencing. The number of reads per individual averaged 2,614,835 and ranged between 318,603 and 5,822,521 (Supplementary Table S2).
The UNEAK bioinformatics pipeline (Tassel 4.0, Glaubitz et al. 2014) was used to align raw sequence reads and call SNPs. UNEAK is a multisample, SNP-calling approach developed for analyzing GBS data from species that lack a reference genome sequence (Lu et al. 2013). GBS and UNEAK have been used to study the genomic signature of adaptation during the range expansion of an invasive rodent (White et al. 2013). In the UNEAK pipeline, reads are trimmed to 64 bp after the barcode, and identical reads are clustered into tags. A 1-bp mismatch between pairwise aligned tags is interpreted as a candidate SNP. This pipeline identified 23,275,556 unique tags, which were sorted into 153,941 tag pairs, each pair comprising two tags that differ by only 1 bp. Final genotypes were called after candidate SNPs passed the following filters according to standard protocols of the BRC: error tolerance rate of tag pairing = 0.03, sequencing error rate per base = 0.01, minimum minor allele frequency = 0.01. After merging duplicate SNPs (i.e., those covered from both forward and reverse reads), 72,517 biallelic SNPs were called.
To limit the incorporation of missing data and erroneous genotypes, analyses focused on the subset of SNPs with a minimum depth of 4× coverage per individual (Perry et al. 2013). This approach excludes a large portion of data, but substantially increases confidence in the accuracy of the SNP genotypes that remain for downstream analyses. Selecting only those reliable sites with at least 4× coverage resulted in a final subset of 8,392 SNPs that were used in all population genomic analyses. The genotypes of these SNPs are available in the supplementary file S1, Supplementary Material online. The average site-wise depth for this final filtered SNP subset was 11.128 and the average coverage across individuals was 10.961.
Population Genomics Analyses
To evaluate the genetic structuring of individuals and populations, three complementary approaches were used: 1) principal components analysis (PCA), a model-free multivariate ordination method implemented in the adegenet package (Jombart 2008) in R (R Core Team 2012); 2) a maximum-likelihood model-based estimation of ancestry implemented in ADMIXTURE (Alexander et al. 2009); and 3) analysis of molecular variance (AMOVA) implemented in GenAlEx (Peakall and Smouse 2006; Peakall and Smouse 2012). To evaluate the genomic diversity within populations, heterozygosity (H) and the inbreeding coefficient (F) were computed using VCFtools (Danecek et al. 2011).
For the PCA, centering and binomial scaling were used to compensate for differences in variance among allele frequencies (Jombart et al. 2009). For the ADMIXTURE analysis, which partitions N samples into K genetic clusters, ten runs were conducted at each value of K ranging from K = 2 through K = 5, after which point additional clusters were no longer informative. Running each value of K 10 times produced a total of 40 Q matrices; this allowed the detection of potential multimodality in the data, the situation when there is more than one way to assign individuals to genetic clusters. Each run was started at a randomly generated seed to explore the full breadth of variation space and determine the major mode present in the data. Multimodality was visualized using pong (Behr et al. 2015). The optimal value of K was chosen based on 5-fold cross-validation procedures implemented during individual ADMIXTURE runs. The optimal value of K is the point at which the addition of more clusters no longer provides a better fit to the data, as judged by minimizing the cross-validation error across runs. Other biologically relevant values of K were also considered, as is recommended by Meirmans (2015). For the AMOVA, population genomic tests for genetic differentiation were conducted between three geographically defined regions: Northeast (NY only), Southwest (OK), and Southeast (NC + SC). ME ticks were excluded from pairwise analyses as they have a low sample size (N = 5) and do not appear to define their own genetic cluster (see Results).
To identify candidate loci that may have been subject to selection during the range expansion of A. americanum, the LOSITAN program (Antao et al. 2008), which employs the FDIST2 algorithm (Beaumont and Nichols 1996), was used to detect SNPs that are FST outliers. This method evaluates the relationship between the expected distribution of FST and heterozygosity assuming an island model of migration. Due to computational constraints of the program, a random subset of 5,000 SNPs was selected and two genetic populations were considered: NY, representing a newly established population, and NC + SC, representing a historic population. OK and ME ticks were excluded because genetic substructure was detected within these samples (see Results). LOSITAN was run for 50,000 simulations, assuming an infinite alleles mutation model, two expected populations, and a conservative false discovery rate of 0.1.
Results and Discussion
Partitioning of Genomic Variation across Space
We observed clear genomic differentiation among the sampled tick populations. The PCA of all 90 specimens revealed a strong distinction between ticks sampled in OK versus other locations (fig. 2A). Moreover, OK ticks formed two distinct clusters in the biplot of the first two principal components, revealing genetic substructure within the lab-reared OK sample. Because the first PCA was dominated by the distinction between OK and non-OK ticks, another PCA was performed excluding OK ticks to further scrutinize the genomic variation in the remaining groups. The PCA of only the northeastern and southeastern samples showed clusters with little overlap, revealing a population structure that broadly corresponds to the geographic locations (fig. 2B). The pairwise distances between the centroids of the three main clusters were as follows: NC to SC = 2.5 units, NC to NY = 2.8 units, and SC to NY = 3.5 units. In this second PCA, the first principal component (which explains the largest fraction of the genetic variance—see fig. 2B inset) partitioned NY as distinct from the Carolinas. These results suggest the next most significant differentiation is between NY and NC/SC ticks. The ME ticks, however, spanned across clusters.
Fig. 2.—
Principal components analysis of genomic diversity in five lone star tick populations: NY, ME, OK, NC, and SC. Panel A includes and panel B excludes the 20 colony-reared OK ticks. The first two components, PC1 and PC2, are shown intersecting at (0, 0). Insets show scree plots of eigenvalues.
The ADMIXTURE cross-validation analysis indicated that lower K values fit the data better. Hence, we focus here on K = 2 to K = 4 and present the major modes at each value of K (fig. 3). At K = 2, the analysis showed that most OK ticks have their own unique ancestry and are distinct from wild-caught ticks in other regions. However, the analysis revealed substructure within OK, congruent with PCA results (fig. 2A). At K = 3, the optimal value as evaluated by minimizing ADMIXTURE cross-validation errors, NY ticks formed a cluster and appeared reasonably distinct from other populations. This pattern is also congruent with PCA results, with the OK population consistently being the most genetically distinct from the others (fig. 2A), followed by the distinction of NY from the Carolinas (PC1 in fig. 2B). The genetic homogeneity of NC and SC ticks is evident at all K values. At K = 4, considerable substructure within OK and ME was detected. For this reason, we do not consider OK and ME to be genetically homogeneous populations. At K = 5, intrastate variation was partitioned into separate clusters of several individuals; therefore, these are no longer biologically meaningful or informative for the hypotheses of interest (data not shown).
Fig. 3.—
Ancestry of individual ticks assuming K clusters of genetic similarity, based on the results of ADMIXTURE analyses. Individuals span across the x axis, sorted by population, and ancestry percentages are visualized as colors on the y axis. The colored proportions for each individual represent the contribution of ancestral genetic clusters. The major mode present across ten runs at each K value is presented along with the agreement in cluster assignment across discordant runs (average pairwise similarity). At K = 2, most OK ticks appear distinct. At K = 3, the optimal K value, the next primary distinction separates historic range (NC and SC) ticks from expanded range (NY) ticks. At K = 4, substructure within OK and ME is detected and multimodality is present. Visualization was created with pong.
We conducted an AMOVA to evaluate population structure among the three regions in table 1. We excluded ME from the analysis because of its small sample size and lack of genomic uniformity. A preliminary AMOVA indicated that there is no significant differentiation between NC and SC tick populations (FST = 0.003, P = 0.197). This result is consistent with the ADMIXTURE results and the shorter geographic distance separating these two populations. We therefore combined NC and SC ticks into one group, calling it the “Southeast” (SE) or “historic range” population in subsequent analyses. A subsequent AMOVA revealed significant population structure (global FST = 0.062, P = 0.001), with 6% of the overall variance occurring among regions. All three pairwise regional comparisons were significant, with FST values ranging from 0.04 (Southeast vs. Southwest) to 0.08 (Northeast vs. Southwest) (table 2).
Table 2.
Pairwise Genetic Differentiation among Three Geographic Regions in the Distribution of Lone Star Tick
| NE | SW | SE | |
| NE | 0.001 | 0.001 | |
| SW | 0.082 | 0.001 | |
| SE | 0.066 | 0.037 | |
Note.—FST values are shown below the diagonal and P values based on 999 AMOVA permutations are shown above diagonal. NE: Northeast (NY only); SW: Southwest (OK); SE: Southeast (NC + SC).
There is marked population structure among ticks collected from different regions. The PCA, ADMIXTURE analysis, and AMOVA all consistently demonstrated clear genetic structuring across geographic areas (figs. 2 and 3, table 2). Our results—based on a broad geographic sampling scheme, many dozens of individuals, and thousands of genome-wide SNPs—provide a fuller picture of tick population structure than what had been presented from earlier, more geographically and genetically limited work. Lone star ticks do not form one genetically uniform, panmictic population across the species range, as had been previously proposed (Hilburn and Sattler 1986). Instead, different geographic regions harbor distinct genomic profiles and show significant genetic differentiation. However, lone star ticks from NC and SC compose one homogeneous population. These two sampling sites are separated by approximately 500 km, a scale comparable to the studies in Georgia and Arkansas based on mitochondrial DNA sequences (Mixson, Lydy, et al. 2006; Trout et al. 2010). Additional genome-wide surveys of A. americanum at smaller spatial extents, especially in the expanded range, are needed to distinguish between fine-scale genetic differentiation and fine-scale panmixia.
We next tested whether particular genomic loci may be subject to selection. The FST-outlier analysis comparing two populations (NY/expanded versus SE/historic) detected numerous outlier sites that had signals compatible with positive and balancing selection. Specifically, 391 SNPs were situated above the 95% confidence interval (CI) and 318 SNPs above the 99% CI (fig. 4). These SNPs are more divergent between geographic regions than expected from a model of island migration between the two regions, and thus may be in genomic regions subject to positive selection. In contrast, the FST-outlier analysis detected 138 SNPs under the 95% CI and 94 SNPs under the 99% CI (fig. 4). These SNPs have FST values significantly smaller than expected under neutrality, and thus may be in genomic regions subject to balancing selection. In total, more than 10% of the 5,000 randomly selected SNPs have signals of recent influence by selection.
Fig. 4.—
FST-outlier analysis of loci under selection. Plotted are FST versus expected heterozygosity (He) calculated for 5,000 SNPs by comparing the recently established NY population to the Historic (NC + SC) population. He is calculated as the probability of choosing two alleles at random that are different, one from each deme. Colored boundaries indicate the 95% CIs obtained through simulations in LOSITAN. The 391 SNPs in the red region are candidates for positive selection coinciding with the range expansion into NY. The 138 SNPs in the yellow region are candidates for balancing selection.
We note that the demography of the lone star tick conforms to a range expansion rather than an island model, and that the FST-outlier test employed by LOSITAN (namely FDIST2) is known to overestimate the number of loci under selection. In their simulations, Lotterhos and Whitlock (2014) found that FDIST2 has a false positive rate of detecting loci under selection that can be as high as 4% in a scenario of range expansion rather than an island model, but the false positive rate declined as the true proportion of selected loci increased. Even if 4% of the 529 SNPs our analysis detected as selected loci were false positives, it remains that more than 10% of SNPs appear to have been influenced by natural selection. This, however, does not mean that 10% of the genome is under selection, as we could not account for correlated coancestry or linkage between sites due to the lack of localization of SNPs in this nonmodel organism.
Rapid range expansions often coincide with rapid evolutionary change (Thomas et al. 2001; Wiens and Graham 2005; Monzón 2012). We aimed to examine the population genetic structure of lone star ticks across different geographic regions representing locations in their historic and expanded range to see if we could detect a signature of a response to natural selection coinciding with range expansion. In the distribution of locus-specific FST values, there were some notable outliers (fig. 4). These highly differentiated sites between regions may have undergone differential selective pressures during or immediately after range expansion. Such sites may represent likely targets of selection for adapting to new environmental pressures in novel habitats or for traits that facilitated the process of range expansion. Although it is impossible to know the specific function of these SNPs with the current lack of a closely related tick reference genome, future studies identifying the genes in which these SNPs lie might shed light on the genetic basis of range expansion in ticks.
Genetic Diversity
Of all populations examined, OK ticks had the highest observed heterozygosity (Ho = 0.108) and the lowest level of inbreeding (F = 0.046). ME ticks had the lowest heterozygosity (Ho = 0.074) and the highest level of inbreeding (F = 0.349). Historic range ticks in the Carolinas and expanded range ticks in NY had intermediate levels of both heterozygosity and inbreeding (table 3). Despite recent range expansion, new populations in NY and OK, which are at most 70 years old (fig. 1; Bishopp and Trembley 1945), are as diverse as historic range populations in NC and SC. Although the degree of genetic diversity is comparable between NY and the Carolinas, the type of diversity is different, based on the FST-outlier analysis described above. The surprisingly high diversity in the NY and OK populations may be due to 1) a single, large founding population of ticks into each newly established area (i.e., no bottleneck); 2) multiple introductions from different parts of its historic range, as in the expansion of the invasive brown anole, Anolis sagrei, (Kolbe et al. 2004); or 3) ticks in previous range margins possessing high genomic diversity that permitted them to survive if they moved into new areas thus further expanding the species range (i.e. expansion through adaptation). Future work should elucidate which of these three hypothetical scenarios best accounts for the relatively high genomic diversity in newly established populations at the species range margins.
Table 3.
Summary Statistics of Genetic Diversity for All Populations Sampled and for the Combined NC + SC Historic Range Population
| Population | N | Ho | F |
|---|---|---|---|
| NY | 26 | 0.086 | 0.246 |
| ME | 5 | 0.074 | 0.349 |
| OK | 20 | 0.108 | 0.046 |
| NC | 22 | 0.093 | 0.180 |
| SC | 17 | 0.089 | 0.216 |
| Historic (NC+SC) | 39 | 0.091 | 0.196 |
Note.—N: sample size; Ho: proportion of observed sites found to be heterozygous, calculated on a per-individual basis; F: coefficient of inbreeding, which estimates the reduction in heterozygosity compared to Hardy–Weinberg expectations, calculated on a per-individual basis.
The OK population of ticks is especially intriguing because it was sampled from a laboratory colony, yet it contains a very high amount of genetic variation—the highest amount of all populations examined in this study—and a negligible measure of inbreeding (table 3). Moreover, both PCA and ADMIXTURE analyses detected population structure within the OK ticks (figs. 2A and 3). This was surprising because all 20 ticks were sampled from the same generation of a laboratory colony. The population structure in OK ticks may be a sampling artifact in that, by chance, some of the sampled ticks were descendants of the inbred line kept since the colony’s origin in 1976, and some were descendants of the wild-caught ticks that are introduced periodically. This possibility is consistent with the colony history in which larvae from two wild egg masses are mixed with larvae from two colony egg masses to produce each successive generation. Hence, the OK ticks may not be representative of a natural tick population, and instead provide an estimate of the genetic variation likely present in the laboratory ticks used for many immunological and disease transmission studies. Our distinct genomic results for the captive OK ticks, compared with the other natural tick populations, present a notable caution to such research on colony-reared ticks. Although presently, there is no evidence that the variation found in the laboratory strain lies in genes involved in the ticks’ competence to transmit pathogens, the possibility that genetic variation influences vector competence or susceptibility to acaricides warrants further investigation. Future immunological and disease transmission experiments should aim to examine ticks from regions that most closely resemble the area of concern, thus ensuring an accurate representation of the natural genetic variation in disease vectors.
Conclusion
The merit of an “integrated genetic epidemiology of infectious diseases” (Tibayrenc 1998) that investigates the genetic diversity of host, vector, and infectious agent has been well established. Many studies have revealed evidence for a genetic basis of a host’s susceptibility to infectious disease (reviewed in Hill 2006), of a vector’s competence to transmit pathogens (reviewed in Gooding 1996), and of a pathogen’s infectivity (e.g., Dykhuizen et al. 2008). Several researchers have emphasized the importance of molecular population genetic studies of arthropod vectors in particular (Tabachnick and Black 1995; Gooding 1996; McCoy 2008). Our study is the first examination of genome-wide genetic variation and population structure throughout the geographic range of A. americanum or any other tick species. Knowledge of the distribution of genetic variability in vectors is critical for furthering our understanding of the ecology, epidemiology, and control of vector-borne diseases. For example, Khatchikian et al. (2015) recently used mitochondrial DNA sequences to infer the demographic history and the rate of a northward expansion of I. scapularis in New York. Additionally, Turissini et al. (2014) used a GBS approach to investigate genomic patterns of polymorphism in a colony-reared inbred line of the mosquito Anopheles gambiae, providing critical insights toward the genetic vector control of malaria.
Climate change is causing expansions and shifts of the geographic ranges of many species, including arthropod vectors of infectious diseases (Lafferty 2009; Monzón et al. 2011). Indeed, the northward expansion of A. americanum is consistent with global warming. However, even with the warming trend of the past century, climatic conditions in the northeastern United States, especially in ME, remain much cooler than in the historic southeastern range of the lone star tick. This suggests the possibility of adaptive evolution causing or coinciding with the range expansion. Consistent with this hypothesis, our analyses identified hundreds of candidate loci (∼8% of SNPs examined) that are divergent between the Northeast population in NY and the Southeast population from the Carolinas. Another possibility is that changes in human land use, which are occurring faster than changes in temperatures, may be facilitating the range expansion of lone star tick. Landscape factors, such as the degree of forest cover and urbanization, appear to be influencing the population growth and range expansion of the Lyme disease tick vector, I. scapularis (Khatchikian et al. 2012, 2015).
Currently, all tick species other than I. scapularis are severely deficient in genomic resources. Here, using next-generation DNA sequencing technology, we discovered thousands of novel polymorphisms and their respective flanking sequences, which may be used to design other low- or high-throughput SNP-based assays of genomic variation in the lone star tick. The development of genomic resources in A. americanum will also enable surveys of neutral and adaptive variation in closely related ixodid ticks, many of which are important disease vectors throughout the world.
Supplementary Material
Acknowledgments
James Coleman, Alvaro Toledo, Indra Jayatilaka, Paul Tesoriero, and Natasha Cambria provided field/lab assistance. Charles Apperson, Lorenza Beati Ziegler, Charles Lubelczyk, and Lisa Coburn provided tick samples. Sharon Mitchell and Charlotte Acharya provided advice and support for GBS. Krishna Veeramah, Dustin Brisson, and three anonymous reviewers provided helpful comments on the manuscript. This work was supported by the National Institutes of Health [grants GM-102778 and AI-027044 to J.L.B.].
Literature Cited
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. 2008. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AW, et al. 2015. County scale distribution of Amblyomma americanum (Ixodida: Ixodidae) in Oklahoma: addressing local deficits in tick maps based on passive reporting. J Med Entomol. 52:269–273. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. 1996. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B: Biol Sci. 263:1619–1626. [Google Scholar]
- Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S. 2015. Pong: fast analysis and visualization of latent clusters in population genetic data. bioRxiv 031815.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp F, Trembley HL. 1945. Distribution and hosts of certain North American ticks. J Parasitol. 31:1–54. [Google Scholar]
- Brown HE, et al. 2011. An acarologic survey and Amblyomma americanum distribution map with implications for tularemia risk in Missouri. Am J Trop Med Hyg. 84:411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpi G, et al. 2011. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6:e25604.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs J, Paddock C. 2003. The ascendancy of Amblyomma americanum as a vector of pathogens affecting humans in the United States. Ann Rev Entomol. 48:307–337. [DOI] [PubMed] [Google Scholar]
- Cooley RA, Kohls GM. 1944. The genus Amblyomma (Ixodidae) in the United States. J Parasitol. 30:77–111. [Google Scholar]
- Cortinas R, Spomer S. 2013. Lone star tick (Acari: Ixodidae) occurrence in Nebraska: historical and current perspectives. J Med Entomol. 50:244–251. [DOI] [PubMed] [Google Scholar]
- Dahlgren FS, Paddock CD, Springer YP, Eisen RJ, Behravesh CB. 2016. Expanding range of Amblyomma americanum and simultaneous changes in the epidemiology of Spotted Fever Group Rickettsiosis in the United States. Am J Trop Med Hyg. 94:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, et al. 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 12:499–510. [DOI] [PubMed] [Google Scholar]
- Dykhuizen D, et al. 2008. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am J Trop Med Hyg. 78:806–810. [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, et al. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg H, Ewing C. 1989. Comparison of flagging, walking, trapping, and collecting from hosts as sampling methods for northern deer ticks, Ixodes dammini, and lone-star ticks, Amblyomma americanum (Acari: Ixodidae). Exp Appl Acarol. 7:313–322. [DOI] [PubMed] [Google Scholar]
- Ginsberg HS, et al. 1991. Increased population densities of Amblyomma americanum (Acari: Ixodidae) on Long Island, New York. J Parasitol. 493–495. [PubMed] [Google Scholar]
- Glaubitz JC, et al. 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:E90346.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J, Varela-Stokes AS. 2009. Role of the lone star tick, Amblyomma americanum (L.), in human and animal diseases. Vet Parasitol. 160:1–12. [DOI] [PubMed] [Google Scholar]
- Gooding R. 1996. Genetic variation in arthropod vectors of disease-causing organisms: obstacles and opportunities. Clin Microbiol Rev. 9:301–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilburn LR, Sattler PW. 1986. Electrophoretically detectable protein variation in natural populations of the lone star tick, Amblyomma americanum (Acari: Ixodidae). Heredity 57:67–74. [DOI] [PubMed] [Google Scholar]
- Hill A. 2006. Aspects of genetic susceptibility to human infectious diseases. Ann Rev Genet. 40:469–486. [DOI] [PubMed] [Google Scholar]
- Hooker WA, Bishopp FC, Hunter WD, Wood HP. 1912. The life history and bionomics of some North American ticks. Washington, DC: U.S; Dept. of Agriculture, Bureau of Entomology. [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Pontier D, Dufour A. 2009. Genetic markers in the playground of multivariate analysis. Heredity 102:330–341. [DOI] [PubMed] [Google Scholar]
- Keirans JE, Lacombe EH. 1998. First records of Amblyomma americanum, Ixodes (Ixodes) dentatus, and Ixodes (Ceratixodes) uriae (Acari: Ixodidae) from Maine. J Parasitol. 84:629–631. [PubMed] [Google Scholar]
- Khatchikian CE, et al. 2012. Geographical and environmental factors driving the increase in the Lyme disease vector Ixodes scapularis. Ecosphere 3:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatchikian CE, et al. 2015. Recent and rapid population growth and range expansion of the Lyme disease tick vector, Ixodes scapularis, in North America. Evolution 69:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe JJ, et al. 2004. Genetic variation increases during biological invasion by a Cuban lizard. Nature 431:177–181. [DOI] [PubMed] [Google Scholar]
- Lafferty KD. 2009. The ecology of climate change and infectious diseases. Ecology 90:888–900. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. 1758. Acarus In: Systema naturae . 10th ed. Stockholm: Salvius; p. 615–618. [Google Scholar]
- Lotterhos KE, Whitlock MC. 2014. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol Ecol. 23:2178–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, et al. 2013. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet. 9:e1003215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy K. 2008. The population genetic structure of vectors and our understanding of disease epidemiology. Parasite 15:444–448. [DOI] [PubMed] [Google Scholar]
- Means R, White D. 1997. New distribution records of Amblyomma americanum (L.) (Acari: Ixodidae) in New York State. J Vector Ecol. 22:133–145. [PubMed] [Google Scholar]
- Meirmans PG. 2015. Seven common mistakes in population genetics and how to avoid them. Mol Ecol. 24:3223–3231. [DOI] [PubMed] [Google Scholar]
- Merten HA, Durden LA. 2000. A state-by-state survey of ticks recorded from humans in the United States. J Vector Ecol. 25:102–113. [PubMed] [Google Scholar]
- Mixson TR, Campbell SR, et al. 2006. Prevalence of Ehrlichia, Borrelia, and Rickettsial agents in Amblyomma americanum (Acari: Ixodidae) collected from nine states. J Med Entomol. 43:1261–1268. [DOI] [PubMed] [Google Scholar]
- Mixson TR, Lydy SL, Dasch GA, Real LA. 2006. Inferring the population structure and demographic history of the tick, Amblyomma americanum Linnaeus. J Vector Ecol. 31:181–192. [DOI] [PubMed] [Google Scholar]
- Monzón J. 2012. Rapid evolution of northeastern coyotes Ecology and Evolution. [PhD dissertation]. New York: Stony Brook University. [Google Scholar]
- Monzón J, Kays R, Dykhuizen D. 2014. Assessment of coyote–wolf–dog admixture using ancestry-informative diagnostic SNPs. Mol Ecol. 23:182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzón J, Moyer-Horner L, Palamar MB. 2011. Climate change and species range dynamics in protected areas. Bioscience 61:752–761. [Google Scholar]
- Peakall R, Smouse PE. 2006. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 6:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, et al. 2013. Aye-aye population genomic analyses highlight an important center of endemism in northern Madagascar. Proc Natl Acad Sci U S A. 110:5823–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reichard M, et al. 2005. Sequence variation of the ribosomal DNA second internal transcribed spacer region in two spatially distinct populations of Amblyomma americanum (L.) (Acari: Ixodidae). J Parasitol. 91:260–263. [DOI] [PubMed] [Google Scholar]
- Schulze TL, et al. 1984. Amblyomma americanum: a potential vector of Lyme disease in New Jersey. Science 224:601–603. [DOI] [PubMed] [Google Scholar]
- Springer Y, Eisen L, Beati L, James A, Eisen R. 2014. Spatial distribution of counties in the continental United States with records of occurrence of Amblyomma americanum (Ixodida: Ixodidae). J Med Entomol. 51:342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabachnick W, Black IVW. 1995. Making a case for molecular population genetic studies of arthropod vectors. Parasitol Today. 11:27–30. [Google Scholar]
- Thomas C, et al. 2001. Ecological and evolutionary processes at expanding range margins. Nature 411:577–581. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M. 1998. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 28:85–104. [DOI] [PubMed] [Google Scholar]
- Tokarz R, Sameroff S, Leon MS, Jain K, Lipkin WI. 2014. Genome characterization of Long Island tick rhabdovirus, a new virus identified in Amblyomma americanum ticks. Virol J. 11:26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Williams SH, et al. 2014. Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J Virol. 88:11480–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trout R, Steelman C, Szalanski A. 2010. Population genetics of Amblyomma americanum (Acari: Ixodidae) collected from Arkansas. J Med Entomol. 47:152–161. [DOI] [PubMed] [Google Scholar]
- Turissini DA, Gamez S, White BJ. 2014. Genome-wide patterns of polymorphism in an inbred line of the African malaria mosquito Anopheles gambiae. Genome Biol Evol. 6:3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TA, Perkins SE, Heckel G, Searle JB. 2013. Adaptive evolution during an ongoing range expansion: the invasive bank vole (Myodes glareolus) in Ireland. Mol Ecol. 22:2971–2985. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Graham CH. 2005. Niche conservatism: integrating evolution, ecology, and conservation biology. Ann Rev Ecol Evol Syst. 36:519–539. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.