Abstract
The phylogenetic relationships among hemosporidian parasites, including the origin of Plasmodium falciparum, the most virulent malaria parasite of humans, have been heavily debated for decades. Studies based on multiple-gene sequences have helped settle many of these controversial phylogenetic issues. However, denser taxon sampling and genome-wide analyses are needed to confidently resolve the evolutionay relationships among hemosporidian parasites. Genome sequences of several Plasmodium parasites are available but only for species infecting primates and rodents. To root the phylogenetic tree of Plasmodium, genomic data from related parasites of birds or reptiles are required. Here, we use a novel approach to isolate parasite DNA from microgametes and describe the first genome of a bird parasite in the sister genus to Plasmodium, Haemoproteus tartakovskyi. Similar to Plasmodium parasites, H. tartakovskyi has a small genome (23.2 Mb, 5,990 genes) and a GC content (25.4%) closer to P. falciparum (19.3%) than to Plasmodium vivax (42.3%). Combined with novel transcriptome sequences of the bird parasite Plasmodium ashfordi, our phylogenomic analyses of 1,302 orthologous genes demonstrate that mammalian-infecting malaria parasites are monophyletic, thus rejecting the repeatedly proposed hypothesis that the ancestor of Laverania parasites originated from a secondary host shift from birds to humans. Genes and genomic features previously found to be shared between P. falciparum and bird malaria parasites, but absent in other mammal malaria parasites, are therefore signatures of maintained ancestral states. We foresee that the genome of H. tartakovskyi will open new directions for comparative evolutionary analyses of malarial adaptive traits.
Keywords: apicomplexa, haemoproteus, host switching, phylogenomics, Plasmodium, Plasmodium ashfordi, transcriptome
Introduction
One hundred years ago, species in the genus Haemoproteus and related avian hemosporidians were essential model organisms for uncovering the complex life cycle of malaria parasites (Martinsen and Perkins 2013), including the discoveries of exflagellation (MacCallum 1897), sporogony in mosquitoes (Ross 1898), and tissue merogony (Aragao 1908). These important life stages forgo the asexual replication in erythrocytes that is associated with malaria. Malaria parasites (Plasmodium) contain over 150 described species infecting mammals, birds, and reptiles within the mainly parasitic phylum Apicomplexa (Chromalveolata) (Perkins 2014). Closely related to Plasmodium are several genera of other hemosporidian parasites infecting mammals (Hepatocystis, Nycteria, and Polychromophilus), birds (Haemoproteus, Leucocytozoon, and Garnia), and reptiles (Saurocytozoon, Fallisia, Garnia, and Haemocystidium) (Valkiūnas 2005; Perkins 2014). To date, there is no consensus on the phylogenetic relationship between hemosporidian parasites. Conflicting phylogenetic hypotheses result from 1) heterogeneous data sets with respect to the genes investigated, limited taxon sampling, and choice of outgroups and 2) saturation of phylogenetic signals that is further complicated by drastically variable GC contents of the parasite genomes (Dávalos and Perkins 2008; Martinsen and Perkins 2013; Perkins 2014). As a consequence, it is challenging to reach unambiguous understanding of the evolutionary history of genes, metabolic pathways, and adaptive traits within this important group of parasites.
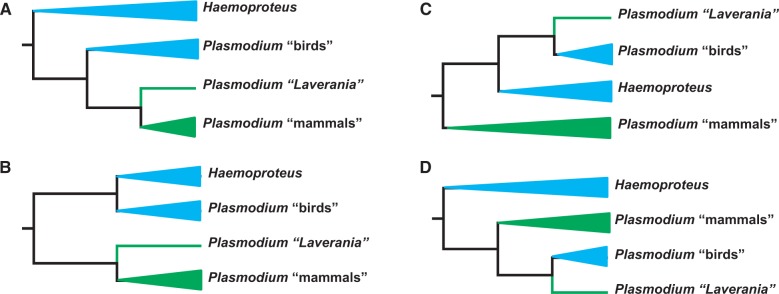
The evolutionary origin of the most virulent malaria parasite of humans, Plasmodium falciparum, has been debated for decades (Hagner et al. 2007). Most of the more recent studies have found it to be closely related to the other mammalian malaria parasites (fig. 1A and B) (Perkins and Schall 2002; Martinsen et al. 2008; Outlaw and Ricklefs 2011; Silva et al. 2011; Schaer et al. 2013; Borner et al. 2016) in contrast to several studies from the 1990s that proposed an origin from a Plasmodium parasite of birds shifting hosts to mammals (fig. 1C and D) (Waters et al. 1991; Escalante et al. 1998; McCutchan et al. 1996). However, more recent studies based on genomic data have also found support for a relationship between P. falciparum and bird Plasmodium (Pick et al. 2011). It is now well established that P. falciparum is one of many closely related species in the subgenus Laverania that infects gorillas and chimpanzees (Prugnolle et al. 2010) and that P. falciparum most likely originated from a parasite of gorillas (Liu et al. 2010). Yet, these important findings of the recent evolutionary history of P. falciparum (Keeling and Rayner 2015) do not answer the question of whether the ancestor of extant Laverania comes from Plasmodium parasites of birds or mammals. Resolving this question will require sequences of multiple genes from a parasite (outgroup) unquestioned as being more distantly related to all the Plasmodium species than those ingroup taxa are to each other, but yet not so distantly related that sequence alignments become problematic.
Fig. 1.—
Four phylogenetic hypotheses (A–D) illustrating the proposed relationships between species of Haemoproteus, Plasmodium of birds (including reptiles), Plasmodium of mammals, and Plasmodium of the subgenus Laverania, which contains the human parasite Plasmodium falciparum. Bird (and reptile) parasite branches are labeled in blue and mammal parasite branches in green. Triangular branch tips contain multiple species (in the range of 100–1,000); the Laverania branch may include about ten species infecting apes. Data presented in this work support phylogenetic hypothesis A.
A potentially useful outgroup to resolve the phylogenetic relationships within Plasmodium could be any species of hemosporidian parasites in the genus Haemoproteus. There are about 150 Haemoproteus parasites described (Valkiūnas 2005), but the genus may contain 10-fold more (cryptic) species as inferred from the high diversity of cytochrome b haplotypes (Outlaw and Ricklefs 2014). Similar to Plasmodium parasites, Haemoproteus spp. produce malarial pigment (hemozoin) in blood cells and their sexual reproduction takes place in dipteran vectors after ingestion of a blood meal. The diploid stage of the parasite is then followed by a series of life stages that eventually result in (haploid) infectious sporozoites that migrate to the salivary gland of the vector and subsequently will be transmitted to a vertebrate host during the next blood meal. The main vectors of Plasmodium parasites are various species of blood-sucking mosquitoes (Culicidae), whereas Haemoproteus parasites are vectored by biting midges (Ceratopogonidae; parasites of the subgenus Parahaemoproteus) and louse flies (Hippoboscidae; parasites of the subgenus Haemoproteus) (Valkiūnas 2005). Another important difference to Plasmodium is that Haemoproteus parasites do not use host red blood cells for asexual replication, only the gametocytes can be found in the blood; instead, the replication takes place in various internal organs of the host.
Using Haemoproteus parasites as outgroups in phylogenetic analyses of Plasmodium parasites is, however, not straightforward as their phylogenetic relationships have been disputed (fig. 1). Plasmodium and Haemoproteus may not be monophyletic when including parasites of other genera, for example, Hepatocystis seem to be nested within Plasmodium, and it is still unknown whether the two subgenera of Haemoproteus are monophyletic (Perkins 2008; Outlaw and Ricklefs 2011; Martinsen and Perkins 2013; Pineda-Catalan et al. 2013; Borner et al. 2016). Although life history data and vector use of the parasites suggest that Haemoproteus and Plasmodium indeed are monophyletic distinct taxa relative to each other, several phylogenetic studies have found support for a common ancestry of Haemoproteus parasites and bird Plasmodium parasites (fig. 1B), separate from Plasmodium parasites of mammals (Perkins and Schall 2002; Outlaw and Ricklefs 2011). Hence, before any Haemoproteus sp. can be used as an outgroup in phylogenetic analyses of Plasmodium, its phylogenetic position requires independent testing and confirmation.
The genomes of malaria and other hemosporidian parasites are relatively small (∼25 Mb) (DeBarry and Kissinger 2011) and should therefore be easily obtained using next-generation sequencing techniques. However, it has proved difficult to acquire sufficiently pure DNA from parasites infecting birds and reptiles for high-quality genome sequencing because their host species (birds and reptiles) have nucleated erythrocytes containing much larger genomes (∼1,300 Mb). There are currently 13 published genome sequences of mammalian malaria parasite species but none from hemosporidians infecting birds and reptiles (Carlton et al. 2013). A partial genome sequence of the bird malaria parasite Plasmodium gallinaceum is available for download but remains unpublished (ftp://ftp.sanger.ac.uk/pub/project/pathogens/Plasmodium/gallinaceum/), last accessed April, 2006.
By using a novel method to purify microgametes from infected bird blood combined with whole-genome amplification (Palinauskas et al. 2013), we present the first genome sequence of a parasite in the sister genus to Plasmodium, the bird parasite Haemoproteus tartakovskyi (subgenus Parahaemoproteus), isolated from a siskin (Spinus spinus). This genome sequence, combined with a novel transcriptome from the bird parasite Plasmodium ashfordi, enabled us to address fundamental questions about the evolution and diversification of malaria parasites.
Materials and Methods
DNA Isolation
We used material from wild-caught Eurasian siskins S. spinus naturally infected with H. tartakovskyi. The birds were captured at the Biological station “Rybachy” of the Zoological Institute of Russian Academy of Sciences on the Curonian Spit in the Baltic Sea (55°50’N, 20°44’E) in 2011. Birds were tested by microscopic examination of thin blood films and a nested polymer chain reaction protocol (Hellgren et al. 2004) targeting a region of the parasite’s cytochrome b (cyt b) gene, to confirm that the specimens were not coinfected with other hemosporidian species. We identified three individual birds with single H. tartakovskyi (cyt b lineage SISKIN1, GenBank accession number AY393806) infections of high parasitemia (between 2% and 5% of erythrocytes infected). The methods of isolating microgametes have been described by Palinauskas et al. (2013). In brief, approximately 200 μl of blood was withdrawn from the brachial vein and placed immediately in a microtube containing 10 μl of sodium citrate solution (3.7%), gently mixed, and exposed to air. The work was performed at 19 ± 1 °C. Four minutes after exposure to air, the sample was centrifuged for 5 min at 7,000 rpm. Approximately 20–50 μl of supernatant (blood plasma) was stored in 150 μl SET-buffer and placed at −20 °C until further processing in the laboratory. DNA was extracted using DNeasy Blood & Tissue kit (Qiagen, Valencia, CA). The amount of DNA was evaluated by 1% agarose gel electrophoresis and in parallel with a serial dilution of lambda-DNA. We obtained approximately 15 μl from each extract with DNA concentrations between 0.06 and 0.26 ng/μl. We used 0.2–1 ng of DNA for whole-genome amplification (Illustra GenomiphiV2 DNA Amplification Kit, GE Healthcare, Waukesha, WI) following the manufacturer’s instructions. Each amplification yielded several micrograms of DNA with a main fragment length of around 10 kb. For the 454-sequencing, we pooled four independent whole-genome amplicons from one of the infected siskins resulting in a total sample of 100 μl at a concentration of 980 ng/μl as determined by a NanoDrop.
Sequencing and Genome Assembly
Genomic shotgun and 3-kb paired-end libraries were each constructed from 5 μg of the above described material. The shotgun library was constructed according to the GS FLX Titanium Rapid Library Preparation method (January 2010 version) (Roche) including fragmentation down to 500 bp by using nebulization. After adaptor ligation and purification, the library was inspected using the DNA High Sensitivity kit on a 2100 BioAnalyzer (Agilent). The shotgun library was then quantified using the RL standard (Roche) as part of the Rapid Preparation method by using a Quantiflour fluorometer (Promega) and was finally diluted to obtain a total of 1 × 107 copies μl − 1.
The 3-kb paired end library was constructed according to the GS FLX Paired End Rapid Library Preparation method (April 2012 version) (Roche) including the use of a HydroShear Plus (Digilab Inc.) to fragmentize DNA. The fragmentation was inspected by the use of the DNA 7,500 kit on a 2100 BioAnalyzer (Agilent). After circularization, the DNA was nebulized for the purpose to size it down to 500 bp. After adaptor ligation and purification, the library was inspected using the DNA High Sensitivity kit on a 2100 BioAnalyzer (Agilent). The 3-kb paired end library was then quantified using the Quant-iT dsDNA assay kit (Invitrogen) and a Quantiflour fluorometer (Promega), and finally diluted to obtain a total of 1 × 107 copies μl − 1. Titrations and library production (aiming at 10–15% enrichment) were performed by emulsion polymer chain reaction and by using the Lib-L kit (Roche). DNA-positive beads were enriched, counted on an Innovatis CASY particle counter (Roche), processed using XLR70 sequencing kit (Roche), and loaded onto picotiter plates for pyrosequencing on a 454 Life Sciences Genome Sequencer FLX+ machine (Roche). Sequencing and library preparation were conducted at Lund University Sequencing Facility (Faculty of Science). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession LSRZ00000000. The version described in this article is version LSRZ01000000. The genome assembly with predicted genes is available for download at http://mbio-serv2.mbioekol.lu.se/Malavi/Downloads, last accessed April, 2016.
The 3.03 million raw reads from the sequencer were initially assembled into 28,500 contigs of a minimum length of 2,000 nt, distributed on 2,243 scaffolds with the Roche runAssembly software, version 2.8. The assembly contained 22.6 million base pairs including 754,000 gaps. To visualize the distribution of sequences originating from the host and parasite, respectively, the contigs and their GC content were calculated and plotted. The GC content showed a bimodal distribution (supplementary fig. S2A, Supplementary Material online). The higher GC content peak matches that of the host, if assuming a similar % GC in the siskin as in the zebra finch Taeniopygia guttata (41%), while the lower peak is more similar to an organism with low GC content, like P. falciparum (19%). The lengths of contigs with a % GC corresponding to the host were typically short (<500 bp, supplementary fig. S2B, Supplementary Material online) as expected for a host genome sequenced at a low coverage (1x, a total of 1.5 Gb sequenced).
Gene prediction was carried out with GeneMark-ES (version 2.3e). As this preliminary assembly contained reads originating from the host, a first filtering step was carried out. A search against UniProtKB, the protein database maintained by EBI, was performed for each gene. The search was conducted with BLASTx (version 2.2.28) using a threshold of 1e-5. This resulted in a list of gene names, which contained genes originating from a bird species according to the top BLAST hit and these were removed. In total, 1.27 million reads were determined to not originate from H. tartakovskyi and were therefore discarded.
A new assembly containing the remaining 1.76 million high-quality reads without host matches was carried out in the same way as described above, with the exception that the minimum scaffold length was reduced to 1,000 (default is 2,000). This assembly contained 5,410 scaffolds with a total length of 26.7 million base pairs. A second filtering step was implemented where 2,375 scaffolds, containing both a length of ≤ 2,000 nt and a GC content % above 33.4, were removed. In addition, 48 short (≤ 3,000 nt) scaffolds were removed after manual inspection (moderate BLAST matches to other vertebrates and suspiciously high GC content). Another four scaffolds were removed after a GenBank Contamination screen. The remaining scaffolds (2,983), with a total length of 23.2 million base pairs (including 459,015 gaps), represents the assembled genome of H. tartakovskyi. Assembly statistics are found in supplementary table S1, Supplementary Material online, and the distribution of the scaffold GC contents in supplementary figure S3, Supplementary Material online. A new gene prediction analysis with the same software and settings as described above was carried out resulting in 5,988 genes.
The Transcriptome of P. ashfordi
Whole blood (20 μl) from three juvenile siskins, experimentally infected with P. ashfordi (cyt b lineage GRW02, GenBank accession number AF254962), was sampled at days 21 and 31 postinfection, corresponding to the peak and decreasing parasitemia levels, respectively. For details regarding the infection experiment, see Palinauskas et al. (2011). Tubes with blood samples were directly put in liquid nitrogen and transferred to −80 °C where stored until RNA extraction. The isolation of RNA and library preparations are described in Videvall et al. (2015). Six infected samples with high parasitemia levels were sequenced as paired-end using an Illumina HiSeq2000. Two of the most highly infected samples (48% and 71% parasitemia) were resequenced, to retrieve more reads from the parasite. After demultiplexing and quality filtering of reads, we ended up with a total of 247 million 90 bp reads and 242 million 65 bp reads.
We performed a de novo assembly of the reads using the RNA-seq assembler Trinity (v. r20131110) (Grabherr et al. 2011). The assembled contigs were initially screened against the zebra finch and chicken (Gallus gallus) genomes with BLASTn (version 2.2.28), and all contigs with matches were removed from the assembly. After a subsequent BLASTx search against the NCBI nonredundant protein database, we retained 8,959 contigs that generated significant hits (e value: 1e-3) against apicomplexan parasites as a preliminary transcriptome of P. ashfordi. Transcripts containing translated open reading frames (ORFs) were scanned using TransDecoder (version rel16JAN2014, http://transdecoder.github.io/, last accessed April, 2016) with default parameters. These 7,953 filtered transcripts were used as input in the ortholog clustering and phylogenetic analyses conducted in this study. Reads used in this study have been uploaded to the NCBI Sequence Read Archive under the accession number PRJNA311546. The assembled transcripts are available for download at http://mbio-serv2.mbioekol.lu.se/Malavi/Downloads (last accessed April, 2016).
Ortholog Clustering, Alignment, and Synteny Analyses
Ortholog clusters (OCs) were generated for 17 apicomplexan species to provide data for phylogenetic analyses. Numbers of clusters for all pairwise combinations of species, and numbers of clusters and genes for specific groups are shown in supplementary tables S2 and S3, Supplementary Material online. All data were from the most current release at the time of analysis (January 21, 2014). Annotated protein-encoding gene sequences, gene IDs, gene coordinates, transcripts, and the numbers, sizes, and IDs, of all chromosomes/contigs/scaffolds for all species were accessed as follows: P. falciparum, Plasmodium vivax, Plasmodium knowlesi, Plasmodium chabaudi, Plasmodium yoelii (strain YM), Plasmodium berghei, and Plasmodium cynomolgi were downloaded from PlasmoDB (Aurrecoechea et al. 2009) (version 9.3). Cryptosprodium parvum and Cryptosprodium muris were downloaded from CryptoDB (Puiu et al. 2004) (version 5.0). Babesia bovis, Babesia microti, Theileria annulata, and Theileria parva were downloaded from PiroplasmaDB (http://piroplasmadb.org/, last accessed June, 2014) (version 3.0). Toxoplasma gondii and Neospora caninum were downloaded from ToxoDB (Gajria et al. 2008) (version 8.2). Orthologs were clustered with WU-BLAST (version 2.2.6) which has become a module within the AB-BLAST package (http://www.advbiocomp.com/, last accessed April, 2016) and OrthoMCL (Li et al. 2003) (version 1.4) as described in (DeBarry and Kissinger 2011). Custom PERL scripts were used to query OrthoMCL output for the groups in table S2. From these groups, we obtained two sets of single-copy gene clusters, 703 unique to Haemoproteus and Plasmodium spp., and 599 shared among all 17 examined apicomplexans. The amino acid sequences for each gene were aligned using ParaAT (Zhang et al. 2012) (version 1.0) with default parameters and the MUSCLE (version 3.8.21) alignment algorithm. Prior to the phylogenetic analyses, the alignments were filtered by GBLOCK (Castresana 2000) (version 91b) with default parameters to remove gaps and poorly aligned regions.
CEGMA orthologs were extracted from the published genomes (Parra et al. 2009) of seven species of Plasmodium, three more distantly related apicomplexan species (To. gondii, B. bovis, and Theileria equi), and our sequenced P. ashfordi transcriptome and the H. tartakovskyi genome.
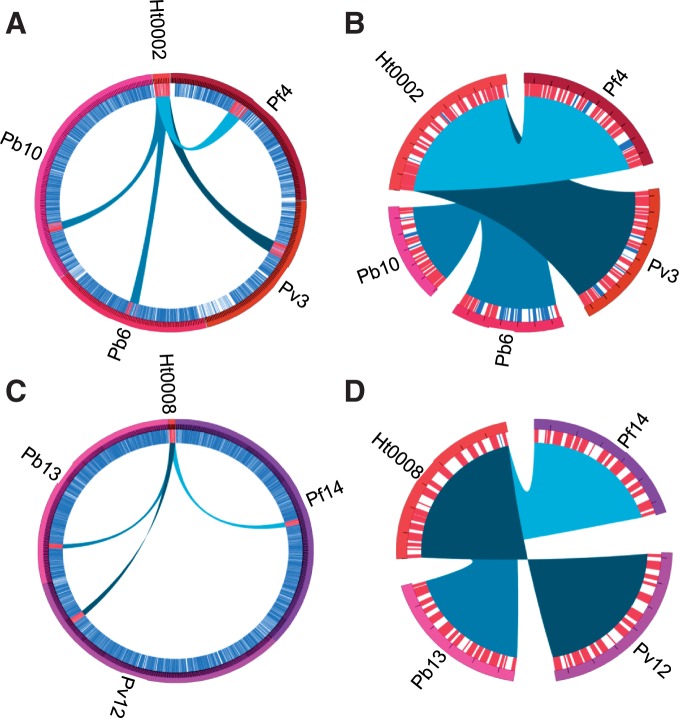
Synteny was calculated with MCScanX (Wang et al. 2012). BLASTp (Altschul et al. 1990) (version 2.2.26) was used to compare annotated protein-encoding gene sequences for P. falciparum, P. vivax, P. berghei, and H. tartakovskyi as input to MCScanX with paralogous synteny detection deactivated, E-value threshold of 1e-5, unit distance set at 1,000, and match size set at 3. Circos (Krzywinski et al. 2009) was used to visualize MCScanX-detected synteny between P. falciparum and H. tartakovskyi scaffolds in figure 2.
Fig. 2.—
Representative synteny between two of the longest Haemoproteus tartakovskyi scaffolds and chromosomes of Plasmodium falciparum, Plasmodium vivax, and Plasmodium berghei. Circles show detected syntenic regions between H. tartakovskyi and Plasmodium spp. Synteny is represented by spans that connect H. tartakovskyi scaffolds (Ht0002 and Ht0008) with chromosome regions of the three Plasmodium species. Circles to the left (A and C) show full sized Plasmodium chromosomes. Circles to the right (B and D) illustrate close ups of the syntenic regions (A and C). Black ticks on scaffolds/chromosomes in (B and D) = 10 kb. Blue and red lines just inside the outer chromosome rings represent nonsyntenic and syntenic protein-encoding genes, respectively.
Phylogenetic Analyses
We used RAxML version 8.0.26 (Stamatakis 2006) to construct bootstrapped (100 iterations) phylogenetic trees on the concatenated protein alignments performed by using GBLOCK (Castresana 2000) (described above). We used the LG + F model for amino acid substitutions as it was found to be the best model for the concatenated alignments and also in 69% of the cases for the individual protein alignments. The individual proteins were analyzed separately to enable the calculation of gene/protein-support frequencies and internode certainties (Salichos and Rokas 2013). This was done using consense from the Phylip package version 3.695 (Felsenstein 1989).
When using GBLOCK default settings, indels are not reported. To identify indels among the nine species of hemosporidians in the 599 + 703 (1,302 total) protein sets used in figure 4, GBLOCK was therefore again run on the original OrthoMCL alignments but now with the b5 option set to “all.” An indel was defined as any number of continuous gaps within a high-quality protein alignment that were flanked by amino acids on both sides (fig. 5B). A value of 1 (regardless of gap length) was given to sequences that contained amino acids for these positions and 0 for the sequences that contained gaps (script available on request). The data were collected in a binary matrix for each protein and then concatenated. All singleton positions (minor indel variant in only one of the nine taxa) were deleted. For the phylogenetically informative indels, we used the IF-function in Microsoft Excel to determine which topology it supported and then to calculate the total number of indels supporting each of the alternative phylogenetic topologies. Maximum parsimony trees were constructed in MEGA (version 6.06) (Tamura et al. 2007) from the concatenated alignment of the phylogenetically informative indels by first replacing the values (0 and 1) to arbitrary nucleotides (C and T).
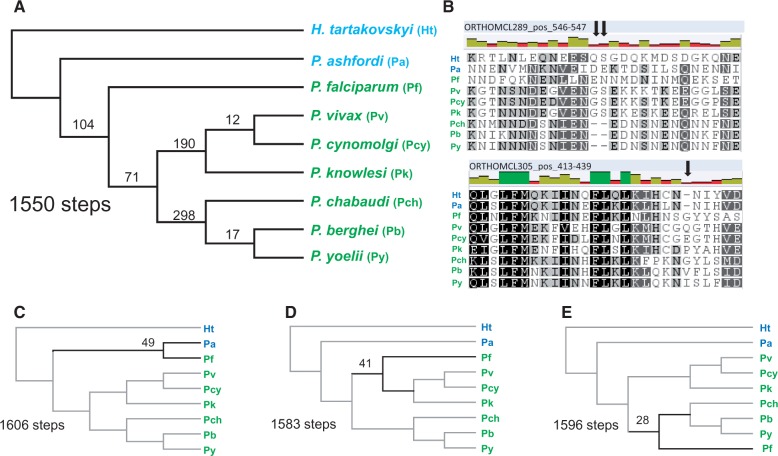
Fig. 4.—
Phylogenetic trees of Plasmodium parasites rooted with Haemoproteus tartakovskyi. The trees were constructed in RAxML using sequences of 599 concatenated genes (192,181 aligned amino acid positions) shared with other apicomplexan species (left) and 703 concatenated genes (128,738 aligned aa positions) unique to parasites of these genera (right). *100% bootstrap support. Individual gene tree support frequencies (in %) and internode certainties are given under the nodes. Taxon names in blue are bird parasites and in green mammal parasites. The scale bar corresponds to branch lengths in the unit of evolutionary distance. H., Haemoproteus; P., Plasmodium. GeneIDs of the included taxa and for each gene alignment are available in supplementary appendices S1 (left panel) and S2 (right panel), Supplementary Material online.
Fig. 5.—
Maximum parsimony tree of 1,116 phylogenetic informative indels of hemosporidian parasites. The tree requiring the lowest number of steps (A) is identical to the tree obtained from sequence-based analyses of amino acids shown in figure 3. (B) Two examples of indels (indicated with arrows) identified within GBLOCKs. The numbers of steps are substantially higher for trees (C) grouping Plasmodium falciparum with the bird parasite Plasmodium ashfordi, (D) grouping P. falciparum with the Plasmodium vivax group, and (E) grouping P. falciparum with Plasmodium parasites infecting rodents. The numbers of indels uniquely shared within clades are shown to the left of the nodes in (A). In (C–E), these numbers indicate the number of indels in conflict with the shortest tree (A). The topology differences between tree (A) and trees (C–E) are indicated by thick branches.
Results
The Genome of H. tartakovskyi
By using 454 pyrosequencing, we obtained approximately 3.03 million reads, which were filtered to exclude sequences originating from the bird host. The remaining approximately 1.73 million reads were used to assemble the genome of H. tartakovskyi into 2,983 scaffolds (supplementary table S1, Supplementary Material online). The apicoplast genome is absent from the assembly, which is expected as we used DNA from microgametes which lack the apicoplast (Valkiūnas 2005). As the microgametes also lack mitochondria (Valkiūnas 2005), it was surprising that one of the scaffolds (HtScaffold0932) contained the complete 5,992 bp mitochondrial genome sequence. However, the sequence depth of this scaffold is lower compared to the average (5X vs. 35X) suggesting that the sequenced mitochondria were from the ruptured microgametocytes or that the DNA isolate also contained some macrogametes.
The size (∼23 Mb) of the H. tartakovskyi nuclear genome and the number of predicted genes (5,988) correspond well to the genomes of Plasmodium species (table 1). The overall GC content (25.4%) is in the lower range and closer to P. falciparum (19.3%) than to P. vivax (42.3%). Of the predicted genes, approximately 60% group into OCs shared with other apicomplexan parasites (supplementary table S3, Supplementary Material online). The remaining genes (∼40%) were identified as unique to H. tartakovskyi. The majority of these genes were single copy (2,199), but clustering analyses demonstrates the presence of 20 expanded gene families with 2–72 copies.
Table 1.
Genome Features of Haemoproteus tartakovskyi in Comparison with Three Other Species of Apicomplexans
| Feature | H. tartakovskyi | Plasmodium falciparum (Pf 3D7) | Plasmodium vivax (Pv Sal1) | Toxoplasma gondii (Tg VEG) |
|---|---|---|---|---|
| Genome | ||||
| Size (Mb) | 23.2 | 23.3 | 27.0 | 64.5 |
| GC content (%)a | 25.4 | 19.3 | 42.3 | 52.4 |
| Genesb | ||||
| Number of genes | 5,988c | 5,542 | 5,286 | 8,410 |
| Mean length (bp) | 2,333 | 2,538 | 2,401 | 4,669 |
| Longest gene (bp) | 29,935 | 30,864 | 34,519 | 53,558 |
| Genes with introns (%) | 69.1 | 55.1 | 53.7 | 80.2 |
| Exonsd | ||||
| Mean number per gene | 2.5 | 2.6 | 2.6 | 5.6 |
| GC content (%)a | 28.3 | 23.8 | 46.2 | 57.9 |
| Mean length (bp) | 661 | 861 | 820 | 420 |
| Introns | ||||
| GC content (%)a | 23.1 | 12.9 | 48.3 | 48.8 |
| Mean length (bp) | 287 | 164 | 182 | 469 |
| Intergenic regions | ||||
| GC content (%)a | 22.6 | 14.4 | 38.0 | 48.8 |
| No. 5.8S/18S/28S rRNA units | 2 | 7 | 7 | ? |
G + C content is defined by the total number of C:s and G:s divided by the number of standard nucleotides (i.e., not N).
Including introns but not untranslated regions. Only protein encoding genes are included.
Predicted genes ≥100 bp. The total number was 6,431 including short genes (10–99 bp).
Only exons with coding sequences are reported. Untranslated regions are not included.
The largest of the expanded gene families (Ht cluster 1) contained 72 predicted genes of variable lengths (375–6,990 bp) and number of exons (between 1 and 11 exons; 66% < 3 exons). These were located on 48 scaffolds with up to five copies appearing in tandem. The pairwise amino acid identities between the copies were ≤ 81%, with a mean of 21%. The next largest expanded gene family (Ht cluster 2) contained 13 predicted genes (lengths: 810–1,437 bp, number of exons: 1–4, mean pairwise amino acid identity 37%). The GC contents of the genes in both these clusters were significantly higher than the mean value for the remaining H. tartakovskyi genes (Table 2).
Table 2.
The GC Content of the Two Largest Expanded Gene Families Unique to Haemoproteus tartakovskyi Compared to Three Expanded Gene Families of Plasmodium falciparum (Pf 3D7)
|
H. tartakovskyi |
P. falciparum |
||||||
|---|---|---|---|---|---|---|---|
| Genes | n | Mean GC (%) | SD | Genes | n | Mean GC (%) | SD |
| Cluster1 | 72 | 30.26a | 2.65 | rif | 185 | 32.65b | 2.06 |
| Cluster2 | 13 | 34.35a | 1.42 | var | 83 | 33.78b | 3.41 |
| stevor | 42 | 29.89b | 2.37 | ||||
| All other genes | 5,905 | 28.09 | 4.80 | All other genes | 5,232 | 24.50 | 3.98 |
Significantly higher than the mean for all other genes, P < 10−6 (Wilcoxon rank sum test).
Significantly higher than the mean for all other genes, P < 10−13 (Wilcoxon rank sum test).
We identified 790 OCs shared among all 17 analyzed apicomplexan species and 35 OCs that are absent in H. tartakovskyi but present in all other apicomplexans (supplementary table S3, Supplementary Material online). By conservatively assuming that these 35 OCs exist in the genome of H. tartakovskyi but are missing in the scaffolds, we can estimate that the assembly includes a minimum of approximately 96% of the existing genes (790/(790 + 35)). Synteny appears to be similar to species of Plasmodium, although this can only be partially evaluated due to the relatively short scaffold lengths (fig. 2).
The mammal Plasmodium species have five to seven copies of the typical eukaryotic ribosomal RNA gene set (18S-5.8S-28S). In P. falciparum, complete copies of these genes are located on chromosomes 1, 5, 7, 11, and 13 and two partial gene sets (5.8S-28S) on chromosome 8 (Rooney 2004). Within the H. tartakovskyi genome, we identified two scaffolds (HtScaffold0062, HtScaffold0602) containing these genes. The sequence similarities between the two H. tartakovskyi 18S copies were 82.0% and between 28S copies 58.7%. The two 28S copies in H. tartakovskyi are more different from each other than the 28S copies of P. falciparum are to each other (between chromosomes 5 and 13; 18S: 86.1%, 28S: 79.1%).
The TRAP-family proteins and proteins that share functional domains with TRAP are of central importance in the host cell invasion process for several life stages (sporozoites, ookinetes, and merozoites) of apicomplexan parasites (Morahan et al. 2009). Of the 10 TRAP and TRAP-like genes described in P. falciparum, we identified nine with high confidence in H. tartakovskyi (supplementary table S4, Supplementary Material online). The pairwise amino acid identities (Ht vs. Pf) were generally low (21–46%), as expected for genes directly involved in host-pathogen evolution.
In the erythrocytic life stages, Plasmodium parasites use hemoglobin as a major nutrient source. This requires a series of enzymes to detoxify the digestion product into hemozoin (malarial pigment), an insoluble microcrystalline form of heme that subsequently remains as pigment granules in the cell. Gametocytes of Haemoproteus parasites also exhibit malarial pigment. We identified orthologs of all the five enzymes (supplementary table S5, Supplementary Material online) shown to be involved in the hemoglobin detoxification pathway in P. falciparum (Chugh et al. 2013). However, we could only detect one H. tartakovskyi copy of the cysteine proteinase falcipain and plasmepsin genes, whereas P. falciparum has two and three copies, respectively. Their pairwise amino acid identities (Ht vs. Pf) were somewhat higher than for the TRAP genes (35–75%).
We identified two OCs exclusively shared between H. tartakovskyi and P. falciparum that were absent in all the other analyzed mammalian Plasmodium species. One of these was an unknown protein (PF3D7_1004100), whereas the other cluster contained the reticulocyte binding protein homolog 1 (RH1) from P. falciparum (PF3D7_0402300) and seven H. tartakovskyi copies. The RH1 protein is involved in the merozoite binding and invasion of erythrocytes (Triglia et al. 2009) and it has a homolog in Plasmodium reichenowi (PRCDC_0005400). Whether the different copies of this expanded gene family in H. tartakovskyi have similar functions in the ligand–receptor interaction between merozoites and host cells remains to be investigated.
Phylogenetic Relationships of Apicomplexan Parasites
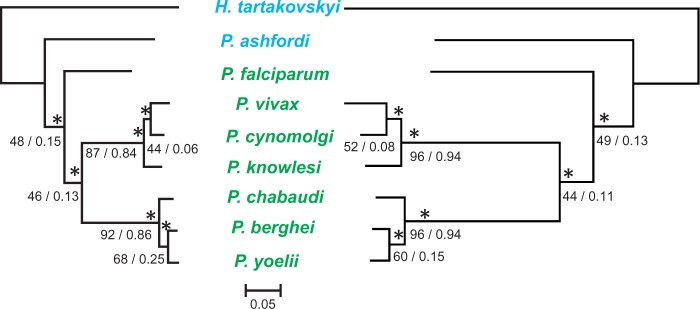
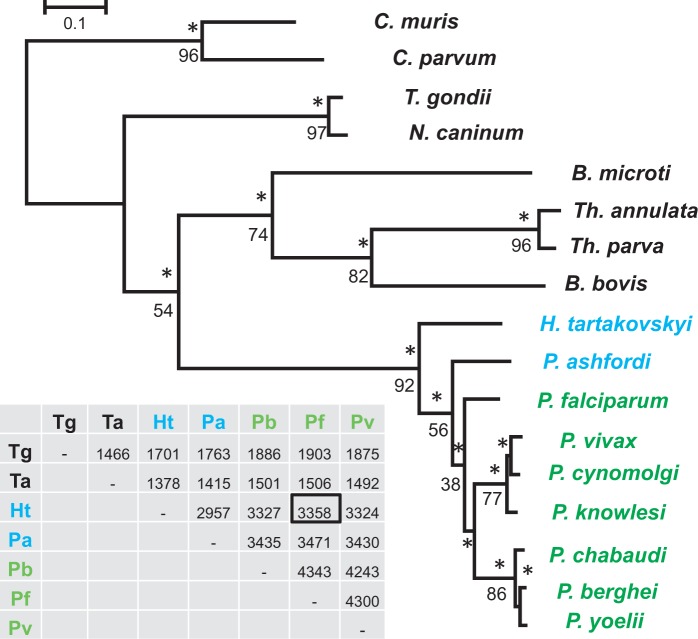
Phylogenetic analyses were executed in two steps. First, we constructed phylogenies from alignments of translated orthologous genes present in 17 apicomplexan species (supplementary table S3, Supplementary Material online) and rooted with C. muris and C. parvum. After GBLOCK (Castresana 2000) filtering of poorly aligned regions, high-quality alignments remained for 593/790 OCs. The maximum-likelihood consensus tree resulted in a topology in general agreement with the phylogeny of apicomplexans (Kuo et al. 2008; DeBarry and Kissinger 2011) and placed H. tartakovskyi as a sister taxon to a clade containing all Plasmodium parasites including the bird parasite P. ashfordi (fig. 3). This topology is supported by 56% of the individual gene trees, whereas the alternative hypothesis of H. tartakovskyi grouping with bird Plasmodium is supported by 16% of gene trees. Together, these results reject the hypotheses depicted in figure 1B and C.
Fig. 3.—
Phylogenetic tree of hemosporidian parasites and other more distantly related apicomplexans. The tree was constructed from 593 concatenated genes shared with other apicomplexan species in RAxML (133,965 aligned aa positions) and rooted with Cryptosporidium muris and Cryptosporidium parvum. *100% bootstrap support. Individual gene tree frequencies that support the topology are indicated below the node (in %). Names of bird hemosporidian parasites are presented in blue and mammal hemosporidian parasites in green. The scale bar corresponds to branch lengths in the unit of evolutionary distance. The table indicates the number of pairwise orthologous gene clusters with the black square marking the number shared between Haemoproteus tartakovskyi and Plasmodium falciparum. The placement of H. tartakovskyi as a sister taxon to a clade of all Plasmodium parasites was supported by 96 CEGMA genes (supplementary fig. S1a, Supplementary Material online) and, with lesser support, by DNA-based analyses of the three mitochondrial genes (supplementary fig. S1b, Supplementary Material online). B., Babesia; C., Cryptosporidium; H. ,Haemoproteus; N., Neospora; P., Plasmodium; Th., Theileria; T., Toxoplasma. GeneIDs of the included taxa and for each gene alignment are available in supplementary appendix S1, Supplementary Material online.
Second, to firmly establish the phylogenetic position of Laverania parasites (here represented by P. falciparum) and to test between the alternative hypotheses in figure 1A and D, we used H. tartakovskyi as an outgroup in two data sets consisting of only hemosporidian species. The first data set consisted of high-quality alignments from 599/790 OCs examined in figure 3 and contained 50% as many aligned amino acid positions. The second data set consisted of 703/1,040 OCs unique to hemosporidians (supplementary table S3, Supplementary Material online). The two resulting phylogenetic trees (fig. 4) show identical topologies to the hypothesis in figure 1A, that is, they reject the hypothesis that Laverania parasites, including P. falciparum, originated as a secondary host shift from a bird-infecting parasite. A monophyletic group of mammalian Plasmodium parasites is supported by 48% and 49% of the individual gene trees, respectively. The alternative hypothesis in figure 1D is the second most common pattern in the individual gene trees (18% and 20%, respectively). Both protein sets place P. (Laverania) falciparum basal to all other mammalian Plasmodium, supported by 46% and 44% of the individual trees.
The topology in figure 4 agrees with four previously identified indels in the mitochondrial genome supporting a monophyletic clade of mammalian Plasmodium parasites (Roy and Irimia 2008). We therefore examined the distribution of informative indels (insertions or deletions of 1–7 amino acids shared between ≥2 taxa) in the protein alignments used for the phylogenetic analyses in figure 4, to ensure that the results from the sequence-based phylogeny were not biased due to, for example, signal saturation, convergent evolution, or bias resulting from base composition. Within high-quality aligned regions identified by GBLOCK (Castresana 2000), there were 1,116 phylogenetically informative indels. These indels were used in maximum parsimony analyses (Tamura et al. 2007) and the tree requiring the fewest steps (fig. 5A) resulted in a topology identical to the tree in figure 4. The competing phylogenetic hypotheses using these indels required 56, 33, and 46 additional steps, respectively (fig. 5C–E).
Discussion
Understanding the process of adaptive evolution requires accurate phylogenies, which in turn depend on proper rooting (Rich and Xu 2011). Our two-step approach, to first establish that Haemoproteus spp. are appropriate outgroups for rooting trees of Plasmodium spp., and second, perform phylogenies with improved parameters, allowed us to obtain longer high-quality alignments and use a larger set of genes to explore the detailed relationships within the genus Plasmodium. High variation in GC content within the ingroup taxa combined with substantial sequence divergence may each have contributed to our finding that many individual gene trees are in conflict with the main topology. However, all nodes in the consensus tree were supported by more than twice as many individual gene trees when compared to the next best alternative. The topology of the best supported sequence-based phylogenetic tree agreed with our analyses of indels, arguably providing parallel, independent phylogenetic support that the phylogeny has been resolved correctly.
Our first finding shows that Haemoproteus is phylogenetically placed outside the clade of the investigated Plasmodium parasites. This disagrees with results in several earlier studies that used mitochondrial cytochrome b gene sequences combined with distant taxa (To. gondii and Th. annulata) for tree rooting (Escalante et al. 1998; Perkins and Schall 2002) but also disputes the conclusions of a recent phylogenetic study based on four genes and an outgroup-free approach for rooting trees (Outlaw and Ricklefs 2011). Our whole-genome phylogenetic analyses of Haemoproteus sp. combined with several distantly related apicomplexan parasites provide strong support for a common origin of Plasmodium parasites in birds and mammals (fig. 1A). This result is in perfect agreement with a recent multigene study that to date covers the broadest taxon sampling of hemosporidians (Borner et al. 2016).
Our second finding is a common origin of mammalian Plasmodium parasites. This phylogenetic topology has previously been recovered in several other studies (Perkins 2008; Outlaw and Ricklefs 2011) but is in conflict with Pick et al. (2011) who used the hitherto largest number of genes (218 orthologous exported proteins) to construct Plasmodium phylogenies. This contradictory result may stem from the large evolutionary distance between the outgroups and the ingroup in the study by Pick et al. (2011), a conclusion corroborated by Borner et al. (2016).
Our results also show, in agreement with several other studies (Perkins 2008; Outlaw and Ricklefs 2011; Borner et al. 2016), that the species of the subgenus Laverania, including P. falciparum, form a basal group relative to the Plasmodium species of primates and rodents. This split was recently dated to 60–120 Ma (Silva et al. 2015). That the mammal Plasmodium parasites form a monophyletic clade implies that the previously reported similarities in circumsporozoite (McCutchan et al. 1996) and chitinase genes (Li et al. 2005) between P. falciparum and P. gallinaceum result from maintained ancestral states rather than a more recently shared ancestry.
On the basis of the phylogeny supported by our analyses, we can conclude that the low GC content of the P. falciparum genome is a trait shared with the ancestors of mammalian Plasmodium species, parsimoniously suggesting that the higher GC content of P. vivax, P. knowlesi, and P. cynomolgi has evolved more recently. A study that analyzed GC/AT composition relative codon positions in P. falciparum and P. vivax reached the same conclusion (Nikbakht et al. 2014), that is, that the high GC content in P. vivax evolved from an ancestral GC poor genome since the split from P. falciparum. A low GC content is commonly found in parasites and endosymbionts with reduced genome sizes. It has been suggested that low GC can result from biased mutations and inefficient selection in small and mainly nonrecombining populations (Moran 1996) or loss of particular DNA repair systems (Lind and Andersson 2008). Thus, it is intriguing that the clade of P. vivax, P. knowlesi, and P. cynomolgi has restored the GC content to a level typical for nonparasitic eukaryotes. Nikbakht et al. (2014) suggested that GC-biased heteroduplex repair (Brown and Jiricny 1988) could explain the resurrected GC in P. vivax but unravelling the mechanisms behind this restoration requires more research.
We found that about 40% of the predicted genes in the genome of H. tartakovskyi did not share orthologs with other apicomplexan species. Such a high proportion of lineage-specific genes is not unusual in comparisons across apicomplexan genera (Kissinger and DeBarry 2011). The reduced genomes of apicomplexan parasites can be explained by rampant and somewhat idiosyncratic gene losses in the course of their evolution from a free-living organism (Woo et al. 2015). Hence, many of the genes here classified as unique to H. tartakovskyi might be traceable to the common ancestor of apicomplexan parasites; a task, however, complicated by strongly divergent sequences and possibly functional changes after hundreds of millions of years of evolution.
A characteristic feature of Plasmodium parasite genomes is the presence of species-specific expanded gene families in the subtelomeric chromosome regions, encoding for proteins expressed on the surface of infected host cells (Scherf et al. 2004), for example, var, rif, and stevor in P. falciparum (Gardner et al. 2002) and members of the pir gene family (Hall et al. 2005); vir in P. vivax (Carlton et al. 2008) and bir, cir, and yir in the rodent parasites (Janssen et al. 2002). By altering the expression of these surface proteins during the course of an infection, the parasite can escape the host immune response. Although the var, rif, and stevor genes in P. falciparum consist of two exons (Gardner et al. 2002), the vir gene family of P. vivax is more heterogeneous (1–5 exons and length variation between 156 and 2,316 bp) (Carlton et al. 2008). We found that the two expanded gene families unique to H. tartakovskyi are highly variable in length and number of exons. The relatively short scaffolds prevent us from establishing their chromosomal locations; however, like the var, rif, and stevor genes, these two gene families have significantly higher GC content compared to the rest of the genes in the genome (Table 2). It is indeed tempting to speculate that these expanded gene families in H. tartakovskyi provide similar functions as the species-specific antigen families in other Plasmodium species; however, testing this hypothesis will require functional validation and data on when and where these genes are expressed.
A caveat of our phylogenetic study is the low level of taxon sampling compared to the existing thousands of species of hemosporidians (Perkins 2014). Based on mtDNA sequences, bird and saurian Plasmodium parasites are highly diverse, and although we here identified them to belong to a monophyletic clade, it would be premature to rule out the possibility that none of these species belong to the clade of mammal Plasmodium parasites. However, our mtDNA result shows that P. ashfordi is closely related to P. gallinaceum (supplementary fig. S1, Supplementary Material online), supporting the hypothesis that the parasite originally suggested to share ancestry with P. falciparum (Waters et al. 1991) rather belongs to the clade of nonmammal Plasmodium parasites.
One trait used to define the genus Plasmodium (in combination with malarial pigment present in blood stages) is “merogony in blood,” that is, the asexual replication in red blood cells that in several species is synchronized with a periodicity of 18–72 h (Mideo et al. 2013) and leads to fever at their coordinated ruptures. However, before we can evaluate whether blood merogony is a gain in Plasmodium spp. since the split from Haemoproteus spp., or a more recent loss in Haemoproteus spp., genomic data from parasites in the other genera within Haemosporida must be generated. For this task, the genome of H. tartakovskyi and our developed method to enrich for parasite DNA (Palinauskas et al. 2013) will be instrumental when recovering and identifying genome sequences from species of hemosporidian genera for which data are lacking. With such data, we can explore how hemosporidian parasites have adapted to different vertebrate hosts (birds, reptiles, or mammals) and vectors (dipterans of the Culicidae, Ceratopogonidae, Simuliidae, Hippoboscidae, Phlebotomidae, Tabanidae, and Nycteribiidae) and investigate the genomic signatures of selection associated with these parasites’ frequent shifts of hosts and vectors (Ricklefs et al. 2014; Outlaw et al. 2015).
Supplementary Material
Supplementary tables S1–S5, Appendices 1 and 2, and figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Swedish Research Council (to S.B. and O.H.), by CAnMove (a Linnaeus research excellence environment financed by Swedish Research Council and Lund University), by the Crafoord Foundation (to O.H.), by the European Social Fund under the Global Grant measure (to G.V.), and in part by resources and technical expertise from the University of Georgia, Georgia Advanced Computing Resource Center. We thank the director of the Biological Station “Rybachy,” Casimir V. Bolshakov, for generously providing facilities for the experimental research and Susan Perkins for comments on the manuscript.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Aragao HB. 1908. Über den entwicklungsgang and die übertragung von Haemoproteus columbae. Arch Protistenkd. 12:154–167. [Google Scholar]
- Aurrecoechea C, et al. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37:D539–D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner J, et al. 2016. Phylogeny of haemosporidian blood parasites revealed by a multi-gene approach. Mol Phyl Evol. 94:221–231. [DOI] [PubMed] [Google Scholar]
- Brown TC, Jiricny J. 1988. Different base/base mispairs are corrected with different efficiencies and specificities in mokey kidney cells. CELL 54:705–711. [DOI] [PubMed] [Google Scholar]
- Carlton JM, et al. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Sullivan SA, Le Roch KG. 2013. Plasmodium genomics and the art of sequencing malaria parasite genomes In: Carlton JM, Perkins SL, Deitsch KW, editors. Malaria parasites: comparative genomics, evolution and molecular biology, Caister Academic Press; p. 35–58 [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chugh M, et al. 2013. Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc Natl Acad Sci U S A. 110:5392–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávalos LM, Perkins SL. 2008. Saturation and base composition bias explain phylogenomic conflict in Plasmodium. Genomics 91:433–442. [DOI] [PubMed] [Google Scholar]
- DeBarry J, Kissinger JC. 2011. Jumbled Genomes: missing apicomplexan synteny. Mol Biol Evol. 28:2855–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. 1998. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci U S A. 95:8124–8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166. [Google Scholar]
- Gajria B, Bahl A, Brestelli J, et al. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36:D553–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagner SC, Misof B, Maier WA, Kampen H. 2007. Bayesian analysis of new and old malaria parasite DNA sequence data demonstrates the need for more phylogenetic signal to clarify the descent of Plasmodium falciparum. Parasitol Res. 101:493–503. [DOI] [PubMed] [Google Scholar]
- Hall N, et al. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307:82–86. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Waldenström J, Bensch S. 2004. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol. 90:797–802. [DOI] [PubMed] [Google Scholar]
- Janssen CS, Barrett MP, Turner CMR, Phillips RS. 2002. A large gene family for putative variant antigens shared by human and rodent malaria parasites. Proc R Soc B. 269:431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Rayner JC. 2015. The origins of malaria: there are more thinhs in heaven and earth. Parasitology 142:S16–S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger JC, DeBarry J. 2011. Genome cartography: charting the apicomplexan genome. Trends Parasitol. 27:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-H, Wares JP, Kissinger JC. 2008. The apicomplexan whole-genome phylogeny: an analysis of incongruence among gene trees. Mol Biol Evol. 25:2689–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Patra KP, Vinetz JM. 2005. An anti-chitinase malaria transmission-blocking single-chain antibody as an effector molecule for creating a Plasmodium falciparum-refractory mosquito. J Infect Dis 192:878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI. 2008. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci U S A. 105:17878–17883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li Y, et al. 2010. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum WG. 1897. On the flagellated form of the malarial parasite. Lancet 11:1240–1241. [Google Scholar]
- Martinsen ES, Perkins SL. 2013. The diversity of Plasmodium and other haemosporidians: the intersection of taxonomy, phylogenetics and genomics In: Carlton JM, Perkins SL, Deitsch KW, editors. Malaria parasites: comparative genomics, evolution and molecular biology. Norfolk, UK: Caister Academic Press; p. 1–15 [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. 2008. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 47:261–273. [DOI] [PubMed] [Google Scholar]
- McCutchan TF, et al. 1996. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci U S A. 93:11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mideo N, Reece SE, Smith AL, Metcalf CJE. 2013. The Cinderella syndrome: why do malaria-infected cells burst at midnight? Trends Parasitol. 29:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan BJ, Wang L, Coppel RL. 2009. No TRAP, no invasion. Trends Parasitol. 25:77–84. [DOI] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93:2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikbakht H, Xia X, Hickey DA. 2014. The evolution of genomic GC content undergoes a rapid reversal within the genus Plasmodium. Genome 57:507–511. [DOI] [PubMed] [Google Scholar]
- Outlaw DC, Ricklefs RE. 2011. Rerooting the evolutionary tree of malaria parasites. Proc Natl Acad Sci U S A. 108:13183–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outlaw DC, Ricklefs RE. 2014. Species limits in avian malaria parasites (Haemosporida): how to move forward in the molecular era. Parasitology 141:1223–1232. [DOI] [PubMed] [Google Scholar]
- Outlaw RK, Counterman B, Outlaw DC. 2015. Differential patterns of molecular evolution among Haemosporidian parasite groups. Parasitology 142:612–622. [DOI] [PubMed] [Google Scholar]
- Palinauskas V, Krizanauskiene A, Iezhova TA, et al. 2013. A new method for isolation of purified genomic DNA from haemosporidian parasites inhabiting nucleated red blood cells. Exp Parasitol. 133:275–280. [DOI] [PubMed] [Google Scholar]
- Palinauskas V, Valkiūnas G, Bolshakov CV, Bensch S. 2011. Plasmodium relictum (lineage SGS1) and Plasmodium ashfordi (lineage GRW2): the effects of the co-infections on experimentally infected passerine birds. Exp Parasitol. 127:527–533. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning ZM, Keane T, Korf I. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res. 37:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SL. 2008. Molecular systematics of the three mitochondrial protein-coding genes of malaria parasites: corroborative and new evidence for the origins of human malaria. Mitochondrial DNA 19:471–478. [DOI] [PubMed] [Google Scholar]
- Perkins SL. 2014. Malaria’s many mates: past, present, and future of systematics of the order Haemosporida. J Parasitol. 100:11–25. [DOI] [PubMed] [Google Scholar]
- Perkins SL, Schall JJ. 2002. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 88:972–978. [DOI] [PubMed] [Google Scholar]
- Pick C, Ebersberger I, Spielmann T, Bruchhaus I, Burmester T. 2011. Phylogenetic analyses of malaria parasites and evolution of their exported proteins. BMC Evol Biol. 11:167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Catalan O, et al. 2013. Revision of hemoproteid genera and description and redescription of two species of chelonian hemoproteid parasites. J Parasitol. 99:1089–1098. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, et al. 2010. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci U S A. 107:1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puiu D, Enomoto S, Buck GA, Abrahamsen MS, Kissinger JC. 2004. CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res. 32:D329–D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SM, Xu G. 2011. Resolving the phylogeny of malaria parasites. Proc Natl Acad Sci U S A. 108:12973–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs RE, et al. 2014. Species formation by host shifting in avian malaria parasites. Proc Natl Acad Sci U S A. 111:14816–14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney AP. 2004. Mechanisms underlying the evolution and maintenance of functionally heterogeneous 18S rRNA genes in apicomplexans. Mol Biol Evol. 21:1704–1711. [DOI] [PubMed] [Google Scholar]
- Ross R. 1898. Report on the cultivation of proteosoma, labbé, in grey mosquitoes. Indian Med Gaz. 33:401–408. [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M. 2008. Origins of human malaria: rare genomic changes and full mitochondrial genomes confirm the relationship of Plasmodium falciparum to other mammalian parasites but complicate the origin of Plasmodium vivax. Mol Biol Evol. 25:1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497:327–331. [DOI] [PubMed] [Google Scholar]
- Schaer J, et al. 2013. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc Natl Acad Sci U S A. 110:17415–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Figueiredo LM, Freitas-Junior LH. 2004. Chromosome structure and dynamics of Plasmodium subtelomeres In: Waters AP, Janse CJ, editors. Malaria parasites: genomes and molecular biology. Wymondham: Caister Academic Press; p. 187–203. [Google Scholar]
- Silva JC, Egan A, Arze C, Spouge JL, Harris DG. 2015. A new method for estimating species age supports the co-existence of malaria parasites and their mammalian hosts. Mol Biol Evol. 32:1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Egan A, Friedman R, et al. 2011. Genome sequences reveal divergence times of malaria parasite lineages. Parasitology 138:1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- Triglia T, Tham WH, Hodder A, Cowman AF. 2009. Reticulocyte binding protein homologues are key adhesins during erythrocyte invasion by Plasmodium falciparum. Cell Microbiol. 11:1671–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkiūnas G. 2005. Avian malaria parasites and other haemosporidia. CRC Press, Boca Raton. [Google Scholar]
- Videvall E, Cornwallis CK, Palinauskas V, Valkiūnas G, Hellgren O. 2015. The avian transcriptome response to malaria infection. Mol Biol Evol. 32:1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Tang HB, DeBarry JD, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AP, Higgins DG, McCutchan TF. 1991. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc Natl Acad Sci U S A. 88:3140–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YH, Ansari H, Otto TD, et al. 2015. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Elife 4: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 419:779–781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.