Abstract
Virtually all aphids (Aphididae) harbor Buchnera aphidicola as an obligate endosymbiont to compensate nutritional deficiencies arising from their phloem diet. Many species within the Lachninae subfamily seem to be consistently associated also with Serratia symbiotica. We have previously shown that both Cinara (Cinara) cedri and Cinara (Cupressobium) tujafilina (Lachninae: Eulachnini tribe) have indeed established co-obligate associations with both Buchnera and S. symbiotica. However, while Buchnera genomes of both Cinara species are similar, genome degradation differs greatly between the two S. symbiotica strains. To gain insight into the essentiality and degree of integration of S. symbiotica within the Lachninae, we sequenced the genome of both Buchnera and S. symbiotica endosymbionts from the distantly related aphid Tuberolachnus salignus (Lachninae: Tuberolachnini tribe). We found a striking level of similarity between the endosymbiotic system of this aphid and that of C. cedri. In both aphid hosts, S. symbiotica possesses a highly reduced genome and is found exclusively intracellularly inside bacteriocytes. Interestingly, T. salignus’ endosymbionts present the same tryptophan biosynthetic metabolic complementation as C. cedri’s, which is not present in C. tujafilina’s. Moreover, we corroborate the riboflavin-biosynthetic-role take-over/rescue by S. symbiotica in T. salignus, and therefore, provide further evidence for the previously proposed establishment of a secondary co-obligate endosymbiont in the common ancestor of the Lachninae aphids. Finally, we propose that the putative convergent split of the tryptophan biosynthetic role between Buchnera and S. symbiotica could be behind the establishment of S. symbiotica as an obligate intracellular symbiont and the triggering of further genome degradation.
Keywords: Buchnera aphidicola, Serratia symbiotica, Lachninae, co-obligate, aphid endosymbiont, symbiont settlement
Introduction
Aphids (Aphididae Latreille, 1802) are a highly diverse group of insects, with over 4,000 species currently organized into 24 distinct extant subfamilies (http://aphid.speciesfile.org, last accessed January 12, 2016). As many phloem feeders, aphids possess obligate intracellular symbionts, generally Buchnera, to complement their nutrient deficient diet (Mittler 1971; Sandstrom and Moran 1999). It has been calculated, through the use of molecular clocks, that this bacterium has been co-evolving with aphids for ∼84–164 Myr (von Dohlen and Moran 2000). Buchnera is mainly in charge of producing essential amino acids and some vitamins (Douglas 1996; Nakabachi and Ishikawa 1999; Akman Gündüz and Douglas 2009; Russell et al. 2014), which has been further corroborated through the whole genome sequencing of eight different Buchnera strains belonging to three distinct aphid subfamilies (Aphidinae Latreille, 1802, Lachninae Herrich-Schaeffer, 1854, and Eriosomatinae: Fordini tribe). This role has also been confirmed through a series of transcriptomics (Moran, Dunbar, et al. 2005; Nakabachi et al. 2005; Hansen and Moran 2011) and proteomics experiments (Poliakov et al. 2011; Russell et al. 2014). Buchnera aphidicola, like many obligate endosymbionts from insects, is vertically transmitted, harbored in specialized host cells called bacteriocytes (which make up a specialized organ-like structure called the bacteriome), and has a reduced genome with a high A + T content and no mobile elements.
While virtually all aphids host Buchnera as an obligate symbiont, they are frequently associated with additional bacteria belonging to different taxa (Fukatsu 2001; Tsuchida et al. 2002; Lamelas et al. 2008; Burke et al. 2009; Henry et al. 2013; Arneodo and Ortego 2014), referred to as secondary symbionts. Some of these are of facultative nature, meaning they are not required for the correct development, reproduction and survival of their host. However, these can provide a desirable trait under certain environmental or ecological conditions. Examples of these include Regiella insecticola (Degnan, Leonardo, et al. 2009), Hamiltonella defensa (Degnan, Yu, et al. 2009), Rickettsia sp. (Sakurai et al. 2005), Rickettsiella viridis (Tsuchida et al. 2014), Wolbachia sp. (Augustinos et al. 2011), the so-called “X-type” (Guay et al. 2009), Spiroplasma sp. (Fukatsu et al. 2001; Łukasik et al. 2013), and Serratia symbiotica (Moran, Russell, et al. 2005). On the other hand, secondary endosymbionts can also be of obligate nature, meaning they are essential for their host. To date, the establishment of a secondary symbiont as an obligate one in aphids has been proposed (through genome-based metabolic inference) for at least two species of aphids in the subfamily Lachninae (Cinara [Cinara] cedri and Cinara [Cupressobium] tujafilina). In these aphids, S. symbiotica strains have apparently established co-obligate associations with both Buchnera and the aphid host (Lamelas, Gosalbes, Manzano-Marín, et al. 2011; Manzano-Marín and Latorre 2014). The comparative genome analyses of S. symbiotica from the two Cinara hosts, along with consistent evidence for the presence of S. symbiotica in various species within the Lachninae (Lamelas et al. 2008; Burke et al. 2009), has previously led us to propose that these co-obligate symbioses were triggered by an ancient pseudogenization of the genes involved in the riboflavin biosynthetic pathway in the Buchnera common ancestor from the Lachninae aphids (Manzano-Marín and Latorre 2014). Nevertheless, S. symbiotica presents very different genomic and phenotypic characteristics in C. cedri and C. tujafilina, reflecting the different stages of integration in their symbiotic systems, the later representing an “earlier” obligate symbiont. While in the C. cedri S. symbiotica it is located obligatorily inside distinct bacteriocytes, presents a pleomorphic shape, and possesses a highly reduced and A + T rich genome (1.76 Mb and 29.2% G + C content) with no traces of mobile elements, in C. tujafilina it is located extracellularly and intracellularly (co-infecting or sequestering Buchnera bacteriocytes), presents a filamentous shape, and possesses a mildly reduced genome (2.49 Mb and 52.0% G + C content) with a high number and variety of mobile elements. A striking metabolic difference between these two endosymbiotic systems was found to be in the genes involved in the biosynthesis of the essential amino acid (EAA) tryptophan. While in C. tujafilina both Buchnera and S. symbiotica retain intact pathways for the biosynthesis of this EAA, in C. cedri, Buchnera has retained only the plasmidic tryptophan biosynthetic genes trpE and trpG (coding for the anthranilate synthase), and has of the rest of the genes in the pathway (trpA, trpB, trpC and trpD) (Pérez-Brocal et al. 2006; Lamelas, Gosalbes, Moya, et al. 2011). These retentions and losses of genes in Buchnera complement those in the S. symbiotica genome, resulting in the biosynthesis of tryptophan now being shared between Buchnera and Serratia (Gosalbes et al. 2008; Lamelas, Gosalbes, Manzano-Marín, et al. 2011).
Both C. cedri and C. tujafilina belong to the same tribe within the Lachninae, the Eulachnini tribe. However, other members of the Lachninae subfamily have also been found to be consistently associated to S. symbiotica strains (Lamelas et al. 2008; Burke et al. 2009; Jousselin et al. 2016), which form at least two phylogenetically distinct clades. Following Lamelas et al. (2008) naming scheme, “cluster A” of S. symbiotica displays short branches and consists of mainly facultative strains of S. symbiotica from the Aphidinae and some Lachninae members (as C. tujafilina’s), while “cluster B” consists solely of strains associated with the Cinara (Cinara) subgenus and other Lachninae (from both Eulachini, Lachnini, and Tuberolachnini tribes). “Cluster B” displayed long branches as well as some phylogenetic congruence with Buchnera (Lamelas et al. 2008; Burke et al. 2009), and thus, also the hosts’ (Munson et al. 1991; Moran et al. 1993; Jousselin et al. 2009). Therefore, it was hypothesized that the S. symbiotica strains from “cluster B” represented obligate-like endosymbionts putatively originating from an infection by a S. symbiotica previous to the split of the Eulachnini and Lachnini tribes (Lamelas et al. 2008). Based on this, to corroborate the loss-of-riboflavin-biosynthesis hypothesis for the establishment of S. symbiotica as a co-obligate endosymbiont in the Lachninae, and to infer the degree of dependence of Buchnera upon S. symbiotica, we have sequenced whole genomes for both Buchnera and S. symbiotica from the aphid Tuberolachnus salignus (Lachninae: Tuberolachnini tribe, cluster B). Tuberolachnus salignus was selected as the S. symbiotica strain it harbors belongs to the “cluster B” of obligate-like symbionts. Additionally, this aphid belongs to the distantly related Tuberolachnini tribe, and would therefore provide a broader evolutionary perspective into the Buchnera–S. symbiotca relationship within the Lachninae aphids, as well as into the establishment of endosymbiotic microbial consortia. We have conducted fluorescent in situ hybridization (FISH) on aphid embryos to locate S. symbiotica. Also, we have performed a complete reannotation of both Buchnera (INSDC:CP000263) and S. symbiotica (INSDC:CP002295) endosymbionts from the aphid C. cedri (hereafter BCc and SCc, respectively). Through comparative genomics and metabolic inference, we have described both the putative genetic and the metabolic convergence found between the endosymbiotic systems of C. cedri and T. salignus. We have inferred presumably convergent losses through the comparison against the endosymbiotic system of C. tujafilina, as well as through comparisons against Buchnera-only systems. Through the use of molecular dating and phylogenetic analyses, we propose a timing and scenario for S. symbiotica’s establishment and further independent genome shrinkage within the Lachninae following putative convergent losses in the branches leading to T. salignus and C. cedri. Finally, and most relevantly, we provide further evidence for the aforementioned loss-of-riboflavin-biosynthesis hypothesis and propose that a convergent secondary biosynthetic-capability-loss by Buchnera in the aphids C. cedri and T. salignus could be behind the triggering of the intracellular establishment inside bacteriocytes and further genomic reduction of S. symbiotica in these members of the Lachninae.
Materials and Methods
Aphid Collection, DNA Extraction, and Sequencing
Tuberolachnus salignus aphids were collected from a single Salix sp. tree on September 5, 2013. The collection site is located at the Pacé (Pazieg, in Breton) commune in the Ille-et-Vilaine department of Brittany in north-western France (48.135161 N 1.786938 W). 15 aphids were dissected under a microscope to remove cuticle, gut, legs and head, in order to enrich bacteriome tissues. Mostly embryos, bacteriocytes and other organs that were dragged along were used for DNA extraction using the JetFlex Genomic DNA Purification Kit (GENOMED, Löhne, Germany). Extracted DNA was sent to Macrogen Inc. (Republic of Korea), where one lane of HiSeq2000 2×100 base pairs (bp) paired-end library was sequenced (mean insert size of 417 bp).
Genome Assembly
HiSeq2000 paired-end reads were first right-tailed trimmed (using a minimum quality threshold of 20) using FASTX-Toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/, last accessed January 12, 2016) and reads shorter than 75 bp where removed. Additionally, PRINSEQ v0.20.4 (Schmieder and Edwards 2011) was used to remove reads containing undefined nucleotides (N) and to separate the resulting paired-end reads from the ones left without a pair. The remaining paired-ends reads where then used to perform a de novo assembly with SOAPdenovo r240 (Luo et al. 2012). The resulting contigs were then filtered by minimum length of 300 and coverage of 50× and where taxonomically assigned using PhymmBL v4.0 (Brady and Salzberg 2011). From these, we separated the contigs into two sets: (1) the ones that were assigned to Buchnera and (2) the ones that were assigned to the rest of Gammaproteobacteria (excluding Buchnera). These two sets were used for paired-end read mapping on Bowtie v2.2.0 (Langmead and Salzberg 2012). The reads assigned to each contig set where then separately used to perform de novo assembly on SOAPdenovo r240, with a post-assembly filtering step as explained above. Then, SSPACE v3.0 (Boetzer et al. 2011) was used to scaffold contigs with a minimum number of links of 30. The resulting gaps in scaffolds where filled using GapFiller v1.10 (Boetzer and Pirovano 2012) as well as manual curation. One circular contig was obtained for S. symbiotica and three for B. aphidicola, belonging to the chromosome and the leucine (circular) and tryptophan (linear) plasmids. Finally, to correct base errors in the assembled consensus sequences and manually check for misassembled regions, we used Polisher v2.0.8 (available for academic use from http://jgi.doe.gov/data-and-tools/polisher/, last accessed January 12, 2016) and Tablet (Milne et al. 2013).
Genome Annotation and Metabolic Reconstruction
The genomes of both B. aphidicola BTs and S. symbiotica STs were annotated in the same manner. First, the origin of replication was predicted with originX (Worning et al. 2006), which in both cases was very near to a putative DnaA-box (used to define the origin). Next, the genomes underwent a round of open reading frame (ORF) prediction using Prodigal v2.6.1 (Hyatt et al. 2010), and were then annotated using the BASys server (Van Domselaar et al. 2005). Second, a step of manual curation of the annotation was done on UGENE v1.18.0 (Okonechnikov et al. 2012) through BLASTx (Altschul et al. 1997) searches of the intergenic regions as well as through BLASTp and DELTA-BLAST (Boratyn et al. 2012) searches of the predicted ORFs against NCBI’s nr database. Priority for the BLAST searches was as follows: (1) against Escherichia coli K-12 substrain MG1655, (2) against Yersinia pestis CO92, and (3) against the whole nr database. The resulting coding sequences (CDSs) were considered to be putatively functional if all essential domains for the function were found or if a literature search supported the truncated version of the protein as functional (details of the literature captured in the annotation file). Third, RNAs were annotated using tRNAscan-SE v1.3.1 (Lowe and Eddy 1997) (tRNAs), ARAGORN v1.2.36 (Laslett and Canback 2004) (tmRNAs) and Infernal v1.1.1 (Nawrocki and Eddy 2013) (rRNAs and other noncoding RNAs usign the Rfam 12.0 database [Nawrocki et al. 2015]). Fourth, ribosomal binding sites (RBSs) were predicted with RBSfinder v1.0 (Suzek et al. 2001) to aid with the proper prediction of the translation start site.
Metabolic reconstruction was performed in Pathway Tools v19.0 (Karp et al. 2015) using the EcoCyc (Keseler et al. 2013) and MetaCyc (Caspi et al. 2014) databases, followed by manual curation. Visual plotting of the inferred metabolism was done by hand using Inkscape v0.91 (https://inkscape.org/en/, last accessed January 12, 2016).
Construction of Homologous Groups of Proteins and Rearrangement Analysis
For both performing phylogenetic inferences and understanding the genetic differences in both Buchnera and Serratia from the different aphids, we first ran a homologous protein clustering analysis using OrthoMCL v2.0.9 (Li et al. 2003; Chen et al. 2007) using the afore-mentioned endosymbiotic bacteria and a set of closely related free-living bacteria (supplementary table S2, Supplementary Material online). These protein clusters were then manually curated to join in a single group proteins such as flagellum and outer membrane proteins which do not tend to cluster together given their low protein identity. Based on these manually curated groups we then created two subsets: (1) single copy-core proteins of currently available Buchnera genomes and free-living relatives and (2) single copy-core proteins of currently available S. symbiotica genomes and free-living relatives. Using the latter set, we recoded the gene arrangements in each S. symbiotica genome for use with MGR v2.03 (Bourque and Pevzner 2002) to infer the phylogeny that absolutely minimizes (no heuristics) the number of rearrangements undergone among the strains. For the rearrangement analysis and the shared unshared genes one between STs and SCc, we added two pseudogenes to the S. symbiotica set from SCc which are present in STs as protein coding genes. No pseudogenes in SCc are present as genes/pseudogenes in STs.
Phylogenetic Reconstruction and Divergence Time Estimations
For phylogenetic reconstruction of both Buchnera and S. symbiotica, we aligned the two subsets, group by group, independently using MAFFT v7.220 (Katoh and Standley 2013) (L-INS-i algorithm). We then removed divergent and ambiguously aligned blocks using Gblocks v0.91b (Talavera and Castresana 2007) and concatenated the resulting alignments into a single one (per subset) (supplementary files 3 and 4, Supplementary Material online) for following phylogenetic inference. For both Buchnera plus its free-living relatives and S. symbiotica plus its free-living relatives, we used the LG + I+G amino acid substitution model, which incorporates the variability of evolutionary rates across sites in the matrix estimation (Le and Gascuel 2008). Bayesian phylogenetic inference was performed in MrBayes v3.2.5 (Ronquist et al. 2012) running two independent analyses with four chains each. In order to alleviate long-branch attraction artefacts commonly seen in endosymbionts (Husník et al. 2011; Philippe and Roure 2011), Phylobayes v4.1 (Lartillot et al. 2009), the analysis was also run under the CAT + GTR + G (four discrete categories) (under four independent runs) using dayhoff6-recoded concatenated amino acid alignments. Chains were run and compared using the tracecomp and bpcomp programs, and were considered converged at a maximum discrepancy of <0.16 and minimum effective size of 50. The resulting trees were visualized and exported with FigTree v1.4.1 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed January 12, 2016) and edited in Inkscape.
For divergence time estimations, we performed two likelihood tests implemented in MEGA v6 (Tamura et al. 2013) on the 24 Buchnera conserved genes that had been previously identified to follow the molecular clock hypothesis (Pérez-Brocal et al. 2006). From these, we identified eight single-copy conserved genes shared by all Buchnera genomes available to date (ssb, rbfA, cspE, rpsS, dapF, leuB, infA, and rpmJ) which still follow this hypothesis under fig. 1A topology. The amino acid sequences from these genes were aligned independently using MAFFT v7.220 and then back-translated to their nucleotide sequences. Concatenated alignments were used for divergence time estimations in Phylobayes v4.1 running two independent analyses with two chains each under the GTR nucleotide substitution model and an underlying across-site rate variations sampled from a discrete gamma distribution (four categories) of mean 1 and variance 1/alpha (exponential of mean 1) (Phylobayes options -gtr -ln -dgam 4). A fixed topology derived from the Bayesian reconstruction done with the single-copy core genes of the currently available Buchnera genomes and free-living relatives was used as input (as Phylobayes requires a fixed topology). A log normal autocorrelated relaxed clock was chosen with a root prior of 100 Myr±100, this last based on previous dating hypothesis of the origin of the family Aphididae (von Dohlen and Moran 2000) (between 84 and 164 Ma), which should be close to that of the splitting of Aphidinae and Lachninae subfamilies (Nováková et al. 2013). We used the time divergence estimate between the tribes Aphidini and Macrosiphini (50–70 Ma) (Clark et al. 1999) as the calibration point for the molecular dating. In addition, a chain was run under the prior (-prior option) and was checked to assess if the resulting distributions were sufficiently wide, finding them to be so. Briefly, regarding the phylogenetic analyses of the trpD, trpC, trpB, and trpA genes, we aligned them using MAFFT, then fed the alignments into Gblocks, followed by phylogenetic reconstruction in MrBayes, as described above.
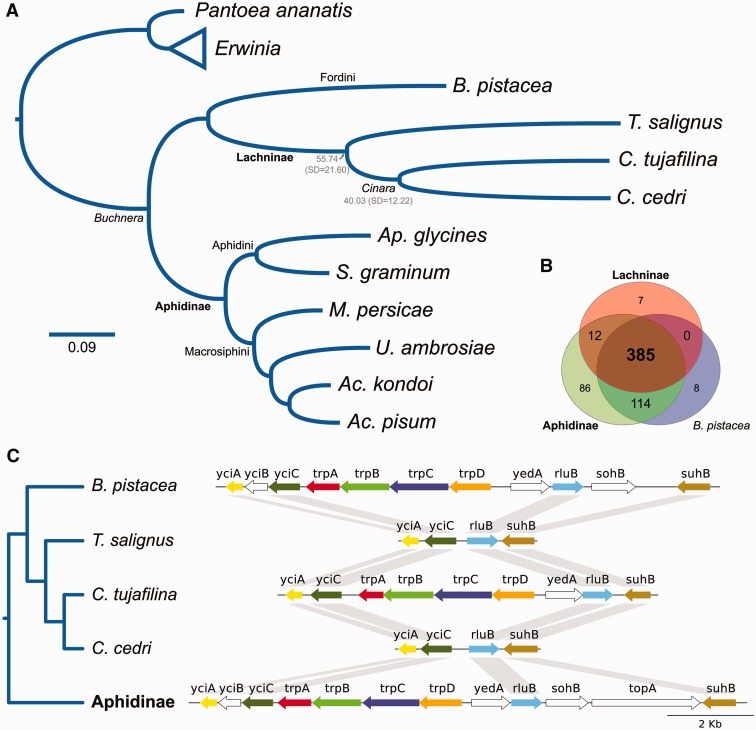
Fig. 1.—
Buchnera phylogenomic reconstruction, genetic-repertoire reduction in the Lachninae and convergent loss of tryptophan biosynthetic genes. (A) Bayesian phylogenomic reconstruction and divergence time estimates between Tuberolachnus salignus–Cinara cedri, and C. tujafilina–C. cedri. Subfamily names are displayed in bold lettering. Mean divergence time estimates are shown in gray under the subfamily or genus clade name. Erwinia spp. and Pantoea ananatis are the outgroups. As all posterior probabilities were equal to 1, they have been excluded from the tree (B) Venn-like diagram displaying the results of the clustering of orthologous proteins. The Lachninae and Aphidinae are represented as pangenomes reconstructed from currently available sequences. (C) Genetic maps of the chromosomal trp genes of different Buchnera strains displaying the convergent loss of the trpD, trpC, trpB, and trpA genes in T. salignus and C. cedri. Nonconserved genes among all strains are displayed in white, while the conserved ones are displayed in different coloring. The Aphidinae plot was done using Buchnera strain APS from the aphid Acyrthosiphon pisum. It was chosen as it would represent the ancestral gene order and content for the Aphidini–Macrosiphini common ancestor (shared by all Aphidinae but Buchnera from Aphis glycines, which has lost both the sohB and topA genes).
Fluorescent In Situ Hybridization
Collected individuals from T. salignus were dissected in absolute ethanol to extract embryos. These embryos were directly transferred to modified Carnoy’s fixative (six chloroform: three absolute ethanol: one glacial acetic acid) and left overnight, following Koga et al. protocol (Koga et al. 2009). Briefly, fixed embryos were then washed with absolute ethanol and transferred into 6% solution of H2O2 diluted in absolute ethanol and were then left in this solution for 2 weeks (changing the solution every 3 days). Then, embryos were washed twice with absolute ethanol. Hybridization was performed overnight at 28 °C in standard hybridization buffer (20 mM Tris–HCl [pH 8.0], 0.9 M NaCl, 0.01% SDS, and 30% formamide) and then washed (20 mM Tris–HCl [pH 8.0], 5 mM EDTA, 0.1 M NaCl, and 0.01% SDS) before slide preparation. Embryos belonging to 20 individuals were viewed under an Olympus FV1000 confocal microscope. We used an eubacterial probe (EUB338-6-FAM [5′-6-FAM-GCTGCCTCCCGTAGGAGT-3′]) plus a specific one targeting S. symbiotica 16S RNA molecules based on (Gómez-Valero et al. 2004), which also makes a perfect match against S. symbiotica from T. salignus (STs-DY-405 [5′-DY-405-CCGCCGCTCGTCACCCAAA-3′]). RNase digestion and no-probe control experiments were done on embryos to confirm the specificity of the detection.
Results
Genomic Features of B. aphidicola and S. symbiotica from T. salignus
The genomes of both B. aphidicola strain BTs isolate Pazieg (hereafter BTs) and S. symbiotica strain STs isolate Pazieg (hereafter STs) from T. salignus. have been assembled to one circular chromosome of 421 kb (375 protein-coding genes) with two plasmids (coding for genes involved in the biosynthesis of tryptophan [plasmid pTrp, ∼2.5 kb] and leucine [plasmid pLeu, 6.5 kb], respectively) for the former, and one circular chromosome of 650 kb (492 protein-coding genes) for the latter, with a mean Illumina HiSeq2500 coverage of 1,473× and 182×, respectively (supplementary fig. S1, Supplementary Material online). The genomes have been deposited in the European Nucleotide Archive under the accession numbers LN890285, LN890286, LN890287, and LN890288. As for many other Buchnera, the pTrp plasmid of BTs was not closed, as it presents long stretches of tandem repeats flanking the trpE and trpG genes (Lai et al. 1994, 1995, 1996; Rouhbakhsh et al. 1996; van Ham et al. 1999; Gil et al. 2006; Gosalbes et al. 2008). The two major tandem repeats present in the plasmid pTrp are located upstream the trpE gene (18 nucleotides in length) and downstream the trpG gene (55 nucleotides in length), both consisting of at least 3 units. The latter repeat contains in its reverse complement strand a putative DnaA-box with the sequence “TTATCCACA.” This repeat could act as an origin of replication for the plasmid. Using the originX software for bioinformatic prediction, together with the identification of the conserved nucleotide motif described by Gil et al. (2006), we located the putative origin of replication of the pLeu plasmid between the genes repA1 and leuB. The location of the origin of replication for this plasmid is similar to the one found in the pLeu plasmids of Buchnera from the aphids Thelaxes suberi (Thelaxinae Baker, 1920) (van Ham et al. 1997) and C. cedri (Gil et al. 2006). The pLeu plasmid also presents a putative regulatory element partially overlapping the 5′ of the repA1 gene consisting of an inverted repeat which could be forming a long secondary structure. This would be consistent with the presence of a small inverted repeat found in the pLeu plasmid of BCc. Both chromosomes of BTs and STs exhibited a putative DnaA-box in the close vicinity of the origin predicted by the aforementioned software. This motif was used as the putative origin of replication for both genomes. No transposable elements of any kind have been found in any of the four replicons (both BTs and STs chromosomes and the pLeu and pTrp plasmids).
Evolutionary Relationships and Genome Reduction in Buchnera
Previous studies have shown that the smallest known Buchnera genomes were those harbored by C. cedri and C. tujafilina (hereafter BCt), both members of the Lachninae subfamily (Pérez-Brocal et al. 2006; Lamelas, Gosalbes, Moya, et al. 2011). Consistent with previous observations for the Buchnera from Lachninae representatives, BTs displays a greatly reduced genome (table 1) and the second smallest after that of BCc. Through a bayesian phylogenetic reconstruction, we found that, as expected, Buchnera from the Lachninae indeed form a monophyletic group, with a sister relationship to the sole representative from the Fordini tribe (Eriosomatinae subfamily) (Buchnera from Baizongia pistacea [hereafter BBp]) (fig. 1A). At the same time, this Fordini/Lachninae clade maintains a sister relationship with the Aphidinae clade, which is further divided into the Aphidini (Latreille, 1802) and Macrosiphini (Wilson, 1910) tribes. This result, along with an analysis on the clustering of orthologous proteins for currently available Buchnera genomes, confirmed previous findings that have proposed a great genetic-repertoire reduction in the Buchera common ancestor of the Lachninae (fig. 1B). We found that the largest Buchnera pangenome was that of the Aphidinae (597 protein-coding genes), which has at least 86 unique protein-coding genes. Thus, the Buchnera common ancestor of the Lachninae would have suffered a loss of at least 208 protein-coding genes (86 unique to the Aphidinae, 114 shared by the Aphidinae and BBp, and 8 unique to BBp). Most importantly, we found a convergent tryptophan biosynthetic gene loss in BTs and BCc, which is not shared by BCt (fig. 1C). This gene loss corresponds to the genomic deletion of the genes trpA, trpB, trpC, and trpD, which code for the enzymes required to synthesize tryptophan from anthranilate. Besides, a phylogenetic analysis of the aforementioned genes showed no indication of these being acquired from another source in BCt (supplementary fig. S2, Supplementary Material online).
Table 1.
Genomic Characteristics of Different Selected Strains of Buchnera
| Subfamily | Aphidinae |
Eriosomatinaea | Lachninae |
|||
|---|---|---|---|---|---|---|
| Tribe | Macrosiphini | Aphidini | Fordini | Tuberolachnini | Eulachnini |
|
| Buchnera strain | BAp | BSg | BBp | BTs | BCt | BCc |
| Genome size (kb)b | 656c | 654c | 618 | 430c | 453 | 425c |
| Chromosome G+C content (%) | 26.3 | 25.3 | 25.3 | 21.6 | 23.0 | 20.1 |
| CDSs (chromosome+plasmid[s]) | 562+7+2c | 547+7+2c | 504+3 | 375+5+2c | 362+6 | 360+5+2c |
| tRNAs | 32 | 32 | 32 | 31 | 31 | 31 |
| rRNAs | 3 | 3 | 3 | 3 | 3 | 3 |
Note.—Comparison of genomic characteristics of Buchnera genomes from distinct aphid subfamilies. The genomic and genetic reduction undergone by the Lachninae’s bacteria is evident. Bap, Buchnera from Acyrthosiphon pisum strain APS; BSg, Buchnera from Schizaphis graminum.
Recent work by Zhang and Chen (1999) and Li et al. (2014) support the split of the Eriosomatinae into up to three subfamilies (Eriosomatinae, Pemphiginae, and Fordinae), given their paraphyly. Baizongia pistacea would then belong to the Eriosomatinae (if divided into two subfamilies) or the Fordinae (if divided into three subfamilies).
Reported genome sizes include both the plasmids.
pTrp plasmid numbers are reported from the assembled contigs, as they are estimated to contain repeated units of the trpE and trpG genes.
To date both the gene loss in S. symbiotica and the maximum age for the loss of the tryptophan biosynthetic capabilities of Buchnera, we used a molecular clock approach, using a subset of Buchnera genes previously inferred to follow this hypothesis (Pérez-Brocal et al. 2006), in combination with the time estimate of 50–70 Ma for the divergence of the Aphidini and Macrosiphini tribes (Clark et al. 1999). We obtained a mean date of 55.74(SD = 21.60) (replicate: 52.62[SD = 16.50]) Ma for the split of T. salignus and Cinara lineage, and 40.03(SD = 12.22) (replicate: 37.98[SD = 12.16]) Ma for the split of C. tujafilina and C. cedri (fig. 1A). These estimates are highly congruent with the dates calculated by Messeguer et al. (2015) using a combination of aphid nuclear and Buchnera gene amplicons. The full table of mean divergence times, standard deviations and other statistics, along with the replicate, can be found in supplementary table S1, Supplementary Material online.
Evolutionary Relationships and Genome Reduction in Serratia
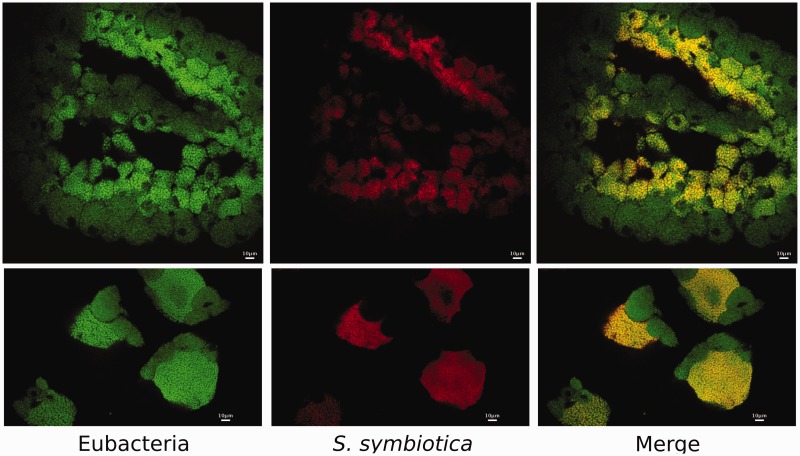
As mentioned before, Buchnera has established a co-obligate association with a S. symbiotica bacterium in the Lachninae aphids C. cedri and C. tujafilina (hereafter SCt) (Lamelas, Gosalbes, Manzano-Marín, et al. 2011; Manzano-Marín and Latorre 2014). This change in lifestyle was probably triggered by an ancient loss of the Buchnera riboflavin biosynthetic genes and, in the case of C. cedri, it was followed by a loss of the tryptophan biosynthetic genes trpA, trpB, trpC, and trpD, resulting in this last role now being split between BCc and SCc (Gosalbes et al. 2008). However, SCc and SCt display very contrasting genomic characteristics, representing two different stages of genome reduction (Lamelas, Gosalbes, Manzano-Marín, et al. 2011; Manzano-Marín and Latorre 2014) (table 2). While SCt presents a cellular shape and size as well as a tissue tropism similar to that of the facultative S. symbiotica from Acyrthosiphon pisum (hereafter SAp), as well as other facultative endosymbionts (Fukatsu et al. 2000; Moran, Russell, et al. 2005; Lamelas et al. 2008; Manzano-Marín and Latorre 2014), SCc (as Buchnera) presents a pleomorphic shape and is confined to specific bacteriocytes (Lamelas et al. 2008). Through the use of FISH using specific probes against STs’s 16S rRNA, we determined that, similarly to the endosymbiotic system present in C. cedri, STs is present in distinct bacteriocytes are surrounded by Buchnera’s bacteriocytes (fig. 2). Strikingly, STs holds an even more degraded genome than that of SCc (table 2), and hence represents a further stage in genome shrinkage, having already lost all the “junk” DNA that is still present in the reduced genome of SCc.
Table 2.
Genomic Characteristics of Serratia Strains with Different Lifestyles
| Subfamily | Aphidinae |
Lachninae |
||||
|---|---|---|---|---|---|---|
| Tribe | Aphidini | Macrosiphini | Eulachnini |
Tuberolachnini | ||
| Serratia strain | Db11 | SAf | SAp | SCt | SCc | STs |
| Lifestyle | Free-living | Facultative endosymbiont | Facultative endosymbiont | Co-obligate endosymbiont | Co-obligate endosymbiont | Co-obligate endosymbiont |
| Genome size (Mb) | 5.11 | 3.58a | 2.76a | 2.49a | 1.76 | 0.65 |
| Plasmids | – | Unknown | Unknown | Unknown | Unknown | Unknown |
| Chromosome G+C content (%) | 59.5 | 52.1 | 48.4 | 52.0 | 29.2 | 20.7 |
| CDSs | 4,709 | 3,398 | 2,098 | 1,601 | 677 | 492 |
| Coding density (%) | 87.9 | 78.2 | 56.8 | 53.4 | 38.8 | 77.2 |
| Pseudogenes | 12 | 126 | 550 | 916 | 98 | 7 |
| tRNAs | 88 | 74 | 44 | 47 | 36 | 33 |
| rRNAs | 22 | 22 | 15 | 13 | 3 | 3 |
| Mobile elements | Yes (few) | Yes | Yes | Yes | No | No |
Note.—Comparison of genomic characteristics of Serratia genomes from the free living Serratia marcescens strain Db11 and endosymbionts from distinct aphid subfamilies. The various degrees of genomic reduction are evident and ordered from left to right. Db11, S. marcescens strain Db11 isolated as a pathogen from Drosophila melanogaster (Flyg et al. 1980); SAf, S. symbiotica strain CWBI-2.3T isolated from Aphis fabae (Aphidinae: Aphidini tribe) (Sabri et al. 2011); Sap, S. symbiotica strain Tucson from Ac. pisum (Burke and Moran 2011).
Genome size is reported as the total size of the assembled scaffolds for unclosed genomes.
Fig. 2.—
Serratia symbiotica localization in Tuberolachnus salignus bacteriomes of early embryos. Whole-mount fluorescence in situ hybridization of early T. salignus embryos using 16S rRNA-directed probes. (Left) Eubacterial staining using EUB338 (Amann et al. 1990) 6-FAM labeled probe. (Center) S. symbiotica staining using STs (see Materials and Methods) DY-405 labeled probe. (Right) Merged image of both eubacterial and S. symbiotica staining, showing double-labeling of S. symbiotica and single-labeling of Buchnera with eubacterial probe in bacteriocytes of T. salignus in (Bottom) early and (Top) later embryos.
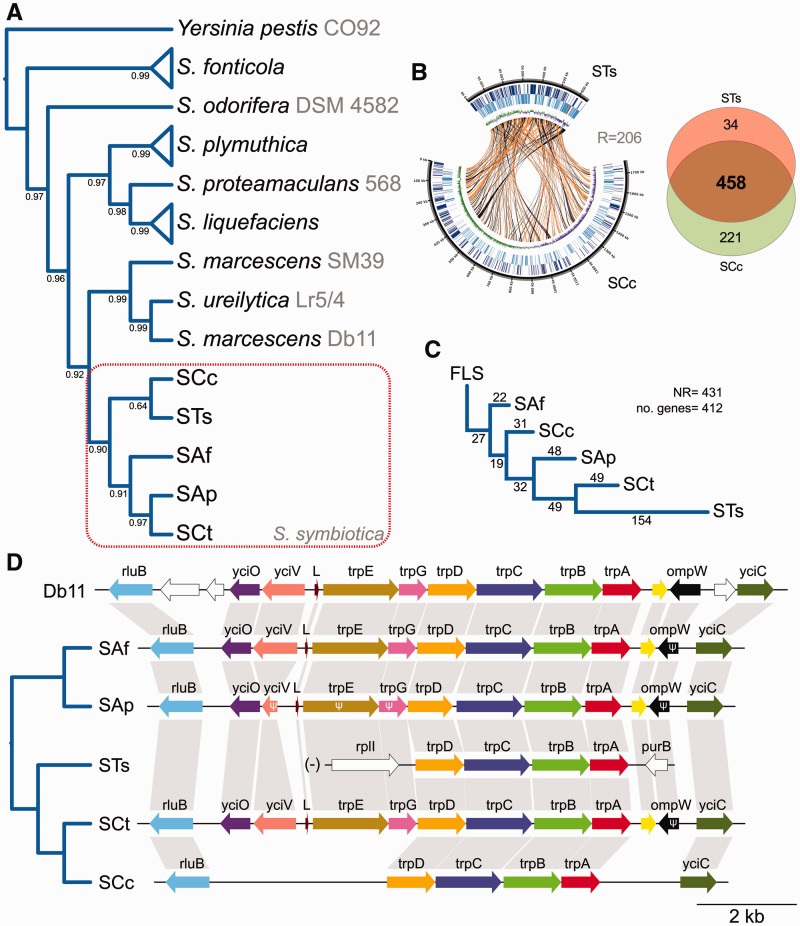
Through a bayesian phylogenomic reconstruction under MrBayes, using single-copy conserved genes, we found S. symbiotica to form a monophyletic clade with a sister relation to the S. marcescens clade (which includes S. ureilytica nested within S. marcescens) (supplementary fig. S3, Supplementary Material online). However, given the very long branches leading to STs and SCc and their apparent sister relation, we also performed a more “sophisticated” phylogenetic reconstruction under Phylobayes using dayhoff6-recoded alignments under the CAT + GTR + G (four discrete categories) model (in order to minimize the effects of long-branch attraction) (fig. 3A). Contrasting the MrBayes reconstruction, the sister relation of STs and SCc is weakly supported, and the relations among S. symbiotica strains become less clear. It is worth mentioning that the “early” co-obligate SCt clusters together with facultative S. symbiotica endosymbionts from Aphis fabae and Ac. pisum, indicating a lack of high sequence divergence, as that seen in STs and SCc. To infer whether both STs and SCc have indeed undergone an early common genomic reduction, as seen in Buchnera, or if both have undergone independent events of genome shrinkage before the loss of mobile genetic elements, we performed a minimal number of rearrangements analysis. If both STs and SCc would have undergone an early drastic common genome reduction and loss of mobile elements (as is the case of Buchnera), it would be expected that they both show a remarkable degree of synteny and phylogenetic congruency. On the contrary, if STs and SCc would have undergone independent events of genome reduction and loss of mobile elements, we would expect to see much genome reordering between the two, as seen among SCc, SCt and SAp (Manzano-Marín and Latorre 2014). We found that at least 206 rearrangements have occurred between STs and SCc (fig. 3B), supporting the hypothesis of early independent events genome reduction. Through a minimum rearrangement phylogeny using a subset of these genes (shared among S. marcescens Db11 and currently available S. symbotica strains as single-copy protein-coding genes), we found that that the rearrangements undergone in STs is highest for all S. symbiotica strains (fig. 3C). In contrast, S. symbiotica strain CWBI-2.3T (hereafter SAf), facultative and culturable endosymbiont from the aphid Ap. fabae, displays the least rearrangements from the ancestral gene order, which is conserved among free-living Serratia strains (Manzano-Marín et al. 2012; Manzano-Marín and Latorre 2014). Disagreeing with the concatenated gene phylogeny, SCc is, rearrangement-wise, more closely related to facultative S. symbiotica strains. Additionally, we have observed the same pseudogenization of the trpE and trpG genes seen in SCc in STs (fig. 3D). In the case of STs, an inversion and translocation of the trp locus has occurred. On the contrary, in SCc, this locus has retained the flanking genes and large “junk” DNA regions putatively corresponding to the degenerated gene sequences, suggesting a closer relationship to SCt.
Fig. 3.—
Serratia phylogenomic reconstruction, gene-order rearrangements and shared genetic repertoire between STs and SCc. (A) Bayesian phylogenomic reconstruction of different Serratia strains using Yersinia pestis strain CO92 as outgroup. Serratia symbiotica forms a monophyletic clade sister to the clade mainly composed of S. marcescens. Strain names are shown after species name in gray. Asterisks at nodes stand for a posterior probability equal to 1 (B) On the left, circular plot displaying the chromosomes of STs and SCc. From outermost to innermost ring, the features on the direct strand, the reverse strand, and GC-skew plot. Lines going from one genome to another represent orthologous genes in direct (orange) or reverse (black) orientation. On the right, Venn-like diagram displaying the shared (core) and unshared protein-coding genes between STs and SCc. (C) Minimum number of rearrangement phylogeny for Serratia strain as calculated by MGR. Numbers on top of branches indicate the number of inferred rearrangements undergone in each branch. NR, number of rearrangements. (D) Genetic maps of the chromosomal trp genes of different Serratia strains displaying the convergent loss of the trpE and trpG genes in Tuberolachnus salignus and Cinara cedri. Dendogram on the left displays phylogenetic relationships of the aphid hosts associated to each S. symbiotica strain. Nonconserved genes among all strains are displayed in white, while the conserved ones are displayed in different coloring. The trp locus of STs is marked with a “(-)” to indicate it has been reverse-complemented (from its original orientation in the genome) to facilitate comparison.
Metabolic Capabilities of BTs and STs Endosymbionts
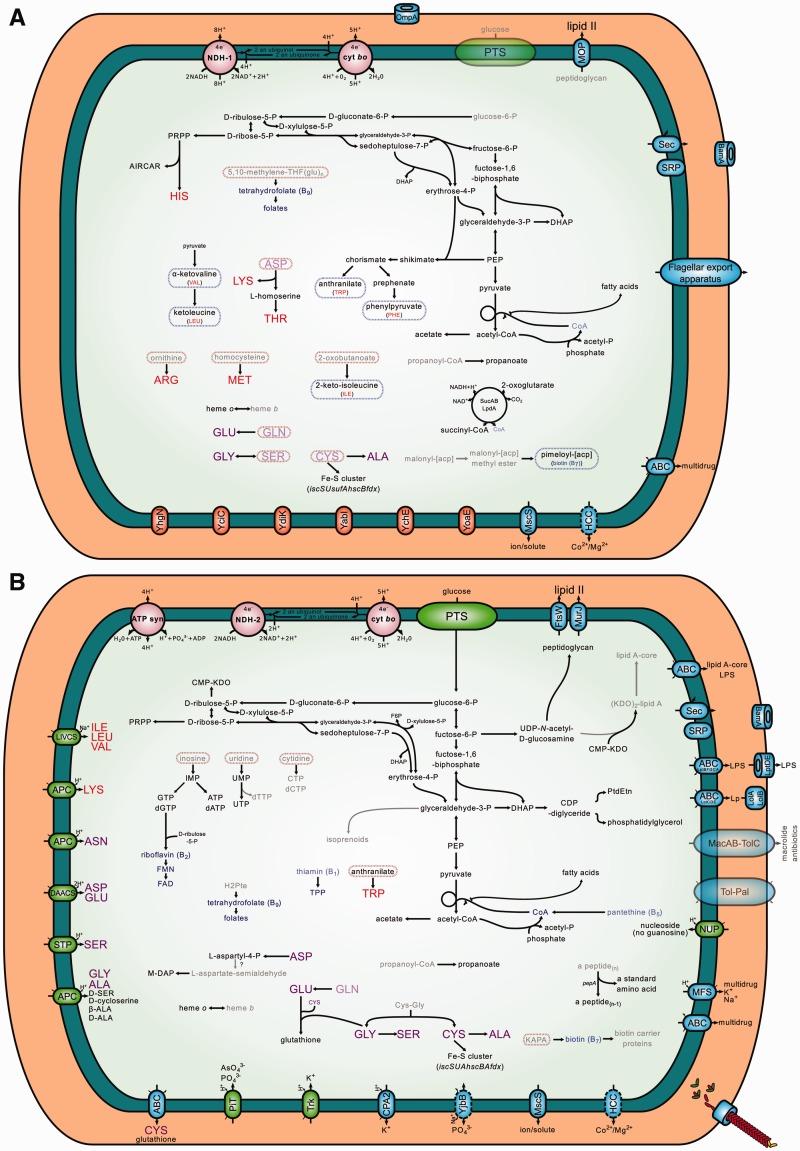
In order to gain insight into the metabolic capabilities of the BTs-STs endosymbiotic consortium, we conducted a metabolic reconstruction using Pathway tools (Karp et al. 2015) (fig. 4A and B). Regarding the biosynthesis of EAA, the main role undertaken by Buchnera in aphids, BTs preserves the capability of synthesizing the same EAAs (and precursors) as Buchnera from Ac. pisum (hereafter BAp) (Hansen and Moran 2011). However, as it is the case for C. cedri’s endosymbiotic system, the capacity of Buchnera to de novo synthesize tryptophan has been partially lost, and this role would now be shared by both members of T. salignus’ endosymbiotic consortium (BTs and STs). Also, differently from BAp, but similarly to both BCc and BCt, the pathway leading to the biosynthesis of arginine has been truncated, and would depend solely on the import of ornithine and glutamine from the host. Regarding glycolysis, we observed an interesting convergence of BTs with BCt, which unlike BCc, have lost the pgi gene. This gene codes for a phosphoglucose isomerase, therefore, BTs would either use glucose-6P (and force glycolytic flux through the pentose phosphate pathway, as has been proposed for an E. coli pgi knock-out mutant [Canonaco et al. 2001; Hua et al. 2003; Charusanti et al. 2010]) or fructose-6P as substrate for glycolysis. This last is probably the case of BCt (Lamelas, Gosalbes, Moya, et al. 2011), which has also lost the genes pgl and zwf of the oxidative pentose pathway. Finally, as BCc and BCt, BTs has lost the capacity to de novo synthesize the essential vitamin riboflavin (role now undertaken by STs) and has lost the genes bioB, bioD, and bioA, involved in the biosynthesis of biotin. This last vitamin could be putatively synthesized by the BTs-STs consortium, where BTs would synthesize pimeloyl-[acp] from an unknown precursor, and then this would in turn be converted to KAPA by the action of an unknown enzyme, and finally, KAPA would be imported into STs to synthesize biotin (fig. 4A and B).
Fig. 4.—
Metabolic reconstruction of Tuberolachnus Salignus’s endosymbiotic consortium. Metabolic reconstruction of (A) Buchnera and (B) Serratia symbiotica of T. salignus. Intact pathways are represented with solid black lines, and unclear ones (missing a specific gene or having it pseudogenized by a frameshift) with solid gray lines. Importers are displayed using green ovals, while exporters and exporters/importers are displayed using blue ovals. The name inside each oval states the family/superfamily they belong to (following TCDB’s classification [Saier et al. 2014]), otherwise the protein name is used. Essential and nonessential amino acids are shown in red and purple lettering, respectively, while cofactors and vitamins are shown in blue. Blurred compounds represent those for which biosynthesis or import cannot be accounted for based on genomic data. Blurred transporters represent those for which a part of the transporter is missing, therefore recently pseudogenized.
With regard to STs (fig. 4B), it preserves a glucose PTS permease and it is able to start glycolysis from glucose-6P as well as go through the pentose phosphate pathway to produce PRPP, ribulose-5P, and erythrose-4P. Regarding the biosynthesis of vitamins, as SCc and SCt, it would be able to synthesize riboflavin (de novo) and tetrahydrofolate (from dihydropterate). STs has completely lost the ability to synthesize thiamin and also the associated ABC transporter (coded by the genes thiB, thiP, and thiQ). However, it keeps the genes thiK and thiL, which catalyze the conversion of thiamin to thiamin diphosphate. STs preserves the capacity to synthesize peptidoglycan from UDP-N-acetyl-d-glucosamine and an almost complete pathway to lipid A-core. This last pathway is missing some steps, whose responsible enzymes are not yet identified in Serratia. Concerning the biosynthesis of nucleotides, it could import inosine, uridine, and cytidine and synthesize all but dTTP, CTP, and dCTP. These last three would require the action of nonspecific kinases yet to be identified, as STs has lost ndk (dTTP from dTDP, CTP from CDP, and dTTP from dTDP) and pyrG (CTP from UTP). ndk has been shown not to be essential for growth in E. coli, but mutants generated high levels of substitutions and frameshifts (Lu et al. 1995; Miller et al. 2002). In addition, we propose that the biosynthesis of coenzyme A might be occurring through the uptake of pantethine. This compound has recently been described to be the most advanced precursor that an E. coli coaBC-deletion mutant strain can utilize to bypass a CoaBC deficiency, in the absence of the pantothenate permease-encoding gene panF (Balibar et al. 2011). This explanation would be consistent with the pseudogenization of the coaBC gene in both STs and SCc, as well as the complete absence of the panF gene in both of these strains. Regarding the biosynthesis of amino acids, similarly to SCc, it has lost the capability of synthesizing almost all essential EAAs, with the notable exception of the retention of the genes trpA, trpB, trpC, and trpD, which identically to SCc, would enable STs to perform the last steps in the biosynthesis of tryptophan following the import of anthranilate, synthesized by BTs. It preserves specific importers for isoleucine, leucine, valine, lysine, asparagine, aspartate, glutamate, serine, glycine, and alanine and could synthesize various nonessential amino acids given the required inputs.
Metabolic and Transporter-Function Convergence between T. salignus’ and C. cedri’s Co-obligate Endosymbiotic Systems
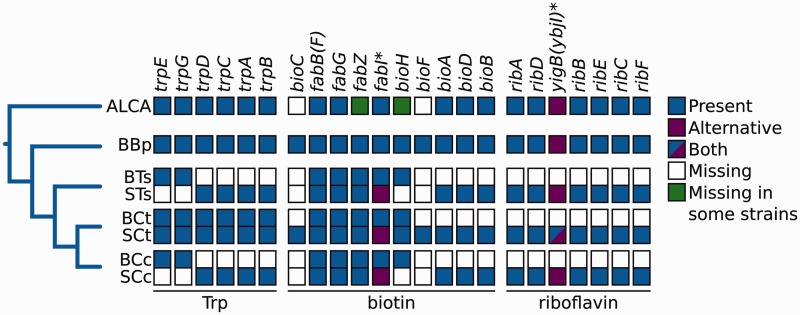
As shown in fig. 3B, the genetic content of STs is mainly a subset of SCc’s, although it has retained 34 genes that are not present in SCc, neither as CDSs nor pseudogenes. It is important to remark the genetic repertoire of both STs and SCc is merely a subset of that of S. marcescens and other S. symbiotica strains (supplementary fig. S4, Supplementary Material online), indicating any retained gene could be inferred to be in the last common ancestor of S. symbiotica. Interestingly, STs’s and SCc’s corresponding Buchnera partners are also very similar in terms of genetic content (supplementary fig. S5, Supplementary Material online). The reduced endosymbiotic systems of T. salignus and C. cedri showed an impressive degree of genetic and metabolic convergence. Firstly, we found that the proposed tryptophan metabolic complementation for SCc and BCc seems to also be true for STs and BTs, as well as the putative biotin pathway split and the aforementioned riboflavin biosynthesis take-over/rescue by S. symbiotica (fig. 5). Regarding the biosynthesis of this last vitamin, two proteins that could catalyze the elusive 5-amino-6-(5-phospho-d-ribitylamino)uracil phosphatase reaction (yigB and ybjI) have just recently been identified (Haase et al. 2013). These two belong to the Haloacid Dehalogenase (HAD) superfamily. Neither STs nor SCc have any of the two aforementioned proteins, however they both preserve the gene yigL, which codes for a putative pyridoxal phosphate phosphatase. YigL (the protein coded by the yigL gene) and YbjI belong to the HAD-superfamily hydrolase, subfamily IIB (Cof-subfamily), and both share all identifiable domains. Thus, YigL could putatively be functioning as the 5-amino-6-(5-phospho-d-ribitylamino)uracil phosphatase in both STs and SCc. It is also important to note that the yigL gene is present in the Buchnera from the mono-endosymbiotic systems of currently sequenced Aphidinae and B. pistacea (Eriosomatinae), and as expected, is missing in the Buchnera from the di-symbiotic systems of currently sequenced Lachninae.
Fig. 5.—
Proposed metabolic complementations found in the obligate endosymbiotic systems of different aphids and riboflavin biosynthesis take-over/rescue by Serratia symbiotica in the Lachninae. Diagram representing the proposed metabolic complementations in the biosynthesis of tryptophan and biotin in the currently available endosymbiotic systems of aphids as well as the riboflavin biosynthesis take-over/rescue by S. symbiotica in the endosymbiotic systems of the Lachninae aphids. The gene names are used as column names, while the abbreviations for the different endosymbiotic bacteria are used as row names, except for Aphididae last common ancestor (ALCA). Asterisks after gene names indicate a putative alternative enzyme could be performing the enzymatic function.
In terms of differential genome reduction and preservation of equivalent enzymes or transporters, convergent patterns can be noticed between STs and SCc. For example, within the lysine biosynthetic pathway, which is intact in SCc but apparently recently impaired in STs given the loss of the asd gene, there exists a most interesting differential degradation pattern. While STs has only retained the lysC gene, which codes for one of three isozymes in E. coli able to catalyze the aspartate kinase reaction, SCc has retained a truncated version of another one of these three isozymes, the bifunctional thrA gene. This truncated gene could preserve its aspartate kinase activity, as has been tested in vivo with a similarly truncated version of the ThrA protein carried in a plasmid using E. coli as a host (Omori and Komatsubara 1993). Additionally, we analyzed the 34 genes that are present in STs but not in SCc, and found that 16 of them actually have equivalent functions to genes present in SCc but that are missing in STs (table 3). This indicates that common functions have been maintained, despite the two independent events of genome reduction. Lastly, we found a putative convergent retention of an apparently complete type-1 fimbriae-like system between STs and SCc. These fimbriae-like systems would be composed of four subunits: (1) A major fimbrin (STSPAZIEG_0410 and SCc_0576), (2) a fimbriae assembly chaperone (STSPAZIEG_0411 and SCc_0575), (3) an outer membrane usher protein (STSPAZIEG_0412 and SCc_0574), and (4) a fimbrial adhesin (STSPAZIEG_0413 and SCc_0167). However, while the first three components seem to be orthologous, the fimbrial adhesins might be of different evolutionary origin, or have already diverged from each other beyond the limits for orthology detection (see table 3, last row).
Table 3.
Metabolic and Transporter-Functional Convergence between STs and SCc
| Genes |
|||
|---|---|---|---|
| STs | SCc | Function | EC Number |
| lysC | thrA | Aspartate kinase | 2.7.2.4 |
| rdgB | mazG (pseudo) | dITP/XTP pyrophosphatase | 3.6.1.19 |
| mdfA,mdlA-mdlB | mdtK,emrAB-tolC | Multidrug efflux | – |
| kefB-kefG | cvrA | Potassium/H+ antiporter | – |
| mltC | mltA,mltD | Membrane-bound lytic murein transglycosylase | – |
| mutY | mutM | Rescue 8-oxo-guanine:cytosine pre (mutM) or post-replication (mutY) | – |
| iscUAhscBAfdx | sufABCDSE | Iron-sulfur cluster biosynthesis | – |
| pgpB | pgpA | Phosphatidylglycerophosphatase | 3.1.3.27 |
| STSPAZIEG_0413 | SCc_0167 | Fimbrial adhesin | – |
Note.—The table displays the genes differentially retained between STs and SCc but that confer similar functions to their corresponding bacterium.
Discussion
A critical issue in evolutionary biology is finding traits in organisms that provide clues into the adaptations and metabolic changes underlying the transition of one lifestyle to another. For many currently available highly reduced endosymbionts like Buchnera or Blochmannia (the primary obligate endosymbiont of Carpenter ants), we lack genomes of organisms that represent their free-living counterparts or intermediate stages needed to infer the adaptations undergone during their establishment as obligate intracellular reduced endosymbionts. Recently, various examples of symbiotic bacteria representing different intermediate steps of the aforementioned lifestyle switch have become available, including S. symbiotica (Burke and Moran 2011; Lamelas, Gosalbes, Manzano-Marín, et al. 2011; Manzano-Marín and Latorre 2014), various Sodalis spp. (Toh et al. 2005; Clayton et al. 2012; Oakeson et al. 2014), Candidatus Providencia siddallii (Manzano-Marín et al. 2015), Candidatus Pantoea carbekii (Kenyon et al. 2015), among others (Gottlieb et al. 2015; Smith et al. 2015). The genomes of these bacteria are proving very interesting insights into the processes behind genomic reduction and settlement of free-living bacteria as obligate endosymbionts.
Aphids from the Lachninae subfamily have been consistently found to be associated with secondary endosymbionts, mainly S. symbiotica (Lamelas et al. 2008; Burke et al. 2009). Strains of this bacterium have also been found in other species belonging to the Aphidinae subfamily, but always with a facultative lifestyle (Fukatsu et al. 2000; Sabri et al. 2011). In fact, the genome sequencing of S. symbiotica from Ac. pisum revealed that this strain found itself in an early stage of the integration process to the intracellular life, as well as confirmed that it was metabolically not necessary for the Buchnera found in Ac. pisum (Burke and Moran 2011). In a previous study, we proposed that an ancient loss of the essential riboflavin biosynthetic genes in Buchnera had putatively triggered the establishment of S. symbiotica as a co-obligate symbiont along with Buchnera in the Lachninae aphids, or at least in the Cinara genus (Manzano-Marín and Latorre 2014). Subsequently, the loss of the tryptophan biosynthetic genes in Buchnera from C. cedri would have accelerated the genomic decay, while in the closely related C. tujafilina, S. symbiotica would have remained in a more “ancient” state, though apparently more degraded than the facultative S. symbiotica from both Ac. pisum. Nevertheless, our previous proposal was based only on S. symbiotica 16S rDNA phylogenetic data and full genomes for both Buchnera and S. symbiotica from aphids belonging to the Eulachnini tribe. In Lamelas et al. (2008) and Burke et al. (2009), the S. symbiotica obligate-like phylogenetic cluster was solely composed of S. symbiotica strains associated to Lachninae aphids, Cinara (Cinara) genus, and the Lachnini and Tuberolachnini tribes. These nonCinara (Cinara) aphids belong to Lachnus roboris, and the Pterochloroides and Tuberolachnus genera. Therefore, it was expected that the Bucnera–S. symbiotica symbiotic systems present in these would resemble the co-obligate endosymbiotic system of C. cedri, where both Buchnera and S. symbiotica reside intracellularly inside bacteriocytes and have established a deeply rooted co-obligate symbiosis. Following the phylogenetic reconstruction of aphids by Novakova et al. (2013), the common ancestor of Cinara genus and T. salignus would coincide with the split of the Lachninae into the two major tribes, the Eulachini and the Lachnini. However, following a very recent phylogentic reconstruction by Chen, Favret, et al. (2015), the aforementioned common ancestor would coincide with the split of the clades Stomaphidini/Tramini/Tuberolachnini and Eulachnini. Regardless, the study of the endosymbiotic system within T. salignus, was expected to first, provide clues into the history of S. symbiotica within the Lachninae, and second, to provide insights into the metabolic adaptations that Buchnera, the obligate symbiont of most aphids, has undergone in the continuous presence of an extra player in the symbiosis.
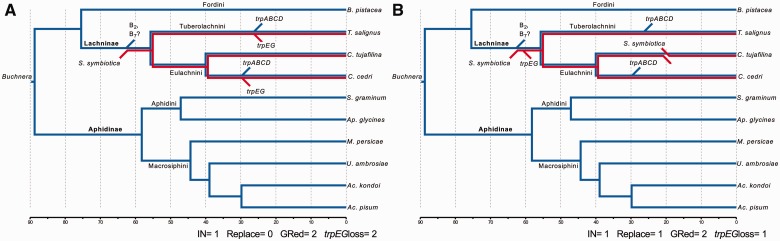
In this work, we have compared the Buchnera–S. symbiotica endosymbiotic systems in two different aphids belonging to two different tribes within the Lachninae subfamily, T. salignus (Tuberolachnini) and C. cedri (Eulachnini), and have found functional and genome size reduction convergence, as well as putative metabolic convergence in the complementation established between the endosymbiotic partners of both systems. However, STs holds a greatly reduced genome, even when compared with SCc, and would thus represent a later stage of genome reduction, where the leftover “junk” DNA and pseudogenes are purged from the genome. We have been able to determine that STs and SCc indeed diverged early in their establishment as obligate intracellular endosymbionts. Thus, two equally parsimonious scenarios (in terms of number of steps) can be proposed in light of the results: the “convergent” (fig. 6A) and the “nonconvergent” one (fig. 6B), in terms of the biotin and tryptophan gene losses. Although we cannot entirely discard replacement by a different S. symbiotica strain in SCt, taking into account the high incidence of obligate-like S. symbiotica strains within the Lachninae (across all five tribes) (Lamelas et al. 2008; Burke et al. 2009; Chen, Wang, et al. 2015; Jousselin et al. 2016), the “fragile” phylogenetic sister relation of STs and SCc (fig. 3A), the S. symbiotica rearrangement phylogeny showing SCc being more closely related to the facultative strains (fig. 3C), and the existence of S. symbiotica strains with both intact and pseudogenized/absent copies of the trpEG genes (fig. 3D), we believe the most likely explanation to be a single infection some 55.74(SD = 21.60) (replicate: 52.62[SD = 16.50]) Ma in the T. salignus/Eulachnini common ancestor, followed by divergence (fig. 6A). The alternative nonconvergent scenario (fig. 6B) would require at least two independent infections of S. symbiotica strains: one in the branch leading to T. salignus/Eulachnini, and one in the branch leading to the C. tujafilina one. Nevertheless, with no extra information from more S. symbiotica genomes from Lachninae aphids, the nonconvergent scenario for the loss of biotin or tryptophan genes cannot be ruled out. Buchnera, which has been co-evolving with its aphid host for ∼84–164 Ma (von Dohlen and Moran 2000), putatively underwent a massive genome reduction and loss of mobile elements before the diversification of aphids. This is evidenced by the high degree of synteny and phylogenetic congruence with the respective hosts (Baumann et al. 1995; Funk et al. 2000; van Ham et al. 2003; Jousselin et al. 2009). Contrasting this, STs and SCc do not display co-cladogenesis with Buchnera, but rather cluster together, although with a low support (fig. 3A). This could be explained by the highly accelerated branches leading to this, possibly causing a long-branch attraction artefact, as is seen in other highly accelerated endosymbiotic lineages (Husník et al. 2011). The contrary case would be exemplified by SCt, whose lack of niche-change (compared with the facultative SAp) (Manzano-Marín and Latorre 2014) would be reflected in the absence of strong evolutionary forces acting on its gene sequences, manifested in its short branch (fig. 3A). STs and SCc’s gene order is indeed quite different (fig. 3B). This contrasts the great genome stasis seen in Buchnera and other “ancient” obligate symbioses, where the bacterial partner’s genome infecting the host’s ancestor, was already greatly reduced (putatively having lost all mobile elements) before the diversification of their hosts (Tamas et al. 2002; van Ham et al. 2003; Degnan et al. 2004; Patiño-Navarrete et al. 2013). This lack of synteny is also displayed among the genomes of SAp, SCt and SCc (fig. 3C) (Manzano-Marín and Latorre 2014). Furthermore, STs display the largest number of rearrangements when compared with all S. symbiotica strains and S. marcescens Db11 (fig. 3C). On the contrary, SCc has a closer rearrangement relation to the other S. sy,mbiotica, supporting the early divergence from STs. To date the maximum age of the presumed independent genome erosions, we used Buchnera genes that follow the molecular clock hypothesis and found that T. salignus and C. cedri’s endosymbiotic systems diverged ∼55.74 Ma, and that the genetic reduction seen in SCc started maximally ∼40.03(SD = 12.22) (replicate: 37.98[SD = 12.16]) Ma. Regarding the putative phylogenetic origin of the S. symbiotica clade, using the currently available genomic sequences for bacteria in the Serratia genus, we have established that S. symbiotica forms a sister clade to one composed mainly of S. marcescens strains (fig. 3A). This would provide phylogenetic evidence that the S. symbiotica strains from aphids originated from a S. marcescens-like bacterium.
Fig. 6.—
Proposed evolutionary scenarios for the establishment of Serratia symbiotica as a co-obligate endosymbiont in currently sequenced Lachninae aphids. Dendogram representing the evolutionary history of Buchnera within aphids and the proposed (A) convergent and (B) nonconvergent scenarios for the establishment of S. symbiotica as a co-obligate endosymbiont in the Lachninae. Blue and red lines represent Buchnera and S. symbiotica, respectively. The blue diagonal lines represent losses in Buchnera, while the red diagonal lines represent those in S. symbiotica. Names on top and below of branches indicate the aphid familiar name (bold) and tribal name. Scale bar at the bottom represents timing in Ma for the branching of the different aphids. IN, infection events; Replace, symbiont replacement events; GRed, Genome reduction events; trpEGloss, trpEG gene-loss events.
As stated before, through comparative genomics and metabolic inference, we have found a high level of similarity between the endosymbiotic systems of C. cedri and T. salignus. Not only have their S. symbiotica evolved to preserve a large common genetic repertoire (fig. 3B), but also part of the observed differential genome reduction undergone by this endosymbiont in both aphids can be attributed to the retention of alternative enzymes/transporters to perform similar or equivalent functions (table 3). This would further evidence of the need of this specific functions/systems, regardless to the gene which performs this function. This is especially evident by the retention of different genes to perform the iron–sulfur cluster biosynthesis, where STs has retained the Isc system, while SCc has retained the Suf one (table 3). As the main role of Buchnera is the provision of EAAs to the host, we would expect that these genes would tend to be lost in a similar fashion in settled down secondary obligate endosymbionts in partnership with Buchnera. We have shown that STs, as SCc (Lamelas, Gosalbes, Manzano-Marín, et al. 2011) but contrary to SCt (Manzano-Marín and Latorre 2014), has evolved to completely lose intact pathways involved in the production of EAAs (fig. 4B). As SCc, it mainly preserves some genes involved in the truncated pathway leading to lysine (from aspartate), as well as genes involved in synthesizing tryptophan from anthranilate. As SCt has been shown to still retain the ability to de novo synthesize chorismate, tryptophan, phenylalanine, and threonine, in addition to retaining pseudogenes for the majority of EAAs biosynthetic pathways (Manzano-Marín and Latorre 2014), it could be inferred that the shared losses between STs and SCc would consequently be examples of evolutionary convergence. However, if SCt represents a case of symbiont replacement, and SCc and STs underwent a common rapid gene loss, while still undergoing massive genome rearrangements (fig. 6B), it is possible that some of these shared losses might not be cases of evolutionary convergence. However, the further exploration of Buchnera–S. symbiotica pairs from the Lachninae should help clarify this.
On the other hand, by comparing the pseudogenizations in the production of vitamins and the EAA tryptophan in the Buchnera from the Lachninae, Aphidinae and Fordini (BBp) (these last two being systems where Buchnera is the sole obligate endosymbiont) (fig. 5), we have corroborated the complete loss of the riboflavin biosynthetic pathway in Buchnera from the Lachninae, role now overtaken by S. symbiotica. We have also identified a second Lachninae-specific loss in Buchnera of the biotin biosynthetic genes bioA, bioD and bioB, which have been interestingly retained in their S. symbiotica co-obligate partners. This pathway is not entirely complete in currently sequenced Buchnera strains from the Aphidinae subfamily, as most have lost the bioC, bioH, and bioF genes, but have preserved, over evolutionary time, all other genes in the pathway (fig. 5). However, in BBp, the sole Buchnera from the Fordini sequenced to date, the biotin pathway remains intact. Therefore, it can be inferred that this pathway could function in a yet-unknown way in most Buchnera and would represent an additional metabolic loss in the Buchnera common ancestor of the Lachninae. For the pea aphid Ac. pisum, it has been proven that riboflavin production by Buchnera is indeed essential for correct development of the aphid (Nakabachi and Ishikawa 1999), but in the case of biotin, to our knowledge, no experimental evidence has yet been put forward to prove the supply or production of this vitamin by the symbiont. Regarding the host need for this last vitamin, to our knowledge, only experiments on artificial diets have been performed in two Macrosiphini aphids. In Myzus persicae, a diet lacking biotin did not prevent larvae from developing into adults, but did adversely affected growth (Dadd et al. 1967), and in Neomyzus circumflexus, the author claimed to have been able to rear aphids “without influencing normal development” for at least five generations on a diet where both biotin and riboflavin were omitted (Ehrhardt 1968). Also, we have found an interesting putative convergent retention of a type-1 fimbriae-like system in both STs and SCc. These types of fimbriae are used by other pathogenic bacteria for infection and surface attachment (reviewed in Korea et al. 2011; Scheller and Cotter 2015), and could thus reflect their need for this mechanism in order to maintain their obligate-intracellular lifestyles inside bacteriocytes. In this line, it is important to mention that neither the facultative SAp nor the “early” co-obligate SCt, which are not found obligatorily inside bacteriocytes, preserve intact fimbriae-related systems. Finally, a most remarkable putative metabolic convergence between the BTs-STs and BCc-SCc endosybmiotic systems, not shared by the BCt-SCt system (where the pathway is intact in both symbionts [Manzano-Marín and Latorre 2014]), is the loss of the genes trpE and trpG genes by both STs and SCc S. symbiotica (figs. 3D and 6), and the genes trpA, trpB, trpC, and trpD by Buchnera (figs. 5 and 6). These losses have led to a metabolic co-dependence with their S. symbiotica partner for the production of tryptophan, an essential compound supplied by the Buchnera in mono-endosymbiotic systems in other aphids (Douglas and Prosser 1992). Interestingly, this pathway is also found split in a similar fashion in some Carsonella-secondary di-symbiotic systems found in psyllids (Sloan and Moran 2012), which could indicate that anthranilate is a metabolite able to pass through the different membranes of the two endosymbionts and hosts. This could be reflected in the apparent selection of this pathway to split up in the same fashion, when splitting actually occurs. However, this pathway is not found split in the same fashion, or even at all, in other di-symbiotic consortia such as the Sulcia-secondary (from sharpshooters, leafhopers, and cicadas) (McCutcheon and Moran 2010), the Carsonella-Profftella (psyllid) (Nakabachi et al. 2013), and the Tremblaya-secondary (mealybugs) ones (Husnik and McCutcheon 2016). This would point toward this feature being putatively somewhat specific to the Buchnera–S. Symbiotica (and the two Carsonella-secondary) endosymbiotic consortia. In the case of most Buchnera strains, the trpE and trpG genes are present in a multi-copy plasmid, and in some cases presenting multiple copies of the two genes within the same replicon (Lai et al. 1994, 1996; Rouhbakhsh et al. 1996; van Ham et al. 1999). This amplification phenomenon has been explained as a mechanism for Buchnera to overproduce anthranilate (Lai et al. 1994). Within Buchnera strains with a trpEG plasmid, BCt is unique, in that it has undergone a fusion of this into the leucine plasmid, putatively losing this amplification (Gil et al. 2006). This would be consistent with SCt still preserving a full gene repertoire for the biosynthesis of tryptophan. Therefore, it could be speculated that when the secondary symbiont adapts to a certain point to the presence of Buchnera (a “better” anthranilate producer), a strong relaxation of selection in the trpEG genes would occur given the high availability of this compound. However, the fact of Buchnera being a “better” anthranilate producer remains to be experimentally tested. In summary, in terms of biosynthetic production of tryptophan, riboflavin, and putatively biotin, the di-symbiotic endosymbiotic systems present in Lachninae members (and more evidently in T. salignus and C. cedri) have evolved to be equivalent to those of the mono-symbiotic systems found in currently available Aphidinae and Fordini aphids (fig. 5), and would thus have established a metabolic-based consortium.
Finally, we speculate that the secondary loss of the tryptophan biosynthetic capability by Buchnera and its putative complementation with S. symbiotica could have occurred independently in the lineages leading to T. salignus and C. cedri. This loss could be behind the establishment (obligatorily inside bacteriocytes) of S. symbiotica in, at least, these two Lachninae aphids. Since that moment, the genome reduction could have been accelerated due to drift, as a consequence of the bottlenecks undergone by these symbionts in each generation, similar to what has happened in ancient obligate endosymbionts (reviewed in Moran and Bennett 2014). We expect further sequencing and morphological studies of symbionts housed by aphids of this peculiar subfamily to shed light on the history of metabolic rewiring as well as the timing, lifestyle switching and genomic changes that bacteria undergo during the settling down process to reduced obligate endosymbionts.
Supplementary Material
Supplementary tables S1 and S2, figs. S1–S5 and files S3 and S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors would like to acknowledge Christelle Buchard, and Evelyne Turpeau for their invaluable help in the collection of aphid samples. This work has been funded by the Ministerio de Economía y Competitividad (Spain) co-financed by FEDER funds (BFU2012-39816-C02-01 to A.L.); the European Commission (Marie Curie FP7 PITN-GA-2010-264774-SYMBIOMICS to A.M.M.); and the Consejo Nacional de Ciencia y Tecnología (Mexico) (Doctoral scholarship CONACYT 327211/381508 to A.M.M.). The Plant Health and Environment department of INRA is also acknowledged for financial support to J.C.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Literature Cited
- Akman Gündüz E, Douglas AE. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 276:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 172:762–770. abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneodo JD, Ortego J. 2014. Exploring the bacterial microbiota associated with native South American species of Aphis (Hemiptera: Aphididae). Environ Entomol. 43:589–594. [DOI] [PubMed] [Google Scholar]
- Augustinos AA, et al. 2011. Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PLoS One 6:e28695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balibar CJ, Hollis-Symynkywicz MF, Tao J. 2011. Pantethine rescues phosphopantothenoylcysteine synthetase and phosphopantothenoylcysteine decarboxylase deficiency in Escherichia coli but not in Pseudomonas aeruginosa. J Bacteriol. 193:3304–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, et al. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 49:55–94. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13:R56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, et al. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct. 7:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA. 2002. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 12:26–36. . [PMC free article] [PubMed] [Google Scholar]
- Brady A, Salzberg S. 2011. PhymmBL expanded: confidence scores, custom databases, parallelization and more. Nat Methods. 8:367.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Moran NA. 2011. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 3:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Normark BB, Favret C, Moran NA. 2009. Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol. 75:5328–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canonaco F, et al. 2001. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 204:247–252. [DOI] [PubMed] [Google Scholar]
- Caspi R, et al. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42:D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charusanti P, et al. 2010. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 6:e1001186.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. 2007. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One 2:e383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Favret C, Jiang L, Wang Z, Qiao G. 2015. An aphid lineage maintains a bark-feeding niche while switching to and diversifying on conifers. Cladistics Advance Access published September 29, 2015, doi:10.1111/cla.12141. [DOI] [PubMed] [Google Scholar]
- Chen R, Wang Z, Chen J, Qiao G-X. 2015. Avoidance and potential remedy solutions of chimeras in reconstructing the phylogeny of aphids using the 16S rRNA gene of Buchnera: a case in Lachninae (Hemiptera). Int J Mol Sci. 16:20152–20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Moran NA, Baumann P. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 16:1586–1598. [DOI] [PubMed] [Google Scholar]
- Clayton AL, et al. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 8:e1002990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadd RH, Krieger DL, Mittler TE. 1967. Studies on the artificial feeding of the aphid Myzus persicae (Sulzer)—IV. Requirements for water-soluble vitamins and ascorbic acid. J Insect Physiol. 13:249–272. [Google Scholar]
- Degnan P, Lazarus A, Brock C, Wernegreen J. 2004. Host–symbiont stability and fast evolutionary rates in an ant–bacterium association: cospeciation of Camponotus species and their endosymbionts, Candidatus Blochmannia. Syst Biol. 53:95–110. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Leonardo TE, et al. 2009. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 12:2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci. 106:9063–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 1996. Reproductive failure and the free amino acid pools in pea aphids (Acyrthosiphon pisum) lacking symbiotic bacteria. J Insect Physiol. 42:247–255. [Google Scholar]
- Douglas AE, Prosser WA. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J Insect Physiol. 38:565–568. [Google Scholar]
- Ehrhardt P. 1968. Der Vitaminbedarf einer siebröhrensaugenden Aphide, Neomyzus circumflexus Buckt. (Homoptera, Insecta). Z Vgl Physiol. 60:416–426. [Google Scholar]
- Flyg C, Kenne K, Boman HG. 1980. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to cecropia immunity and a decreased virulence to Drosophila. Microbiology 120:173–181. [DOI] [PubMed] [Google Scholar]
- Fukatsu T. 2001. Secondary intracellular symbiotic bacteria in aphids of the genus Yamatocallis (Homoptera: Aphididae: Drepanosiphinae). Appl Environ Microbiol. 67:5315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Nikoh N, Kawai R, Koga R. 2000. The secondary endosymbiotic bacterium of the pea Aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol. 66:2748–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Tsuchida T, Nikoh N, Koga R. 2001. Spiroplasma symbiont of the pea Aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol. 67:1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Helbling L, Wernegreen JJ, Moran NA. 2000. Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc Biol Sci. 267:2517–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Sabater-Muñoz B, Perez-Brocal V, Silva FJ, Latorre A. 2006. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene 370:17–25. [DOI] [PubMed] [Google Scholar]
- Gómez-Valero L, et al. 2004. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the Aphid Cinara cedri. J Bacteriol. 186:6626–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes MJ, Lamelas A, Moya A, Latorre A. 2008. The striking case of tryptophan provision in the cedar Aphid Cinara cedri. J Bacteriol. 190:6026–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb Y, Lalzar I, Klasson L. 2015. Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-like endosymbionts in ticks. Genome. Biol Evol. 7:1779–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay J-F, Boudreault S, Michaud D, Cloutier C., 2009. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J Insect Physiol. 55:919–926. [DOI] [PubMed] [Google Scholar]
- Haase I, et al. 2013. Enzymes from the haloacid dehalogenase (HAD) superfamily catalyse the elusive dephosphorylation step of riboflavin biosynthesis. ChemBioChem. 14:2272–2275. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci. 108:2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry LM, et al. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol. 23:1713–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q, Yang C, Baba T, Mori H, Shimizu K. 2003. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol. 185:7053–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husník F, Chrudimský T, Hypša V. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. biorXiv. Available from: http://dx.doi.org/10.1101/042267. [DOI] [PMC free article] [PubMed]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin E, et al. 2016. Assessment of a 16S rRNA amplicon Illumina sequencing procedure for studying the microbiome of a symbiont-rich aphid genus. Mol. Ecol. Resour. 16:628–640. [DOI] [PubMed] [Google Scholar]
- Jousselin E, Desdevises Y, Coeur d’acier A. 2009. Fine-scale cospeciation between Brachycaudus and Buchnera aphidicola: bacterial genome helps define species and evolutionary relationships in aphids. Proc Biol Sci. 276:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, et al. 2015. Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform. bbv079.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon LJ, Meulia T, Sabree ZL. 2015. Habitat visualization and genomic analysis of ‘Candidatus Pantoea carbekii,’ the primary symbiont of the brown marmorated stink bug. Genome Biol Evol. 7:620–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, et al. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Tsuchida T, Fukatsu T. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl Entomol Zool. 44:281–291. [Google Scholar]
- Korea C-G, Ghigo J-M, Beloin C. 2011. The sweet connection: solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. Bioessays 33:300–311. [DOI] [PubMed] [Google Scholar]
- Lai C-Y, Baumann L, Baumann P. 1994. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci. 91:3819–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C-Y, Baumann P, Moran NA. 1995. Genetics of the tryptophan biosynthetic pathway of the prokaryotic endosymbiont (Buchnera) of the aphid Schlechtendalia chinensis. Insect Mol Biol. 4:47–59. [DOI] [PubMed] [Google Scholar]
- Lai C-Y, Baumann P, Moran NA. 1996. The endosymbiont (Buchnera sp.) of the aphid Diuraphis noxia contains plasmids consisting of trpEG and tandem repeats of trpEG pseudogenes. Appl Environ Microbiol. 62:332–339. abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, et al. 2008. Evolution of the secondary symbiont ‘Candidatus Serratia symbiotica’ in Aphid species of the subfamily Lachninae. Appl Environ Microbiol. 74:4236–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, Gosalbes MJ, Manzano-Marín A, et al. 2011. Serratia symbiotica from the Aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 7:e1002357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, Gosalbes MJ, Moya A, Latorre A. 2011. New clues about the evolutionary history of metabolic losses in bacterial endosymbionts, provided by the genome of Buchnera aphidicola from the Aphid Cinara tujafilina. Appl Environ Microbiol. 77:4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Y, Jiang L-Y, Qiao G-X. 2014. Is the subfamily Eriosomatinae (Hemiptera: Aphididae) monophyletic? Turkish J Zool. 38:285–297. [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. 1995. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 254:337–341. [DOI] [PubMed] [Google Scholar]
- Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett. 16:214–218. [DOI] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Lamelas A, Moya A, Latorre A. 2012. Comparative genomics of Serratia spp.: two paths towards endosymbiotic life. PLoS One 7:e47274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Latorre A. 2014. Settling down: the genome of Serratia symbiotica from the Aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol. 6:1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Oceguera-Figueroa A, Latorre A, Jiménez-García LF, Moya A. 2015. Solving a bloody mess: B-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the leech Haementeria officinalis. Genome Biol Evol. 7:2871–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseguer AS, Coeur d’acier A, Genson G, Jousselin E. 2015. Unravelling the historical biogeography and diversification dynamics of a highly diverse conifer-feeding aphid genus. J Biogeogr. 88:1482–1492. [Google Scholar]
- Miller JH, et al. 2002. Escherichia coli Strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics 162:5–13. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, et al. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202. [DOI] [PubMed] [Google Scholar]
- Mittler TE. 1971. Some effects on the aphid Myzus persicae of ingesting antibiotics incorporated into artificial diets. J Insect Physiol. 17:1333–1347. [Google Scholar]
- Moran NA, Bennett GM. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68:195–215. [DOI] [PubMed] [Google Scholar]
- Moran NA, Dunbar HE, Wilcox JL. 2005. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 187:4229–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc B Biol Sci. 253:167–171. [Google Scholar]
- Moran NA, Russell JA, Koga R, Fukatsu T. 2005. Evolutionary relationships of three new species of enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 71:3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson MA, et al. 1991. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 173:6321–6324. abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2005. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci U S A. 102:5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 23:1478–1484. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H. 1999. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 45:1–6. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43:D130–D137. 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E, et al. 2013. Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol Phylogenet Evol. 68:42–54. [DOI] [PubMed] [Google Scholar]
- Oakeson KF, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome. Biol Evol. 6:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- Omori K, Komatsubara S. 1993. Role of serine 352 in the allosteric response of Serratia marcescens aspartokinase I-homoserine dehydrogenase I analyzed by using site-directed mutagenesis. J Bacteriol. 175:959–965. [cited 2015 Jun 4]. Available from: http://jb.asm.org/content/175/4/959.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño-Navarrete R, Moya A, Latorre A, Peretó J. 2013. Comparative genomics of Blattabacterium cuenoti: the frozen legacy of an ancient endosymbiont genome. Genome Biol Evol. 5:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Brocal V, et al. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314:312–313. [DOI] [PubMed] [Google Scholar]
- Philippe H, Roure B. 2011. Difficult phylogenetic questions: more data, maybe; better methods, certainly. BMC Biol. 9:91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, et al. 2011. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol Cell Proteomics. 10:M110.007039.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhbakhsh D, et al. 1996. The tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the aphididae. J Mol Evol. 42:414–421. [DOI] [PubMed] [Google Scholar]
- Russell CW, et al. 2014. Matching the supply of bacterial nutrients to the nutritional demand of the animal host. Proc R Soc B Biol Sci. 281:20141163–20141163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri A, et al. 2011. Isolation, pure culture and characterization of Serratia symbiotica sp. nov., the R-type of secondary endosymbiont of the black bean aphid Aphis fabae. Int J Syst Evol Microbiol. 61:2081–2088. [DOI] [PubMed] [Google Scholar]
- Saier MH, Reddy VS, Tamang DG, Västermark A. 2014. The transporter classification database. Nucleic Acids Res. 42:D251–D258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Koga R, Tsuchida T, Meng X-Y, Fukatsu T. 2005. Rickettsia symbiont in the pea Aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the, essential symbiont Buchnera. Appl Environ Microbiol. 71:4069–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom J, Moran N. 1999. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl. 91:203–210. [Google Scholar]
- Scheller EV, Cotter PA. 2015. Bordetella filamentous hemagglutinin and fimbriae: critical adhesins with unrealized vaccine potential. Pathog Dis. 73:ftv079.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 29:3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TA, Driscoll T, Gillespie JJ, Raghavan R. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol. 7:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123–1130. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, et al. 2005. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Koga R, Fujiwara A, Fukatsu T. 2014. Phenotypic effect of ‘Candidatus Rickettsiella viridis’, a facultative symbiont of the pea aphid (Acyrthosiphon pisum), and its interaction with a coexisting symbiont. Appl Environ Microbiol. 80:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol Ecol. 11:2123–2135. [DOI] [PubMed] [Google Scholar]
- van Ham RCHJ, et al. 2003. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci. 100:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RCHJ, Martínez-Torres D, Moya A, Latorre A. 1999. Plasmid-encoded anthranilate synthase (TrpEG) in Buchnera aphidicola from aphids of the family Pemphigidae. Appl Environ Microbiol. 65:117–125. [cited 2012 Mar 23]. Available from: http://aem.asm.org/cgi/content/abstract/65/1/117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RCHJ, Moya A, Latorre A. 1997. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola (endosymbiont of aphids). J Bacteriol. 179:4768–4777. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=179323&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Domselaar GH, et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33:W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dohlen CD, Moran NA. 2000. Molecular data support a rapid radiation of aphids in the Cretaceous and multiple origins of host alternation. Biol J Linn Soc. 71:689–717. [Google Scholar]
- Worning P, Jensen LJ, Hallin PF, Stærfeldt H-H, Ussery DW. 2006. Origin of replication in circular prokaryotic chromosomes. Environ Microbiol. 8:353–361. [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen X. 1999. Study on the phylogeny of Pemphigidae (Homoptera:Aphidinea). Acta Entomol Sin. 42:176–183. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.