Abstract
After decades of debate, a mostly satisfactory resolution of relationships among the 11 recognized holometabolan orders of insects has been reached based on nuclear genes, resolving one of the most substantial branches of the tree-of-life, but the relationships are still not well established with mitochondrial genome data. The main reasons have been the absence of sufficient data in several orders and lack of appropriate phylogenetic methods that avoid the systematic errors from compositional and mutational biases in insect mitochondrial genomes. In this study, we assembled the richest taxon sampling of Holometabola to date (199 species in 11 orders), and analyzed both nucleotide and amino acid data sets using several methods. We find the standard Bayesian inference and maximum-likelihood analyses were strongly affected by systematic biases, but the site-heterogeneous mixture model implemented in PhyloBayes avoided the false grouping of unrelated taxa exhibiting similar base composition and accelerated evolutionary rate. The inclusion of rRNA genes and removal of fast-evolving sites with the observed variability sorting method for identifying sites deviating from the mean rates improved the phylogenetic inferences under a site-heterogeneous model, correctly recovering most deep branches of the Holometabola phylogeny. We suggest that the use of mitochondrial genome data for resolving deep phylogenetic relationships requires an assessment of the potential impact of substitutional saturation and compositional biases through data deletion strategies and by using site-heterogeneous mixture models. Our study suggests a practical approach for how to use densely sampled mitochondrial genome data in phylogenetic analyses.
Keywords: mitochondrial phylogenomics, compositional bias, rate variation, PhyloBayes, Holometabola phylogeny, tree-of-life
Introduction
Mitochondrial genomes have been widely applied to infer intraordinal relationships across insects, which in most instances were found to be congruent with other sources of data and provided convincing levels of support (Cameron et al. 2007; Timmermans et al. 2010, 2015; Wiegmann et al. 2011; Li et al. 2012, 2013; Cameron 2014; Gillett et al. 2014; Wu et al. 2014; Bourguignon et al. 2015). However, for deep relationships mitochondrial genomes are considered ineffective, for example, for the study of interordinal and intraordinal relationships of Insecta and Arthropoda, due to base compositional heterogeneity and variable evolutionary rate that potentially mislead tree inference and frequently result in strong topological conflicts with morphological and nuclear data and high inconsistency among different inference methods (Nardi et al. 2003; Cameron et al. 2004; Sheffield et al. 2009; Talavera and Vila 2011; Simon and Hadrys 2013). These compositional and mutational biases in mitochondrial genomes are in violation of the most widely used substitution models, which accommodate among-site rate variation by applying a GTR (general-time-reversible) model with uniform gamma distribution to all characters and thus accommodate differences in rates (fast or slow sites), but neglect variation in other parameters that may differ among characters (Kolaczkowski and Thornton 2004; Lartillot and Philippe 2004). Accelerated evolutionary rates can lead to systematic errors in the inference of relationships due to long-branch attraction (LBA) (Siddall and Whiting 1999; Brinkmann et al. 2005; Yang and Rannala 2012). In addition, evolutionary models that do not account for compositional heterogeneity can lead to false groupings of unrelated taxa with similar base composition (Tarrío et al. 2001; Sheffield et al. 2009). Recent evolutionary models such as the site-heterogeneous mixture model establish a number of rate categories that each are defined by different equilibrium frequencies of nucleotide or amino acid characters estimated from the empirical data (Lartillot and Phillippe 2004). This approach permits exchangeabilities that differ over sites, and thereby relax the assumption of homogeneity across sites of the standard models. These models apparently reduce the negative effects of compositional and mutational bias (Lartillot et al. 2009; Husník et al. 2011; Morgan et al. 2013; Li et al. 2015; Timmermans et al. 2015).
The use of the most accurate models will reduce the probability of systematic errors, as they are able to extract the genuine phylogenetic signals (Delsuc et al. 2005; Jeffroy et al. 2006; Philippe et al. 2011; Morgan et al. 2013; Liu et al. 2014). However, improved models might not hold all the answers to the inconsistency problem, especially when the genuine phylogenetic signal is weak (e.g., for ancient phylogenetic relationships) or the nonphylogenetic signal is predominant (Delsuc et al. 2005; Jeffroy et al. 2006; Philippe et al. 2011). Fast-evolving sites are particularly challenging for phylogenetic reconstruction because they are likely to have experienced multiple substitutions, eroding the genuine signal and misleading phylogenetic inference (Brinkmann and Philippe 1999; Pisani 2004; Delsuc et al. 2005; Goremykin et al. 2013). Various strategies have been suggested to reduce the impact of nonphylogenetic signal, for example, 1) sampling more species to correctly infer multiple substitutions at a site (Baurain et al. 2007; Dunn et al. 2008; Pick et al. 2010; Philippe et al. 2011); 2) excluding the third codon position of protein-coding genes or using RY-coding to decrease saturation and compositional bias (Delsuc et al. 2003; Phillips et al. 2004; Breinholt and Kawahara 2013); and 3) identifying and removing the fast-evolving sites, for example, the slow-fast (SF) method (Kostka et al. 2008) and the observed variability (OV) sorting method (Zhong et al. 2011; Goremykin et al. 2013; Liu et al. 2014).
The debate over the utility of mitochondrial genomes in deep relationships of insects is ongoing (Cameron et al. 2004; Talavera and Vila 2011; Simon and Hadrys 2013). However, most previous attempts to reconstruct interordinal relationships using mitochondrial genomes have used site-homogeneous models that do not account for variation in composition or exchange rates across the data (Cameron et al. 2004; Simon and Hadrys 2013). Bayesian site-heterogeneous mixture models have been employed in a few studies, with apparently good success (Talavera and Vila 2011), but it is not clear if they can compensate for the effect of compositional heterogeneity and saturation among sites on phylogenetic inference of deep relationships of insect mitochondrial genomes.
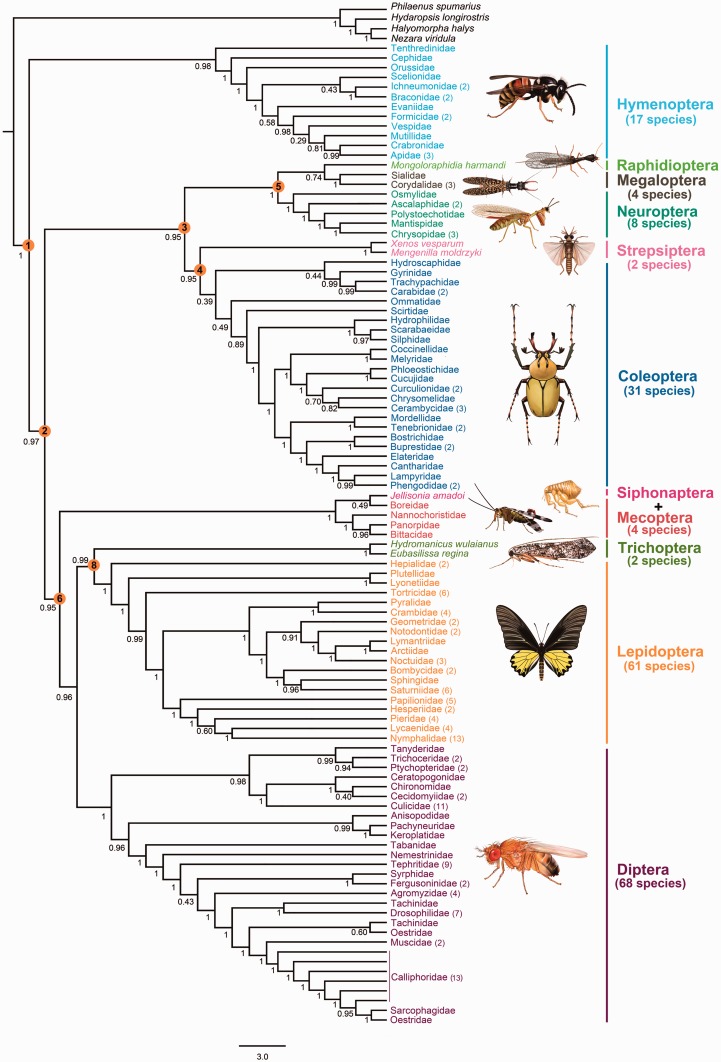
Holometabola are the most species-rich lineage of animals on Earth (Kristensen 1999; Trautwein et al. 2012), containing approximately 780,000 described species. Most of the species richness is concentrated in four super-radiations: Coleoptera (beetles), Hymenoptera (bees, ants, and wasps), Lepidoptera (moths and butterflies), and Diptera (true flies) (Grimaldi and Engel 2005), which combined represent about 50% of all known metazoan species (Wilson 1992). A credible resolution of relationships among the 11 recognized holometabolan orders is now in sight, as integrative systematics and independent data types corroborate an increasingly well-supported tree topology (see discussion in Trautwein et al. 2012). Progress has come especially from several recent phylogenetic studies based on multiple nuclear protein-coding genes (Wiegmann et al. 2009; Ishiwata et al. 2010) and large numbers of single-copy orthologs from expressed sequence tags (ESTs) (Meusemann et al. 2010; Letsch et al. 2012), transcriptomes (Misof et al. 2014; Peters et al. 2014), and whole genomes (Niehuis et al. 2012). According to the molecular evidence, and largely consistent with traditional morphological hypotheses (Kristensen 1999; Beutel and Pohl 2006; Wiegmann et al. 2009; Ishiwata et al. 2010; Misof et al. 2014; Peters et al. 2014), Holometabola is divided into three main lineages: Hymenoptera, Neuropteroidea (Neuropterida, Coleoptera, Strepsiptera), and Mecopterida (Lepidoptera, Trichoptera, Diptera, Mecoptera, Siphonaptera) (fig. 1). The basal split of Hymenoptera from all others has been confirmed by most molecular studies (Wiegmann et al. 2009; Ishiwata et al. 2010; Meusemann et al. 2010; Peters et al. 2014; Misof et al. 2014) and recently by morphological analyses (Beutel et al. 2011). The longstanding controversial position of Strepsiptera has also been resolved in favor of a close relationship with Coleoptera by both nuclear genes (Wiegmann et al. 2009; Ishiwata et al. 2010; Longhorn et al. 2010; Niehuis et al. 2012; Misof et al. 2014; Peters et al. 2014) and morphological data (Beutel et al. 2011).
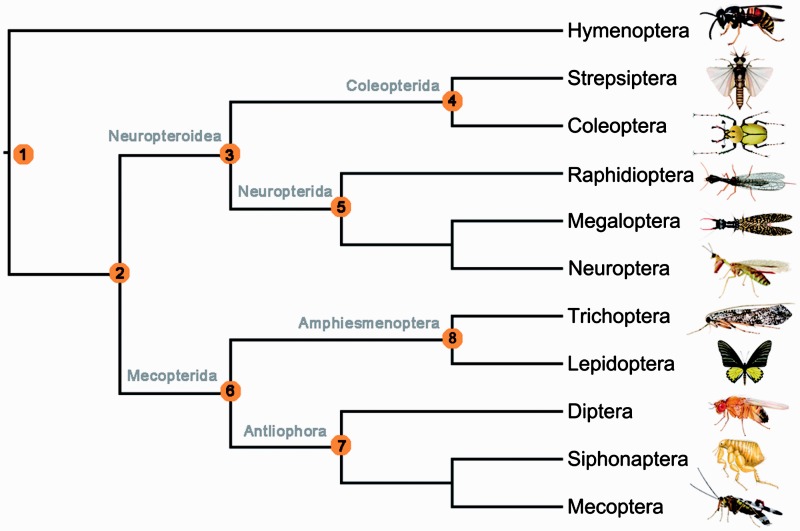
Fig. 1.—
Current view of higher level relationships of Holometabola. This tree represents the best recent estimate of holometabolan insect relationships based on nuclear genes (Wiegmann et al. 2009; Misof et al. 2014; Peters et al. 2014). Eight nodes were selected to assess the quality of trees under the different methodological strategies. These uncontroversial relationships are labeled by orange circles with number: 1, the basal split of Hymenoptera from all others; 2, Neuropteroidea + Mecopterida; 3, Neuropteroidea; 4, Coleopterida; 5, Neuropterida; 6, Mecopterida; 7, Antliophora; 8, Amphiesmenoptera.
Unlike the recent successes of resolving holometabolan relationships with nuclear genes, interordinal studies with mitochondrial genomes are limited, and most of them omit too many orders to be of much comparative value and recovered very few of the widely accepted clades (Cameron et al. 2009; Wei et al. 2010; Talavera and Vila 2011; Kaltenpoth 2012; Wang et al. 2014). Furthermore, mitochondrial genomes of holometabolan insects showed significant lineage-specific variation in base composition and mutational rate, creating phylogenetic error in superimposed substitutions, but these confounding effects may be more manageable as taxon sampling increases. Thus, holometabolan insects constitute an excellent model to evaluate the impact of systematic errors in mitochondrial phylogenomics and to test challenging questions of phylogenetic methodology.
To investigate the potential and pitfalls of the mitochondrial phylogenomic data under dense taxon sampling, we conduct a phylogenetic analysis of mitochondrial genome sequences of 199 representative taxa from all 11 holometabolan orders. Eight uncontroversial relationships of deep holometabolan relationships are selected as indicators to test the performance of different phylogenetic methodological strategies (fig. 1). We find that high compositional heterogeneity and saturation of the sequences can lead to strongly supported but incorrect phylogenies under standard “site-homogeneous” models, especially when the phylogenetic signal for a given branch is significantly weaker than the nonphylogenetic signal. We show that nonphylogenetic signal can be reduced by using 1) the site-heterogeneous mixture model, 2) the inclusion of rRNA (ribosomal RNA) genes, and 3) removal of fast-evolving sites. To address the difficult question of the phylogenetic relationships among major insects, and specifically in the Holometabola, all these strategies are required at the same time.
Materials and Methods
Taxon Sampling and Sequence Alignment
We downloaded nucleotide sequences of the 13 protein-coding genes and 2 rRNA genes for 197 Holometabola insects from the NCBI (National Center of Biotechnology Information). To this initial data set, we added newly generated mitochondrial genomes for two Trichoptera species (GenBank accession numbers KF717094 and KF717095) and added 4 Hemiptera species as outgroups, thus generating a data set of 203 taxa (199 Holometabola species and 4 outgroups; see supplementary table S1, Supplementary Material online). The annotated mitochondrial genomes of the two newly sequenced caddisflies were presented in supplementary figure S1, Supplementary Material online.
Each protein-coding gene was aligned individually based on codon-based multiple alignments by using the MAFFT algorithm implemented in TranslatorX with the L-INS-i strategy (Abascal et al. 2010). Two rRNA genes were individually aligned using the MAFFT 7.0 online server with the G-INS-i strategy (Katoh and Standley 2013).
Base Composition and Substitution Rate of Protein-Coding Genes
For each species of Holometabola, we concatenated the 13 mitochondrial protein-coding genes in MEGA 6.0 (Tamura et al. 2013) and calculated 1) the overall nucleotide GC% (Guanine-Cytosine) using all 3 codon positions and 2) the frequency of amino acids encoded by GC-rich codons (glycine, alanine, arginine, and proline [GARP]). The nonsynonymous substitution rate (Ka) was calculated using the DnaSP v.5.0 (Librado and Rozas 2009). We also extracted branch length estimates from the most likely tree after standard Bayesian inference (BI) and maximum-likelihood (ML) analysis.
Phylogenetic Analyses Using Site-Homogeneous Models
Alignments of individual genes were concatenated to generate five 203-taxa data sets: 1) the PCG matrix, including all three codon positions of protein-coding genes (total of 11,169 bp); 2) the PCG12 matrix, including the first and second codon positions of protein-coding genes (total of 7,446 bp); 3) the PCGR matrix, including all three codon positions of protein-coding genes and two rRNA genes (total of 13,799 bp); 4) the PCG12R matrix, including the first and second codon positions of protein-coding genes and two rRNA genes (total of 10,076 bp); and 5) the AA matrix, including amino acid sequences of protein-coding genes (total of 3,723 amino acids). The heterogeneity of sequence divergence within data sets was analyzed using AliGROOVE (Kück et al. 2014) with the default sliding window size. Indels in nucleotide data set were treated as ambiguity and a BLOSUM62 matrix was used as default amino acid substitution matrix. The metric establishes pairwise sequence comparisons of individual terminals or subclades with terminals outside of the focal group. The obtained scoring distance between sequences is then compared with similarity over the entire data matrix. Values can vary between −1 if comparisons are very different from the remainder of the matrix to +1 for comparisons whose score match the average for the entire matrix. This provides an indirect measure of heterogeneity of a given sequence or clade with respect to the full data set.
We first analyzed five datasets under both BI and ML frameworks using MrBayes 3.2.3 (Ronquist et al. 2012), PhyloBayes MPI 1.4f (Lartillot et al. 2013), and RAxML-HPC2 8.1.11 (Stamatakis 2006), respectively. Separate partitions were created for each gene in the data set. Bootstrap ML analyses with 1,000 replicates were performed with the fast ML method implemented in RAxML using the GTRGAMMA model for nucleotide data and the MtArt (Abascal et al. 2007) model for amino acid data. For MrBayes analyses, the best-fit model of nucleotide sequences of each gene was determined with jModelTest 0.1.1 (Posada 2008) according to the Akaike Information Criterion (AIC) (supplementary table S2, Supplementary Material online). Two simultaneous runs of 6–20 million generations were conducted for the matrix and trees were sampled every 1,000 generations, with the first 25% discarded as burn-in. Stationarity was considered to be reached when the average standard deviation of split frequencies was below 0.01 (Huelsenbeck et al. 2001). The amino acid data set was analyzed using PhyloBayes MPI with the MtArt model. Two independent chains starting from a random tree were run for 30,000 cycles, with trees being sampled at every cycle. The initial 7,500 trees of each Markov chain Monte Carlo (MCMC) run were discarded as burn-in. A consensus tree was computed from the remaining 45,000 trees combined from two runs.
Three data sets (PCG, PCGR and AA) were also used to test the different partitioning schemes for ML and BI methods. The optimal partitioning scheme and substitution model was selected by PartitionFinder v1.1.1 (Lanfear et al. 2012). We created input configuration files that contained different predefined partitions for each data set: 1) 13 gene partitions for PCG; 2) 39 codon partitions for PCG; 3) 15 gene partitions for PCGR; 4) 41 partitions for PCGR (39 codon positions for PCGs and 2 partitions for rRNAs); and 5) 13 gene partitions for AA. We used the “greedy” algorithm with branch length estimated as “unlinked” and AIC criteria to search for the best-fit partitioning scheme and substitution model. The best selected partitioning schemes and models of three data sets for ML analyses were listed in supplementary table S3, Supplementary Material online. Partitioned ML analyses were conducted using RAxML.
Phylogenetic Analyses based on Site-Heterogeneous Models
Ten-fold Bayesian cross-validation was performed to test the fit of the site-heterogeneous mixture models (CAT and CAT + GTR) and “site-homogeneous” models (GTR and MtArt) to our complete nucleotide (PCG and PCGR) and amino acid (AA) data sets using PhyloBayes 3.3f (Lartillot et al. 2009) (see PhyloBayes manual). The result of cross-validation showed that the CAT + GTR model was the best fitting model for all data sets (supplementary table S4, Supplementary Material online).
We then inferred phylogenies from the AA, PCG, PCG12, PCG-RY (protein-coding genes with third codon positions R-Y coded), PCGR, PCG12R, and PCGR-RY (the third codon position of protein-coding genes R-Y coded) data sets using PhyloBayes MPI 1.4f (Lartillot et al. 2013), with the CAT + GTR model. In each individual analysis, two independent chains starting from a random tree were run for 30,000 cycles, with trees being sampled at every cycle. The initial 7,500 trees of each MCMC run were discarded as burn-in. A consensus tree was computed from the remaining 45,000 trees combined from 2 runs. All phylogenetic analyses were carried out on the CIPRES Science Gateway (Miller et al. 2010) and the High Performance Computing Cluster at the University of Kentucky Analytics and Technologies.
Model-based Saturation Plots and Posterior Predictive Analyses of Sequence Homoplasy
For the saturation plots, the overall best fitting CAT + GTR model was selected as a reference model. Patristic distances derived from trees obtained under site-homogeneous models or using the observed distances (uncorrected P-distances) were plotted against the CAT + GTR distances, and the slope of the regression line in the plot was used as a measure of saturation (relative to the divergences estimated from the rate-heterogeneous model). Patristic distances were generated using PATRISTIC (Fourment and Gibbs 2006). Posterior predictive analysis implemented in PhyloBayes 3.3f (Lartillot et al. 2009) was also used to compare the ability of alternative models to estimate the homoplasy in our data sets.
Exploration of the Signal in the Nucleotide and Amino Acid Data Sets
To explore the phylogenetic signal in the nucleotide and amino acid data sets, we excluded the fast-evolving sites using SlowFaster (Kostka et al. 2008). To assign substitution rates to individual positions, eight widely recognized groups (Hymenoptera, Megaloptera, Neuroptera, Strepsiptera, Coleoptera, Trichoptera, Lepidoptera, and Diptera) were chosen. Positions with the highest rates were gradually excluded and new restricted sub-data sets were produced. The nucleotide and amino acid sub-data sets were analyzed with PhyloBayes under the CAT + GTR model.
Phylogenetic Error Reduction by OV-Sorting Method
By using OV sorting (Goremykin et al. 2010, 2013), the full data set was ordered from the most variable sites to the most conserved sites, and a series of alignments was generated by successively shortening the OV-sorted alignment in steps of 500 sites. At each round of data removal, two data partitions were obtained: 1) an “A” partition, which includes sites from the conserved end of the alignment that are left after the iterative removal of the fastest sites, and 2) a “B” partition, which includes the variable sites removed in the current round. After model fitting was applied to each partition using ModelTest (Posada and Crandall 1998), the ML distance and uncorrected p distance were calculated using PAUP* (Swofford 2002). Correlation analyses were conducted at each shortening step to establish 1) the correlation of the ML and uncorrected p distances for partition B, testing for the change in this correlation as uncorrected p distances increasingly fail to capture the true divergence in the most variable data, unlike the model-based estimates; and 2) the correlation of the ML distances on pairs of taxa for partition A and B, in the expectation that both partitions produce roughly similar distances, unless the two partitions differ in their rates, in which case the B partition should be removed. The stopping point for site removal was determined as the point at which the two correlations showed a significant improvement (also named the GNB criterion in Goremykin et al. 2013). To reduce computation time for PAUP tree-building analyses on a 24-core Linux server, a small taxon selection (36-taxa) from the full 203-taxa PCGR data set was used in the OV-sorting analyses. The 36-taxa data set included species from all 11 holometabolan insect orders, and we fully considered the diversity of their branch length and A + T content to simulate the complicacy of the full 203-taxa data set. Finally, the OV-sorted data set selected by the GNB criterion was analyzed with PhyloBayes under the CAT + GTR model.
Results and Discussion
High Degree of Compositional Heterogeneity
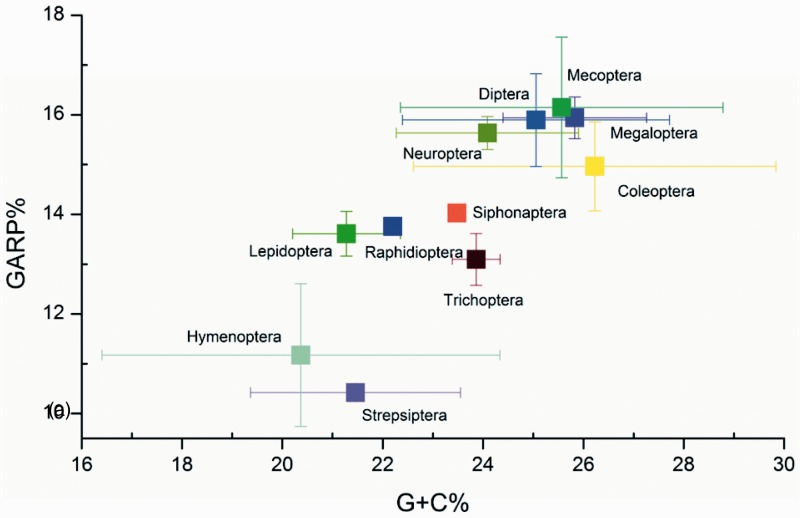
We explored the compositional diversity of both nucleotides and amino acids of mitochondrial protein-coding genes across holometabolan orders (fig. 2). Sequences of Hymenoptera and Strepsiptera were more extremely A + T rich and low in the GC-encoding GARP amino acids than other orders. Among the three orders of Neuropterida, sequences of Raphidioptera were more A + T rich than Neuroptera and Megaloptera. In Mecopterida, sequences of Lepidoptera, Trichoptera, and Siphonaptera were more A + T rich than Diptera and Mecoptera. Five orders (Hymenoptera, Strepsiptera, Diptera, Mecoptera, and Coleoptera) showed high compositional bias at the intraordinal level; for example, sequences of two gall midges (Diptera: Cecidomyiidae) were more A + T rich than other species of Diptera (supplementary table S5, Supplementary Material online). Our observation showed a high degree of compositional heterogeneity among holometabolan mitochondrial genomes in both nucleotide and amino acid level and such variability is known as the source of systematic errors in phylogenetic reconstructions (Sheffield et al. 2009; Rota-Stabelli et al. 2010).
Fig. 2.—
Compositional properties of holometabolan mitochondrial protein-coding genes. The G + C content of the concatenated alignment is plotted against the percentage of amino acids encoded by G- and C-rich codons (GARP). Values are averaged for orders, with standard deviations indicated.
Contrasting Rates of Evolution in the Mitochondrial Genome of Holometabola
We measured Ka for each taxon included in our study in comparison with the outgroup (supplementary table S5, Supplementary Material online). These comparisons showed that Ka is low for Mecoptera (0.24–0.27), Neuroptera (0.25–0.26), Megaloptera (0.25–0.27), Siphonaptera (0.28), Trichoptera (0.29 and 0.30), Raphidioptera (0.29), Coleoptera (0.24–0.33), Diptera (0.23–0.32), and Lepidoptera (0.23–0.31), and generally higher for Hymenoptera (0.30–0.49) and Strepsiptera (0.37 and 0.45). In Hymenoptera, the Ka of a sawfly Monocellicampa pruni (Wei et al. 2015) (Tenthredinidae) (0.30) was also markedly lower than the other species. Comparison of branch lengths in the phylogenetic tree showed a similar trend, and a positive correlation was observed between Ka and branch length (R2 = 0.88) (supplementary table S5, Supplementary Material online). Overall, these findings demonstrate the lineage-specific substitution rate and contrasting rate of mitochondrial genome evolution both across and within Holometabola orders, especially a significantly accelerated evolutionary rate in Hymenoptera and Strepsiptera. Accelerated substitution rates may play a role in masking and eroding phylogenetic signal through unrecognized homoplasy and lead to increased susceptibility to systematic bias, such as LBA (Bergsten 2005; Rota-Stabelli et al. 2010; Talavera and Vila 2011).
Heterogeneous Sequence Divergence within Holometabola Mitochondrial Genomes
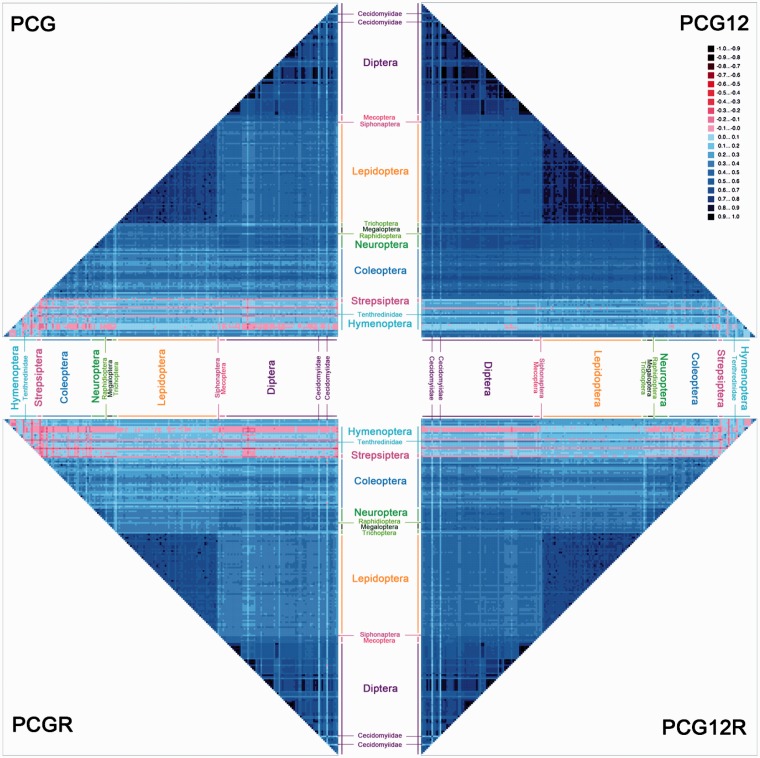
The AliGROOVE procedure provides a measure of heterogeneity of sequence divergence by conducting pairwise comparisons of nucleotide divergences for each terminal or set of terminals defined by an internal node against all other sequences in a multiple sequence alignment (Kück et al. 2014). This analysis found strong heterogeneity in sequence divergence for a subset of taxa (fig. 3 and supplementary fig. S2, Supplementary Material online). In particular, pairwise sequence comparisons involving Hymenoptera (excluding Tenthredinidae), Strepsiptera, and two gall midges (Diptera: Cecidomyiidae) received lower similarity scores than pairwise comparisons between other sequences. The divergence of these taxa may indicate that species in these groups cannot be robustly placed in the phylogenetic tree or may be misplaced. The heterogeneity was strongest for data sets PCG and PCGR that include all nucleotide positions, compared with the PCG12, PCG12R, and AA data sets, indicating that third codon positions are greatly more rate-heterogeneous than the first and second codon positions and consistently scored negative in the AliGROOVE pairwise comparisons (supplementary fig. S2, Supplementary Material online). Data sets with the third codon position excluded (PCG12 and PCG12R) therefore reduced the degrees of random sequence similarity and sequence heterogeneity.
Fig. 3.—
AliGROOVE analysis for four data sets. The mean similarity score between sequences is represented by a colored square, based on AliGROOVE scores from −1, indicating great difference in rates from the remainder of the data set, that is, heterogeneity (red coloring), to +1, indicating that rates match all other comparisons (blue coloring).
Systematic Errors in Standard Bayesian Inference and Maximum-Likelihood Analysis
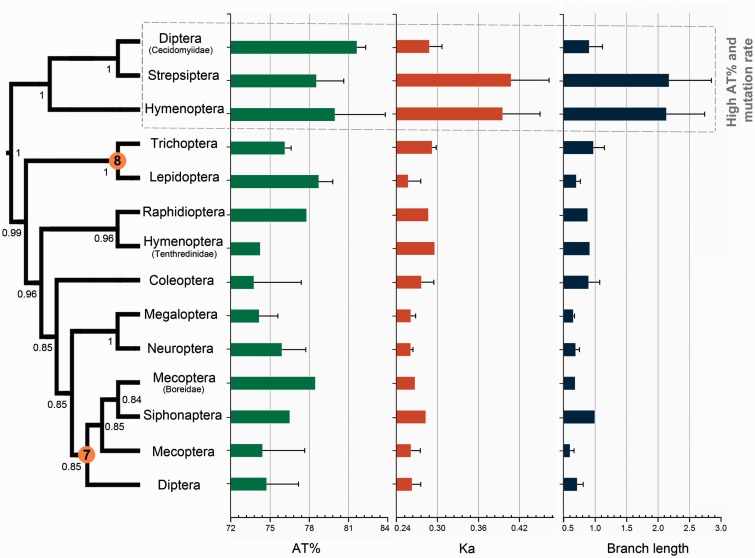
Standard BI and ML analysis were conducted for each of the five 203-taxa data sets (PCG, PCG12, PCGR, PCG12R, and AA). These analyses using site-homogenous models based on empirical frequencies of amino acid or nucleotide substitutions (including MtArt or GTR-based models) produced contradictory topologies (supplementary figs. S3 and S4, Supplementary Material online). Bayesian analyses of three data sets (BI-PCG, BI-PCG12, and BI-PCG12R) recovered two uncontroversial relationships, Amphiesmenoptera and Antliophora, and only Amphiesmenoptera was supported by other standard BI and ML analyses. ML analyses of three data sets (PCG, PCGR, and AA) with optimized partition schemes could not improve the results recovering one uncontroversial relationship at most (supplementary fig. S5, Supplementary Material online). There is no significant difference in topology or nodal support between different partitioning schemes. In general, the unexpected grouping of two gall midges (Diptera: Cecidomyiidae), Strepsiptera and Hymenoptera (excluding Tenthredinidae), was highly supported by most methods (fig. 4 and supplementary figs. S3 and S4, Supplementary Material online). After mapping the values of Ka, branch lengths, and A + T content onto the phylogenetic tree, we found that species in this clade showed higher values for these parameters than other holometabolan insects (fig. 4), and the similarity scores showed high divergence to all other species displayed in the AliGROOVE analyses (fig. 3 and supplementary fig. S2, Supplementary Material online). These results suggest that phylogenetic artifacts are responsible for recovery of this spurious clade.
Fig. 4.—
Systematic errors in the standard phylogenetic analyses under site-homogeneous model. The tree is obtained by Bayesian analysis of nucleotide sequences of protein-coding genes (BI-PCG) under site-homogeneous models. Orange circles with number indicate recovered uncontroversial relationships in figure 1. The unexpected clade caused by accelerated substitution rates and compositional heterogeneity of holometabolan mitochondrial genomes is highlighted by a dotted line box. Error bars represent standard deviations from data of multiple species.
Phylogenetic Results under Site-Heterogeneous Models
Bayesian (PhyloBayes) analyses under the CAT + GTR model from the AA and PCG data sets recovered three uncontroversial relationships, Neuropterida (BPP [Bayesian posterior probabilities] = 0.65 and 0.99), Amphiesmenoptera (BPP = 1.0), and Antliophora (BPP = 0.98 and 0.77) (supplementary fig. S6, Supplementary Material online). Artifactual clades similar to those in the standard Bayesian and ML analyses were also found in these two analyses, for example, the grouping of two gall midges (Diptera: Cecidomyiidae), Strepsiptera, and most Hymenoptera in the AA data set, and the sister relationship between Coleoptera and Strepsiptera plus most Hymenoptera in the PCG data set. Removal of third codon positions (PCG12) or RY-coding (PCG-RY) could break up these artifactual groupings, but the positions of Strepsiptera, Amphiesmenoptera, and Hymenoptera were still unresolved (supplementary fig. S6, Supplementary Material online). These results indicate that when the CAT + GTR model is used, RY-coding and removal of third codon positions can reduce the effect of systematic bias. However, the effect of these strategies was insufficient to recover the correct relationships.
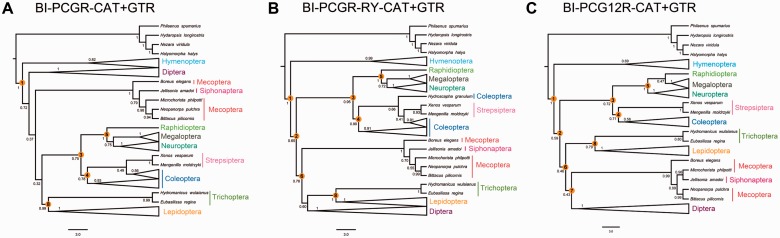
Further improvements were obtained when also including the rRNA genes (BI-PCGR), which recovered the basal split of Hymenoptera from all others (BPP = 1.0), and the monophyly of Neuropteroidea (BPP = 0.75), Coleopterida (BPP = 0.78), Neuropterida (BPP = 1.0), and Amphiesmenoptera (BPP = 0.99) (fig. 5A). PhyloBayes analysis of the two rRNA genes alone (BI-RNA) recovered the monophyly of Amphiesmenoptera (BPP = 0.96), Coleopterida (BPP = 0.89; although Strepsiptera was placed within Coleoptera), and Diptera (BPP = 0.95), and the artifactual clades common to the analyses of AA and PCG data sets were not found (supplementary fig. S6, Supplementary Material online). Thus, the inclusion of the rRNA genes reduced the impact of systematic errors to a certain extent. In combination with RY-coding (PCGR-RY), we found the above five relationships (BPP ≥ 0.75) and further recovered the monophyly of Mecopterida (BPP = 0.76) and the sister relationship of Neuropteroidea and Mecopterida (BPP = 0.65) (fig. 5B). Finally, when applying BI-PCG12R, we obtained the best tree topology of deep relationships of Holometabola consistent with accepted nuclear data and morphology-based hypotheses: (Hymenoptera + ((Coleopterida + Neuropterida) + (Amphiesmenoptera + Antliophora))) (fig. 5C and supplementary fig. S7, Supplementary Material online).
Fig. 5.—
Holometabolan phylogenies inferred from the combined protein-coding genes and rRNA gens using PhyloBayes with the CAT + GTR model. (A) Bayesian tree from the data set PCGR under the CAT + GTR model. (B) Bayesian tree from the data set PCGR-RY under the CAT + GTR model. (C) Bayesian tree from the data set PCG12R under the CAT + GTR model. We show a schematic version of the Bayesian trees with some lineages collapsed for clarity. Supports at nodes are Bayesian posterior probabilities. Orange circles with number indicate recovered uncontroversial relationships in figure 1.
Improvements Using Site-Heterogeneous Models
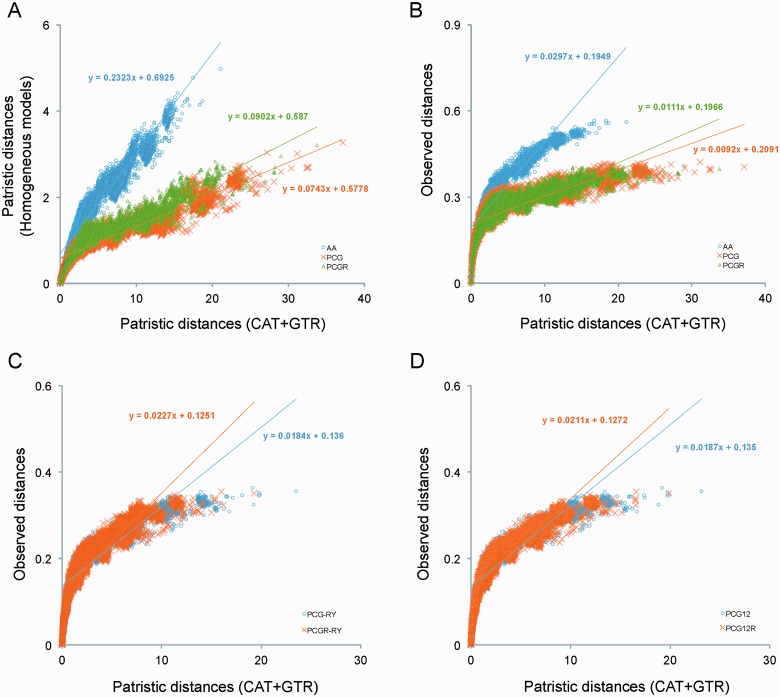
Sequence saturation and compositional heterogeneity are two frequent causes of phylogenetic artifacts (Philippe et al. 2011; Rota-Stabelli et al. 2013). Inferences of ancient nodes are hampered by multiple superimposed substitutions (homoplasy) and therefore depend on the accuracy of the model of sequence evolution, to avoid spurious convergences (Lartillot and Philippe 2004; Sperling et al. 2009; Morgan et al. 2013). We measured how well various models estimate sequence saturation and homoplasy. Posterior predictive analysis showed that MtArt and GTR models inferred a much lower level of homoplasy in the amino acid (AA) and nucleotide (PCG and PCGR) data sets compared to the CAT + GTR model (supplementary table S6, Supplementary Material online). Using the CAT + GTR model, predicted homoplasies of the data sets PCG (251.59) and PCGR (220.51) were much higher than after removal of third positions (PCG12, 92.25; PCG12R, 119.76) or RY-coding (PCG-RY, 117.46; PCGR-RY, 111.62) and amino acid coding (AA, 92.32) (supplementary table S6, Supplementary Material online).
These results were confirmed based on the level of sequence divergence assessed by plotting patristic distances in standard and CAT + GTR models. The slope of these plots was extremely low for data sets that include all nucleotide positions under the standard GTR model (0.0743 for PCG and 0.0902 for PCGR) (fig. 6A) and amino acids under the MtArt model (0.2323 for AA) (fig. 6B). The plots of uncorrected p-distance against the distances from the CAT-GTR model still showed high saturation after removal of third positions and RY-coding (slope of PCG12 = 0.0187 and PCG-RY = 0.0184), whereby the inclusion of the rRNA genes clearly reduced the level of saturation shown for PCGR-RY (0.0227; fig. 6C) and PCG12R (0.0211; fig. 6D). Thus, removal of third positions or RY-coding greatly reduced saturation against the full PCG data set (0.0092), while AA recoding had an even greater effect. These results indicate the difficulty of estimating the correct rates due to saturation of nucleotide changes under the GTR and MtArt models, which may account for spurious groups exhibiting high A + T content and fast evolutionary rate (e.g., the grouping of Cecidomyiidae, Strepsiptera, and most Hymenoptera), while the site-heterogeneous model are less affected and resolve the tree correctly, in particular once the third positions have been removed.
Fig. 6.—
Model-based saturation plots for the amino acid and nucleotide data sets. (A) Plots of the patristic distances of all data (AA, PCG, and PCGR) estimated from the CAT + GTR tree compared with the distances from the “site-homogeneous” MtArt and GTR-based models. Plots of the observed distances (uncorrected P-distances) against distance estimated from the CAT + GTR tree, using (B) all data, (C) all data after RY coding, and (D) first and second positions only.
Phylogenetic Signal in the Nucleotide and Amino Acid Data Sets of Mitochondrial Genomes
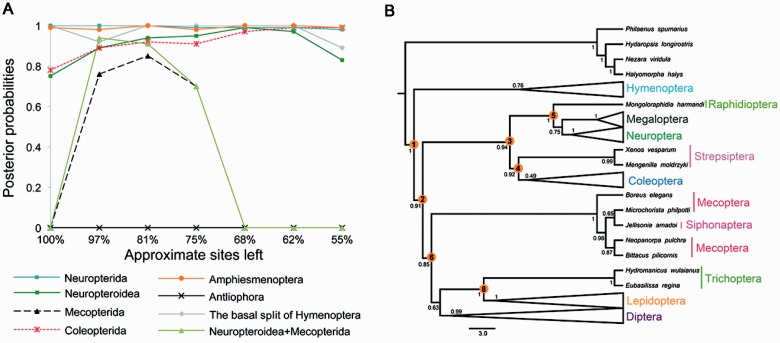
As homoplasy and degree of saturation varied greatly among classes of nucleotides and had great effects on the phylogenetic trees even under the CAT + GTR model, we attempted to dissect the role of variable rates among nucleotides by gradually excluding an ever-larger proportion of the fastest evolving sites using SlowFaster (Kostka et al. 2008). Based on the best fitting CAT + GTR model and sub-data sets of PCGR, this analysis showed that signal supporting the basal split of Hymenoptera, and the monophyly of Coleopterida, Neuropterida, and Amphiesmenoptera was stable under any level of data removal (fig. 7A). However, the monophyly of Mecopterida and the sister relationship of Neuropteroidea and Mecopterida were recovered only over a small window of data exclusion, after approximately 19% of the fastest evolving sites were excluded, but lost again after exclusion of approximately 32% or more of sites (fig. 7B). Signal for Neuropteroidea was also lost after exclusion of approximately 53% of fastest evolving sites. Seven of eight selected uncontroversial relationships were recovered with high support (BPP > 0.85) and only the monophyletic Antliophora was not recovered by SF analyses of the PCGR data set. The best SF analyses of AA and PCG recovered few selected uncontroversial relationships, with three relationships recovered in the complete AA data and four relationships supported by the PCG after excluding approximately 47% of fastest evolving sites (supplementary fig. S8, Supplementary Material online). Overall, the SF analyses obtained better topologies with the PCGR rather than the PCG and AA data sets, as inclusion of the rRNA genes apparently increased the amount of phylogenetic signal and the removal of fastest evolving sites further decreased the proportion of spurious variation. However, phylogenetic signal supporting different deep relationships was derived from sites with various evolutionary rates, and therefore the removal of the fastest evolving sites also caused the loss of phylogenetic signal and the increased effect of competing signal for some nodes, which resulted in incorrect topology or low support levels (e.g., the low support for nodes Neuropteroidea + Mecopterida, Mecopterida, and Antliophora in the PhyloBayes tree of PCG12R; fig. 5C).
Fig. 7.—
Slow-fast analyses of the nucleotide data set of the combined protein-coding genes and rRNA genes. (A) Posterior probabilities using Bayesian CAT + GTR model for various sub-data sets deprived of classes of fast-evolving sites in the data set PCGR (as indicated by the amount of sites left in the data sets). Eight uncontroversial relationships in figure 1 (orange circles) are selected as indicators to test the phylogenetic signals in the data sets. (B) Holometabolan phylogeny inferred from the data set PCGR with approximately 19% fastest evolving sites excluded using PhyloBayes under the CAT + GTR model. We show a schematic version of the Bayesian trees with some lineages collapsed for clarity.
Applying an A Priori Criterion for the Removal of Most Fast-Evolving Sites
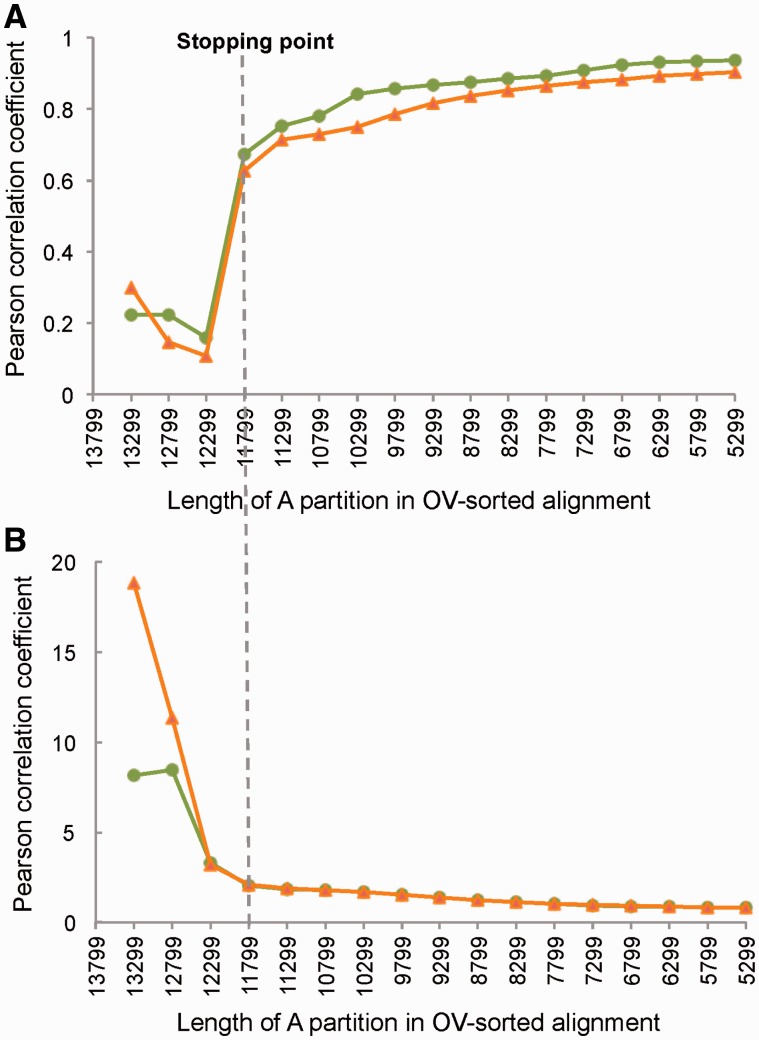
The cut-off for removal of fast sites is a subjective choice unless an a priori criterion can be applied for determining what sites should be targeted. The OV-sorting method reorders the sites in the alignment according to their observed rates, which is assessed by a statistical analysis identifying the most deviating sites in the matrix relative to the rates in the other sites and thus provides an objective stopping point for data removal (Zhong et al. 2011, 2014; Goremykin et al. 2013). This method was applied to the 13,799 sites of the PCGR data set by removing sites in increments of 500 nt. The optimal number of sites retained (GNB criterion) was 11,799 (2,000 sites removed from the full data set), identified by significant improvement in the correlation analysis that compares the variability in the removed fraction (partition B) and the retained data (partition A) in the iterative data reduction procedure (fig. 8). When the OV-sorted data set was subjected to PhyloBayes, seven of the eight selected uncontroversial relationships were recovered with high support (BPP ≥ 0.95) although the monophyletic Antliophora was not recovered (fig. 9 and supplementary fig. S9, Supplementary Material online). The topology of interordinal relationships was similar to the best SF analysis of PCGR data set, but has higher support levels for the recovered nodes. Within insect orders, many well-resolved relationships were also recovered, for example, 1) the paraphyletic Symphyta and the monophyletic Apocrita in Hymenoptera (Mao et al. 2015); 2) a robust intraordinal relationships in Lepidoptera that is in line with the recent phylogenomic study based on 2,696 nuclear genes (Kawahara and Breinholt 2014); 3) the monophyletic groups including Cyclorrhapha, Brachycera, and Neodiptera (placing Bibionomorpha as a sister group to Brachycera) in Diptera (Wiegmann et al. 2011); and 4) the monophyly of Polyphaga and Adephaga, and the sister relationship between Scirtidae and the remaining Polyphaga in Coleoptera (Timmermans et al. 2015). This indicates that phylogenetic error can be reduced by OV sorting, and the GNB criterion is suitable for identifying erroneous signal from fast-evolving sites and for selecting the appropriate data for mitochondrial phylogenomic studies. Although this criterion is applied in insect mitochondrial genomic data for the first time, we suggest that more studies in different insect groups are necessary to test the reliability of this method.
Fig. 8.—
Results of OV analysis. (A) Plot showing results of Pearson correlation analyses. The green dotted line indicates the Pearson correlation coefficients (r) of ML distances for A partitions (the more conserved) and B partitions (less conserved). The orange dotted line represents r value of uncorrected p-distances and ML distances for B partitions. The r values begin to increase sharply at the forth OV-shortening step of the PCGR data set (11,799 position remained). (B) Plot showing mean deviations between ML and p distances for B partitions. In calculating ML distances, the best-fitting ML model for each partition was first determined under the AIC using ModelTest (Posada and Crandall 1998). The orange dotted line indicates results from analyses using a neighbor-joining tree to fit ML model parameters. The green dotted line indicates results obtained when an ML tree is used to fit substitution model parameters.
Fig. 9.—
Holometabolan phylogenies inferred from the OV-sorted PCGR data set using PhyloBayes with the CAT + GTR model. The OV-sorted PCGR data set (11,799 bp) was selected by the GNB criterion (fig. 8). We show a schematic version of the Bayesian trees with some lineages collapsed for clarity and the full tree with branch lengths can be inspected in supplementary figure S9, Supplementary Material online. Bracket with number indicates the number of sampled species in a family. Supports at nodes are Bayesian posterior probabilities. Orange circles with number indicate recovered uncontroversial relationships in figure 1.
Methodological Implications for Mitochondrial Phylogenomics of Deep Insect Relationships
It is well known that insect mitochondrial genomes display strong base compositional and mutational rate heterogeneity, variation among different genes, among codon positions within a gene, and among different taxonomic levels (Sheffield et al. 2009; Castellana et al. 2011; Bernt et al. 2013; Li et al. 2013; Cameron 2014), as shown for the holometabolan insects in this study. Such heterogeneities violate the stationarity assumption of the widely used site-homogeneous models of nucleotide substitution (Rosenberg and Kumar 2003; Kolaczkowski and Thornton 2004; Lartillot and Philippe 2004; Hassanin 2006). Site-homogeneous models assume the same evolutionary process for every site of the data set (only the evolutionary rate can be modeled as heterogeneous across sites, usually through a gamma distribution of rates), although evolutionary processes are known to be heterogeneous across positions (Philippe et al. 2011). The CAT + GTR in PhyloBayes is better suited to accommodate this variation in the evolutionary process across sites (Lartillot and Philippe 2004; Philippe et al. 2011). This model establishes k profiles of equilibrium frequencies combined with general exchange rates (using a Dirichlet process to describe the likelihood of the distribution of discrete categories) (Lartillot and Philippe 2004; Lartillot et al. 2009). In this study, the better fit of the site-heterogeneous mixture model to holometabolan mitochondrial phylogenomic data sets was confirmed by analyses of cross-validation, model-based saturation plots, and posterior predictive simulation. The method also seems well suited to deal with different types of data composed of protein-coding and rRNA genes. Other studies have also demonstrated that these models provide a better fit to phylogenomic data and tend to reduce tree reconstruction artifacts (Sperling et al. 2009; Rota-Stabelli et al. 2010; Husník et al. 2011; Morgan et al. 2013; Li et al. 2015; Timmermans et al. 2015). We show the power of this approach for resolving the tree of Holometabola, indicating that model adequacy is critical for accurate tree reconstruction in mitochondrial phylogenomic studies.
Yet, the site-heterogeneous model is still affected by the biases in the data driven by saturation of variation and high levels of homoplasy. The rapidly evolving sites in mitochondrial genomes and in particular the third codon position of protein-coding genes are expected to be the most heterogeneous in composition and saturated in substitutions, and often contribute to various phylogenetic artifacts (Pisani 2004; Rota-Stabelli et al. 2010). Removal of these sites or the use of amino acid data is considered an effective method dealing with systematic errors (Kostka et al. 2008; Zhong et al. 2011; Cameron 2014; Liu et al. 2014). Our analyses confirmed that these strategies could significantly reduce the sequence saturation of different data sets (e.g., in AA, PCG, and PCGR). However, the genuine phylogenetic signal for ancient phylogenetic relationships is always weak and differs between protein-coding and rRNA genes. The inclusion of two rRNA sequences is helpful in increasing the amounts of phylogenetic signal and the signal-to-noise ratio. Furthermore, the site-heterogeneous models demand more data to correctly estimate the distribution of site-specific effects and to discriminate among competing phylogenetic hypotheses (Quang et al. 2008; Morgan et al. 2013). Thus adding the two rRNA genes to the protein-coding genes improved the topology of the tree by adding more phylogenetic information.
Based on the analyses with the site-heterogeneous model, we show that a part of fast-evolving sites should be discarded from phylogenetic analyses to resolve the deepest nodes in holometabolan phylogeny. Exclusion of these sites significantly reduced the phylogenetic errors and actually generated the best topological resolution. However, nodal support of some clades was also reduced. These results indicate that these fast-evolving sites include not only the majority of misleading signal, but also contribute valuable phylogenetic information. Accordingly, our results suggest that mitochondrial phylogenomic studies of basal relationships of insects require rigorous analyses with suitable evolutionary models and careful evaluation of which data to include. Therefore, an accurate method for detecting and removing the part of data containing a high level of nonphylogenetic signal is important for mitochondrial phylogenomic studies. Our exploratory analysis indicates that the GNB criterion of the OV-sorting method (Goremykin et al. 2010, 2013) is suitable for identifying sites most affected by multiple substitutions and it could be useful for other insect groups and data sets with similar properties.
Phylogenomics, the inference of phylogenetic relationships using genome scale data (from EST, transcriptome, and whole-genome sequences), have shown their power for assembling the tree-of-life (Philippe et al. 2009; Jarvis et al. 2014; Misof et al. 2014). However, systematic error resulting from nonphylogenetic signal is not expected to disappear with the addition of large amounts of data (Delsuc et al. 2005; Jeffroy et al. 2006; Philippe et al. 2011). In addition, the requirement for living materials is still a major limitation to transcriptomic analyses. With next-generation sequencing technology, hundreds or even thousands of mitochondrial genomes can be efficiently and economically obtained from a pooled mixture of DNA extracts (Gillett et al. 2014; Tang et al. 2014). As little as 1 ng of genomic DNA from each species is sufficient for pooling and many degraded voucher specimens of rare species in museum collections can also be suitable for sequencing (Timmermans et al. 2016). The ease of sequencing, the feasibility of large taxon sampling, and other advantages of mitochondrial genomes (easy alignment, straightforward gene orthology, etc.) remain major reasons for its continued use in the phylogenomic era.
In order to effectively use mitochondrial genome data to correctly resolve difficult phylogenetic questions, particularly the ancient divergences, inferences from mitochondrial genomic data should always assess the possible impact of substitutional saturation and compositional biases whose effect is not independent but potentially correlated. We therefore suggest that phylogenetic hypotheses inferred from mitochondrial genomic data be interpreted with caution, even when highly supported, until the effects of systematic errors are fully assessed through data deletion strategies (e.g., OV-sorting method), and by using site-heterogeneous mixture models.
Supplementary Material
Supplementary tables S1–S6 and figures S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Yang Liu (University of Connecticut, USA) for assistance with phylogenetic analysis and Dr Peter Foster (Natural History Museum, London, UK) for discussing likelihood models. This work was supported by grants from the National Basic Research Program of China (grant no. 2013CB127600), the National Natural Science Foundation of China (grant nos. 31420103902, 31372229, and 31401991), the Beijing Natural Science Foundation (grant nos. 6144027 and 6152016), and the Chinese Universities Scientific Fund (grant nos. 2015NX001, 2016QC025, and 2016QC072).
Literature Cited
- Abascal F, Posada D, Zardoya R. 2007. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 24:1–5. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Philippe H. 2007. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Mol Biol Evol. 24:6–9. [DOI] [PubMed] [Google Scholar]
- Bergsten J. 2005. A review of long-branch attraction. Cladistics 21:163–193. [DOI] [PubMed] [Google Scholar]
- Bernt M, et al. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69:252–364. [DOI] [PubMed] [Google Scholar]
- Beutel RG, Pohl H. 2006. Endopterygote systematics—where do we stand and what is the goal (Hexapoda, Arthropoda)? Syst Entomol. 31:202–219. [Google Scholar]
- Beutel RG, et al. 2011. Morphological and molecular evidence converge upon a robust phylogeny of the megadiverse Holometabola. Cladistics 26:1–15. [DOI] [PubMed] [Google Scholar]
- Bourguignon T, et al. 2015. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol Biol Evol. 32:406–421. [DOI] [PubMed] [Google Scholar]
- Breinholt JW, Kawahara AY. 2013. Phylotranscriptomics: saturated third codon positions radically influence the estimation of trees based on next-gen data. Genome Biol Evol. 5:2082–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann H, Giezen M, Zhou Y, Raucourt GP, Philippe H. 2005. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol. 54:743–757. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Philippe H. 1999. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 16:817–825. [DOI] [PubMed] [Google Scholar]
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. [DOI] [PubMed] [Google Scholar]
- Cameron SL, Lambkin CL, Barker SC, Whiting MF. 2007. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol. 32:40–59. [Google Scholar]
- Cameron SL, Miller KB, D’Haese CA, Whiting MF, Barker SC. 2004. Mitochondrial genome data alone are not enough to unambiguously resolve the relationships of Entognatha, Insecta and Crustacea sensu lato (Arthropoda). Cladistics 20:534–557. [DOI] [PubMed] [Google Scholar]
- Cameron SL, Sullivan J, Song H, Miller KB, Whiting MF. 2009. A mitochondrial genome phylogeny of the Neuropterida (lacewings, alderflies and snakeflies) and their relationship to the other holometabolous insect orders. Zool Scr. 38:575–590. [Google Scholar]
- Castellana S, Vicario S, Saccone C. 2011. Evolutionary patterns of the mitochondrial genome in Metazoa: exploring the role of mutation and selection in mitochondrial protein-coding genes. Genome Biol Evol. 3:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 6:361–375. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Phillips MJ, Penny D. 2003. Comment on ‘Hexapod origins: monophyletic or paraphyletic?’. Science 301:1482. [DOI] [PubMed] [Google Scholar]
- Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749. [DOI] [PubMed] [Google Scholar]
- Fourment M, Gibbs MJ. 2006. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol. 6:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett CPDT, et al. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31:2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Nikiforova SV, Bininda-Emonds OR. 2010. Automated removal of noisy data in phylogenomic analyses. J Mol Evol. 71:319–331. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, et al. 2013. The evolutionary root of flowering plants. Syst Biol. 62:51–62. [DOI] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. 2005. Evolution of the insects. Melbourne (VIC: ): Cambridge University Press. [Google Scholar]
- Hassanin A. 2006. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol. 38:100–116. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314. [DOI] [PubMed] [Google Scholar]
- Husník F, Chrudimský T, Hypša V. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata K, Sasaki G, Ogawa J, Miyata T, Su Z. 2010. Phylogenetic relationships among insect orders based on three nuclear protein coding gene sequences. Mol Phylogenet Evol. 58:169–180. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22:225–231. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, et al. 2012. Accelerated evolution of mitochondrial but not nuclear genomes of Hymenoptera: new evidence from crabronid wasps. PLoS One 7:e32826.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara AY, Breinholt JW. 2014. Phylogenomics provides strong evidence for relationships of butterflies and moths. Proc R Soc B. 281:20140970.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. 2004. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature 431:980–984. [DOI] [PubMed] [Google Scholar]
- Kostka M, Uzlíková M, Čepička I, Flegr J. 2008. SlowFaster, a user-friendly program for slow-fast analysis and its application on phylogeny of Blastocystis. BMC Bioinformatics 9:341.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen NP. 1999. Phylogeny of endopterygote insects, the most successful lineage of living organisms. Eur J Entomol. 96:237–253. [Google Scholar]
- Kück P, Meid SA, Groß C, Wägele JW, Misof B. 2014. AliGROOVE—visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinformatics 15:294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Letsch HO, et al. 2012. Insect phylogenomics: results, problems and the impact of matrix composition. Proc R Soc B. 279:3282–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2012. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLoS One 7:e29419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2013. Mitochondrial genomes of two barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLoS One 8:e61685.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2015. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci Rep. 5:8527.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cox CJ, Wang W, Goffinet B. 2014. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst Biol. 63:862–878. [DOI] [PubMed] [Google Scholar]
- Longhorn SJ, Pohl HW, Vogler AP. 2010. Ribosomal protein genes of holometabolan insects reject the Halteria, instead revealing a close affinity of Strepsiptera with Coleoptera. Mol Phylogenet Evol. 55:846–859. [DOI] [PubMed] [Google Scholar]
- Mao M, Gibson T, Dowton M. 2015. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol Phylogenet Evol. 84:34–43. [DOI] [PubMed] [Google Scholar]
- Meusemann K, et al. 2010. A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol. 27:2451–2464. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans (LA): Institute of Electrical and Electronics Engineers (IEEE). p. 1-8.
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Morgan CC, et al. 2013. Heterogeneous models place the root of the placental mammal phylogeny. Mol Biol Evol. 30:2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi F, et al. 2003. Hexapod origins: monophyletic or paraphyletic? Science 299:1887–1889. [DOI] [PubMed] [Google Scholar]
- Niehuis O, et al. 2012. Genomic and morphological evidence converge to resolve the enigma of Strepsiptera. Curr Biol. 22:1309–1313. [DOI] [PubMed] [Google Scholar]
- Peters RS, et al. 2014. The evolutionary history of holometabolous insects inferred from transcriptome-based phylogeny and comprehensive morphological data. BMC Evol Biol. 14:52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, et al. 2009. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 19:706–712. [DOI] [PubMed] [Google Scholar]
- Philippe H, et al. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9:e1000602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Delsuc F, Penny D. 2004. Genome-scale phylogeny and the detection of systematic biases. Mol Biol Evol. 21:1455–1458. [DOI] [PubMed] [Google Scholar]
- Pick KS, et al. 2010. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 27:1983–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D. 2004. Identifying and removing fast-evolving sites using compatibility analysis: an example from the Arthropoda. Syst Biol. 53:983–994. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818. [DOI] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Quang LS, Gascuel O, Lartillot N. 2008. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24:2317–2323. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M, Kumar S. 2003. Heterogeneity of nucleotide frequencies among evolutionary lineages and phylogenetic inference. Mol Biol Evol. 20:610–621. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Lartillot N, Philippe H, Pisani D. 2013. Serine codon usage bias in deep phylogenomics: pancrustacean relationships as a case study. Syst Biol. 62:121–133. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, et al. 2010. Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda. Genome Biol Evol. 2:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF. 2009. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol. 58:381–394. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Whiting MF. 1999. Long-branch abstractions. Cladistics 15:9–24. [Google Scholar]
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69:393–403. [DOI] [PubMed] [Google Scholar]
- Sperling EA, Peterson KJ, Pisani D. 2009. Phylogenetic-signal dissection of nuclear housekeeping genes supports the paraphyly of sponges and the monophyly of Eumetazoa. Mol Biol Evol. 26:2261–2274. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates.
- Talavera G, Vila R. 2011. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol. 11:315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrío R, Rodríguez-Trelles F, Ayala FJ. 2001. Shared nucleotide composition biases among species and their impact on phylogenetic reconstructions of the Drosophilidae. Mol Biol Evol. 18:1464–1473. [DOI] [PubMed] [Google Scholar]
- Timmermans MJTN, Viberg C, Martin G, Hopkins K, Vogler AP. 2016. Rapid assembly of taxonomically validated mitochondrial genomes from historical insect collections. Biol J Linn Soc. 117:83–95. [Google Scholar]
- Timmermans MJTN, et al. 2010. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic Acids Res. 38:e197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJTN, et al. 2015. Family-level sampling of mitochondrial genomes in Coleoptera: compositional heterogeneity and phylogenetics. Genome Biol Evol. 8:161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein MD, Wiegmann BM, Beutel R, Kjer KM, Yeates DK. 2012. Advances in insect phylogeny at the dawn of the postgenomic era. Annu Rev Entomol. 57:449–468. [DOI] [PubMed] [Google Scholar]
- Wang YY, Liu XY, Yang D. 2014. The first mitochondrial genome for caddisfly (Insecta: Trichoptera) with phylogenetic implications. Int J Biol Sci. 10:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX. 2010. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to holometabolous insects. BMC Genomics 11:371.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Wu QL, Liu W. 2015. Sequencing and characterization of the Monocellicampa pruni (Hymenoptera: Tenthredinidae) mitochondrial genome. Mitochondrial DNA 26:157–158. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, et al. 2009. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biol. 7:34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann BM, et al. 2011. Episodic radiations in the fly tree of life. Proc Natl Acad Sci U S A. 108:5690–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO. 1992. The diversity of life. Cambridge: Belknap Press. [Google Scholar]
- Wu LW, Lin LH, Lees DC, Hsu YF. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics 15:468.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. 2012. Molecular phylogenetics: principles and practice. Nat Rev Genet. 13:303–314. [DOI] [PubMed] [Google Scholar]
- Zhong B, et al. 2011. Systematic error in seed plant phylogenomics. Genome Biol Evol. 3:1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, et al. 2014. Streptophyte algae and the origin of land plants revisited using heterogeneous models with three new algal chloroplast genomes. Mol Biol Evol. 31:177–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.