Abstract
We have examined a collection of the free-living marine bacterium Alteromonas genomes with cores diverging in average nucleotide identities ranging from 99.98% to 73.35%, i.e., from microbes that can be considered members of a natural clone (like in a clinical epidemiological outbreak) to borderline genus level. The genomes were largely syntenic allowing a precise delimitation of the core and flexible regions in each. The core was 1.4 Mb (ca. 30% of the typical strain genome size). Recombination rates along the core were high among strains belonging to the same species (37.7–83.7% of all nucleotide polymorphisms) but they decreased sharply between species (18.9–5.1%). Regarding the flexible genome, its main expansion occurred within the boundaries of the species, i.e., strains of the same species already have a large and diverse flexible genome. Flexible regions occupy mostly fixed genomic locations. Four large genomic islands are involved in the synthesis of strain-specific glycosydic receptors that we have called glycotypes. These genomic regions are exchanged by homologous recombination within and between species and there is evidence for their import from distant taxonomic units (other genera within the family). In addition, several hotspots for integration of gene cassettes by illegitimate recombination are distributed throughout the genome. They code for features that give each clone specific properties to interact with their ecological niche and must flow fast throughout the whole genus as they are found, with nearly identical sequences, in different species. Models for the generation of this genomic diversity involving phage predation are discussed.
Keywords: Alteromonas, pangenome, genomic islands, recombination, intraspecies diversity, phages
Introduction
The Neodarwinian evolutionary paradigm (Maynard 1989) has little value for the analysis of the evolution of prokaryotes (Koonin and Wolf 2012). Prokaryotes have two-tiered genomes formed by a core of genes, that have homologs in all the strains within the species, and a flexible set characteristic of each strain that have no homologs in most or none of the others (Welch et al. 2002; Tettelin et al. 2008). The flexible pool is often associated to horizontal gene transfer (HGT) and site-specific recombination catalyzed by mobile genetic elements, whereas the core tends to be identified with vertical transmission and homologous recombination (Wiedenbeck and Cohan 2011; Kuenne et al. 2013; Polz et al. 2013). However, it is better to define these two genomic compartments as evolutionary trends. Thus, the flexible pool provides diversity in the population, e.g., at the level of surface features or transporters that interact with the biotic and abiotic components of the environment, whereas the core provides a stable metabolic and genomic backbone for the population (Rodriguez-Valera and Ussery 2012). A significant part of the flexible genome is concentrated in genomic islands of >10 kb. We use the term flexible genomic island (fGI) to refer the clusters of flexible genes that occupy equivalent genomic locations in different strains within a taxonomic unit (normally the species, although here we have used the term at the genus level) (Gonzaga et al. 2012; Chan et al. 2015) to distinguish them from the unique islands that are found in either one or the other strain. These fGIs can be classified into two major categories (López-Pérez et al. 2013; Chan et al. 2015). Additive fGIs that are site directed recombination hotspots for the integration of gene cassettes that vary in their numbers and nature (Kuenne et al. 2013). Their functional role is variable depending on the combination of cassettes they carry. Replacement fGIs are gene clusters related to polysaccharides or glycoproteins present in the outer layers of the cell. They are coded by completely different gene clusters in different strains, with no evidence of similarity (except for the overall functional annotation) but code for equivalent exposed structures. Replacement fGIs are instrumental for the environmental interactions of the cell that, in addition, acquires through them a surface identity. For example, they allow specific host recognition by phages (Rodriguez-Valera et al. 2009; Avrani et al. 2012). In pathogenic bacteria, in which this diversity has been extensively explored and described (Reeves 1993; Milkman 1997) they are recognized by the vertebrate immune system and generate a characteristic serotype. However, for free-living microbes this terminology seems out of place. In this work, we call the specific glycosydic receptors coded by these sets of genes “glycotype.” They are exchanged by homologous recombination facilitated by the conserved neighboring genes (Milkman et al. 2003; Jeong et al. 2009; Dingle et al. 2012; López-Pérez, Martin-Cuadrado, et al. 2014).
For some years, we have studied the marine bacterium Alteromonas with the focus on understanding its genomic make-up and dynamics (Gonzaga et al. 2012; López-Pérez et al. 2012, 2013; López-Pérez, Gonzaga, et al. 2014). This microbe is a heterotrophic marine gammaproteobacterium that is found in the open ocean and takes advantage of the sporadic inputs of organic matter that appear in this relatively impoverished environment. It is a typical “bloomer” that grows very fast and can be easily retrieved in pure culture. Although initially we focused on a single species (A. macleodii) this group of strains turned out to be made up of two species: Alteromonas macleodii and A. mediterranea, that display <84% average nucleotide identity (ANI) between them (Ivanova et al. 2015). We compared two genomes of A. mediterranea isolated from the same sample and compared them to a fosmid metagenomic library also from the same Mediterranean seawater sample. This study (Gonzaga et al. 2012) showed that the two isolates, members of the same population, had similar cores (over 98.51% ANI) but differed broadly in gene content of their fGIs. The presence of fosmids with overlapping sections indicated that there were at least five coexisting clones in the population. When we added to the comparison isolates from a different Mediterranean location and obtained at a different time (5 years apart) we still detected members of the same clones (López-Pérez et al. 2013). These isolates had evidence of very close common ancestry [<100 single nucleotide polymorphism (SNPs) throughout the core genome], and had identical glycotype determinants, but differed slightly in their additive fGIs. These highly similar genomes belonging to independent isolates were defined as members of the same clonal frame (CF). They are the smallest unit of differentiation of prokaryotes (often referred to as strains, biotypes, or serotypes). This term is also equivalent with “epidemic clone” in clinical microbiology (Cho et al. 2010; Mutreja et al. 2011; Cui et al. 2015).
With the addition of other species, A. macleodii (López-Pérez et al. 2012) and A. australica (López-Pérez, Gonzaga, et al. 2014), the genomes revealed remarkable conservation of synteny, which has facilitated the study of the physical arrangement of the core and flexible regions in the genomes of representatives of this genus (López-Pérez, Gonzaga, et al. 2014). In Alteromonas, four replacement fGIs were identified (López-Pérez et al. 2012, 2013; López-Pérez, Gonzaga, et al. 2014). They are associated to the synthesis of structural polysaccharides in the outer surface of the cell, specifically, flagellum glycosylation, capsular exopolysaccharide (EPS) and O-chain polysaccharide (two separate clusters) of the cell-wall lipopolysaccharide. Each different CF has a different glycotype with different versions for each of the four glycotypic determinants (López-Pérez et al. 2013). In addition, several additive fGIs were identified coding for multiple functions from H2 oxidation to polysaccharide metabolism.
We considered that the availability of this gradient of genomic diversity provided an opportunity to understand its origin and evolution within the genus Alteromonas. The results of this analysis could be extrapolated to other free-living aquatic bacteria. Therefore, we have analyzed Alteromonas genomes through comparative genomics, starting from the closest strains (same CF), and then adding gradually more distant taxonomic entities (strains belonging to different CFs within the same species and strains belonging to different species). Specifically, for this work, we have sequenced eight new genomes; we have also collected strains from strain collections and genomes available in public databases. All the genomes analyzed here are fully sequenced and assembled. They have been compared to dissect the mechanistic processes of their evolution using diverse bioinformatic tools. Finally, we describe a model of diversity fixation in populations of closely related bacteria by bacteriophage predation.
Materials and Methods
Sample Collection, Sequencing, Assembly and Annotation
Details of isolation and origin of the Alteromonas strains using are provided in supplementary table S1, Supplementary Material online. Strains R10SW13, LMG 21861 and LMG 21856 have been obtained from the culture collection of the DSMZ (https://www.dsmz.de/home.html). DNA was extracted by phenol-chloroform as described in (Neumann et al. 1992) and checked for quality on a 1% agarose gel. The quantity was measured using Quant-iT® PicoGreen® dsDNA Reagent (Invitrogen). Sequencing was performed using Illumina Hiseq 2000 (100-bp paired-end read) (BGI Tech Solutions, Hong Kong). The generated reads were trimmed and assembled de novo using the IDBA assembler (Peng et al. 2012). We used a combination of Geneious Pro 5.0.1 (with default parameters) using previously Alteromonas assembled genomes as a reference (Gonzaga et al. 2012; López-Pérez et al. 2012, 2013) and oligonucleotides designed from the sequence of the ends of assembled contigs to obtain one single-closed contig. The genomes were annotated using the NCBI PGAAP annotation pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/). The predicted protein sequences were compared using BLASTP to the NCBI nr protein database (e-value 10 − 5). ORFs <100 bp and without significant homology to other proteins were not considered. Reciprocal BLASTN and TBLASTXs searches between the genomes were carried out, leading to the identification of regions of similarity, insertions and rearrangements. The ANI between strains was calculated using JSpecies software package v1.2.1 using default parameters (Richter and Rosselló-Móra 2009). To allow the interactive visualization of genomic fragment comparisons Artemis v.12 and Artemis Comparison Tool ACTv.9 were used (Carver et al. 2005, 2012). Additional annotation was carried out using the RAST server (Aziz et al. 2008). Additional local BLAST searches against the latest NCBI nr database were performed whenever necessary. Clustering algorithm CD-HIT (Fu et al. 2012) was performed with cut-offs of >65% minimal alignment coverage and >85% sequence identity for each sequence pair in order to assign the sequences to orthologous clusters.
Phylogenetic Analysis
Whole-genome alignments for all 25 strains (plus Pseudoalteromonas atlantica T6c that was used as an outgroup) were constructed using the progressiveMauve algorithm (Darling et al. 2010) of the Mauve software v2.3.1. The resulting alignments were subsequently used in ClonalFrame software v1.2 (Didelot et al. 2010). ClonalFrame is a Bayesian inference method which jointly reconstructs the clonal relationships between the isolates in a sample. The clonal genealogy inferred from the generated alignment of 1.38 Mb of the core genome by ClonalFrame is shown in fig. 1. StripSubsetLCBs script, also including in the Mauve software v2.3.1, was used to extract all the regions of at least 1000 bp found in all the genomes and maximum-likelihood trees using RAxML (version 7.2.6) (Stamatakis 2014) were generated using these regions in order to confirm the low level of recombination detected among different species.
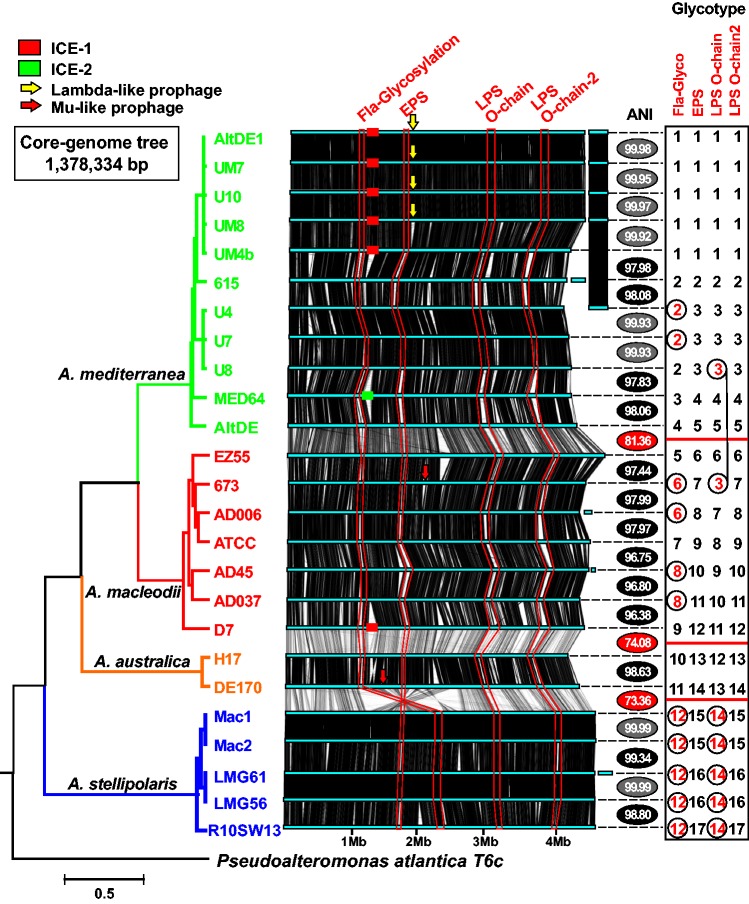
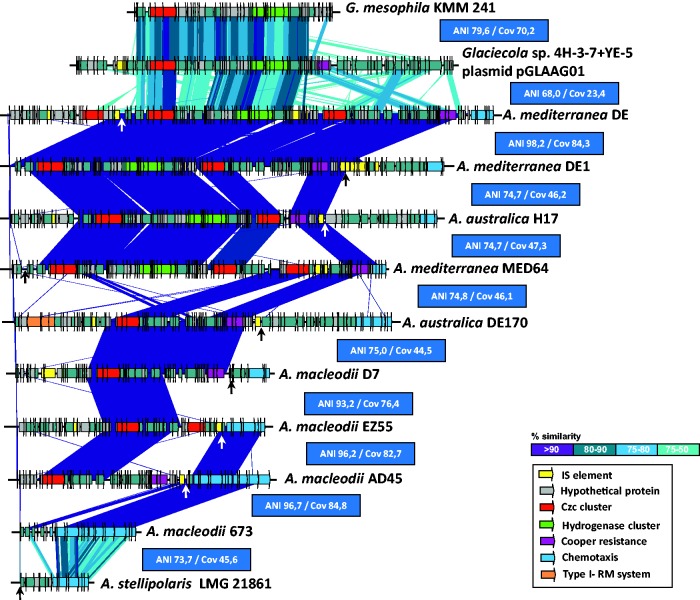
Fig. 1.—
Phylogenomic tree and whole-genome alignment of Alteromonas strains. The genomes have been linearized for the sake of simplicity and are arranged by a phylogenomic tree constructed using a concatenation of the core genome (1.38 Mb). Synteny is indicated by vertical lines connecting the linearized genomes. Plasmids appear at the right hand side. Color of the tree branches indicates members of the same species. Replacement fGIs have been connected by red lines and identified by the inferred function at the top. Arrows show location of phages inserted within the genomes and Integrative and Conjugative Elements (ICEs) are indicated with rectangles, same color indicates same version of the genomic element. The ANI value is intercalated between each genome and the one below (next in the tree). The ANI values with a gray background are between strains belonging to the same clonal frame (CF), black background members of the same species but different CF and red background strains of different species. The panel on the right indicates the type of each replacement fGI present in the corresponding genome. Different numbers correspond to the different versions of each replacement fGI. The combination of numbers for each strain provides a glycotype numerical code. Exchanges between clonal frames or species, i.e., identical or very similar version of the gene cluster in different genomic backgrounds, are highlighted in red.
Evolutionary Rate and SNPs Analysis
ClustalW was used to align the sequence of orthologous proteins and program pal2nal program (Suyama et al. 2006) to obtain the multiple codon alignment from the corresponding aligned protein sequences in order to evaluate the type and rate of nucleotide substitutions. For each sequence pair, dN/dS ratios were calculated based on the codon alignments using SNAP (Korber 2000). A low ratio (dN/dS < 1) indicates purifying selection, whereas a high ratio (dN/dS > 1) is a clear signal of diversifying selection. For each pair of strains, pairwise alignments were performed using progressiveMauve algorithm (Darling et al. 2010). The resulting locally collinear blocks of at least 500 bp were extracted from the output of the program and concatenated to form the core genome alignment. DnaSP 5.10.01 (Librado and Rozas 2009) was used to obtain the total number of SNPs between the genomes and SNPs in windows of 1 kb. Indels and SNPs between small regions of the genome such as genomic islands were identified using nucmer program in the MUMmer3+ package (Kurtz et al. 2004).
Results
Genomes Studied
We have analyzed genome sequences of Alteromonas species isolated over a 50-year span and from all around the world (supplementary table S1, Supplementary Material online), although we do not believe there is any biogeographic signal in this microbe (López-Pérez et al. 2013; López-Pérez, Gonzaga, et al. 2014). In total, we have studied here 25 fully assembled genomes classified into four clear-cut genospecies. We define genospecies as clusters of strains with genomes having >95% ANI (Konstantinidis and Tiedje 2005). Specifically, we have analyzed representatives of A. mediterranea (formerly A. macleodii deep ecotype) (11 strains), A. macleodii (7 strains), “A. stellipolaris” (5 strains) and A. australica (2 strains). For A. mediterranea, we have analyzed several strains that belong to the same CF. They provide a gradient of variation that goes from a few SNPs (ANI 99.98%), for members of the same CF, to borderline values for strains belonging to the same genus (ANI 73.35%) (supplementary fig. S1, Supplementary Material online). Eight strain genomes (U10, UM8, D7, Mac1, Mac2, LMG 21861, LMG 21856, and R10SW13) are reported here for the first time. The Ionian isolate UM8 was formerly considered identical to UM7 (López-Pérez et al. 2013) but has been now shown to be different and is therefore described for the first time. Two strains from polar latitudes A. stellipolaris (LMG 21861 and LMG 21856) (Van Trappen et al. 2004) and A. addita R10SW13 (described as different species based on 16S rRNA) (Ivanova et al. 2005), together with two new Mediterranean isolates described here for the first time (Alteromonas sp. Mac1 and Mac2), belong to a single genospecies that we will call “A. stellipolaris” (the oldest denomination).
Overall Conservation of Synteny
In order to analyze the phylogenetic relationships of all the strains, we have clustered them using a whole-genome phylogeny from a concatenation of shared genes (ca. 1.38 Mb) (fig. 1). The genomes analyzed have relatively similar sizes (ranging from 4.34 to 4.94 Mb, plasmids included). The strains cluster into four genospecies (ANI> 95%) (fig. 1 and supplementary fig. S1, Supplementary Material online). Synteny was quite well conserved throughout the genus and even the positions of the main features of the flexible genome were conserved, as has been observed before in other bacteria within this range of ANI variation (Kuenne et al. 2013; López-Pérez, Gonzaga, et al. 2014) (fig. 1). Replacement fGIs coding for the glycotype are highlighted in fig. 1 and the different versions of the replacement fGIs coding for them assigned a number.
The main variation in synteny was detected in all the “A. stellipolaris” isolates between positions 1–2.5 Mb (affecting to ca. 30% of the genome) (fig. 1 and supplementary fig. S2, Supplementary Material online). These strains had a large rearrangement located in the second quadrant between the second and third ribosomal operon. Within this large rearrangement, we identified a large inversion (ca. 869 kb) that included both the flagellum glycosylation and EPS fGIs (supplementary fig. S2, Supplementary Material online). Although the position of the flagellum glycosylation fGI appeared inverted, the position relative to the origin of replication of the EPS fGI (very close to the center of the inversion) remains similar to that of the other species. Using DE1 as a reference, the rearrangement breakpoint was located at a 4-kb intergenic region at the boundary of the flagellum glycosylation island and in the other side in a region enriched in IS elements. There are two additional small inversions of ca. 74 and 120 kb, respectively (supplementary fig. S2, Supplementary Material online).
The conservation of syntheny and the availability of more than one strain for each species allowed us to assess the patterns of genomic variation within and between species and eventually try to understand the evolutionary mechanisms behind. A summary of the divergence parameters of some of the pairwise genome comparisons is shown in table 1.
Table 1.
Summary of Parameters of Examples of Genomes Pairwise Comparisons with Increasing Levels of Divergence
| Strains Compared | Intra-clonalframe |
Intra-species |
Inter-species |
||||||
|---|---|---|---|---|---|---|---|---|---|
| DE1-UM7 | DE1-UM4b | U4-U8 | DE1-U4 | LMG 21861- R10SW13 | H17-DE170 | ATCC 27126- AD45 | DE1-ATCC 27126 | DE1-LMG 21861 | |
| Core length (bp) | 4,607,484 | 4,281,619 | 4,181,088 | 3,863,565 | 4,402,959 | 3,890,654 | 4,006,297 | 3,347,798 | 2,706,307 |
| ANI (%) | 99.98 | 99.91 | 99.93 | 98.14 | 98.83 | 98.63 | 96.75 | 80.94 | 73.8 |
| Coverage (%) | 99.49 | 95.66 | 95.22 | 85.65 | 94.13 | 88.44 | 85.13 | 65.72 | 43.42 |
| #Total SNPs | 97 | 135 | 507 | 33,279 | 27,456 | 24,792 | 66,301 | 310,462 | 351,405 |
| ΔS (%) | 0.002 | 0.003 | 0.012 | 0.861 | 0.624 | 0.637 | 1.655 | 9.274 | 12.985 |
| Clonal fraction | 0.998 | 0.997 | 0.995 | 0.501 | 0.619 | 0.516 | 0.040 | 0.000 | 0.000 |
| #SNPs Mutation | 48 | 43 | 109 | 2,341 | 3,010 | 3,783 (9,399)* | 358 (41,297)* | 5 (251,792)* | 3 (333,513)* |
| #SNPs Synonymous | 23 | 16 | 49 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| #SNPs NonSynon. | 17 | 19 | 45 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| #SNPs IR | 8 | 8 | 15 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| ΔM (%) | 49.48 | 31.85 | 21.49 | 7.03 | 12.48 | 0 (37.91)* | 0 (62.29)* | 0 (81.10)* | 0 (94.91)* |
| ΔR (%) | 50.52 | 68.15 | 78.51 | 92.97 | 87.52 | 100 (53.69)* | 100 (37.71)* | 100 (18.90)* | 100 (5.09)* |
| r/m | 3.04 | 4.39 | 8.18 | 2.23 | 3.19 | 0.73 | 0.03 | 0.00 | 0.00 |
| dN | – | – | – | 0.02 | 0.01 | 0.01 | 0.03 | 0.19 | 0.30 |
| dS | – | – | – | 0.02 | 0.01 | 0.01 | 0.04 | 0.29 | 0.42 |
| dN/dS | – | – | – | 0.73 | 0.67 | 0.69 | 0.65 | 0.65 | 0.72 |
| LCA (age in years) | 86 | 77 | 196 | 4,210 | 5,414 | 16,905* | 74,275* | 452,863* | 599,844* |
Note.—ANI, average nucleotide identity; ΔS, percentage of SNPs for aligned genomes; Clonal fraction, fraction of segments containing 0–3 SNPs (Dixit et al. 2015); IR, intergenic regions; ΔM, fraction of the SNPs involved in mutation; ΔR, fraction of the SNPs involved in recombination; ΔC, SNP density in the clonal fraction; r/m, ΔR* Clonal fraction/ΔM; LCA, age of last common ancestor determined using the evolutionary rate of Legionella pneumophila (Sanchez-Buso et al. 2014); n.d., not determined.
*Values that deviate from the analysis described in Dixit et al. (2015) are in parentheses.
Evolution of the Core Genome
Intraclonalframe
We have compared the genomes by incremental steps of divergence starting by isolates that belong to a single CF. Alteromonas mediterranea CF1 and CF2 were described previously (López-Pérez et al. 2013). Now we have included more strains and analyzed in more detail the differences among them. Whole genome alignment among the five members of A. mediterranea CF1 (AltDE1, UM7, UM8, UM4b and U10) revealed a total of 151 homologous regions representing a core genome of 4.2 Mb (97% of the average genome size) and a total of 146 variable sites that include both SNPs and insertions or deletions.
Following the approach used for Escherichia coli by (Dixit et al. 2015), we have calculated different parameters based on SNP density to gauge the relative evolutionary divergence among these strains (summarized in table 1). Aligned regions between genomes were divided into 1 kb segments and divergence was estimated as the percentage of SNPs (ΔS). The clonal fraction (nonrecombined) of each pair was identified as the segment fraction containing 0–3 SNPs (Dixit et al. 2015) and the SNPs average in this fraction was associated to mutation (ΔM), while the remaining, at least in part, to recombination (ΔR). The clonal fraction for strains within a single CF was 0.998–0.995. The two most similar strains (DE1 and UM7; ANI 99.98/Coverage 99.49%) had only 48 mutational SNPs (out of a total of 97) (table 1). As previously reported (López-Pérez et al. 2013), most of the genomic variation between the two strains was found at the main integron site and could be attributed to differential loss of gene cassettes. Strains U10 and UM8 have a similar number of mutational SNPs (table 1) but reveal many more changes and synteny variations, even though these four strains have the same lysogenic lambda-like phage still inserted at the same location. UM4b, the most different within CF1 (ANI 99.92/Coverage 95.66% to DE1), appears to have suffered genome reduction, losing the plasmid pAMDE1-300 and ca. 200 kb of the chromosome (including the lysogenic phage). However, only 43 of the SNPs detected in this strain could be ascribed to mutation, indicating a similar divergence time with the other strains. The ratios of SNPs attributable to synonymous and nonsynonymous replacements (table 1) were similar for all of the CF1 strains, ca. 25% were located in intergenic regions. These figures support a recent divergence that has not allowed time for selection to act. To translate mutational SNP values into divergence time, we have used the evolutionary rate of the nonrecombining core genome of Legionella pneumophila (Sanchez-Buso et al. 2014), another aquatic gammaproteobacterium, as reference. We calculated that ca. 80 years (table 1) have elapsed since the last common ancestor of CF1 strains.
Similar figures were found for CF2, the two strains U4 and U8 core genome revealed 109 mutational SNPs in coding regions, of which 49 (45%) were nonsynonymous (table 1). These two strains should be separated by ca. 196 years since their last common ancestor (table 1). A careful examination of the variable sites within members of the same CF indicated that most could be the product of intragenomic recombination during genome replication, typically involving tandem repeats of different lengths, gene duplications and the movement of transposable elements.
Intraspecies
When the comparison was shifted to strains belonging to different CFs within the same species, clonal fractions decreased to 0.624–0.040 as ΔS varied from nearly 0 to 1.66% (table 1). Besides, the total number of SNPs over the core genome varied from a few hundred to a range from 13,461 (LMG 21861-Mac1) to 66,301 (ATCC 27126-AD45). We aligned the core of four genome pairs, one for each Alteromonas genospecies, covering the whole range of intraspecies ANI values (supplementary fig. S3, Supplementary Material online). We plotted the SNP density (in 1 kb segments) (Dixit et al. 2015) for each genospecies pair (supplementary fig. S3, Supplementary Material online). They showed a similar distribution to those found when comparing different strains of E. coli (Dixit et al. 2015), having a maximum at the average frequency of mutational changes per kb, one for pairs within “A. stellipolaris” (LMG 21861-R10SW13) and A. mediterranea (DE1-U4); four for A. australica (H17-DE170) and nine for A. macleodii (ATCC 27126-AD45), respectively. Recently, it has been proposed for E. coli (Dixit et al. 2015) that clonal segments disappear at levels of divergence at or above 1 SNP/kb. Accepting this premise, in the pairs ATCC 27126-AD45 (ANI 96.75) and H17-DE170 (ANI 98.63), all clonal vertically-inherited parts are lost (supplementary fig. S3, Supplementary Material online). Interestingly this happens at about the same overall divergence (ANI 98.8) as in E. coli (Dixit et al. 2015). The most divergent intraspecies pairs (<98% ANI) also followed a Poisson distribution with peak values >1 SNP/kb (supplementary fig. S3, Supplementary Material online). Although after the previously mentioned model (Dixit et al. 2015), these genomes are likely to be completely covered by a mosaic of recombined segments (ΔM = 0%; ΔR = 100%), we posit that fragments with small amounts of SNPs (located to the left of the peak) could be due to mutation and the rest (to the right of the peak) to recombination. If this assumption is correct, ΔR varied from 93% (LMG21861-R10SW13, ANI 98.8%) to 37% (ATCC 27126-AD45, ANI 96.75) (table 1). These values are similar to those found for intraspecies recombination in other Gammaproteobacteria (Vos and Didelot 2009).
In order to analyze the impact of homologous recombination in the evolution of the core genome, we estimated the r/m, parameter that measures the relative weight of recombination to mutation in sequence divergence (table 1). Results showed that for all intraclonalframe and most intraspecies comparisons r/m was >1, i.e., recombination introduced more substitutions than mutation, with the highest value observed between U4 and U8 members of the CF2 (table 1). However, in the two most divergent intraspecies pairs (H17-DE170 and ATCC 27126-AD45), r/m was significantly <1 (more mutation than recombination). These data suggest that recombination rates decrease proportionally to ANI values.
The last common ancestor of the different CFs within each species calculated from ΔM values, as above, was between 4,000 and 74,000 years old (table 1). Annotation of the segments with the highest numbers of SNPs attributed to recombination, showed enrichment in proteins neighboring the replacement fGIs (supplementary fig. S3, Supplementary Material online), known to be frequent targets of recombination (López-Pérez et al. 2013). The same kind of genes has been found to be highly recombinogenic in E. coli (Dixit et al. 2015). Other genes overrepresented in the high recombination tails coded for proteins that are exposed in the outermost cell structures like TonB receptors, porins or pillins. They might also be involved in environmental or phage interaction.
To gauge the selection pressure acting on these strain pairs, we estimated the ratio of nonsynonymous to synonymous substitution rates dN/dS. The values for all the intraspecies comparisons (table 1) were very similar, with a mean of 0.69 ± 0.04, indicating weak stabilizing selection, similar values were obtained for different serovars of the bacterial pathogen Salmonella enterica Agona (Zhou et al. 2013), Typhi (Holt et al. 2008) and Typhimurium (Hawkey et al. 2013) and other pathogenic bacteria. We also examined the dN/dS variation throughout the genome. A whole genome alignment was constructed with the five A. mediterranea genomes belonging to different CFs partitioning the alignment into 10 kb windows. This process generated 348 fragments containing 3.4 Mb of core aligned nucleotides. Among these fragments, 81 (23%) had higher dN than dS. Similar values were obtained for the strains within A. macleodii. Supplementary table S2, Supplementary Material online, lists the genes with the highest dN/dS for A. mediterranea strains. Among the 20 genes associated with highest dN/dS values eight were transporters of different kinds, including TonB receptors. This might reflect a divergence in nutrient ranges utilized by different strains within the same species.
Interspecies
The cores of strains belonging to different genospecies were much more divergent, A. macleodii and A. mediterranea with an ANI ca. 84% had ΔS values ca. 9.2% while A. australica and “A. stellipolaris” (ANI ca. 73%) 13% (table 1). Despite the 10-fold increase in ΔS compared with the intraspecies values, the dN/dS ratio was similar, 0.65 for the first group and ca. 0.72 for the more distant genospecies. Thus, it appears that purifying selection is already saturated at the intraspecies level. The variation of the SNP density plots with increasing ANI distance is shown in fig. 2, from CFs to interspecies, using as reference the genome of A. mediterranea DE1. It is apparent that recombination is much rarer between species (ΔR ca. 5% compared with ca. 60% within species). This reduction has been described in other Gammaproteobacteria such as Vibrio (Urbanczyk et al. 2015). Given that for the most divergent pairs the model predicting recombination from SNP density is not applicable (Dixit et al. 2015), to asses recombination rates at the interspecies level, we generated ML trees of different alignable genomic regions and compared them to the consensus tree generated by the core genome of all the strains (supplementary fig. S4, Supplementary Material online). The results illustrate that recombination events often break the topology within the species but not between species. All these analysis support the decline in homologous recombination between increasingly divergent genomes that, as has been suggested before, might lead to prokaryotic speciation (Shapiro et al. 2012).
Fig. 2.—
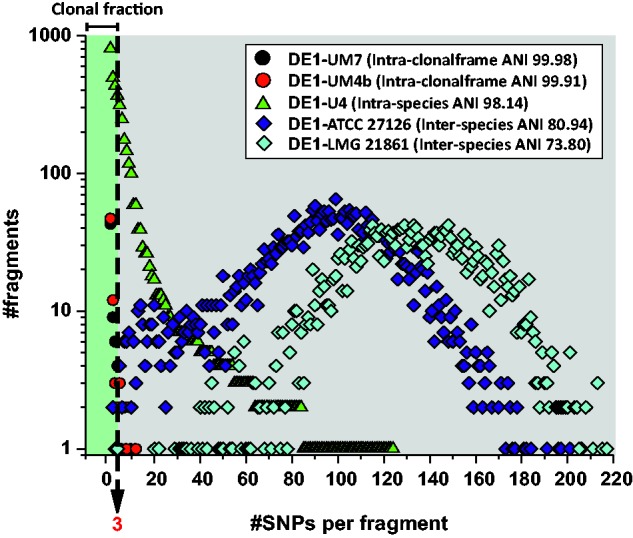
SNP density in homologous 1 kb fragments derived from pairwise genome comparisons using A. mediterranea DE1 as a reference. The analysis has been applied to strains with increasing levels of divergence, from strains belonging to the same clonal frame, to intraspecies (but belonging to different clonal frames), and interspecies. Different symbols are color coded for visibility.
Flexible Genome
Flexible Genome Expansion
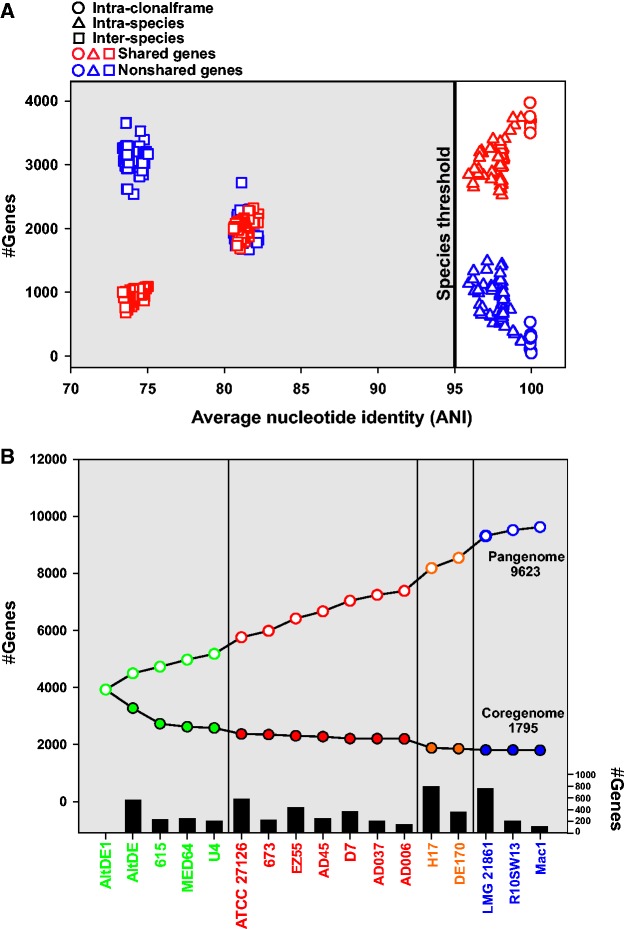
The pairwise variation in shared genes among all the Alteromonas strains (all vs. all) is shown in fig. 3. The shared gene clusters (core) and differential (flexible) for all couples of strains have reciprocal values, as could be expected given that the genome size is relatively constant. In addition, as expected, both values were proportional to the ANI values, with the number of common clusters increasing, and the numbers of not shared clusters decreasing, at higher ANI values (fig. 3A). It is remarkable that most of the expansion of the flexible genome takes place within the species range of variation, i.e., some pairs belonging to the same species have as many different genes as pairs of strains belonging to different species. Furthermore, even when comparing pairs belonging to the same CF the numbers of nonshared genes was quite significant (in the most extreme values close to 500). Figure 3B plot shows the incremental change of core and flexible with each of the genomes added. Typically each new strain within the species added 300 new genes while the species change added between 500 and 700. Together all the strains (considering only one member of each CF) were shown to possess a pangenome of 9,623 genes (1,795 core and 7,828 flexible) (fig. 3B).
Fig. 3.—
Pairwise gene pool variation (shared and nonshared genes) among Alteromonas strains. (A) Number of genes shared by pairs of strains with different core ANI values. Pairwise comparisons among members of the same clonal frame, species or different species are indicated by circles, triangles or squares. Numbers of gene clusters shared are indicated in red and not shared in blue symbols. (B) Pangenome (empty circles) and core genome (full circles) plots of the Alteromonas strains analyzed. Different colors indicate different species. Black bars indicate the number of new genes contributed by each strain.
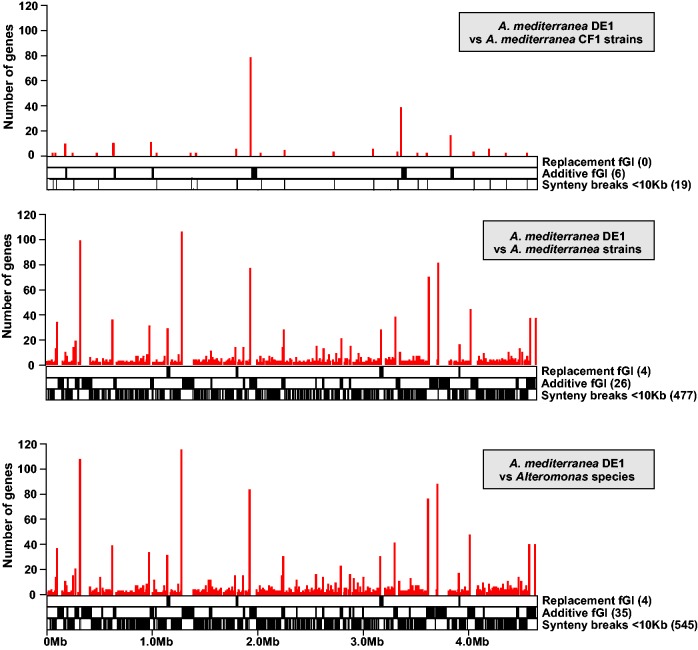
We have analyzed the locations where synteny was lost across the genomes due to gene acquisition and loss for the two species of Alteromonas with a significant number of strains each (A. macleodii and A. mediterranea). Using as reference strain DE1 of A. mediterranea, we identified only 25 synteny breaks when its genome was compared with the five strains belonging to the same CF (fig. 4, upper panel). By contrast, when compared with the other five CFs within A. mediterranea, the number of synteny breaks increased to 507 (fig. 4, middle panel). When the comparison was extended to all the other strains in the genus (including the other species) the number of synteny breaks increased only to 584 (fig. 4, lower panel). It seems that the sites for synteny breaks were largely exhausted within the species boundaries.
Fig. 4.—
Synteny breaks for Alteromonas using A. mediterranea DE1 as a reference and comparing to genomes with increasing levels of divergence. Upper panel indicates synteny breaks within clonal frame 1, middle panel within A. mediterranea species and lower panel considering all the other strains in the genus. Vertical lines indicate gaps in the reference genome for which synteny is lost in any of the other genomes. Height of the line indicates the number of genes present in the reference strain that are not present in any of the others.
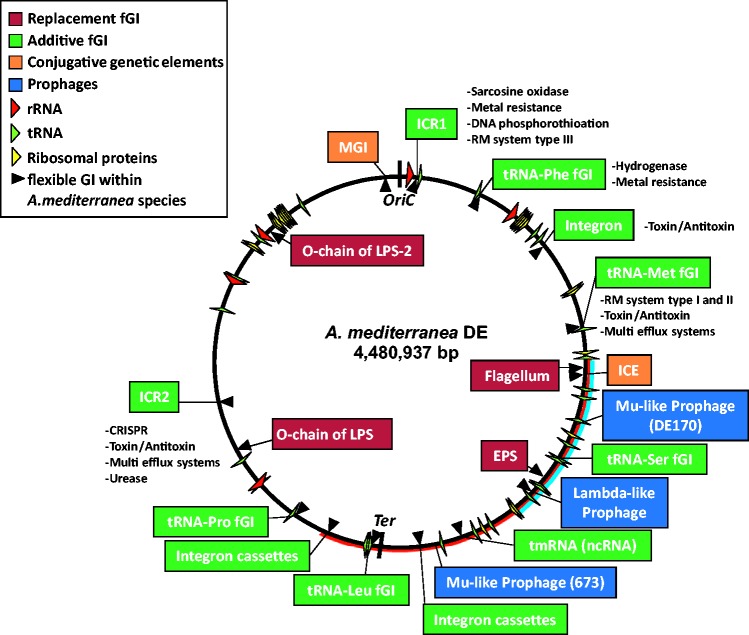
Thus, the real qualitative jump in the flexible genome diversity in Alteromonas happens at the intraspecies level. We found a similar number of syntheny breaks (494) for the seven strains in A. macleodii (supplementary fig. S5, Supplementary Material online). These breaks contain 1,783 and 1,496 genes in A. mediterranea and A. macleodii, respectively. Synteny breaks involving segments of <10 kb were ca. 93% of the total number of synteny breaks in both species, but they contained only around half of the flexible genes (49% and 59%, respectively). The other part of the flexible genome was present in segments >10 kb (largely fGIs), 30 in A. mediterranea and 34 in A. macleodii. The biological role of the larger synteny breaks and their relative location appeared conserved throughout the genus (fig. 5).
Fig. 5.—
A schematic representation of the fGIs detected in all the strains described here using the genome of A. mediterranea DE1 as a reference. Those already present within the A. mediterranea species are labelled with a triangle pointing to the insertion site. Red and blue lines indicate the position of the rearrangement and the main inversion present in all the “A. stellipolaris” isolates. Each location is characterized by a feature found at the site tRNA, Integron, ICR1 immigration control region 1; ICR2 immigration control region 2, MGI mobilizable genetic island, ICE Integrative Conjugative Element. Some of the functions detected in the additive fGIs are indicated to the right or under the colored box.
Exchange of Alteromonas Glycotype Gene Clusters
Figure 1 shows the location and different versions (identified by a number) of the four replacement fGIs (glycotype gene clusters) described in Alteromonas (Gonzaga et al. 2012; López-Pérez et al. 2012, 2013). Each different CF had a totally different set of these islands. The exceptions are highlighted in red in fig. 1. The case of “A. stellipolaris” can be considered special because the flagellum glycosylation island and the O-chain main gene cluster were highly similar although the five strains belong to three different CFs. However, the EPS and the O-chain cluster 2 were different for each CF as is normally the case, their relationship at ANI level was also atypical when compared with the other species with intermediate values between the intraclonalframe and intraspecies level.
It is known that replacement fGIs are exchanged by homologous recombination (Jeong et al. 2009; López-Pérez et al. 2013; López-Pérez, Martin-Cuadrado, et al. 2014). We already had detected exchange between different CFs (a nearly identical version of the flagellum glycosylation island was found in two different strains within A. mediterranea) (López-Pérez et al. 2013). In the collection of strains presented here, more examples of exchange between different CFs, and even species, could be detected (fig. 1). It is important to emphasize, however, that recombination of glycotype determinants in Alteromonas happens at a much lower rate than the exchange of ecologically relevant characters found in additive islands (López-Pérez et al. 2013; López-Pérez, Martin-Cuadrado, et al. 2014). Firstly, in CF1 of A. mediterranea the gene clusters were identical, i.e., likely to remain vertically transmitted, although the strains have diverged for a large number of generations (see above), and were pretty divergent at the level of other components of the flexible genome. Secondly, the frequency of SNPs when comparing the putatively exchanged gene clusters indicates that thousands of generations elapse between these homologous recombination events of exchange. For example, in A. macleodii, the strain pairs 673-AD006 and AD45-AD037 share the same flagellum cluster but with 116 SNPs over 18.4 kb and 926 SNPs over 28.8 kb, respectively. These values are indicative of old exchanges.
Although the most frequently exchanged fGI was the flagellum glycosylation cluster, the members of A. mediterranea CF2 and strain A. macleodii 673 share the main LPS O-chain cluster (fig. 1). In this case, the similarity between the common (putatively exchanged) fGI was relatively low (ca. 95%), but much higher than the average of the two genomes (ca. 81%) what is suggestive of an ancient transfer and significantly posterior to the split of the two species. E. coli and S. enterica have been shown to share O-antigens, but again the genes show low similarity consistent with ancient transfers (Reeves 1993). These examples illustrate the way in which cells can diverge in glycotype by replacing the gene clusters that code for the biosynthesis of these exposed structures. Milkman and co-workers suggested that the enormous diversity of O-antigens in E. coli derives from the capture of exotic clusters coming from distantly related microbes in “isolated and improbable exchanges” (Milkman 1997). This idea of “retransmission” of exotic gene clusters within a species was justified by the drive of escaping host immunity, i.e., they provide antigenic novelty that would be strongly selected for and will be preserved in the species. Along these lines, by searching with all the glycotype gene clusters of Alteromonas, we have found examples of syntenic clusters (albeit at very low similarity) in distantly related microbes. Specifically, a similar O-chain cluster (ca. 60%) to the one of A. macleodii EZ55 was found in Catenovulum agarivorans YM01, an Alteromonaceae isolate from the Yellow Sea (Shi et al. 2012) (supplementary fig. S6, Supplementary Material online). We have found also that the flagellum glycosylation cluster of A. australica H17 have a syntenic cluster in Glaciecola polaris LMG 21857 (supplementary fig. S6, Supplementary Material online). These examples illustrate that glycotype clusters are recruited from a pool of microbes of taxonomic diversity at least at the rank of family. However, it is important to emphasize that Alteromonas is a free-living microbe never exposed to any immune system.
Additive fGIs
Most synteny breaks correspond to additive fGIs that contain different gene cassette combinations, although their functions are sometimes related (fig. 5). The simplest fGI (and also the most common) are integration hotspots, most (6 out of 8) have tRNA and tmRNA genes at the insertion site. The additions of some cassettes were still hall marked by the tell-tale repeat of part of the tRNA gene downstream of the inserted cassette (direct repeat). Nearly all the strains have different versions of these fGIs except for those belonging to the same CF, and even those sometimes differed by several cassettes (López-Pérez et al. 2013). The presence of different cassettes at the same location within the same CF attests to their change in short evolutionary times. The number of additive fGIs was significantly higher in the right replichore (fig. 5) that is also enriched in tRNA genes.
Although most of the fGIs are already detectable at the intraspecies level (fig. 5), a few novel hotspots were detected by interspecies comparison, specifically at the tRNA-Leu and tRNA-Ser. Their function could not be elucidated from annotation (mostly hypothetical proteins). The average number of tRNA genes in the different species was about 70 but the number of tRNA genes used as insertional hotspots was much smaller (fig. 5). Within the Alteromonas genomes the most frequently targeted tRNA genes were tRNA-Phe and tRNA-Met (18 strains), followed by tRNA-Leu (11 strains), tRNA-Pro (9 strains), tmRNA (9 strains) and tRNA-Ser (6 strains).
Additive cassettes were obviously acquired by HGT as shown by their disconnection to the core phylogeny, i.e., the number and order of the shared cassettes is not dependent on the phylogenetic position of the strain, even when this phylogeny indicates very close last common ancestor (see below). Actually, the most remarkable finding when comparing additive fGIs of different species was the frequent detection of nearly identical cassettes. Figure 6 shows an example of cassette conservation in the heavy metal resistance additive fGI that is associated with a tRNA-Phe gene (López-Pérez et al. 2012). In A. mediterranea DE, this island contains genes coding for metal resistance (clusters of czcABC genes and a copper resistance operon) and a hydrogenase. The cassettes were present or absent in strains of the different genospecies with no apparent relationship with the phylogeny of the strain. Furthermore, when cassettes were shared they were always nearly identical. Actually, we found syntenic collections of cassettes related to this cluster, including the Czc and the hydrogenase cluster, in other genera within the family (Glaciecola mesophila KMM 241 and Glaciecola sp. 4H-3-7 + YE-5 plasmid), although in these cases the similarity was much lower. Another example is the postsegregational killing system zeta toxin found in the tRNA-Met (fig. 5). This zeta toxin system was found with low identity in Pseudoalteromonas haloplanktis inserted in the same tRNA. While the core and the replacement fGIs appear to drift apart, additive islands can show very high similarities, likely as a reflection of their fast turnover in genomes of members of the same genus.
Fig. 6.—
Schematic representation of the tRNA-Phe fGI gene cassettes found in different strains representatives of different species of Alteromonas. The conservation of some cassettes present in widely divergent genomic backgounds is apparent. Some cassettes were found still syntenic but at lower similarity in members of the genus Glaciecola.
Conjugative genomic elements such as the Integrative and Conjugative Elements (ICEs) or the mobilizable genomic island (MGI) (Daccord et al. 2013; López-Pérez et al. 2013) can be considered additive fGIs, and always shared the same chromosomal integration site, but their presence was much more sporadic. Lysogenic phages appear to be the only genomic elements that appeared delocalized, i.e., different insertion sites depending on the species. Thus, a Mu-like prophage (fig. 5) appears at different integration sites in A. australica DE170 and A. macleodii 673. The presence of conjugative plasmids has also been previously described (Lopez-Perez et al. 2013) and follows a similar core-genome disconnected presence.
Discussion
Intraspecies Diversity is Key to Bacterial Evolution
The main change in pace in genomic variation appeared at the level of different clones or strains within the same species, i.e., over the 95% ANI threshold. These strains show strong evidence of recombination along the core and present multiple variations in their flexible genomes. They have different glycotype determinants, which provide them with different surface properties and different targets for phage recognition. In Alteromonas, a few examples of shared glycotype islands reveal how these critical gene clusters are exchanged with other clonal lineages sometimes outside of the species boundaries. However, as shown by their stability within CFs, the exchange of these islands is rare enough to keep their linkage with the main determinants of the lineage ecological properties, at least within ecological time frames (López-Pérez et al. 2013; Kashtan et al. 2014), preventing their sweep across populations as some authors have claimed (Cordero and Polz 2014; Takeuchi et al. 2015). The exchange between species appeared to be older (containing more SNPs) than those within species indicating that these exchanges are probably even much less frequent, but they might be essential to renew the stock of glycosydic receptors available within the species. This phenomenon has been known for long in E. coli and Salmonella (Reeves 1993) and might be relevant for antigenic variation in pathogens with large environmental reservoirs such as Vibrio cholerae (Mutreja et al. 2011). Additive fGIs were the most obvious drivers of intraspecies diversity and represent the largest part of the flexible genome. Actually, their gene cassettes seem to flow rapidly through taxonomic entities of at least the rank of families and provide adaptive traits that make each CF different at multiple levels. Those include, metabolism (transporters, degradation pathways, efflux pumps, porins), environmental sensing (two component systems) protection from abiotic (metal resistance) and biotic factors (antibiotic production and resistance, CRISPR, toxin-antitoxin, restriction modification systems) (Gonzaga et al. 2012; López-Pérez et al. 2012, 2013; López-Pérez, Gonzaga, et al. 2014).
By contrast, the comparison of genomes belonging to different species did not bring much more diversity into the picture. Actually, the numbers of new genes found when comparing two strains of A. mediterranea and A. macleodii did not increase significantly over the figures found among CFs of each species. Even dN/dS values increased little, indicating that positive (Darwinian) selection did not increase when considering different species, i.e., there seemed to be little increase in the adaptive evolution of the core. The main difference found in the comparison between strains of different species was that recombination along the core seemed to be much less prevalent. This supports a decline in gene flow correlated with core genome divergence that, as has been suggested before, might lead to prokaryotic speciation (Shapiro et al. 2012). However, it is important to emphasize that the exchange of additive cassettes must happen frequently among different species, as shown by the detection of identical cassettes in distantly related ones. This proves that there is no genetic isolation of the different genospecies.
A decrease in recombination was observed when analyzing the genomes of Vibrio cyprinolyticus associated to different particle sizes (Shapiro et al. 2012). Something similar could be taking place in Alteromonas and actually, the description of the genomes of A. mediterranea DE and A. macleodii ATCC 27126 revealed differences consistent with adaptation to particles of different size. Alteromonas is a typical bloomer that relies on fast growth rates to compete in marine oligotrophic waters, which probably restricts its habitat to the relatively nutrient rich and extremely diverse realm of particulate organic matter. In fact, typically pelagic free-living microbes such as Prochlorococcus, Ca. Pelagibacter or the ammonia oxidizing archaea seem to have less species diversity (Penno et al. 2006; Gilbert et al. 2008; Hatzenpichler 2012).
Niche separation would lead to different phenotypic traits that would fit the classic polyphasic definition of bacterial species (taking into account all available phenotypic and genotypic parameters) (Ramasamy et al. 2014). Another factor to consider would be the barriers to recombination and exchange derived from the decrease in co-infecting phages, i.e., much fewer phage lineages can infect members of two separate species (Dixit et al. 2015) as a consequence of the divergence and lower exchange rates of replacement fGIs coding for cell surface phage targets.
A Model for Pangenome Expansion, from Clones to Species
The ecotype speciation theory sustains that each ecotype exploits a specific niche. Genetic heterogeneity should be purged from the population by periodic selective sweeps (Cohan 2002; Lassalle et al. 2015). However, different lines of evidence from metagenomics, single cell genomics and culture of concurrent clones indicate a large diversity in different populations of aquatic microbes (Gonzaga et al. 2012; Kashtan et al. 2014; Bendall et al. 2016). We would like to posit that the survival of a single clone in nature is unlikely because the restricted genetic information carried by one prokaryotic cell might not be enough to allow the population to adjust efficiently to environmental changes, even in one single niche (e.g., particulate matter in the ocean water column). In addition, a monoclonal population could be preyed to extinction by specific phages (Leggett et al. 2013). On the other hand, the coexistence of several clones with different ecological attributes not only dilutes the selective pressure of phages, it can be good for the population as a whole that can exploit resources more efficiently and share the load of interaction with stressful environmental factors as suggested by the Black Queen hypothesis (Morris et al. 2012). This equilibrium can be maintained with a complement of phages that prevent any single clone from replacing the others (a clonal sweep or periodic selection event). This model was proposed as “constant-diversity” dynamics (Rodriguez-Valera et al. 2009) and the metaclonal population including the phages proposed to be a single selection unit (Rodriguez-Valera et al. 2009; Rodriguez-Valera and Ussery 2012).
With these hypotheses in mind and with our set of strain genomes, it is possible to visualize how this high clonal diversity could be established (fig. 7). As a clone multiplies asexually, it diverges at the level of SNPs as both mutation and recombination take place along the core. At the same time, genomes incorporate different additive cassettes received from other microbes by HGT. This way clonal lineages start to diverge and acquire different traits. We can see how this is starting to happen in the populations represented by the five isolates in CF1. However, as long as they have the same replacement genomic islands (belonging to a single glycotype), phages prey similarly on all the cells, and there is the risk that a clonal sweep or a massive lytic event will make the incipient clone extinct. On the other hand, if one clone manages to persist for long enough, it will eventually acquire a different set of replacement fGIs (phage receptors) becoming a different glycotype, as seems to be starting to happen in strain U4 (fig. 1). The new glycotype with the cognate set of phages (that could be transplanted from the same origin as the glycotype determinant), will then become integrated in the multi-clonal population. Incidentally, phage receptor swapping could increase recombination rates by originating phages with broad infection range that can transfer genes between more different genomic backgrounds. Understanding the nuts and bolts of prokaryotic evolution will require profound changes in our way to think about population genetics and evolution at large. Comparative genomics provides a wonderful tool to start unraveling its intricacy.
Fig. 7.—
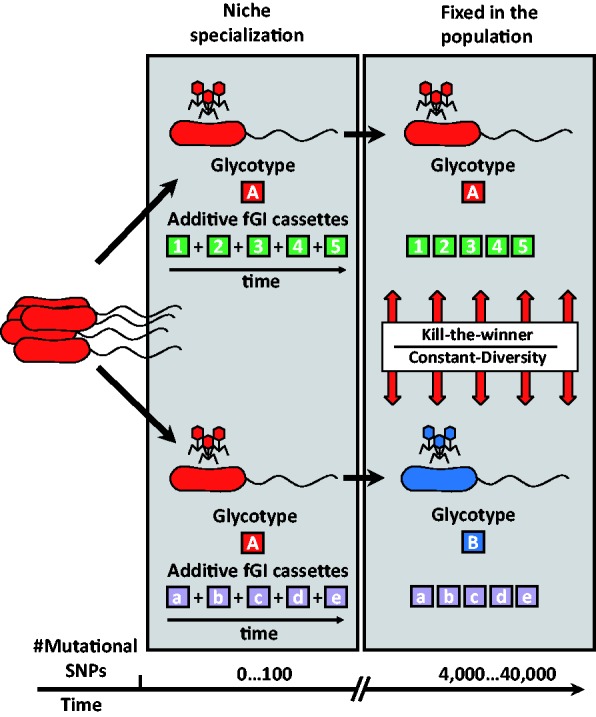
A model for the evolution of the Alteromonas pangenome at the intraspecies level. The figure depicts two clonal lineages that start diverging by accumulation of different cassettes in additive fGIs. The number of different cassettes added to the recombination hot-spot increases with time, they appear represented by letters or numbers. The new flexible genes can modify the clone’s lifestyle. However, as long as both clonal frames belong to the same glycotype (A), they can be subject to a clonal sweep that would reduce diversity. Acquisition of a different replacement fGI by one of the clones, indicated by a change of color, leads to a new glycotype (B) with different phage susceptibility. Then both clones can coexist indefinitely thanks to a predation dependent dynamics (Rodriguez-Valera et al. 2009).
Supplementary Material
Supplementary tables S1 and S2 and figs. S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We are grateful for the suggestions to improve the manuscript of some of the referees during the review process. This work was supported by projects MEDIMAX BFPU2013-48007-P from the Spanish Ministerio de Economía y Competitividad, MaCuMBA Project 311975 of the European Commission FP7 and PROMETEO II/2014/012 project AQUAMET from the Generalitat Valenciana. Strain D7 was kindly provided by Professor Åke Hagström (Linnaeus University, Sweden).
Literature Cited
- Avrani S, Schwartz DA, Lindell D. 2012. Virus-host swinging party in the oceans. Mob Genet Elem. 2:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall ML, et al. 2016. Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. doi: 10.1038/ismej.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. [DOI] [PubMed] [Google Scholar]
- Chan AP, et al. 2015. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 16:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y-J, Yi H, Lee JH, Kim DW, Chun J. 2010. Genomic evolution of Vibrio cholerae. Curr Opin Microbiol. 13:646–651. [DOI] [PubMed] [Google Scholar]
- Cohan FM. 2002. Sexual isolation and speciation in bacteria. Genetica 116:359–370. [PubMed] [Google Scholar]
- Cordero OX, Polz MF. 2014. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol. 12:263–273. [DOI] [PubMed] [Google Scholar]
- Cui Y, et al. 2015. Epidemic clones, oceanic gene pools and eco-LD in the free living marine pathogen Vibrio parahaemolyticus. Mol Biol Evol. 32(6):1396–410 [DOI] [PubMed] [Google Scholar]
- Daccord A, Ceccarelli D, Rodrigue S, Burrus V. 2013. Comparative analysis of mobilizable genomic islands. J Bacteriol. 195:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D. 2010. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186:1435–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, et al. 2012. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large scale whole genome sequencing. J Infect Dis. jis734.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit PD, Pang TY, Studier FW, Maslov S. 2015. Recombinant transfer in the basic genome of Escherichia coli. Proc Natl Acad Sci. 112:9070–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Mühling M, Joint I. 2008. A rare SAR11 fosmid clone confirming genetic variability in the ‘Candidatus Pelagibacter ubique’ genome. ISME J. 2:790–793. [DOI] [PubMed] [Google Scholar]
- Gonzaga A, et al. 2012. Polyclonality of concurrent natural populations of Alteromonas macleodii. Genome Biol Evol. 4:1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R. 2012. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol. 78:7501–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey J, et al. 2013. Evidence of microevolution of Salmonella typhimurium during a series of egg-associated outbreaks linked to a single chicken farm. BMC Genomics 14:800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, et al. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat Genet. 40:987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EP, et al. 2005. Alteromonas addita sp. nov. Int J Syst Evol Microbiol. 55:1065–1068. [DOI] [PubMed] [Google Scholar]
- Ivanova EP, et al. 2015. Ecophysiological diversity of a novel member of the genus Alteromonas, and description of Alteromonas mediterranea sp. nov. Antonie Van Leeuwenhoek 107:119–132. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21 (DE3). J Mol Biol. 394:644–652. [DOI] [PubMed] [Google Scholar]
- Kashtan N, et al. 2014. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 102:2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. 2012. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Frontiers in Cellular and Infection Microbiology 2:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B. 2000. HIV Signature and Sequence Variation Analysis. In: Rodrigo AG, Learn GH, editors. Computational Analysis of HIV Molecular Sequences, Chapter 4. Dordrecht (The Netherlands): Kluwer Academic Publishers. [Google Scholar]
- Kuenne C, et al. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, Muller D, Nesme X. 2015. Ecological speciation in bacteria: reverse ecology approaches reveal the adaptive part of bacterial cladogenesis. Res Microbiol. 166(10):729–741. [DOI] [PubMed] [Google Scholar]
- Leggett HC, Buckling A, Long GH, Boots M. 2013. Generalism and the evolution of parasite virulence. Trends Ecol Evol. 28:592–596. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- López-Pérez M, et al. 2012. Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep. 2:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pérez M, Gonzaga A, Ivanova EP, Rodriguez-Valera F. 2014. Genomes of Alteromonas australica, a world apart. BMC Genomics 15:483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pérez M, Gonzaga A, Rodriguez-Valera F. 2013. Genomic diversity of “deep ecotype” Alteromonas macleodii isolates: evidence for Pan-Mediterranean clonal frames. Genome Biol Evol. 5:1220–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pérez M, Martin-Cuadrado A-B, Rodriguez-Valera F. 2014. Homologous recombination is involved in the diversity of replacement flexible genomic islands in aquatic prokaryotes. Front Genet. 5:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J. 1990. Did Darwin Get it right? Essays on Games, sex and evolution. American Journal of Physical Anthropology 81:456–457. [Google Scholar]
- Milkman R. 1997. Recombination and population structure in Escherichia coli. Genetics 146:745.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R, Jaeger E, McBride RD. 2003. Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics 163:475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3:e00036–e00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja A, et al. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Pospiech A, Schairer HU. 1992. Rapid isolation of genomic DNA from Gram-negative bacteria. Trends Genet. 8:332–333. [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu S-M, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. [DOI] [PubMed] [Google Scholar]
- Penno S, Lindell D, Post AF. 2006. Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N‐regulatory gene ntcA. Environ Microbiol. 8:1200–1211. [DOI] [PubMed] [Google Scholar]
- Polz MF, Alm EJ, Hanage WP. 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 29:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy D, et al. 2014. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 64:384–391. [DOI] [PubMed] [Google Scholar]
- Reeves P. 1993. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 9:17–22. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 106:19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, et al. 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 7:828–836. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Ussery DW. 2012. Is the pan-genome also a pan-selectome?. F1000Research 1:16. [DOI] [PMC free article] [PubMed]
- Sanchez-Buso L, Comas I, Jorques G, Gonzalez-Candelas F. 2014. Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat Genet. 46:1205–1211. [DOI] [PubMed] [Google Scholar]
- Shapiro BJ, et al. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Yu M, Yan S, Dong S, Zhang X-H. 2012. Genome sequence of the thermostable-agarase-producing marine bacterium Catenovulum agarivorans YM01T, which reveals the presence of a series of agarase-encoding genes. J Bacteriol. 194:5484–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Cordero O, Koonin E, Kaneko K. 2015. Gene-specific selective sweeps in bacteria and archaea caused by negative frequency-dependent selection. BMC Biol. 13:20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 11:472–477. [DOI] [PubMed] [Google Scholar]
- Urbanczyk H, Ogura Y, Hayashi T. 2015. Contrasting inter-and intraspecies recombination patterns in the “Harveyi Clade” vibrio collected over large spatial and temporal scales. Genome Biol Evol. 7:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Trappen S, Tan T-L, Yang J, Mergaert J, Swings J. 2004. Alteromonas stellipolaris sp. nov., a novel, budding, prosthecate bacterium from Antarctic seas, and emended description of the genus Alteromonas. Int J Syst Evol Microbiol. 54:1157–1163. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199–208. [DOI] [PubMed] [Google Scholar]
- Welch R, et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci. 99:17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 35:957–976. [DOI] [PubMed] [Google Scholar]
- Zhou Z, et al. 2013. Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet. 9:e1003471.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.