Abstract
The transduction of biological signals often involves structural rearrangements of proteins in response to input signals, which leads to functional outputs. This review discusses the role of regulated partial and complete protein unfolding as a mechanism of controlling protein function and the prevalence of this regulatory mechanism in signal transduction pathways. The principles of regulated unfolding, the stimuli that trigger unfolding, and the coupling of unfolding with other well characterized regulatory mechanism are discussed.
Keywords: regulated unfolding, signal transduction, protein dynamics, unfolding trigger, alternative protein fold, NMR spectroscopy
Introduction
Proteins are the work horses of biological systems, performing a plethora of tasks, including chemical catalysis, signal transmission, molecular transportation, cellular movement and forming the structural framework of cells and tissues. Protein function is dictated by the primary amino acid sequence which, in turn, determines the three-dimensional organization and dynamic behavior of proteins. Through evolution, proteins have achieved a fine balance between thermodynamic stability and dynamic fluctuations to optimally perform their biological functions in the environmental setting of their host [1]. It has long been understood that the three dimensional structure of a protein determines its function. Growing evidence, however, establishes the pervasive roles of disorder and dynamics in mechanisms of protein function [2–6]. In fact, nearly a third of all proteins, in all kingdoms of life, contain disordered regions of at least 30 amino acids [7]. Disorder is manifested in different ways, from short, flexible linkers and long “random coil-like” disordered segments to compact but disordered domains and whole proteins termed intrinsically disordered proteins (IDPs) [8]. Structural flexibility and disorder mediates critical biological functions; consequently, these dynamic features are often evolutionarily conserved [9, 10]. A noteworthy example is the topologically conserved activation loop in kinases [11]. In the inactive state of Serine/Threonine and Tyrosine kinases (e.g. PKA, IRK) the flexible loop is collapsed on the active site, preventing substrate binding. An evolutionary conserved kinase activation mechanism involves phosphorylation of this loop, which results in (i) stabilization of an open conformation, and (ii) rearrangement of key catalytic residues, enabling substrate binding and phospho-transfer, respectively [11]. Classic allostery, which mediates signal transduction through the tertiary and quaternary structure of proteins (e.g., hemoglobin, receptor tyrosine kinases), causes structural rearrangements in one functional domain or subunit in response to ligand binding within a distal domain/subunit of the same protein [12]. This regulatory mechanism depends upon the ability of whole proteins or domains to fluctuate between different defined conformations to regulate function. However, accumulating evidence shows that partial or complete protein unfolding is also utilized as a mechanism of regulating function, particularly in signal transduction pathways. Here we introduce the concept of regulated unfolding as a protein regulatory mechanism, provide illustrative examples, and discuss its future implications.
Protein unfolding as a type of signaling output
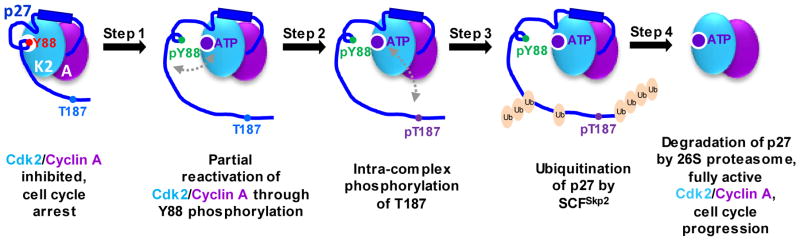
Signaling mechanisms often involve posttranslational modifications and/or protein-ligand (e.g. protein, nucleic acids, lipid, etc.) interactions that couple an upstream input to a conformational change, which alters function and produces a downstream signal. The extent of the conformational change ranges from subtle, local unfolding events to full unfolding of protein domains. For example, the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 (p27) regulates progression through the cell division cycle by interacting with and inhibiting several Cdk/cyclin complexes in the nucleus [13]. Cell cycle progression to S phase is characterized by rapid turnover of p27 via the proteasome pathway, a fate which is signaled by phosphorylation of p27 on Thr187 [14]. Counter intuitively, this posttranslational modification is performed by the Cdk/cyclin complexes for which p27 is a potent inhibitor [14, 15]. Grimmler, et al. [14], demonstrated that non-receptor tyrosine kinases phosphorylate Tyr88 of p27, a residue which occupies the active site of Cdk2 [16]. This modification causes an inhibitory 310 helix containing Tyr88 to be ejected from the ATP binding pocket of Cdk2, partially restoring kinase activity. Intrinsic flexibility of the C-terminal domain of p27 allows Thr187 to fluctuate into close proximity to the Cdk active site and become phosphorylated, creating a phosphodegron that leads to selective p27 ubiquitination and degradation, and ultimately full activation of Cdk/cyclin complexes (Figure 1). Regulated partial unfolding of the inhibitory conformation of p27 through tyrosine phosphorylation triggers this signaling cascade that ultimately drives progression of cells into S phase of the division cycle.
Figure 1.
p27 as a signaling conduit. Tyrosine phosphorylation-dependent partial unfolding of p27 triggers signal propagation through the length of the protein and regulates its degradation. Step 1 involves phosphorylation of Y88 of p27 that is bound to Cdk/cyclin complexes [Cdk2 (K2)/cyclin A (A) here] by non-receptor tyrosine kinases such as BCR-ABL, Src, Lyn, and Jak2, which ejects Y88 from the ATP binding pocket of Cdk2 and restores partial kinase activity. Following Step 1, Step 2 involves phosphorylation of T187 within the flexible C-terminal domain of p27 by partially active Cdk2 through a pseudo uni-molecular mechanism (indicated by gray arrow). Phosphorylation of T187 creates a phosphodegron signal for ubiquitination of Lysine residues within the p27 C-terminus by the E3 ligase, SCFSkp2, during Step 3. Finally, during Step 4, ubiquitinated p27 is selectively degraded by the 26S proteasome, leading to the release of fully active Cdk2/cyclin A, which drives progression into S phase of the cell division cycle.
Regulated unfolding mechanisms are also involved in the control of programmed cell death. Cytoplasmic p53 tumor suppressor initiates apoptosis by binding to and activating pro-apoptotic proteins [17]. This lethal function is inhibited by association of p53 with the anti-apoptotic protein BCL-xL [18]. Release and activation of p53 in response to DNA damage is signaled by a BH3-only protein ligand (PUMA) binding to BCL-xL. A π-stacking interaction between His113 in BCL-xL and Trp71 in PUMA, causes unfolding of BCL-xL at an allosteric site comprising two α-helix structural elements and dissociation of p53 from BCL-xL [19]. This example illustrates a signaling mechanism which combines traditional allosteric, ligand binding-induced structural changes with unfolding to release a binding partner.
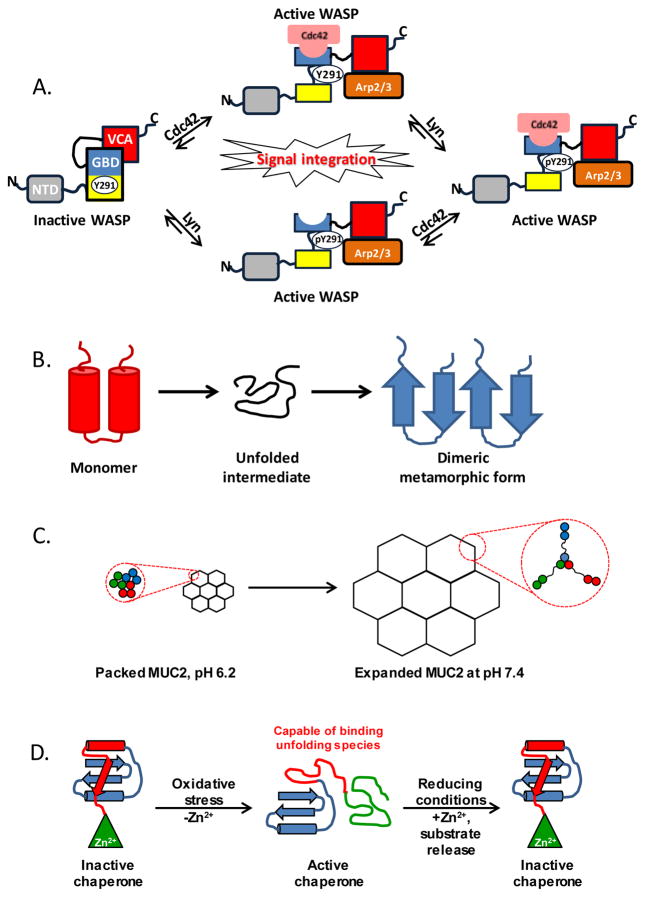
The Wiskott-Aldrich syndrome protein (WASP) provides an example of both posttranslational modification- and ligand binding-induced unfolding involving several protein domains. WASP regulates cytoskeletal actin polymerization through direct interaction of its C-terminal domain with the Arp2/3 and actin complex. However, this domain is auto-inhibited through tertiary interactions with other domains of WASP. Cdc42, a Rho-family GTPase, signals activation of auto-inhibited WASP to initiate actin polymerization. Cdc42 and the C-terminal domain of WASP compete for binding to the WASP GTPase binding domain (GBD). Activation of WASP by Cdc42 involves partial unfolding of the hydrophobic core of the auto-inhibited conformation of WASP and folding of the WASP-Cdc42 complex. Furthermore, the partially unfolded conformation exposes Tyr291, a phosphorylation site for the non-receptor tyrosine kinase Lyn. This modification further relieves inhibition and enables the unfolding required for the structural switch to the Cdc42-bound conformation [20, 21]. This activation mechanism (Figure 2A) is an example of regulated unfolding wherein two input signals, posttranslational modification and ligand binding, synergize to control the three-dimensional organization and function of WASP with switch-like precision. Utilization of two input mechanisms allows WASP to integrate disparate upstream signals [21] and to respond through regulated unfolding.
Figure 2.
Examples of various regulated unfolding mechanisms involved in signaling. (A) Signal integration in the regulatory mechanism of WASP. In the autoinhibited form, the GBD domain (blue and yellow boxes) is bound to the C-terminal VCA domain (red box), inhibiting the binding of VCA to the Arp2/3 complex (orange box). Binding of Cdc42 GTPase (coral object) to the GBD domain of WASP, which requires partial unfolding and remodeling of this binding site, releases VCA and activates WASP. Phosphorylation of Y291 further stabilizes the active form of WASP. WASP integrates disparate signals (Cdc42 binding and phosphorylation by Lyn) to enable Arp2/3 binding and promote actin polymerization. (B) Metamorphic proteins require partial or global unfolding to interconvert between different tertiary and quaternary structures. (C) Formation of colon mucus through regulated unfolding of MUC2. (D) Functional cycle of the Hsp33 oxidative stress-response chaperone. The sensing domain (green) unfolds in response to oxidative stress, leading to exposure of the substrate binding domain (red) that binds and holds partially unfolded substrates. Restoration of reducing conditions causes the structure of the chaperone to revert to the folded, inactive form, releasing substrates to undergo folding under non-stress conditions.
However, these two mechanisms are not the only inputs that propagate biological signals through regulated unfolding. For example, phototropins, a class of Ser/Thr kinases, play critical roles in signal transduction in plants. Their activation is signaled by exposure to blue light, when a covalent bond forms between a flavin chromophore and the light-oxygen-voltage 2 domain (LOV2), causing unfolding of an inhibitory Jα helix and consequently the activation of the kinase domain [22, 23]. A similar mechanism is utilized by a class of bacterial photoactivatable proteins [24]. These examples have illustrated regulated unfolding mechanisms involving relatively subtle alterations of secondary and tertiary structure.
Protein shape-shifters
Other examples of regulated unfolding include a class of so-called ‘metamorphic proteins’ ([25, 26], Figure 2B). The intriguing structural shape-shifting of these proteins mediates multiple cellular functions. For example, the chemokine lymphotactin (Ltn) switches between a monomeric α-helical and dimeric β-sheet sandwich conformation. The monomeric form, which exhibits the classical chemokine fold, binds to the canonical XCRI receptor. In contrast, the dimeric form binds to heparin and localizes to the plasma membrane [27]. The two mutually exclusive functional states exist in equilibrium under physiological conditions and require global unfolding for their inter-conversion [28]. Mad2, a protein involved in regulation of the mitotic spindle assembly, provides another example of metamorphic behavior. This protein undergoes a significant structural reorganization from an inactive to active conformation which requires a partially unfolded intermediate [29]. While the in vitro evidence for the alternative structures of metamorphic proteins supports the observations of functional switching in cells, the exact mechanisms that regulate conformational switching of Ltn and Mad2 in vivo are currently not well understood.
Proteins such as the glycoprotein MUC2, the major colon mucin, constitute the scaffold for formation of extensive biomolecular networks. Trimerization of MUC2 via its N-terminal domain, coupled with dimerization via its C-terminal domain forms planar polymers that assemble as stacked gel sheets on the inner epithelium of the colon [30]. Compact, ring-shaped polymers composed of folded monomers are stabilized in the presence of Ca2+ and at low pH (6.2) and transported by secretory granulae to the epithelial cell layer. At pH 7.4 and in the presence of chelating agents, conditions which mimic those at the epithelial cell layer, the N-terminal rings of MUC2 partially unfold, causing an expansion of the proteinaceous network by greater than 1,000-fold (Figure 2C). This expanded polymer is stabilized by covalent disulfide bonds formed within the N-terminal trimerization domains [30]. The use of regulated unfolding maximizes the surface area that can be engaged by the polymer and likely mediates the physical and mechanical properties required for its function as a protective barrier in the colon. The energy expenditure for delivering MUC2 from the site of synthesis to the epithelial layer via the secretory pathway is significantly reduced though the employment of the compact form in early stages of the functional cycle.
Protein unfolding as a mechanism for revealing occluded signals
While some proteins perform unique, well-defined tasks, many exhibit multiple functions, often performed in multiple subcellular locations. A preponderance of these multi-functional proteins is involved in cellular signaling. Translocation between subcellular compartments is mediated by specialized machinery which recognizes specific signals, such as nuclear localization (NLS) or nuclear export signals (NES), which are encoded by short linear motifs within the primary sequence [31]. The transport machinery is always active; therefore, switchable signals are needed to control when a particular protein is transported from one cellular compartment to another. For example, KSRP, also known as FBP2, a protein involved in various aspects of mRNA metabolism [32], contains a 14-3-3ζ consensus binding sequence which is structurally occluded within an atypical KH1 domain. Phosphorylation by AKT of Ser193 causes the kinase domain of KH1 to unfold, consequently revealing the 14-3-3ζ interaction site [33]. This regulated unfolding event results in re-localization of KSRP to the nucleus in a 14-3-3ζ dependent manner [33] and reduction of the rate of mRNA degradation [34]. A similar mechanism of exposing structurally inaccessible localization signals is employed by the influenza virus to hijack the nuclear import machinery of its host cell. The C-terminal segment of viral polymerase PB2 unfolds in order to reveal a bipartite NLS which binds to importin α5 and allows the parasitic enzyme to enter the host cell nucleus and process newly synthesized viral genomic material [35].
The Crk-like (CRKL) adaptor protein, involved in mediating a variety of signal transduction cascades, including subcellular re-localization and activation of kinases and other signaling molecules [36], is another example of a protein which harbors an occluded recognition sequence [37]. An evolutionarily conserved NES is encoded in SH3C, a functionally important domain of CRKL that is otherwise uninvolved in recruitment of signaling molecules. Through a combination of structural and biophysical analyses, Harkiolaki and colleagues [37] demonstrated that the SH3C domain of CRKL is able to form a domain-swapped dimer that exposes two symmetrically disposed NESs. These signals are structurally occluded in the monomeric form of the protein [37]. Interestingly, domain swapping is also employed by other proteins as a method of regulating function [38, 39]. The ‘hinge loop’, a topologically required region for formation of domain swapped dimers, extends from its collapsed configuration in the monomeric form to an extended conformation in the dimer. This hinge loop is an favorable location for conditional signaling sequences, such as sites of phosphorylation that regulate function, which become solvent exposed upon dimerization. Tyr926, a conserved phosphorylation site in the ‘hinge loop’ of the focal adhesion targeting (FAT) domain of focal adhesion kinase (FAK) is modified by Src with greater efficiency when the protein adopts the domain-swapped conformation, affecting downstream signaling through the Ras-MAPK pathway [40, 41].
Due to its critical role in maintaining DNA integrity and controlling cell fate, the level and activity of the tumor suppressor p53 is controlled by complex signaling networks involving a staggering number of positive and negative feedback systems [42]. Acetylation of tetrameric p53 by the acetyltransferase p300 enhances specific DNA binding [43]. The acetylation site, located in the C-terminal regulatory domain of p53, is sterically occluded when this domain is phosphorylated, but becomes accessible for p300 modification when p53 binds to DNA, as well as under heat-denaturation conditions. These results suggest an allosterically regulated local unfolding mechanism [44].
Central to the conserved, inter-cellular Notch signaling pathway are the Notch family of modular, single-pass transmembrane receptors [45]. In their resting state, Notch receptors adopt an auto-inhibited fold, in which two key proteolytic sites, S2 and S3, located within the negative regulatory region (NRR) are sterically protected from proteolytic cleavage. Binding of Notch on the signal-receiving cell to a transmembrane ligand located on the signal-sending cell causes ligand endocytosis as well as simultaneous endocytosis in trans (into the signal-sending cell) of the ecto-domain of Notch [45]. Since the transmembrane domain of Notch remains anchored in the membrane of the signal-receiving cell, the adjacent NRR domain is subjected to mechanical strain, which exposes the occluded S2 proteolytic site for cleavage [45]. In a molecular dynamics study, Chen and Zolkiewska [46] identify the protease-sensitive conformation of Notch1 as an on-pathway unfolding intermediate, in which two Lin12/Notch repeats dissociate from the heterodimerization domain (HD), causing unfolding of a secondary structure element within HD that contains S2. Furthermore, Stephenson and Avis [47] demonstrated through a combination of atomic force microscopy, biophysical assays and molecular dynamics that a β-strand containing the S2 site within the NRR domain of Notch2 undergoes stepwise unfolding in response to pulling force. Unfolding of the S2 site exhibited a low energy barrier and was an early event on the unfolding pathway. Experimental evidence associated the unfolding of the S2-containing structural element with proteolytic cleavage by the TACE and ADAM10 proteases, linking mechanically-induced unfolding with trans-endocytosis, a critical step in the Notch signaling pathway.
The mechanism of regulated unfolding as a means of exposing hidden signaling sequences is also utilized by a giant amongst proteins, the Van Willebrand factor (VWF), which forms ultra-large multimers. Buried protease recognition sites are revealed via local unfolding generated by tensile force created in response to arterial bleeding. Cleavage by the metalloprotease ADAMTS13 severs the ultra-large VWF multimers into smaller oligomers as part of a regulatory mechanism of hemostasis [48, 49].
Together, these findings demonstrate that regulated unfolding to expose otherwise structurally occluded signaling sequences is a frequently utilized and effective mechanism for controlling the functional repertoire of numerous multitasking proteins.
Protein unfolding as a mechanism for modulating ligand binding affinity
Modulation of protein ligand binding affinity is another prevalent regulatory mechanism utilized in transcriptional regulation [50], signal transduction [51–53], metabolism [54] and other biological processes. The mechanisms employed include allosteric regulation [52–54], assembly of multi-subunit complexes [50], and modulation of binding site affinity via homo- or hetero-oligomerization [50]. Regulated unfolding is also utilized as a means to decrease binding affinity [19, 55], and somewhat counter intuitively, to enhance substrate binding affinity [56].
Chaperones are molecular machines that recognize misfolded proteins and promote their refolding. Interestingly, cellular stress signals that trigger protein misfolding also initiate chaperone activation. For example, redox-regulated chaperones, such as the bacterial holdases Hsp33 [49, 57, 58] and HdeA [59, 60], are activated upon oxidative stress and a drop in cellular pH, respectively. Strikingly, activation of these chaperones is achieved through conditional domain unfolding [56]. The structural transition to the partially unfolded state confers high affinity towards partially unfolded chaperone substrates, to which they bind and ‘hold’ until environmental conditions favor native protein folding. When these normal conditions are restored, substrates are released and allowed to fold independently [60] or are transferred to an ATP-dependent foldase [49]. Exposure of hydrophobic surfaces on the C-terminal substrate binding domain (the so-called ‘sensor’ domain) of the chaperone through regulated unfolding provides selectivity and high binding affinity for unfolding/misfolding intermediates. Utilization of folded-to-unfolded transitions in the functional cycle of these disordered chaperones provides two-fold functional advantages. First, this energy-independent mechanism allows maintenance of proteostasis under stress conditions, when the pool of ATP required by ATP-dependent chaperones is depleted. Second, utilization of a disordered chaperone region for substrate recognition enables binding to a broad pallete of unfolded protein substrates [61].
The unfolding/folding functional cycle of Hsp33 has been elegantly elucidated by Jakob and colleagues ([49] and Figure 2D). Under normal physiological conditions, the ‘sensor domain’ of Hsp33 is stabilized by a Zn2+ ion which coordinates four highly conserved cysteines. In response to oxidative stress, the stabilizing ion is released and, consequently, the C-terminal domain unfolds. Oxidation of the four Zn-coordinating cysteines acts as an allosteric switch that causes unfolding of the previously folded linker connecting the N- and C-terminal domains [62]. This unfolded linker domain serves as the high-affinity binding site for early unfolding intermediates, while selecting against self-recognition for intrinsically disordered regions within the chaperone, as well as against other cellular IDPs [49].
Unfolding is a means to dramatically decrease the binding affinity between two folded biomolecules. A particularly interesting example of this regulatory mechanism involves the sarcoplasmic reticulum (SR) Ca2+-ATPase (SERCA) and the SR integral membrane protein phospholamban (PLN), which regulate cardiac contractility [55]. Activation of SR and plasma-membrane Ca2+ channels in myocytes causes increased cytoplasmic Ca2+ concentration and leads to cellular contraction. SERCA, a SR calcium pump, mediates transport of cytoplasmic Ca2+ into the SR lumen, causing muscle relaxation [63]. PLN binding to SERCA inhibits SERCA-mediated Ca2+ flux from the cytoplasm into the SR. Phosphorylation of PLN at Ser16 by PKA causes unfolding of domains Ia and Ib, positioned in the cytoplasm and the hydrophilic layer of the SR membrane, respectively. This modification reduces the affinity of PLN for SERCA and restores SERCA-mediated uptake of Ca2+ into the SR membrane [55, 63]. Through EPR and NMR-based analyses, Gustavsson, et al. [55], identified several partially disordered, alternative conformational states that exist in equilibrium with the folded form. The equilibrium distribution of conformational states for the conditionally unfolded species can be regulated by phosphorylation and lipid binding, which determines the binding affinity between SERCA and PLN and regulates cardiac contraction. The Ia domain of PLN is also involved in signal transduction by interacting with a number of binding partners. This function is most likely enabled by the conformational dynamics of this conditionally unstructured domain [55].
Triggers of regulated unfolding
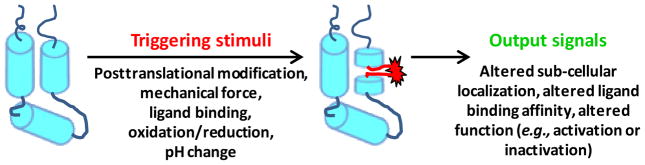
The cellular functions affected by regulated unfolding mechanisms are highly diverse. Furthermore, the extent of disorder induced during signal switching ranges from subtle, local unfolding events [35, 37, 44, 48, 64] to unfolding of entire domains [30, 55, 65, 66]. Similarly diverse is the spectrum of molecular triggers that unleash regulated unfolding events.
Environmental stimuli
Changes in chemical environment, such as alteration of pH [30, 60], redox condition [49], exposure to light [22–24, 67], and metal ion concentrations [30, 55], are signals that trigger cells to activate specific regulatory pathways (Figure 3). These stimuli can affect the physico-chemical properties of proteins, providing a mechanism for coupling them with structural changes (e.g., unfolding) and downstream signaling. For example, oxidative-stress conditions promote disulfide bond formation between Cys residues in Hsp33 and signal activation of the chaperone through conditional unfolding [66] and chelation of stabilizing Ca2+ ions promotes unfolding and physical expansion of the colon mucus [30].
Figure 3.
Several different triggering stimuli mediate protein unfolding and regulate function.
Chemical modification
The state of foldedness of proteins is also controlled by chemical modifications arising from posttranslational modifications [14, 55, 64, 65, 68]. An illustrative example is the cyclic phosphorylation and dephosphorylation of the SERCA/PLN system [55] that modulates the regulated unfolding mechanism responsible for controlling cardiac contractility. Another example is regulation of the Cdk inhibitory activity of p27 by tyrosine phosphorylation, which disrupts the inhibitory conformation and partially activates Cdk activity [14]. A plethora of other posttranslational modifications, including acetylation [44], methylation, ubiquitination, sumoylation, etc., are either known to, or likely to mediate regulated unfolding events within diverse signaling pathways.
Ligand binding
Protein-protein interactions constitute the basis for intracellular signaling. Often, these interactions are triggered by structural rearrangements of one or more of the binding partners, either at the interaction site or at an allosterically regulated site. Several protein re-activation mechanisms employ ligand binding-induced unfolding steps. For instance, the C-terminal domain of inactivated peroxiredoxin must unfold when bound to the repair enzyme sulphiredoxin, in order to allow access to its active site [69]. PUMA binding to BCL-xL induces local unfolding of an allosteric site, thereby signaling release and activation of the tumor suppressor p53 [19]. Local unfolding induced by ligand binding has been observed in mechanisms that regulate the sub-cellular localization of proteins. For example, the C-terminal segment of the influenza virus polymerase unfolds when bound to human importin α5 for efficient nuclear import [35] and the nuclear export signal of CRKL becomes accessible only upon local unfolding of the polypeptide chain, upon self-association into a domain-swapped dimer [37]. Proteins are dynamic entities that sample multiple conformations within their folding landscape [70]. For the protein examples discussed here, intrinsic fluctuations within this landscape are enhanced through regulated unfolding to enable exposure of otherwise occluded binding sites, providing a mechanism for enabling interactions in a tightly controlled manner.
Mechanical force
In addition to chemical modifications and ligand binding, mechanical force is an important regulatory mechanism employed, in particular, in the muscular and vascular systems. Mechanical stress-induced local unfolding of titin in striated muscle is thought to play an important role in regulating its kinase activity [71], while fluid shear stress in blood vessels controls the length and function of the thrombogenic factor VWF, by exposing a buried proteolytic site [48]. Furthermore, trans-endocytosis of the ecto-domain of Notch receptor exerts mechanical strain within its auto-inhibitory domain causing strain-induced local unfolding that exposes otherwise occluded sites for proteolytic cleavage, allowing propagation of Notch signals [46, 47].
The existence of these diverse triggering mechanisms highlights the broad utilization of regulated unfolding in all kingdoms of life and as a response to widely divergent environmental stimuli.
Concluding remarks
The process of protein unfolding is utilized by all organisms to facilitate amino acid recycling [72] and to transport macromolecules through membranes, by threading them through tight pores [73]. Here we show that all kingdoms of life utilize mechanisms involving regulated protein unfolding to mediate signal transduction. Evolutionary conservation of the protein regions involved in regulated unfolding (e.g., the conserved occluded NES in CRKL [37], tyrosine residues within Cdk inhibitors [74], etc.) highlights the biological importance of this type of signaling mechanism. A theme that emerges from the examples discussed above is that, through various triggering mechanisms, regulated unfolding is a means to alter the dynamic properties of proteins, or segments within them, and, in so doing, alter protein function. Enhanced sampling of unfolded, or less structured, states in response to the triggering stimuli discussed above provides physical mechanisms for proteins to transmit biological signals [26].
Partial or global unfolding of proteins or protein domains facilitates interconversion between isoenergetic, alternative conformational states. Often, the structural rearrangement exposes new hydrophobic interaction surfaces and thus promotes the formation of oligomers [26]. For example, the metamorphic proteins Mad2 and Ltn [26] have evolved alternative folds [75], with one of two folds stabilized via dimerization and at least partial unfolding required for the structural transition between these conformational states [26, 28, 29]. Furthemore, the form of CRKL that is exported from the nucleus is a domain swapped dimer with an unfolded segment that contains a NES [37]. Oligomerization is a mechanism for enhancing the functional complexity associated with a particular protein sequence [26, 75] and this complexity can be further enhanced via regulated unfolding to control transitions between different oligomeric states [26–29, 37, 55, 68].
Recent advances in NMR spectroscopy methodology [76] and single molecule techniques (e.g. atomic force microscopy [77], single molecule fluorescence [78, 79]) have allowed detailed characterization of the molecular mechanisms by which structural fluctuations mediate protein function. For example, NMR relaxation studies have shown that enzymes fluctuate between different sets of structural states at different stages of catalytic cycles [80]. Furthermore, Kay and co-workers have characterized lowly populated unfolding intermediates for several proteins using similar NMR methods [81, 82]. In addition, single-molecule FRET techniques identified alterations in the folding pathway of α-synuclein due to mutations associated with Parkinson’s disease [83]. Advances in computational methods in studies of protein folding and unfolding, as well as advances in computing power, provide opportunities to understand regulated unfolding mechanisms in atomistic detail. An illustrative example is the mechanism that controls VWF size in arterial thrombosis (reviewed in [84]) which was elucidated though a combination of molecular dynamics [85, 86], single molecule experiments [48, 87], X-ray crystallography [88] and biophysical assays [86, 89]. Together, these approaches will be valuable tools in future studies into the roles of regulated unfolding—from subtle order-to-disorder transitions to large-scale polypeptide unfolding—in protein function. Importantly, the identification of functionally relevant unfolded states requires monitoring dynamics on multiple time-scales which necessitates the use of complementary experimental and computational techniques.
We anticipate that the list of proteins recognized to utilize regulated unfolding will grow, as conformational states identified in biophysical assays as simple folding/unfolding intermediates are shown to be physiologically relevant. Similar to the example of local unfolding and acetylation of p53 in response to DNA binding [44], these intermediates may be stabilized through the types of triggering modifications discussed above. For example, we proposed that the multifunctional protein nucleophosmin (NPM1), a histone chaperone involved in ribosome biogenesis, cell cycle control and tumor suppression [90], may use regulated unfolding of its N-terminal domain from a folded β-sheet rich pentamer to a disordered monomer in order to switch functions and sub-cellular localization [68]. Identification and characterization of functionally relevant unfolded states for other proteins will require addressing several challenges, such as (i) identification of switching triggers through biochemical, structural and cellular investigations, (ii) elucidation of the functional outcome(s) of the regulated unfolding phenomena, (iii) determination of the lifetimes of the unfolded species in an appropriate functional setting, and (iv) elucidation of the mechanisms by which regulated unfolding signals are reset when triggering stimuli are absent.
Finally, our growing knowledge of the broad utilization of regulated unfolding mediated by diverse triggering mechanisms provides opportunities for applications in protein engineering. In fact, mechanisms that couple protein domain folding and unfolding have been previously explored as general designs for biomolecular switches, with mechanical force [91–93], Ca2+ ion binding [94, 95] and proteolytic cleavage [96] utilized as input signals, and alteration of protein function as the output signal. Understanding the structural and biophysical underpinnings of regulated unfolding mechanisms will advance our knowledge of multifunctional protein regulation. It is likely that understanding the physical principles of evolutionarily selected mechanisms of regulated unfolding will lead protein design efforts in new directions, with possible applications in the biotechnology and pharmaceutical industries.
Acknowledgments
We acknowledge the following funding sources: NIH R01CA082491 and 1R01GM083159 (to R.W.K.), a National Cancer Institute Cancer Center Support Grant P30CA21765 (at St. Jude Children’s Research Hospital), and the American Lebanese Syrian Associated Charities (ALSAC).
Abbreviations
- IDP
Intrinsically disordered protein
- NLS
nuclear localization signal
- NES
nuclear export signal
References
- 1.Feller G. Protein stability and enzyme activity at extreme biological temperatures. Journal of physics Condensed matter: an Institute of Physics journal. 2010;22:323101. doi: 10.1088/0953-8984/22/32/323101. [DOI] [PubMed] [Google Scholar]
- 2.Galea CA, Pagala VR, Obenauer JC, Park CG, Slaughter CA, Kriwacki RW. Proteomic studies of the intrinsically unstructured mammalian proteome. Journal of proteome research. 2006;5:2839–48. doi: 10.1021/pr060328c. [DOI] [PubMed] [Google Scholar]
- 3.Galea CA, High AA, Obenauer JC, Mishra A, Park CG, Punta M, Schlessinger A, Ma J, Rost B, Slaughter CA, Kriwacki RW. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. Journal of proteome research. 2009;8:211–26. doi: 10.1021/pr800308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. Journal of molecular biology. 2004;337:635–45. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. Journal of molecular biology. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 6.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic acids research. 2004;32:1037–49. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome informatics Workshop on Genome Informatics. 2000;11:161–71. [PubMed] [Google Scholar]
- 8.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson J, Grahn M, Wright AP. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome biology. 2011;12:R65. doi: 10.1186/gb-2011-12-7-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong CS, Kim D. Coevolved residues and the functional association for intrinsically disordered proteins. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing; 2012. pp. 140–51. [PubMed] [Google Scholar]
- 11.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–82. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 12.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–90. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 13.Mitrea DM, Yoon MK, Ou L, Kriwacki RW. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biological chemistry. 2012;393:259–74. doi: 10.1515/hsz-2011-0254. [DOI] [PubMed] [Google Scholar]
- 14.Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128:269–80. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 15.Lacy ER, Wang Y, Post J, Nourse A, Webb W, Mapelli M, Musacchio A, Siuzdak G, Kriwacki RW. Molecular basis for the specificity of p27 toward cyclin-dependent kinases that regulate cell division. Journal of molecular biology. 2005;349:764–73. doi: 10.1016/j.jmb.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10453–8. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–30. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–5. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 19.Follis AV, Chipuk JE, Fisher JC, Yun MK, Grace CR, Nourse A, Baran K, Ou L, Min L, White SW, Green DR, Kriwacki RW. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nature chemical biology. 2013 doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, Rosen MK. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–83. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 21.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–8. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 22.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–4. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 23.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–92. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 24.Dux P, Rubinstenn G, Vuister GW, Boelens R, Mulder FA, Hard K, Hoff WD, Kroon AR, Crielaard W, Hellingwerf KJ, Kaptein R. Solution structure and backbone dynamics of the photoactive yellow protein. Biochemistry. 1998;37:12689–99. doi: 10.1021/bi9806652. [DOI] [PubMed] [Google Scholar]
- 25.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–6. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 26.Bryan PN, Orban J. Proteins that switch folds. Current opinion in structural biology. 2010;20:482–8. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5057–62. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyler RC, Murray NJ, Peterson FC, Volkman BF. Native-state interconversion of a metamorphic protein requires global unfolding. Biochemistry. 2011;50:7077–9. doi: 10.1021/bi200750k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner JJ, Wood S, Shorter J, Englander SW, Black BE. The Mad2 partial unfolding model: regulating mitosis through Mad2 conformational switching. The Journal of cell biology. 2008;183:761–8. doi: 10.1083/jcb.200808122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambort D, Johansson ME, Gustafsson JK, Nilsson HE, Ermund A, Johansson BR, Koeck PJ, Hebert H, Hansson GC. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5645–50. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–6. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 32.Briata P, Chen CY, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. KSRP, many functions for a single protein. Frontiers in bioscience: a journal and virtual library. 2011;16:1787–96. doi: 10.2741/3821. [DOI] [PubMed] [Google Scholar]
- 33.Diaz-Moreno I, Hollingworth D, Frenkiel TA, Kelly G, Martin S, Howell S, Garcia-Mayoral M, Gherzi R, Briata P, Ramos A. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nature structural & molecular biology. 2009;16:238–46. doi: 10.1038/nsmb.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS, Briata P. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS biology. 2006;5:e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature structural & molecular biology. 2007;14:229–33. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 36.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–71. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 37.Harkiolaki M, Gilbert RJ, Jones EY, Feller SM. The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure. 2006;14:1741–53. doi: 10.1016/j.str.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Rousseau F, Schymkowitz J, Itzhaki LS. Implications of 3D domain swapping for protein folding, misfolding and function. Advances in experimental medicine and biology. 2012;747:137–52. doi: 10.1007/978-1-4614-3229-6_9. [DOI] [PubMed] [Google Scholar]
- 39.Ding F, Prutzman KC, Campbell SL, Dokholyan NV. Topological determinants of protein domain swapping. Structure. 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Dixon RD, Chen Y, Ding F, Khare SD, Prutzman KC, Schaller MD, Campbell SL, Dokholyan NV. New insights into FAK signaling and localization based on detection of a FAT domain folding intermediate. Structure. 2004;12:2161–71. doi: 10.1016/j.str.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Prutzman KC, Gao G, King ML, Iyer VV, Mueller GA, Schaller MD, Campbell SL. The focal adhesion targeting domain of focal adhesion kinase contains a hinge region that modulates tyrosine 926 phosphorylation. Structure. 2004;12:881–91. doi: 10.1016/j.str.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 42.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 43.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 44.Ceskova P, Chichger H, Wallace M, Vojtesek B, Hupp TR. On the mechanism of sequence-specific DNA-dependent acetylation of p53: the acetylation motif is exposed upon DNA binding. Journal of molecular biology. 2006;357:442–56. doi: 10.1016/j.jmb.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 45.Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Current topics in developmental biology. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Zolkiewska A. Force-induced unfolding simulations of the human Notch1 negative regulatory region: possible roles of the heterodimerization domain in mechanosensing. PloS one. 2011;6:e22837. doi: 10.1371/journal.pone.0022837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2757–65. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–4. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–57. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Funnell AP, Crossley M. Homo- and heterodimerization in transcriptional regulation. Advances in experimental medicine and biology. 2012;747:105–21. doi: 10.1007/978-1-4614-3229-6_7. [DOI] [PubMed] [Google Scholar]
- 51.Luo BH, Springer TA. Integrin structures and conformational signaling. Current opinion in cell biology. 2006;18:579–86. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu G, Wang W, Luo BH. Overview: structural biology of integrins. Methods in molecular biology. 2012;757:81–99. doi: 10.1007/978-1-61779-166-6_7. [DOI] [PubMed] [Google Scholar]
- 53.Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning. PLoS computational biology. 2011;7:e1002154. doi: 10.1371/journal.pcbi.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denisov IG, Sligar SG. A novel type of allosteric regulation: functional cooperativity in monomeric proteins. Archives of biochemistry and biophysics. 2012;519:91–102. doi: 10.1016/j.abb.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G. Lipid-mediated folding/unfolding of phospholamban as a regulatory mechanism for the sarcoplasmic reticulum Ca2+-ATPase. Journal of molecular biology. 2011;408:755–65. doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends in biochemical sciences. 2012 doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graf PC, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. Activation of the redox-regulated chaperone Hsp33 by domain unfolding. The Journal of biological chemistry. 2004;279:20529–38. doi: 10.1074/jbc.M401764200. [DOI] [PubMed] [Google Scholar]
- 58.Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Molecular cell. 2005;17:381–92. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 59.Tapley TL, Korner JL, Barge MT, Hupfeld J, Schauerte JA, Gafni A, Jakob U, Bardwell JC. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5557–62. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC. Protein refolding by pH-triggered chaperone binding and release. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1071–6. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright PE, Dyson HJ. Linking folding and binding. Current opinion in structural biology. 2009;19:31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbirz S, Jakob U, Glocker MO. Mass spectrometry unravels disulfide bond formation as the mechanism that activates a molecular chaperone. The Journal of biological chemistry. 2000;275:18759–66. doi: 10.1074/jbc.M001089200. [DOI] [PubMed] [Google Scholar]
- 63.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nature reviews Molecular cell biology. 2003;4:566–77. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 64.Nardozzi J, Wenta N, Yasuhara N, Vinkemeier U, Cingolani G. Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5. Journal of molecular biology. 2010;402:83–100. doi: 10.1016/j.jmb.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Díaz-Moreno IHD, Frenkiel TA, Kelly G, Martin S, Howell S, García-Mayoral M, Gherzi R, Briata P, Ramos A. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nature structural & molecular biology. 2009;16:238–246. doi: 10.1038/nsmb.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. The Journal of biological chemistry. 2010;285:11243–51. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harper SM, Neil LC, Day IJ, Hore PJ, Gardner KH. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. Journal of the American Chemical Society. 2004;126:3390–1. doi: 10.1021/ja038224f. [DOI] [PubMed] [Google Scholar]
- 68.Mitrea DM, Kriwacki RW. Cryptic disorder: an order-disorder transformation regulates the function of nucleophosmin. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing; 2012. pp. 152–63. [PubMed] [Google Scholar]
- 69.Jonsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plaxco KW, Gross M. Cell biology. The importance of being unfolded. Nature. 1997;386:657–659. doi: 10.1038/386657a0. [DOI] [PubMed] [Google Scholar]
- 71.Stahl SW, Puchner EM, Alexandrovich A, Gautel M, Gaub HE. A conditional gating mechanism assures the integrity of the molecular force-sensor titin kinase. Biophysical journal. 2011;101:1978–86. doi: 10.1016/j.bpj.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochimica et biophysica acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prakash S, Matouschek A. Protein unfolding in the cell. Trends in biochemical sciences. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 74.Yoon MK, Mitrea DM, Ou L, Kriwacki RW. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochemical Society transactions. 2012;40:981–8. doi: 10.1042/BST20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7287–92. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sze KH, Lai PM. Probing protein dynamics by nuclear magnetic resonance. Protein and peptide letters. 2011;18:373–9. doi: 10.2174/092986611794653897. [DOI] [PubMed] [Google Scholar]
- 77.Katan AJ, Dekker C. High-speed AFM reveals the dynamics of single biomolecules at the nanometer scale. Cell. 2011;147:979–82. doi: 10.1016/j.cell.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 78.Ferreon AC, Deniz AA. Protein folding at single-molecule resolution. Biochimica et biophysica acta. 2011;1814:1021–9. doi: 10.1016/j.bbapap.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gambin Y, Deniz AA. Multicolor single-molecule FRET to explore protein folding and binding. Molecular bioSystems. 2010;6:1540–7. doi: 10.1039/c003024d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weikl TR, Boehr DD. Conformational selection and induced changes along the catalytic cycle of Escherichia coli dihydrofolate reductase. Proteins. 2012;80:2369–83. doi: 10.1002/prot.24123. [DOI] [PubMed] [Google Scholar]
- 81.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–90. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 82.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nature chemical biology. 2009;5:808–14. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 83.Ferreon AC, Moran CR, Ferreon JC, Deniz AA. Alteration of the alpha-synuclein folding landscape by a mutation related to Parkinson’s disease. Angewandte Chemie. 2010;49:3469–72. doi: 10.1002/anie.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Di Stasio E, De Cristofaro R. The effect of shear stress on protein conformation: Physical forces operating on biochemical systems: The case of von Willebrand factor. Biophysical chemistry. 2010;153:1–8. doi: 10.1016/j.bpc.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 85.Baldauf C, Schneppenheim R, Stacklies W, Obser T, Pieconka A, Schneppenheim S, Budde U, Zhou J, Grater F. Shear-induced unfolding activates von Willebrand factor A2 domain for proteolysis. Journal of thrombosis and haemostasis: JTH. 2009;7:2096–105. doi: 10.1111/j.1538-7836.2009.03640.x. [DOI] [PubMed] [Google Scholar]
- 86.Schneider SW, Nuschele S, Wixforth A, Gorzelanny C, Alexander-Katz A, Netz RR, Schneider MF. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7899–903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ying J, Ling Y, Westfield LA, Sadler JE, Shao JY. Unfolding the A2 domain of von Willebrand factor with the optical trap. Biophysical journal. 2010;98:1685–93. doi: 10.1016/j.bpj.2009.12.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Q, Zhou YF, Zhang CZ, Zhang X, Lu C, Springer TA. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9226–31. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Auton M, Cruz MA, Moake J. Conformational stability and domain unfolding of the Von Willebrand factor A domains. Journal of molecular biology. 2007;366:986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 90.Lindstrom MS. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochemistry research international. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ha JH, Butler JS, Mitrea DM, Loh SN. Modular enzyme design: regulation by mutually exclusive protein folding. Journal of molecular biology. 2006;357:1058–62. doi: 10.1016/j.jmb.2006.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Radley TL, Markowska AI, Bettinger BT, Ha JH, Loh SN. Allosteric switching by mutually exclusive folding of protein domains. Journal of molecular biology. 2003;332:529–36. doi: 10.1016/s0022-2836(03)00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng Q, Kong N, Wang HC, Li H. Designing redox potential-controlled protein switches based on mutually exclusive proteins. Protein science: a publication of the Protein Society. 2012;21:1222–30. doi: 10.1002/pro.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stratton MM, McClendon S, Eliezer D, Loh SN. Structural characterization of two alternate conformations in a calbindin D(9)k-based molecular switch. Biochemistry. 2011;50:5583–9. doi: 10.1021/bi102040g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stratton MM, Mitrea DM, Loh SN. A Ca2+-sensing molecular switch based on alternate frame protein folding. ACS chemical biology. 2008;3:723–32. doi: 10.1021/cb800177f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitrea DM, Parsons LS, Loh SN. Engineering an artificial zymogen by alternate frame protein folding. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2824–9. doi: 10.1073/pnas.0907668107. [DOI] [PMC free article] [PubMed] [Google Scholar]