Abstract
Fibroblast growth factor (FGF)2, FGF4, FGF7 and FGF20 are representative paracrine FGFs binding to heparan-sulfate proteoglycan and fibroblast growth factor receptors (FGFRs), whereas FGF19, FGF21 and FGF23 are endocrine FGFs binding to Klotho and FGFRs. FGFR1 is relatively frequently amplified and overexpressed in breast and lung cancer, and FGFR2 in gastric cancer. BCR-FGFR1, CNTRL-FGFR1, CUX1-FGFR1, FGFR1OP-FGFR1, MYO18A-FGFR1 and ZMYM2-FGFR1 fusions in myeloproliferative neoplasms are non-receptor-type FGFR kinases, whereas FGFR1-TACC1, FGFR2-AFF3, FGFR2-BICC1, FGFR2-PPHLN1, FGFR3-BAIAP2L1 and FGFR3-TACC3 fusions in solid tumors are transmembrane-type FGFRs with C-terminal alterations. AZD4547, BGJ398 (infigratinib), Debio-1347 and dovitinib are FGFR1/2/3 inhibitors; BLU9931 is a selective FGFR4 inhibitor; FIIN-2, JNJ-42756493, LY2874455 and ponatinib are pan-FGFR inhibitors. AZD4547, dovitinib and ponatinib are multi-kinase inhibitors targeting FGFRs, colony stimulating factor 1 receptor (CSF1R), vascular endothelial growth factor (VEGF)R2, and others. The tumor microenvironment consists of cancer cells and stromal/immune cells, such as cancer-associated fibroblasts (CAFs), endothelial cells, M2-type tumor-associating macrophages (M2-TAMs), myeloid-derived suppressor cells (MDSCs) and regulatory T cells. FGFR inhibitors elicit antitumor effects directly on cancer cells, as well as indirectly through the blockade of paracrine signaling. The dual inhibition of FGF and CSF1 or VEGF signaling is expected to enhance the antitumor effects through the targeting of immune evasion and angiogenesis in the tumor microenvironment. Combination therapy using tyrosine kinase inhibitors (FGFR or CSF1R inhibitors) and immune checkpoint blockers (anti-PD-1 or anti-CTLA-4 monoclonal antibodies) may be a promising choice for cancer patients. The inhibition of FGF19-FGFR4 signaling is associated with a risk of liver toxicity, whereas the activation of FGF23-FGFR4 signaling is associated with a risk of heart toxicity. Endocrine FGF signaling affects the pathophysiology of cancer patients who are prescribed FGFR inhibitors. Whole-genome sequencing is necessary for the detection of promoter/enhancer alterations of FGFR genes and rare alterations of other genes causing FGFR overexpression. To sustain the health care system in an aging society, a benefit-cost analysis should be performed with a focus on disease-free survival and the total medical cost before implementing genome-based precision medicine for cancer patients.
Keywords: cholangiocarcinoma, circulating tumor cells, clinical sequencing, CXCR4, FOXP3, gatekeeper mutation, infigratinib, glioblastoma, intra-tumor heterogeneity, ovarian cancer, peritoneal dissemination
1. Introduction
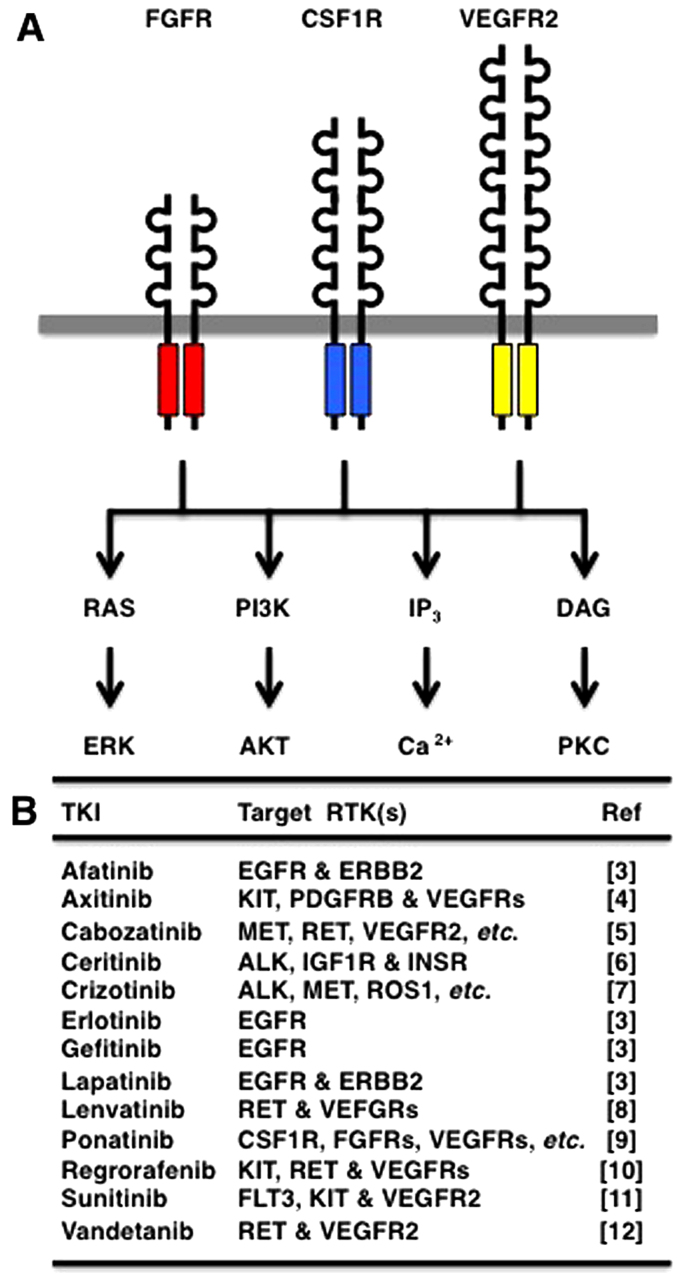
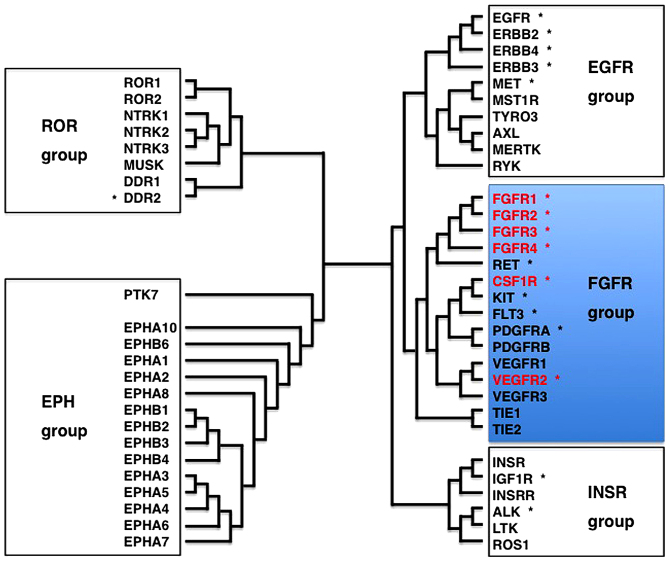
Receptor tyrosine kinases (RTKs) are transmembrane-type receptors with cytoplasmic tyrosine kinase domains (1), which transduce extracellular signals to a variety of intracellular signaling cascades, such as RAS-ERK, PI3K-AKT, IP3-Ca2+ and DAG-PKC (Fig. 1A). Phylogenetic analyses of 518 protein kinases revealed that RTKs are clustered with non-receptor-type tyrosine kinases (2), and analyses of 54 human RTKs using the Clustal Omega program revealed that RTKs are classified into the epidermal growth factor receptor (EGFR) group (EGFR, ERBB2, MET, RYK, etc.), the fibroblast growth factor receptor (FGFR) group [FGFRs, colony stimulating factor 1 receptor (CSF1R), vascular endothelial growth factor (VEGF)R2, etc.], the insulin receptor (INSR) group (INSR, IGF1R, ALK, ROS1, etc.), the RAR-related orphan receptor (ROR) group (ROR1, ROR2, DDR2, NTRK1, etc.) and the EPH receptor (EPH) group (EPHA1, EPHB1, PTK7, etc.) (Fig. 2).
Figure 1.
Receptor tyrosine kinase (RTK) signaling cascades and RTK inhibitors. (A) RTK signaling cascades. Multiple RTKs, such as fibroblast growth factor receptors (FGFRs), CSF1R and VEGFR2, redundantly activate the RAS-ERK, PI3K-AKT, IP3-Ca2+ and DAG-PKC signaling cascades. (B) Representative tyrosine kinase inhibitors (TKIs) and their targets.
Figure 2.
Receptor tyrosine kinase (RTK) superfamily. Phylogenetic tree of 54 human RTKs are shown. *, 18 RTKs in the Oncomine Comprehensive Panel (137). RTKs are classified into the EGFR, FGFR, INSR ROR and EPH groups. FGFR1, FGFR2, FGFR3, FGFR4, CSF1R and VEGFR2, shown in red, belong to the FGFR group.
Since the aberrant activation of RTKs is a driving force of human carcinogenesis, small-molecule inhibitors targeting RTKs have been developed for cancer therapy (3–12). For example, erlotinib and gefetinib target EGFR; afatinib and lapatinib target EGFR and ERBB2; and ponatinib (AP24534) targets multiple RTKs, such as CSF1R, FGFRs, PDGFRs, RET and VEGFRs (Fig. 1B).
FGFR1, FGFR2, FGFR3 and FGFR4 constitute the FGFR family of RTKs with three immunoglobulin-like domains in the extracellular region (13–16). FGF1 (acidic FGF), FGF2 (basic FGF), FGF3-FGF10, FGF16, FGF17, FGF18, FGF20 and FGF22 bind to heparin-sulfate proteoglycan for paracrine signaling through FGFRs, whereas FGF19, FGF21 and FGF23 bind to Klotho proteins for endocrine signaling through FGFRs. FGFRs are involved in the regulation of cell survival, proliferation, differentiation and motility during embryogenesis, adult-tissue homeostasis and carcinogenesis (17–20).
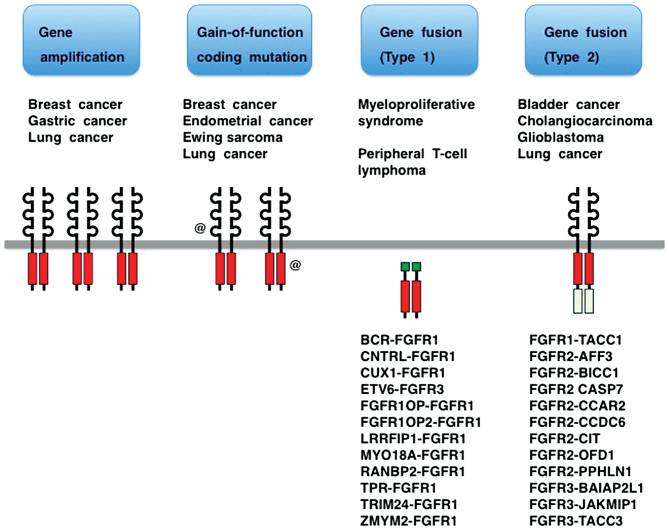
Gene amplification, gain-of-function coding mutation and gene fusion are three major classes of FGFR alterations in human cancer (Fig. 3) (14,21–24). Clinical trials of several tyrosine kinase inhibitors (TKIs) targeting FGFRs are ongoing (25–28), while TKI resistance and tumor-stromal interaction related to FGFRs are hot issues (29–32). Knowledge of FGFRs has been exponentially growing as a result of the advancement of massively parallel sequencing technology combined with the global trend toward translational medicine. In this review, recent progress in the field of FGFR medicine is reviewed with emphases on FGFR alterations in human cancer, the classification of small-molecule FGFR inhibitors and the effects of FGFR inhibitors on the tumor microenvironment and whole-body homeostasis.
Figure 3.
Fibroblast growth factor receptor (FGFR) alterations in human cancer. FGFR genes are activated in human cancer as a result of gene amplification, coding mutation and gene fusion. FGFR gene fusions are further classified into two groups. Type 1 FGFR fusions in hematological malignancies encode non-transmembrane-type FGFR kinases. Type 2 FGFR fusions in solid tumors encode transmembrane-type FGFRs with C-terminal substitution to the region of fusion partners.
2. FGFR genetic alterations in human cancer
FGFR alterations in major cancers
Lung cancer is the most common malignancy worldwide (33), which is histologically classified into adenocarcinoma, large cell carcinoma, small cell carcinoma and squamous cell carcinoma. FGFR1 amplification preferentially occurs in squamous cell lung cancer (34); 9.3% of stage I cases, 22% of stage II cases (35) and 19% of stage IV cases with brain metastasis (36). FGFR2-CCAR2, FGFR2-CIT and FGFR3-TACC3 fusions and gain-of-function mutations of FGFR2 (A266_S267insSTVVGGD and 290_291WI>C) are also observed in lung cancer (37–39). Aberrant FGFR signaling may promote the proliferation and survival of tumor cells in the early stage, and invasion and metastasis in the later stage. As smoking is a risk factor of squamous cell lung cancer, the smoking-induced DNA damage of bronchial epithelial cells may cause lung carcinogenesis in part through FGFR1 gene amplification.
Breast cancer is the second most common malignancy worldwide (33). Breast cancers have been classified based on immunohistochemical analyses of estrogen receptor (ER), progesterone receptor (PR) and ERBB2 (HER2) receptor. The prognosis for luminal breast cancer (ER+) is better than that for non-luminal HER2+ breast cancer (ER−/PR−/HER2+) and triple-negative breast cancer (ER−/PR−/HER2−) (40). Endocrine therapy using tamoxifen or aromatase inhibitors is the standard therapy for patients with luminal breast cancer without lymph node metastasis; however, recurrence as a result of resistance to endocrine therapy is a serious issue in clinical practice (41). FGFR1 gene amplifications and FGFR2 gain-of-function missense mutations are detected in circulating tumor cells of two cases and one case of breast cancer patients, respectively (42,43). The FGFR1 gene is more frequently amplified in metastatic breast cancer than the FGFR2 and FGFR3 genes (44), which leads to FGFR1 overexpression and resistance to endocrine therapy (45). The preclinical studies mentioned above indicate that FGFR1-targeted therapy is applicable for a subset of breast cancer patients.
Gastric cancer is the fifth most common malignancy worldwide, although its incidence and mortality have been decreasing (33,46). The amplifications of genes encoding RTKs, such as EGFR, ERBB2, FGFR2 and MET, occur in gastric cancer (47,48). Gastric cancer with FGFR2 amplification is significantly associated with lymphatic invasion and a poor prognosis (49,50); however, the molecular mechanisms through which FGFR2 amplification promotes lymph node metastasis remain unclear. Preclinical studies using small-molecule FGFR2 inhibitor and patient-derived cancer xenografts revealed that FGFR2 amplification in human gastric cancer is a promising therapeutic target (51,52).
Gain-of-function and loss-of-function mutations
Somatic FGFR1 mutation (N546K) in Ewing sarcoma (53), FGFR2 mutations (R203C, N549K and K659N) in breast cancer (43,54), FGFR2 mutations (S252W and N549K) in endometrial cancer (55), FGFR2 coding in/del mutations in lung cancer (39) and FGFR3 mutations (R248C and S249C) in bladder cancer (56) are gain-of-function mutations (Fig. 3), whereas somatic FGFR2 mutations (D530N, I642V and A648T) in melanoma are loss-of-function mutations (57).
Missense mutations of FGFRs involved in congenital disorders rather than cancer have been well characterized. Gremlin FGFR1 mutations (P252R and Y372C) and FGFR2 mutations (S252W, P253R, K526E, N549K, K641R, etc.) in patients with craniosynostosis and FGFR3 mutations (R248C, S249C, G380R, N540K, K650E, etc.) in patients with skeletal dysplasia are gain-of-function mutations (17–20); however, germline FGFR1 mutations (R254W and V429E) in patients with congenital hypogonadotropic hypogonadism are loss-of-function mutations (58,59).
These facts clearly indicate that there are gain-of-function, as well as loss-of-function mutations in FGFRs. Therefore, validation of gain-of-function based on kinase or cell-based assay is mandatory before prescribing FGFR inhibitors to cancer patients with FGFR coding mutations.
Two types of FGFR fusions
FGFR fusions in human cancers are classified into type 1 fusions caused by chromosomal translocations in hematological malignancies, and type 2 fusions caused by chromosomal rearrangements in solid tumors (Fig. 3). Chromosomal translocations involving the FGFR1 gene occur in patients with myeloproliferative neoplasms initially presenting with eosinophilia and lymphadenopathy and then developing acute myeloid or mixed-lineage leukemia (60,61); chromosomal translocations involving the FGFR3 gene have been shown to occur in patients with peripheral T cell lymphoma (62). Type 1 FGFR fusion proteins, including BCR-FGFR1, CNTRL-FGFR1, CUX1-FGFR1, FGFR1OP-FGFR1, FGFR1OP2-FGFR1, LRRFIP1-FGFR1, MYO18A-FGFR1, RANBP2-FGFR1, TPR-FGFR1, TRIM24-FGFR1, ZMYM2-FGFR1 and ETV6-FGFR3 are non-receptor-type FGFR kinases (Fig. 3). CNTRL-FGFR1 fusion kinase promotes the expansion of the stem cell population and causes myeloid and lymphoid malignancies (63). By contrast, type 2 FGFR fusion proteins in solid tumors, including FGFR1-TACC1, FGFR2-AFF3, FGFR2-BICC1, FGFR2-CASP7, FGFR2-CCAR2, FGFR2-CCDC6, FGFR2-CIT, FGFR2-OFD1, FGFR2-PPHLN1, FGFR3-BAIAP2L1, FGFR3-JAKMIP1 and FGFR3-TACC3, are transmembrane-type FGFRs with C-terminal substitution to the region of fusion partners (37,38,56,64,65) (Fig. 3).
Both types of FGFR fusion proteins are endowed with oncogenic potential through the acquisition of protein-protein-interaction modules from fusion partners for ligand-independent dimerization and/or recruitment of aberrant substrates. Type 1 FGFR fusion proteins acquire oncogenic potential through altered subcellular localization as a result of the loss of the extracellular and transmembrane domains of wild-type FGFRs. Type 2 FGFR fusion proteins lose the PLC-γ-binding tyrosine (Tyr or Y) residue (Y766 in FGFR1, Y769 in FGFR2 or Y760 in FGFR3) owing to the C-terminal alterations. To understand the mechanisms of carcinogenesis caused by the FGFR fusions, substrates and downstream signaling cascades of FGFR fusion proteins need be elucidated.
3. FGFR inhibitors targeting cancer cells
Classification of FGFR inhibitors
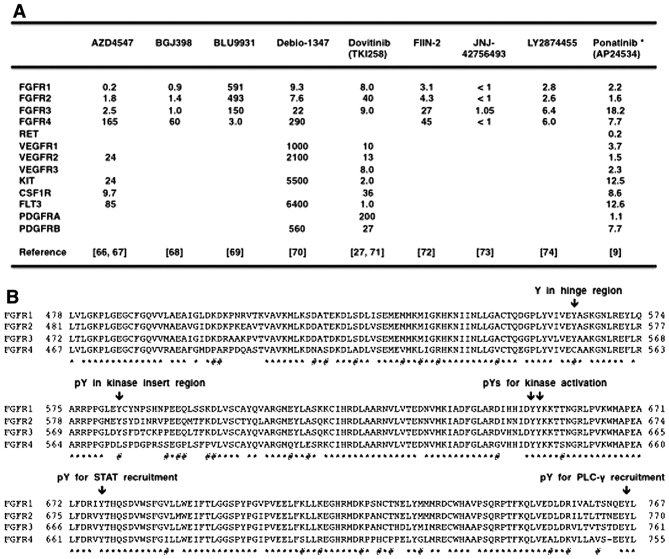
AZD4547 (66,67), BGJ398 (infigratinib) (68), BLU9931 (69), Debio-1347 (70), dovitinib (TKI258) (27,71), FIIN-2 (72), JNJ-42756493 (73), LY2874455 (74) and ponatinib (9) are small molecule compounds that inhibit FGFRs at low nM levels (IC50 value <20 nM) (Fig. 4A). AZD4547, BGJ398, Debio-1347, dovitinib, JNJ-42756493 and LY2874455 are reversible FGFR inhibitors that occupy the ATP-binding pocket in the kinase domain, whereas BLU9931 and FIIN-2 are irreversible FGFR inhibitors that covalently bind to each specific cysteine (Cys or C) residue in the kinase domain. These FGFR inhibitors reduce phosphorylation of FGFRs themselves and their direct targets, FRS2 and PLC-γ, and inactivate downstream signaling, such as the RAS-ERK, PI3K-AKT, IP3-Ca2+ and DAG-PKC signaling cascades.
Figure 4.
Small-molecule fibroblast growth factor receptor (FGFR) inhibitors and FGFRs. (A) Small-molecule FGFR inhibitors. Enzymatic IC50 values for FGFRs and other substrates are listed. (B) Alignment of the tyrosine kinase domain of FGFR1, FGFR2, FGFR3 and FGFR4. Amino-acid position is shown on both sides of the alignment. Amino-acid residues conserved in all members of FGFRs are shown by asterisk, whereas amino-acid residues conserved in FGFR1, FGFR2 and FGFR3 but not in FGFR4 are shown by sharp. FGFR4 is relatively divergent from FGFR1, FGFR2 and FGFR3. Tyrosine residue in the hinge region (Y563 in FGFR1, Y566 in FGFR2 and Y557 in FGFR3) is substituted to C552 in FGFR4, which divergence is involved in the selectivity of FGFR inhibitors for FGFR1/2/3 and FGFR4.
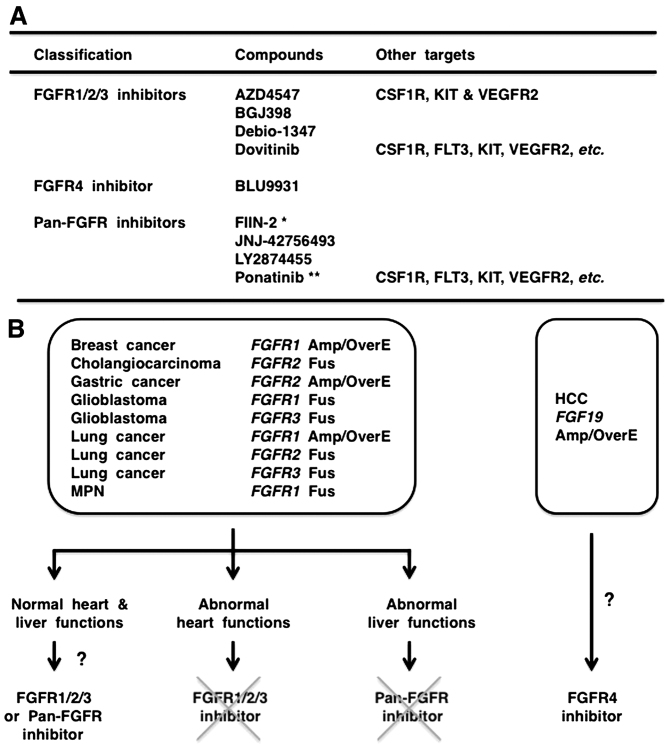
AZD4547, BGJ398, Debio-1347 and dovitinib are FGFR1/2/3 inhibitors that are less effective on FGFR4; BLU9931 is a selective FGFR4 inhibitor; FIIN-2, JNJ-42756493, LY2874455 and ponatinib are pan-FGFR inhibitors (Fig. 5A). Phylogenetic analysis on 54 RTKs revealed diversification of FGFR4 from other FGFRs (Fig. 2), and amino-acid alignment of the tyrosine kinase domains in the FGFR family members revealed relatively frequent amino-acid substitutions specifically in FGFR4 (Fig. 4B). Phospho-tyrosine residues involved in catalytic activation (Y653 and Y654 in FGFR1), STAT recruitment (Y677 in FGFR1) and PLC-γ recruitment (Y766 in FGFR1) are conserved in all members of the FGFR family. By contrast, one tyrosine residue in the hinge region (Y563 in FGFR1, Y566 in FGFR2 and Y557 in FGFR3) is changed to C552 in FGFR4, and phospho-tyrosine residue in the kinase insert region (Y583 in FGFR1, Y586 in FGFR2 and Y577 in FGFR3) is changed to L572 in FGFR4 (Fig. 4B). Y563 in FGFR1 is necessary for the interaction with Debio-1347 (70), whereas C552 in FGFR4 is necessary for the covalent binding with BLU9931 (69). As the diversification of FGFR4 significantly affects the biding affinities of TKIs, it is reasonable to functionally classify FGFR-targeting TKIs into FGFR1/2/3 inhibitors, FGFR4 inhibitor and pan-FGFR inhibitors.
Figure 5.
Selection of fibroblast growth factor receptor (FGFR) inhibitor for precision medicine. (A) Classification of FGFR inhibitors based on substrate specificities. *, FIIN-2 is for experimental use only. **, Severe adverse effects of ponatinib have been reported in patients with chronic myeloid leukemia. (B) Flow-chart for the choice of FGFR inhibitor in clinic. MPN, myeloproliferative neoplasm; Amp, gene amplification; OverE, overexpression; Fus, fusion. FGFR1/2/3 inhibitors and pan-FGFR inhibitors are applicable for human cancer with genetic alteration in FGFR1, FGFR2 or FGFR3. FGFR1/2/3 inhibitors are not the choice for patients with heart diseases, whereas pan-FGFR inhibitors are not the choice for patients with liver dysfunction. FGFR4 inhibitor is applicable for HCC with genetic alteration in FGF19.
Clinical trials and adverse effects
AZD4547, BGJ398, Debio-1347, dovitinib, JNJ-42756493 and ponatinib are currently being investigated in clinical trials (https://clinical-trials.gov): phase II studies of AZD4547 in patients with breast, gastric and squamous-cell lung cancer (FGFR1 or FGFR2 amplification) and metastatic breast or non-small-cell lung cancer (FGFR genetic alterations, umbrella trial); phase II studies of BGJ398 in patients with solid tumors or hematological malignancies (FGFR genetic alterations); phase II study of dovitinib in patients with gastric cancer (FGFR2 amplification); phase II study of JNJ-42756493 in patients with urothelial cancer (FGFR genetic alterations); phase II study of ponatinib in patients with advanced biliary cancer (FGFR2 fusion) or refractory metastatic solid tumors (genetic alterations in FGFRs and other targets).
TKIs have been approved for cancer therapy by regulatory authorities in expectation of an improved risk/benefit ratio; however, adverse effects on viral organs, such as the cardiovascular system and liver, are serious issues that may occur in the clinic (75). Hypertension, bleeding and thrombosis are adverse effects of anti-angiogenic therapy targeting the VEGF signaling pathway (76), while cardiovascular events are serious adverse effects of ponatinib for the treatment of chronic myeloid leukemia (77). AZD4547, dovitinib and ponatinib are representative multi-kinase inhibitors targeting FGFRs and other tyrosine kinases (Fig. 5A). Selective FGFR targeting is expected to reduce adverse effects, whereas the dual targeting of FGFR and VEGFR/CSF1R is expected to enhance the anti-tumor effects indirectly through the normalization of tumor microenvironment.
4. FGFR inhibitors targeting the tumor microenvironment
Tumor microenvironment and paracrine signaling
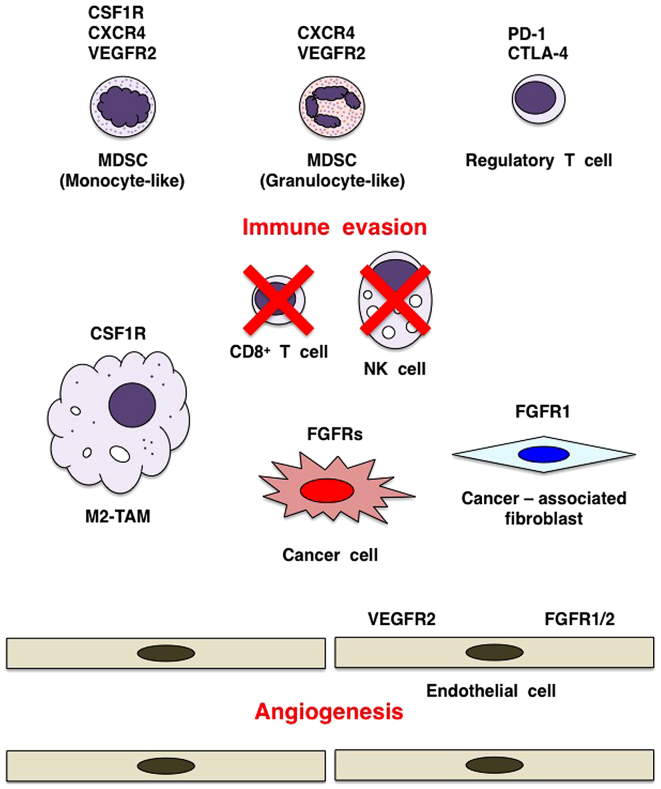
The tumor microenvironment consists of cancer cells and stromal/immune cells, such as fibroblasts, endothelial cells, lymphocytes, macrophages, monocytes and neutrophils (Fig. 6). The interactions of tumor cells and stromal cells are involved in almost all stages of tumor development, including neoplastic transformation, proliferation, invasion and metastasis, through the regulation of various cellular processes in a context-dependent manner (78–80). FGFs derived from cancer cells, as well as stromal cells play a key role in the tumor microenvironment.
Figure 6.
Cancer therapy targeting tumor microenvironment. Cancer cells, cancer-associated fibroblasts (CAFs), endothelial cells, myeloid-derived suppressor cells (MDSCs), tumor-associating macrophages of M2 type (M2-TAMs) and regulatory T cells are representative components of tumor microenvironment. Interactions between cancer cells and stromal/immune cells are involved in almost all steps of carcinogenesis. CSF1 signaling through CSF1R induces proliferation and differentiation of MDSCs and M2-TAMs. Dual inhibition of fibroblast growth factor receptor (FGFR) and CSF1R/VEGFR2 is expected to increase antitumor effects through targeting immune evasion and angiogenesis in the tumor microenvironment. Combination of FGFR/CSF1R inhibitor targeting cancer cells and stromal/immune cells and anti-PD-1/CTLA-4 monoclonal antibody targeting regulatory T cells and de-repressing CD8+ T cells may be a promising choice for cancer patients.
Cancer-associated fibroblasts (CAFs) are activated stromal fibroblasts that support tumorigenesis (Fig. 6). FGF2 activates human dermal fibroblasts through transcriptional downregulation of the TP53 gene, whereas BGJ398 or ponatinib treatment induces their senescence through the upregulation and activation of TP53 (81). By contrast, FGF2 signaling through FGFR1 causes resistance to EGFR inhibitor in lung cancer cells, and combination therapy using EGFR inhibitor and AD4547 is effective to overcome drug resistance (82). Multiple myeloma cells induce FGF23 secretion from osteocytes, and then FGF23 signaling through FGFR3 to multiple myeloma cells promotes proliferation and induces heparanase upregulation, which explains the pathogenesis of osteolytic 'punched-out lesion' in patients with multiple myeloma. BGJ398 treatment inhibits FGF23-dependent growth and heparanase expression of multiple myeloma cells (83). These results indicate the rational for the application of FGFR inhibitors to target paracrine FGF signaling in the tumor microenvironment.
Angiogenesis
Tumor angiogenesis is largely classified into sprouting angiogenesis and vasculogenesis (84). Sprouting angiogenesis is the formation of new blood vessels as a result of endothelial sprouting from preexisting blood vessels, whereas vasculogenesis is the de novo formation of blood vessels owing to endothelial differentiation of progenitor cells and endothelial-like differentiation of cancer cells. Tumor angiogenesis is involved in the supply of oxygen and nutrient (mature blood vessels), as well as the formation of hypoxic environment (immature blood vessels).
VEGF, FGF, angiopoietin (ANGPT) and Notch signaling cascades are major players of tumor angiogenesis (85,86). VEGF signaling through VEGFR2 promotes endothelial cell proliferation via the DAG-PKC signaling cascade, endothelial cell survival via the PI3K-AKT signaling cascade, endothelial cell migration via the FAK-Paxillin signaling cascade as well as vascular permeability and vasodilatation via the IP3-eNOS (NOS3) signaling cascade (76,87,88). Pro-angiogenic FGF2 also promotes the proliferation and migration of endothelial cells directly through FGFR1 (or FGFR2) signaling activation (89,90) and indirectly through the induction/secretion of VEGF and ANGPT2 from endothelial cells (91,92). ANGPT1 is secreted from pericytes and maintains endothelial quiescence or stabilization through TIE2 signaling activation. ANGPT2 is secreted from endothelial cells and promotes the endothelial activation or sprouting through TIE2 signaling inhibition (92,93). VEGF signaling in endothelial tip cells induces DLL4 expression, which subsequently activates Notch signaling in endothelial stalk cells for vascular quiescence through VEGFR downregulation (94–96). VEGF, FGF2 and ANGPT2 are involved in endothelial activation, whereas ANGPT1 and Notch are involved in endothelial quiescence. VEGFR2 and FGFR1/2 on endothelial cells are representative RTKs that promote tumor angiogenesis (Fig. 6).
VEGF signaling is targeted using anti-VEGF monoclonal antibody (mAb) or small-molecule VEGFR inhibitors in cancer patients; however, some tumors do not respond to the VEGF blockade therapy and other tumors recur after transient response [(Gacche and Meshram (76); Jain (78)]. As FGF signaling activation in endothelial cells is one of the mechanisms responsible for intrinsic and acquired resistance to the VEGF blockade therapy (97), FGFR inhibitors may be applicable to overcome the resistance to the VEGF blockade therapy. There are two options for the dual blockade of FGF and VEGF signaling cascades. Combination therapy using FGFR inhibitor and anti-VEGF mAb is a preferable choice to reduce adverse effects, whereas monotherapy using small-molecule FGFR/VEGFR2 dual inhibitors, such as AZD4547 and dovitinob, may be a preferable choice to reduce medical cost. FGF/VEGF dual blockade therapy should be optimized in consideration of safety issues and medical costs.
Immune evasion
Cancer immunity and immune tolerance in the tumor microenvironment are regulated by the interaction between cancer cells and immune cells (98). CD8+ T cells, NK cells and NKT cells are immune effector cells involved in tumor elimination (98), whereas myeloid-derived suppressor cells (MDSCs) (99), tumor-associating macrophages of M2 type (M2-TAMs) (100) and regulatory T (Treg) cells (101) are immune modifier cells involved in immune evasion and tumor growth (Fig. 6).
MDSCs are heterogeneous populations of immature myeloid cells, including monocyte-like MDSCs (CD14+, CXCR4+, CSF1R+ and VEGFR2+), granulocyte-like MDSCs (CD15+, CXCR4+, KIT+ and VEGFR2+) and endothelial progenitor cells (CD31+, CXCR4+, KIT+ and VEGFR2+) (99). CSF1 (M-CSF), CSF2 (GM-CSF) and CSF3 (G-CSF) are secreted from the tumor microenvironment and stimulate the growth and survival of MDSCs and other myeloid-lineage cells, while CXCL12 (SFD-1α) and VEGF promote the recruitment of MDSCs to the tumor microenvironment (102–106). MDSCs activate M2-TAMs and Treg cells, but inhibit CD8+ T cells and NK cells, leading to immune evasion in the tumor microenvironment. In addition, endothelial progenitor cell-like MDSCs are involved in tumor angiogenesis (104). MDSC infiltration and tumor angiogenesis during mammary tumorigenesis in MMTV-Wnt1/iFGFR1 bi-genic mice are significantly enhanced in comparison with MMTV-Wnt1 transgenic mice, and BGJ398 treatment results in tumor regression and disappearance of MDSCs from the residual mammary gland (107). By contrast, AZD4547 treatment inhibits the proliferation and lung metastasis of 4T1 mouse mammary tumor cells, and reduces MDSCs in the tumor microenvironment and systemic circulation (108). FGFR inhibitors induce the reduction or disappearance of MDSCs from the tumor microenvironment, partly by targeting cytokine-producing CAFs.
CSF1 signaling through CSF1R on monocyte/macrophage-lineage cells are involved in their proliferation, survival and differentiation (109–111). CSF1R inhibitors (GW2580 and PLX3397) and anti-CSF1R mAb (RG7155) have been developed as therapeutics for CSF1 signaling blockade in monocyte-like MDSCs and M2-TAM (112–115). Combination therapy of CSF1R inhibitor PLX3397 and paclitaxel inhibits tumor-infiltration of MDCSs and M2-TAM and suppresses mammary tumorigenesis (113,114). IC50 value of PLX3397 to CSF1R is 20 nM (113), whereas IC50 values of AZD4547 (67), ponatinib (9) and dovitinib (71) to CSF1R are 9.7, 8.7 and 36 nM, respectively. By contrast, PI3K is one of common signaling effectors CSF1R and FGFRs (Fig. 1A), and PI3K activation enhances immune suppressor and pro-angiogenic potentials of M2-TAMs (116). Therefore, as CSF1 and FGF signals are both involved in the accumulation of tumor-infiltrating/promoting MDSCs and M2-TAMs, the dual inhibition of CSF1R and FGFRs may be more effective for cancer therapy than selective CSF1R inhibition.
5. FGFR inhibitors affecting whole-body homeostasis
Endocrine FGF signaling
FGF19, FGF21 and FGF23 are endocrine FGFs that transduce signals to target organs through FGFRs and the Klotho family of co-receptors, α-Klotho (KL) and β-Klotho (KLB) (117). FGF19 is upregulated by bile acid in the intestine to transduce endocrine signaling through FGFR4 and β-Klotho in the liver. FGF21 is upregulated by fasting in the liver and adipose tissue to transduce paracrine signaling through FGFR1 and β-Klotho locally and endocrine signaling to the pancreas and brain. FGF23 is upregulated by serum phosphate, vitamin D and parathyroid hormone in bone to transduce endocrine signaling through FGFR1 and α-Klotho in the kidneys and negative feedback signaling through FGFR3 and α-Klotho in the parathyroid gland (118). As endocrine FGFs are involved in the maintenance of whole-body homeostasis, FGFR inhibitors elicit endocrine or metabolic abnormalities. This section will be focused on adverse effects of FGFR inhibitors on endocrine FGF signaling in cancer patients.
FGF19-FGFR4 signaling in liver homeostasis
FGF19 signaling through FGFR4 in the liver stimulates hepatocyte proliferation and glycogen synthesis but reduces bile acid synthesis and triglyceride synthesis (69,119). FGF19-FGFR4 signaling blockade in cynomolgus monkeys using anti-FGF19 monoclonal antibody causes hepatotoxicity, increased bile acid secretion and severe diarrhea (120). Fgfr4 knockout in mice also causes increased bile acid secretion in the liver, which leads to induction of Fgf15 (mouse ortholog of human FGF19) in the intestine and subsequent improvement of insulin resistance and glucose metabolism (121). The selective FGFR4 inhibitor, BLU9931, may be applied for the treatment of patients with hepatocellular carcinoma depending on FGF19-FGFR4 signaling. By contrast, as FGFR4 blockade is associated with a risk of liver toxicity, FGFR1/2/3 inhibitors rather than pan-FGFR inhibitors are preferable for the treatment of cancer patients with genetic alterations in FGFR1, FGFR2 or FGFR3, especially those with liver dysfunction (Fig. 5B).
FGF23-FGFR4 signaling in heart homeostasis
Serum FGF23 elevation is a biomarker indicating on-target effects of FGFR1/2/3 and pan-FGFR inhibitors in cancer patients, whereas serum FGF23 levels are also elevated in patients with non-cancerous diseases, such as hypophosphatemic rickets and chronic kidney diseases (118). Physiological FGF23 signaling through FGFR1 and α-Klotho in the kidneys decreases the serum phosphate level through the downregulation of phosphate reabsorption. FGFR inhibitors, hindering FGF23 signaling in the kidneys, promote hyperphosphatemia and subsequent FGF23 secretion from bone and soft-tissue mineralization. Pathological FGF23 upregulation is associated with endothelial dysfunction and arterial stiffness (122). Pathological FGF23 signaling through FGFR4 in cardiac myocytes then induces phosphorylation of PLC-γ and activation of the IP3-Ca2+ signaling cascade, which results in cardiac remodeling, such as cardiac hypertrophy and cardiac fibrosis (123). As FGF23-FGFR4 signaling activation is associated with a risk of cardiac toxicity, pan-FGFR inhibitors rather than FGFR1/2/3 inhibitors may be selected for the treatment of cancer patients with FGFR genetic alterations, particularly those with heart dysfunction (Fig. 5B).
6. Conclusion
FGFR genetic alterations in human cancer are classified into gene amplification, gain-of-function coding mutation and two types of fusions. AZD4547, BGJ398, Debio-1347 and dovitinib are FGFR1/2/3 inhibitors; BLU9931 is a selective FGFR4 inhibitor; FIIN-2, JNJ-42756493, LY2874455 and ponatinib are pan-FGFR inhibitors. FGFR inhibitors elicit antitumor effects directly on cancer cells as well as indirectly through normalization of tumor microenvironment, especially paracrine signaling, angiogenesis and immune evasion. FGF19-FGFR4 signaling inhibition is associated with a risk of liver toxicity, whereas FGF23-FGFR4 signaling activation is associated with a risk of heart toxicity. Endocrine FGF signaling affects pathophysiology of cancer patients with FGFR-targeting therapy.
7. Perspectives
Massively parallel sequencing technology for the whole-exome or whole-genome sequencing has been used to clarify genomic landscapes in various types of human cancer (124). The over-expression of FGFR occurs in human cancers through gene amplification, as well as other types of aberrations. Rearrangement in the distal enhancer region and point mutation in the proximal promoter region are both able to induce FGFR overexpression. Repression of FGFR-targeting microRNA (miRNA) precursor gene or upregulation of long-non-coding RNA (lncRNA) sequestering FGFR-targeting miRNA leads to FGFR overexpression. SWI/SNF mutation dysregulating chromatin remodeling, as well as cancer-associated fusion transcription factor also cause FGFR overexpression. For example, lung cancer cells with FGFR1 upregulation rather than FGFR1 copy number gain are sensitive to ponatinib (125). Rhabdoid tumor cells with FGFR overexpression as a result of SMARCB1 (SNF5) deletion are sensitive to BGJ398 (126). Myxoid liposarcoma cells with FGFR2 upregulation owing to FUS-DDIT3 or EWS1R-DDIT3 fusion are sensitive to BGJ398 and dovitinib (127). Taken together, these facts clearly indicate that FGFR inhibitors are applicable for the treatment of cancers with FGFR overexpression in the absence of gene amplification, particularly rare cancers with their specific alterations inducing FGFR overexpression. However, development of biomarkers for FGF dependence is necessary before clinical application of FGFR inhibitors for the treatment of cancers with FGFR overexpression.
Genomic heterogeneity is the major mechanism of tumor evolution for recurrence after cancer therapy (128). FGFR2-BICC1 and FGFR2-PPHLN1 fusions in mostly distinct, but some overlapping cases of cholangiocarcinomas (65) suggest convergent evolution and intra-tumor heterogeneity, respectively. Resistance to EGFR-targeted therapy occurs based on paracrine FGF signaling from tumor-stromal cells (82) or activating FGFR alterations, such as FGFR1 amplification and FGFR3 mutation, in cancer cells (129,130). On the other hand, resistance to FGFR-targeted therapy occurs based on paracrine signaling through EGFR/HER2/MET (29,107) or secondary FGFR alterations (131). FGFR1 V561M (34), FGFR2 V564M (72), FGFR3 V555M (131) and FGFR4 V550M (132) are gatekeeper mutations that cause resistance to ATP-competitive FGFR inhibitors, such as AZD4547 and BGJ398, whereas FGFR2 V564M is sensitive to a covalent pan-FGFR inhibitor, FIIN-2 (72). Because antitumor effects of FIIN-2 are limited to cell-based assays, orally bioavailable derivatives of FIIN-2 should be developed for the treatment of cancer resistant to ATP-competitive FGFR inhibitors.
Immune-checkpoint blockade therapy is a frontier in the field of clinical oncology. PD-1 ligand (PD-L1) is expressed on cancer cells and stromal/immune cells, whereas PD-1 and CTLA-4 are expressed on CD8+ T cells and Treg cells (133–136). As PD-1 signaling and CTLA-4 signaling are both involved in functional suppression of cytotoxic T cells directly or indirectly through Treg cells, anti-PD-1 mAb, anti-PD-L1 mAb and anti-CTLA-4 mAb are clinically applied for cancer immunotherapy, which leads to sustainable remission in a fraction of patients. By contrast, FGFR and CSF1R inhibitors are shown to target immune cells, such as MDSCs and M2-TAMs, in the tumor microenvironment (Fig. 6) and are expected to indirectly repress PD-L1 expression on tumor cells and stromal/immune cells through normalization of tumor microenvironment. Therefore, combination therapy using TKI (FGFR or CSF1R inhibitor) and immune checkpoint blocker (anti-PD-1 or anti-CTLA-4 mAb) may be a promising choice for cancer patients.
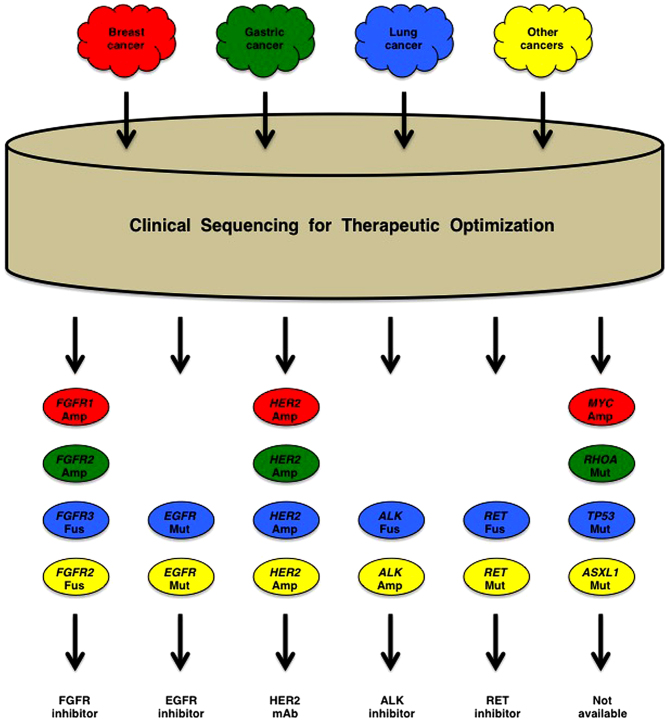
Cancer patients are prescribed appropriate drug based on their genetic alterations to reduce the costs of diagnosis and to increase the amounts of knowledge (Fig. 7). Partial-exome sequencing of a panel of cancer-associated genes are utilized for therapeutic optimization of cancer patients in the field of clinical oncology, which is relatively inexpensive but unable to detect cis-acting enhancer/promoter alterations and transacting rare coding alterations. By contrast, integrative genomic analyses based on whole-genome sequencing are utilized for precise characterization of human cancers in the field of basic oncology, which is expensive but comprehensive and informative. As a benefit-cost ratio is a critical issue to sustain health care system of aging society, it is necessary to discuss the benefit-cost issue with a focus on disease-free survival and total medical cost before implementation of genome-based precision medicine for cancer patients.
Figure 7.
Genome-based precision medicine for cancer patients. Amp, amplification; Fus, fusion; Mut, mutation. Patients with breast cancer (red), gastric cancer (green), lung cancer (blue) and other cancers (yellow) are reorganized into the groups of cancer patients with specific genetic alterations. Patients with fibroblast growth factor receptor (FGFR), EGFR, HER2, ALK and RET alterations are prescribed FGFR inhibitor, EGFR inhibitor, HER2 monoclonal antibody (mAb), ALK inhibitor and RET inhibitor, respectively. However, at present, there is no targeted therapy for cancer patients with MYC Amp, RHOA Mut, TP53 Mut, ASXL1 Mut, etc.
Acknowledgments
This study was financially supported in part by a Grant-in-Aid for the Knowledgebase Project from the M. Katoh's Fund.
References
- 1.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Rugo HS, Herbst RS, Liu G, Park JW, Kies MS, Steinfeldt HM, Pithavala YK, Reich SD, Freddo JL, Wilding G. Phase I trial of the oral antiangiogenesis agent AG-013736 in patients with advanced solid tumors: Pharmacokinetic and clinical results. J Clin Oncol. 2005;23:5474–5483. doi: 10.1200/JCO.2005.04.192. [DOI] [PubMed] [Google Scholar]
- 5.Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P, Qian F, Chu F, Bentzien F, Cancilla B, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- 6.Marsilje TH, Pei W, Chen B, Lu W, Uno T, Jin Y, Jiang T, Kim S, Li N, Warmuth M, et al. Synthesis, structure-activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulfonyl)phenyl) pyrimidine-2,4-diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J Med Chem. 2013;56:5675–5690. doi: 10.1021/jm400402q. [DOI] [PubMed] [Google Scholar]
- 7.Cui JJ, Tran-Dubé M, Shen H, Nambu M, Kung PP, Pairish M, Jia L, Meng J, Funk L, Botrous I, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54:6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 8.Tohyama O, Matsui J, Kodama K, Hata-Sugi N, Kimura T, Okamoto K, Minoshima Y, Iwata M, Funahashi Y. Antitumor activity of lenvatinib (e7080): An angiogenesis inhibitor that targets multiple receptor tyrosine kinases in preclinical human thyroid cancer models. J Thyroid Res. 2014;2014:638747. doi: 10.1155/2014/638747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm SM, Dumas J, Adnane L, Lynch M, Carter CA, Schütz G, Thierauch KH, Zopf D. Regorafenib (BAY 73-4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer. 2011;129:245–255. doi: 10.1002/ijc.25864. [DOI] [PubMed] [Google Scholar]
- 11.Chow LQ, Eckhardt SG. Sunitinib: From rational design to clinical efficacy. J Clin Oncol. 2007;25:884–896. doi: 10.1200/JCO.2006.06.3602. [DOI] [PubMed] [Google Scholar]
- 12.Wells SA, Jr, Gosnell JE, Gagel RF, Moley J, Pfister D, Sosa JA, Skinner M, Krebs A, Vasselli J, Schlumberger M. Vandetanib for the treatment of patients with locally advanced or metastatic hereditary medullary thyroid cancer. J Clin Oncol. 2010;28:767–772. doi: 10.1200/JCO.2009.23.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Katoh M, Nakagama H. FGF receptors: Cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 15.Coleman SJ, Bruce C, Chioni AM, Kocher HM, Grose RP. The ins and outs of fibroblast growth factor receptor signalling. Clin Sci (Lond) 2014;127:217–231. doi: 10.1042/CS20140100. [DOI] [PubMed] [Google Scholar]
- 16.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh M. FGFR2 abnormalities underlie a spectrum of bone, skin, and cancer pathologies. J Invest Dermatol. 2009;129:1861–1867. doi: 10.1038/jid.2009.97. [DOI] [PubMed] [Google Scholar]
- 18.Turner N, Grose R. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 19.Kelleher FC, O'Sullivan H, Smyth E, McDermott R, Viterbo A. Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis. 2013;34:2198–2205. doi: 10.1093/carcin/bgt254. [DOI] [PubMed] [Google Scholar]
- 20.Helsten T, Schwaederle M, Kurzrock R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: Biologic and clinical implications. Cancer Metastasis Rev. 2015;34:479–496. doi: 10.1007/s10555-015-9579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 22.Chang J, Liu X, Wang S, Zhang Z, Wu Z, Zhang X, Li J. Prognostic value of FGFR gene amplification in patients with different types of cancer: A systematic review and meta-analysis. PLoS One. 2014;9:e105524. doi: 10.1371/journal.pone.0105524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker BC, Zhang W. Fusion genes in solid tumors: An emerging target for cancer diagnosis and treatment. Chin J Cancer. 2013;32:594–603. doi: 10.5732/cjc.013.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S, Zhou L, Nice EC, Huang C. Fibroblast growth factor receptors: Multifactorial-contributors to tumor initiation and progression. Histol Histopathol. 2015;30:13–31. doi: 10.14670/HH-30.13. [DOI] [PubMed] [Google Scholar]
- 25.Liang G, Chen G, Wei X, Zhao Y, Li X. Small molecule inhibition of fibroblast growth factor receptors in cancer. Cytokine Growth Factor Rev. 2013;24:467–475. doi: 10.1016/j.cytogfr.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 26.André F, Cortés J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res Treat. 2015;150:1–8. doi: 10.1007/s10549-015-3301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porta C, Giglione P, Liguigli W, Paglino C. Dovitinib (CHIR258, TKI258): Structure, development and preclinical and clinical activity. Future Oncol. 2015;11:39–50. doi: 10.2217/fon.14.208. [DOI] [PubMed] [Google Scholar]
- 28.Carter EP, Fearon AE, Grose RP. Careless talk costs lives: Fibroblast growth factor receptor signalling and the consequences of pathway malfunction. Trends Cell Biol. 2015;25:221–233. doi: 10.1016/j.tcb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Chang J, Wang S, Zhang Z, Liu X, Wu Z, Geng R, Ge X, Dai C, Liu R, Zhang Q, et al. Multiple receptor tyrosine kinase activation attenuates therapeutic efficacy of the fibroblast growth factor receptor 2 inhibitor AZD4547 in FGFR2 amplified gastric cancer. Oncotarget. 2015;6:2009–2022. doi: 10.18632/oncotarget.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Mikse O, Liao RG, Li Y, Tan L, Janne PA, Gray NS, Wong KK, Hammerman PS. Ligand-associated ERBB2/3 activation confers acquired resistance to FGFR inhibition in FGFR3-dependent cancer cells. Oncogene. 2015;34:2167–2177. doi: 10.1038/onc.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronca R, Giacomini A, Rusnati M, Presta M. The potential of fibroblast growth factor/fibroblast growth factor receptor signaling as a therapeutic target in tumor angiogenesis. Expert. Opin Ther Targets. 2015;19:1361–1377. doi: 10.1517/14728222.2015.1062475. [DOI] [PubMed] [Google Scholar]
- 32.Salazar L, Kashiwada T, Krejci P, Meyer AN, Casale M, Hallowell M, Wilcox WR, Donoghue DJ, Thompson LM. Fibroblast growth factor receptor 3 interacts with and activates TGFβ-activated kinase 1 tyrosine phosphorylation and NFκB signaling in multiple myeloma and bladder cancer. PLoS One. 2014;9:e86470. doi: 10.1371/journal.pone.0086470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 34.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cihoric N, Savic S, Schneider S, Ackermann I, Bichsel-Naef M, Schmid RA, Lardinois D, Gugger M, Bubendorf L, Zlobec I, et al. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br J Cancer. 2014;110:2914–2922. doi: 10.1038/bjc.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preusser M, Berghoff AS, Berger W, Ilhan-Mutlu A, Dinhof C, Widhalm G, Dieckmann K, Wöhrer A, Hackl M, von Deimling A, et al. High rate of FGFR1 amplifications in brain metastases of squamous and non-squamous lung cancer. Lung Cancer. 2014;83:83–89. doi: 10.1016/j.lungcan.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanizaki J, Ercan D, Capelletti M, Dodge M, Xu C, Bahcall M, Tricker EM, Butaney M, Calles A, Sholl LM, et al. Identification of oncogenic and drug-sensitizing mutations in the extracellular domain of FGFR2. Cancer Res. 2015;75:3139–3146. doi: 10.1158/0008-5472.CAN-14-3771. [DOI] [PubMed] [Google Scholar]
- 40.Maxmen A. The hard facts. Nature. 2012;485:S50–S51. doi: 10.1038/485S50a. [DOI] [PubMed] [Google Scholar]
- 41.Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 2013;3:27–34. doi: 10.1158/2159-8290.CD-12-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoo BL, Lee SC, Kumar P, Tan TZ, Warkiani ME, Ow SG, Nandi S, Lim CT, Thiery JP. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget. 2015;6:15578–15593. doi: 10.18632/oncotarget.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.André F, Bachelot T, Commo F, Campone M, Arnedos M, Dieras V, Lacroix-Triki M, Lacroix L, Cohen P, Gentien D, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: A multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15:267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 45.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality 1980–2011 with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 47.Holbrook JD, Parker JS, Gallagher KT, Halsey WS, Hughes AM, Weigman VJ, Lebowitz PF, Kumar R. Deep sequencing of gastric carcinoma reveals somatic mutations relevant to personalized medicine. J Transl Med. 2011;9:119. doi: 10.1186/1479-5876-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung EJ, Jung EJ, Min SY, Kim MA, Kim WH. Fibroblast growth factor receptor 2 gene amplification status and its clinicopathologic significance in gastric carcinoma. Hum Pathol. 2012;43:1559–1566. doi: 10.1016/j.humpath.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Su X, Zhan P, Gavine PR, Morgan S, Womack C, Ni X, Shen D, Bang YJ, Im SA, Ho Kim W, et al. FGFR2 amplification has prognostic significance in gastric cancer: Results from a large international multicentre study. Br J Cancer. 2014;110:967–975. doi: 10.1038/bjc.2013.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X, Xu Y, Gao Z, Liu K, Zhou M, et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res. 2013;19:2572–2583. doi: 10.1158/1078-0432.CCR-12-3898. [DOI] [PubMed] [Google Scholar]
- 52.Zhang T, Zhang L, Fan S, Zhang M, Fu H, Liu Y, Yin X, Chen H, Xie L, Zhang J, et al. Patient-derived gastric carcinoma xenograft mouse models faithfully represent human tumor molecular diversity. PLoS One. 2015;10:e0134493. doi: 10.1371/journal.pone.0134493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agelopoulos K, Richter GH, Schmidt E, Dirksen U, von Heyking K, Moser B, Klein HU, Kontny U, Dugas M, Poos K, et al. Deep sequencing in conjunction with expression and functional analyses reveals activation of FGFR1 in Ewing sarcoma. Clin Cancer Res. 2015;21:4935–4946. doi: 10.1158/1078-0432.CCR-14-2744. [DOI] [PubMed] [Google Scholar]
- 54.Reintjes N, Li Y, Becker A, Rohmann E, Schmutzler R, Wollnik B. Activating somatic FGFR2 mutations in breast cancer. PLoS One. 2013;8:e60264. doi: 10.1371/journal.pone.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byron SA, Gartside M, Powell MA, Wellens CL, Gao F, Mutch DG, Goodfellow PJ, Pollock PM. FGFR2 point mutations in 466 endometrioid endometrial tumors: Relationship with MSI, KRAS, PIK3CA, CTNNB1 mutations and clinicopathological features. PLoS One. 2012;7:e30801. doi: 10.1371/journal.pone.0030801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ross JS, Wang K, Al-Rohil RN, Nazeer T, Sheehan CE, Otto GA, He J, Palmer G, Yelensky R, Lipson D, et al. Advanced urothelial carcinoma: Next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2014;27:271–280. doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 57.Gartside MG, Chen H, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL, Bengston A, Yudt LM, Eliseenkova AV, Ma J, et al. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol Cancer Res. 2009;7:41–54. doi: 10.1158/1541-7786.MCR-08-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koika V, Varnavas P, Valavani H, Sidis Y, Plummer L, Dwyer A, Quinton R, Kanaka-Gantenbein C, Pitteloud N, Sertedaki A, et al. Comparative functional analysis of two fibroblast growth factor receptor 1 (FGFR1) mutations affecting the same residue (R254W and R254Q) in isolated hypogonadotropic hypogonadism (IHH) Gene. 2013;516:146–151. doi: 10.1016/j.gene.2012.12.041. [DOI] [PubMed] [Google Scholar]
- 59.Villanueva C, Jacobson-Dickman E, Xu C, Manouvrier S, Dwyer AA, Sykiotis GP, Beenken A, Liu Y, Tommiska J, Hu Y, et al. Congenital hypogonadotropic hypogonadism with split hand/foot malformation: A clinical entity with a high frequency of FGFR1 mutations. Genet Med. 2015;17:651–659. doi: 10.1038/gim.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: A review. Hum Pathol. 2010;41:461–476. doi: 10.1016/j.humpath.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Kumar KR, Chen W, Koduru PR, Luu HS. Myeloid and lymphoid neoplasm with abnormalities of FGFR1 presenting with trilineage blasts and RUNX1 rearrangement: A case report and review of literature. Am J Clin Pathol. 2015;143:738–748. doi: 10.1309/AJCPUD6W1JLQQMNA. [DOI] [PubMed] [Google Scholar]
- 62.Yagasaki F, Wakao D, Yokoyama Y, Uchida Y, Murohashi I, Kayano H, Taniwaki M, Matsuda A, Bessho M. Fusion of ETV6 to fibroblast growth factor receptor 3 in peripheral T-cell lymphoma with a t(4;12)(p16;p13) chromosomal translocation. Cancer Res. 2001;61:8371–8374. [PubMed] [Google Scholar]
- 63.Ren M, Qin H, Kitamura E, Cowell JK. Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood. 2013;122:1007–1016. doi: 10.1182/blood-2013-03-489823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, et al. St Jude Children's Research Hospital-Washington University Pediatric Cancer Genome Project: Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K, Bonal D, Miltiadous O, Zhang Z, Hoshida Y, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun. 2015;6:6087. doi: 10.1038/ncomms7087. [DOI] [PubMed] [Google Scholar]
- 66.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, Rooney C, Coleman T, Baker D, Mellor MJ, et al. AZD4547: An orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 67.Kwak Y, Cho H, Hur W, Sim T. Antitumor effects and mechanisms of AZD4547 on FGFR2-deregulated endometrial cancer cells. Mol Cancer Ther. 2015;14:2292–2302. doi: 10.1158/1535-7163.MCT-15-0032. [DOI] [PubMed] [Google Scholar]
- 68.Guagnano V, Kauffmann A, Wöhrle S, Stamm C, Ito M, Barys L, Pornon A, Yao Y, Li F, Zhang Y, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–1133. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- 69.Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, Bifulco N, Kim JL, Hodous B, Brooijmans N, et al. First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 2015;5:424–437. doi: 10.1158/2159-8290.CD-14-1029. [DOI] [PubMed] [Google Scholar]
- 70.Nakanishi Y, Akiyama N, Tsukaguchi T, Fujii T, Sakata K, Sase H, Isobe T, Morikami K, Shindoh H, Mio T, et al. The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Mol Cancer Ther. 2014;13:2547–2558. doi: 10.1158/1535-7163.MCT-14-0248. [DOI] [PubMed] [Google Scholar]
- 71.Lee SH, Lopes de Menezes D, Vora J, Harris A, Ye H, Nordahl L, Garrett E, Samara E, Aukerman SL, Gelb AB, et al. In vivo target modulation and biological activity of CHIR-258, a multitargeted growth factor receptor kinase inhibitor, in colon cancer models. Clin Cancer Res. 2005;11:3633–3641. doi: 10.1158/1078-0432.CCR-04-2129. [DOI] [PubMed] [Google Scholar]
- 72.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, Zhu SJ, Zhang Y, Ercan D, Liao RG, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci USA. 2014;111:E4869–E4877. doi: 10.1073/pnas.1403438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tabernero J, Bahleda R, Dienstmann R, Infante JR, Mita A, Italiano A, Calvo E, Moreno V, Adamo B, Gazzah A, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2015;33:3401–3408. doi: 10.1200/JCO.2014.60.7341. [DOI] [PubMed] [Google Scholar]
- 74.Zhao G, Li WY, Chen D, Henry JR, Li HY, Chen Z, Zia-Ebrahimi M, Bloem L, Zhai Y, Huss K, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10:2200–2210. doi: 10.1158/1535-7163.MCT-11-0306. [DOI] [PubMed] [Google Scholar]
- 75.Shah RR, Morganroth J. Update on cardiovascular safety of tyrosine kinase inhibitors: With a special focus on QT interval, left ventricular dysfunction and overall risk/benefit. Drug Saf. 2015;38:693–710. doi: 10.1007/s40264-015-0300-1. [DOI] [PubMed] [Google Scholar]
- 76.Gacche RN, Meshram RJ. Angiogenic factors as potential drug target: Efficacy and limitations of anti-angiogenic therapy. Biochim Biophys Acta. 2014;1846:161–179. doi: 10.1016/j.bbcan.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Douxfils J, Haguet H, Mullier F, Chatelain C, Graux C, Dogné JM. Association between BCR-ABL tyrosine kinase inhibitors for chronic myeloid leukemia and cardiovascular events, major molecular response, and overall survival: A systematic review and meta-analysis. JAMA Oncol. 2016 Feb 4; doi: 10.1001/jamaoncol.2015.5932. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 78.Jain RK. Normalizing tumor microenvironment to treat cancer: Bench to bedside to biomarkers. J Clin Oncol. 2013;31:2205–2218. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 81.Procopio MG, Laszlo C, Al Labban D, Kim DE, Bordignon P, Jo SH, Goruppi S, Menietti E, Ostano P, Ala U, et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol. 2015;17:1193–1204. doi: 10.1038/ncb3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, Cummings CT, Graham DK, Astling D, Tan AC, et al. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suvannasankha A, Tompkins DR, Edwards DF, Petyaykina KV, Crean CD, Fournier PG, Parker JM, Sandusky GE, Ichikawa S, Imel EA, et al. FGF23 is elevated in multiple myeloma and increases heparanase expression by tumor cells. Oncotarget. 2015;6:19647–19660. doi: 10.18632/oncotarget.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weis SM, Cheresh DA. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 85.Bridges E, Oon CE, Harris A. Notch regulation of tumor angiogenesis. Future Oncol. 2011;7:569–588. doi: 10.2217/fon.11.20. [DOI] [PubMed] [Google Scholar]
- 86.Katoh M. Therapeutics targeting angiogenesis: Genetics and epigenetics, extracellular miRNAs and signaling networks (Review) Int J Mol Med. 2013;32:763–767. doi: 10.3892/ijmm.2013.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmitt J, Matei D. Targeting angiogenesis in ovarian cancer. Cancer Treat Rev. 2012;38:272–283. doi: 10.1016/j.ctrv.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 88.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. 2013;13:871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oladipupo SS, Smith C, Santeford A, Park C, Sene A, Wiley LA, Osei-Owusu P, Hsu J, Zapata N, Liu F, et al. Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proc Natl Acad Sci USA. 2014;111:13379–13384. doi: 10.1073/pnas.1324235111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao L, Yang S, Hao J, Yuan X, Luo W, Jiang L, Hu Y, Fu Z, Zhang Y, Zou C. Endostar attenuates melanoma tumor growth via its interruption of b-FGF mediated angiogenesis. Cancer Lett. 2015;359:148–154. doi: 10.1016/j.canlet.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 91.Choi HJ, Armaiz Pena GN, Pradeep S, Cho MS, Coleman RL, Sood AK. Anti-vascular therapies in ovarian cancer: Moving beyond anti-VEGF approaches. Cancer Metastasis Rev. 2015;34:19–40. doi: 10.1007/s10555-014-9538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hilbert T, Klaschik S. The angiopoietin/TIE receptor system: Focusing its role for ischemia-reperfusion injury. Cytokine Growth Factor Rev. 2015;26:281–291. doi: 10.1016/j.cytogfr.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 93.Fagiani E, Christofori G. Angiopoietins in angiogenesis. Cancer Lett. 2013;328:18–26. doi: 10.1016/j.canlet.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 94.Zhou W, Wang G, Guo S. Regulation of angiogenesis via Notch signaling in breast cancer and cancer stem cells. Biochim Biophys Acta. 2013;1836:304–320. doi: 10.1016/j.bbcan.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rostama B, Peterson SM, Vary CP, Liaw L. Notch signal integration in the vasculature during remodeling. Vascul Pharmacol. 2014;63:97–104. doi: 10.1016/j.vph.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang P, Yan X, Chen Y, Yang Z, Han H. Notch signaling in blood vessels: From morphogenesis to homeostasis. Sci China Life Sci. 2014;57:774–780. doi: 10.1007/s11427-014-4716-0. [DOI] [PubMed] [Google Scholar]
- 97.Bertolini F, Marighetti P, Martin-Padura I, Mancuso P, Hu-Lowe DD, Shaked Y, D'Onofrio A. Anti-VEGF and beyond: Shaping a new generation of anti-angiogenic therapies for cancer. Drug Discov Today. 2011;16:1052–1060. doi: 10.1016/j.drudis.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 98.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: Integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 99.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mantovani A, Sica A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 101.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 102.Balkwill FR. The chemokine system and cancer. J Pathol. 2012;226:148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 103.Lippitz BE. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013:e218–e228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 104.Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med. 2015;66:97–110. doi: 10.1146/annurev-med-051013-052304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rivera LB, Bergers G. Intertwined regulation of angiogenesis and immunity by myeloid cells. Trends Immunol. 2015;36:240–249. doi: 10.1016/j.it.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Y, Ramjiawan RR, Reiberger T, Ng MR, Hato T, Huang Y, Ochiai H, Kitahara S, Unan EC, Reddy TP, et al. CXCR4 inhibition in tumor microenvironment facilitates anti-programmed death receptor-1 immunotherapy in sorafenib-treated hepatocellular carcinoma in mice. Hepatology. 2015;61:1591–1602. doi: 10.1002/hep.27665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Holdman XB, Welte T, Rajapakshe K, Pond A, Coarfa C, Mo Q, Huang S, Hilsenbeck SG, Edwards DP, Zhang X, et al. Upregulation of EGFR signaling is correlated with tumor stroma remodeling and tumor recurrence in FGFR1-driven breast cancer. Breast Cancer Res. 2015;17:141. doi: 10.1186/s13058-015-0649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu L, Ye TH, Han YP, Song H, Zhang YK, Xia Y, Wang NY, Xiong Y, Song XJ, Zhu YX, et al. Reductions in myeloid-derived suppressor cells and lung metastases using AZD4547 treatment of a metastatic murine breast tumor model. Cell Physiol Biochem. 2014;33:633–645. doi: 10.1159/000358640. [DOI] [PubMed] [Google Scholar]
- 109.Hume DA, MacDonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 2012;119:1810–1820. doi: 10.1182/blood-2011-09-379214. [DOI] [PubMed] [Google Scholar]
- 110.Sieweke MH, Allen JE. Beyond stem cells: Self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 111.Hamilton JA, Achuthan A. Colony stimulating factors and myeloid cell biology in health and disease. Trends Immunol. 2013;34:81–89. doi: 10.1016/j.it.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 112.Moughon DL, He H, Schokrpur S, Jiang ZK, Yaqoob M, David J, Lin C, Iruela-Arispe ML, Dorigo O, Wu L. Macrophage blockade using CSF1R inhibitors reverses the vascular leakage underlying malignant ascites in late-stage epithelial ovarian cancer. Cancer Res. 2015;75:4742–4752. doi: 10.1158/0008-5472.CAN-14-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu J, Escamilla J, Mok S, David J, Priceman S, West B, Bollag G, McBride W, Wu L. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer Res. 2013;73:2782–2794. doi: 10.1158/0008-5472.CAN-12-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 116.Rivera LB, Meyronet D, Hervieu V, Frederick MJ, Bergsland E, Bergers G. Intratumoral myeloid cells regulate responsiveness and resistance to antiangiogenic therapy. Cell Rep. 2015;11:577–591. doi: 10.1016/j.celrep.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beenken A, Mohammadi M. The FGF family: Biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Degirolamo C, Sabbà C, Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat Rev Drug Discov. 2016;15:51–69. doi: 10.1038/nrd.2015.9. [DOI] [PubMed] [Google Scholar]
- 119.Liu WY, Xie DM, Zhu GQ, Huang GQ, Lin YQ, Wang LR, Shi KQ, Hu B, Braddock M, Chen YP, et al. Targeting fibroblast growth factor 19 in liver disease: A potential biomarker and therapeutic target. Expert. Opin Ther Targets. 2015;19:675–685. doi: 10.1517/14728222.2014.997711. [DOI] [PubMed] [Google Scholar]
- 120.Pai R, French D, Ma N, Hotzel K, Plise E, Salphati L, Setchell KD, Ware J, Lauriault V, Schutt L, et al. Antibody-mediated inhibition of fibroblast growth factor 19 results in increased bile acids synthesis and ileal malabsorption of bile acids in cynomolgus monkeys. Toxicol Sci. 2012;126:446–456. doi: 10.1093/toxsci/kfs011. [DOI] [PubMed] [Google Scholar]
- 121.Ge H, Zhang J, Gong Y, Gupte J, Ye J, Weiszmann J, Samayoa K, Coberly S, Gardner J, Wang H, et al. Fibroblast growth factor receptor 4 (FGFR4) deficiency improves insulin resistance and glucose metabolism under diet-induced obesity conditions. J Biol Chem. 2014;289:30470–30480. doi: 10.1074/jbc.M114.592022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Silswal N, Touchberry CD, Daniel DR, McCarthy DL, Zhang S, Andresen J, Stubbs JR, Wacker MJ. FGF23 directly impairs endothelium-dependent vasorelaxation by increasing superoxide levels and reducing nitric oxide bioavailability. Am J Physiol Endocrinol Metab. 2014;307:E426–E436. doi: 10.1152/ajpendo.00264.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grabner A, Amaral AP, Schramm K, Singh S, Sloan A, Yanucil C, Li J, Shehadeh LA, Hare JM, David V, et al. Activation of cardiac fibroblast growth factor receptor 4 causes left ventricular hypertrophy. Cell Metab. 2015;22:1020–1032. doi: 10.1016/j.cmet.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, Edwards MG, Böhm D, Perner S, Helfrich BA, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20:3299–3309. doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wöhrle S, Weiss A, Ito M, Kauffmann A, Murakami M, Jagani Z, Thuery A, Bauer-Probst B, Reimann F, Stamm C, et al. Fibroblast growth factor receptors as novel therapeutic targets in SNF5-deleted malignant rhabdoid tumors. PLoS One. 2013;8:e77652. doi: 10.1371/journal.pone.0077652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Künstlinger H, Fassunke J, Schildhaus HU, Brors B, Heydt C, Ihle MA, Mechtersheimer G, Wardelmann E, Büttner R, Merkelbach-Bruse S. FGFR2 is overexpressed in myxoid liposarcoma and inhibition of FGFR signaling impairs tumor growth in vitro. Oncotarget. 2015;6:20215–20230. doi: 10.18632/oncotarget.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, Sausen M, Phallen J, Hruban CA, Tokheim C, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, Frias RL, Gainor JF, Amzallag A, Greninger P, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chell V, Balmanno K, Little AS, Wilson M, Andrews S, Blockley L, Hampson M, Gavine PR, Cook SJ. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene. 2013;32:3059–3070. doi: 10.1038/onc.2012.319. [DOI] [PubMed] [Google Scholar]
- 132.Ang D, Ballard M, Beadling C, Warrick A, Schilling A, O'Gara R, Pukay M, Neff TL, West RB, Corless CL, et al. Novel mutations in neuroendocrine carcinoma of the breast: Possible therapeutic targets. Appl Immunohistochem Mol Morphol. 2015;23:97–103. doi: 10.1097/PDM.0b013e3182a40fd1. [DOI] [PubMed] [Google Scholar]
- 133.Okazaki T, Honjo T. PD-1 and PD-1 ligands: From discovery to clinical application. Int Immunol. 2007;19:813–824. doi: 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 134.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 135.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 137.Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, Bandla S, Bien G, Choppa P, Hyland F, et al. Development and validation of a scalable next-generation sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]