Abstract
Background
The molecular mechanisms linking environmental exposures to earlier pubertal development are not well characterized. Epigenetics may play an important role, but data on the relationship between epigenetic marks and puberty, particularly in humans, is limited.
Methods
We used pyrosequencing to measure Alu and long interspersed nucleotide elements (LINE-1) methylation in DNA isolated from whole blood samples collected from newborns and 9-year-old children (n=266). Tanner staging was completed six times between ages 9 and 12 years to determine pubertal status, and hormone levels were measured in 12-year-old boys.
Results
Among girls, we observed a suggestive trend of increased odds of breast and pubic hair development with higher Alu and LINE-1 methylation in 9-year-old blood, respectively. The strongest association identified was an inverse association of LINE-1 methylation in 9-year-old girls with odds of experiencing menarche by age 12 (OR(95%CI): 0.63(0.46,0.87); p=0.005). We observed a consistent inverse relationship for Alu and LINE-1 methylation at 9 years with luteinizing hormone (LH), testosterone and follicle stimulating hormone levels in boys but it was only significant between LINE-1 and LH.
Conclusion
DNA methylation of Alu and LINE-1 may be involved in puberty initiation and development. This relationship should be confirmed in future studies.
Introduction
The average age of puberty development in girls, as marked by onset of breast development and menarche(1,2), has been decreasing in recent years and there is some evidence that age at puberty may also be decreasing in boys (3). These trends have public health significance since timing of puberty can affect health later in life. For instance, earlier puberty in girls has been associated with increased risk of behavior problems, breast cancer, and all-cause mortality among other adverse health outcomes (4,5). In girls, risk factors for earlier puberty include increased body weight (6), paternal absence, and family stressors (5,7). Though data is more limited, a few studies suggest body mass index (BMI) may also be associated with earlier puberty in boys (8,9). Genome-wide association studies (GWAS) have identified several loci related to pubertal timing in girls and more recently also in boys, but these genetic variants explain only a small percentage of the variation in timing of menarche, breast, and genital development (8,10). Exposure to endocrine disrupting chemicals (EDCs) like the pesticide dichlorodiphenyltrichloroethane (DDT), bisphenol-A (BPA), and phthalates have also been associated with puberty timing, though findings have been inconsistent (11). The molecular mechanisms through which these factors can affect pubertal timing are not well characterized and there is a growing consensus that epigenetics may play an important role. Epigenetic modifications regulate gene expression without DNA sequence modifications and can be heritable. Some examples include DNA methylation, histone modifications, and noncoding RNAs. Since epigenetic marks are malleable to environmental conditions like nutrition, chemical exposure, and social stressors, researchers have been particularly interested in them as potential biological mechanisms through which the environment can affect health (12). Furthermore, early development in utero and during the neonatal period is considered a critical window in which the epigenome may be particularly sensitive to the environment (13,14). DNA methylation of repetitive elements, Alu, and long interspersed nucleotide elements (LINE-1), have been extensively utilized in population studies to assess relationships of methylation profiles with exposures and/or health outcomes because this method is informative and cost effective (15,16).
Methylation of Alu and LINE-1 repeats represents up to 50% of global genomic methylation (17), with approximately 1.4 million Alu repetitive elements and half a million LINE-1 elements interspersed throughout the human genome. Hypomethylation of these retrotransposable elements has been associated with genomic instability and is therefore biologically relevant (18,19). Furthermore, methylation changes in response to environmental exposures can vary between Alu and LINE-1 elements (16,20), supporting the concept that LINE-1 and Alu methylation each represent distinct measures of methylation in different parts of the methylome (21,22).
Recent animal studies observed that the release of gonadotropin releasing hormone (GnRH), which coincides with the initiation of puberty, may be influenced by changes in DNA methylation, providing evidence of epigenetic control on pubertal development (23). Only a few studies have examined associations of repetitive element methylation with pubertal status or hormone levels. LINE-1 and Alu methylation were not associated with hormone levels in healthy female adults (24). Among postmenopausal women, a significant inverse association of LINE-1 methylation with hormone levels was observed only in women with low folate levels (25). To our knowledge, no studies of repetitive element methylation and pubertal timing or hormone levels have been reported in adolescent children. The purpose of the present study is to examine the relationship of Alu and LINE-1 repetitive elements in fetal and child blood with puberty status and hormone levels in participants of the Center for Health Assessment of Mothers and Children of Salinas (CHAMACOS), a longitudinal birth cohort study.
Results
Table 1 summarizes CHAMACOS maternal and child characteristics. The majority of mothers were low-income and relatively young. Most did not smoke or drink alcohol during pregnancy and many either worked in agriculture (41%) or lived with someone that worked in agriculture (82%). The sex distribution of children included in this analysis was relatively even. Among girls, 39% had initiated puberty as measured by breast development (Tanner Stage >1) at age 9 and 58% had begun menses by age 12. Among 10.5 year old boys, 51% had experienced pubertal onset as indicated by genital development (Tanner Stage > 1).
Table 1.
Demographics and pubertal status of CHAMACOS participantsa
| N | % | |
|---|---|---|
| Child sex | ||
| Boy | 126 | 47.4 |
| Girl | 140 | 52.6 |
| Child Obesity Status at 9 years | ||
| Normal (≤85th percentile) | 118 | 45.2 |
| Overweight (>85th, < 95th percentile) | 39 | 14.9 |
| Obese (≥95th percentile) | 104 | 39.8 |
| Breast Development (9 yr Girls)b | ||
| Tanner Stage≤1 | 81 | 61.4 |
| Tanner Stage > 1 | 51 | 38.6 |
| Pubic Hair Development (10.5 yr Girls)b | ||
| Tanner Stage≤1 | 58 | 42.3 |
| Tanner Stage > 1 | 79 | 57.7 |
| Menarche at Age 12b | ||
| No | 59 | 42.1 |
| Yes | 81 | 57.9 |
| Genital Development (10.5 yr Boys)b | ||
| Tanner Stage≤1 | 61 | 49.2 |
| Tanner Stage > 1 | 63 | 50.8 |
| Pubic Hair Development (12 yr Boys)b | ||
| Tanner Stage≤1 | 53 | 49.1 |
| Tanner Stage > 1 | 55 | 50.9 |
| Paternal Presence at 10.5 years | ||
| No | 76 | 29 |
| Yes | 186 | 71 |
| Maternal Menarche at Age 12 | ||
| No | 112 | 42.1 |
| Yes | 154 | 57.9 |
Total number of observation vary due to missing data.
Pubertal development is shown at the age at which close to half of the children have begun development. For girls, it was ages 9, 10.5, and 12 years for breast, pubic hair, and menarche, respectively. For boys, it was 10.5 and 12 years for genital and pubic hair development, respectively.
Hormone Levels in 12 Year Old Boys
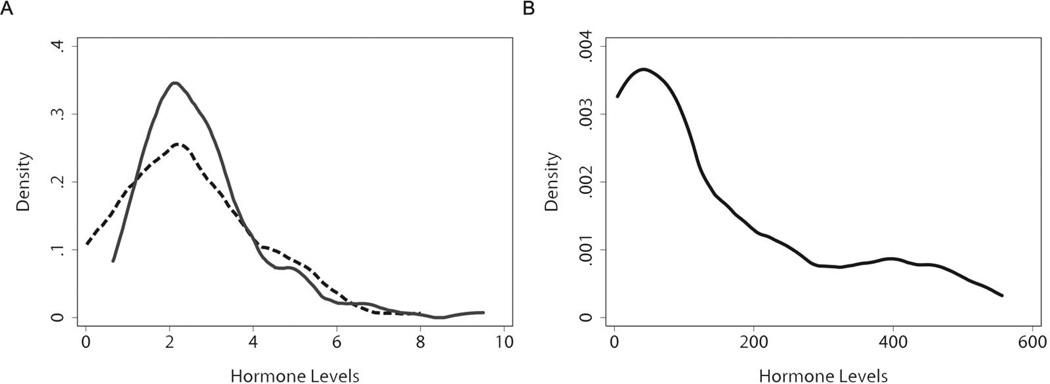
Figure 1 shows the distribution of FSH, LH, and T among 12-year-old boys. Levels of all three hormones were well within published reference ranges. The mean ± standard deviation for FSH, LH, and T were 2.79 ± 1.36 mIU/mL, 2.47 ± 1.54 mIU/mL, and 157.89 ± 167.32 ng/dl, respectively. LH was significantly correlated with both T (r=0.62, p<0.0005) and FSH (0.42, p<0.0005). T and FSH, however, were only weakly correlated with each other (r=0.18, p=0.09). As expected, boys who had entered more advanced stages of pubertal development by age 10.5 years also had higher mean hormone levels at age 12 years as summarized in Table 2. This was particularly strong for T, where levels more than tripled in boys who had reached or surpassed G2 or PH2.
Figure 1.
Distribution of hormone levels in 12 year old CHAMACOS boys (n=91). In panel A, LH (miU/mL) and FSH (miU/mL) in CHAMACOS boys are shown using dashed and solid lines, respectively. Testosterone in CHAMACOS boys is shown in panel B using a solid line. Reference levels of LH range from 0.3–6.0 mIU/mL in prepubertal boys and 1.6–12 mIU/mL in adult males. FSH reference levels range from 0.5–10.5 mIU/mL in 14 year old boys. Reference levels of testosterone range from 7–800 ng/dL at age 12 and from 100–1200 ng/dL at age 15–18 years in males.
Table 2.
Mean (SD) hormone levels by pubertal status in 12 year old boys (n=91)
| FSH (mIU/mL) |
LH (mIU/mL) |
Testosterone (ng/dL) |
|
|---|---|---|---|
| Genital Development at 10.5 Yr | |||
| Tanner Stage≤1 | 2.56(1.24) | 1.92(1.62) | 68.43(113.92) |
| Tanner Stage > 1 | 2.99(1.45) | 3.02(1.24) | 242.94(166.01) |
| Pubic Hair Development at 12 Yr | |||
| Tanner Stage≤1 | 2.49(1.15) | 1.99(1.42) | 64.57(91.5) |
| Tanner Stage > 1 | 3.11(1.59) | 3.08(1.48) | 264.16(172.86) |
DNA Methylation
LINE-1 methylation ranged 74.4 to 82.4 %5mC in cord blood and from 75.4 to 82.0 in 9-year old children. Means and standard deviations (Mean ± SD) were 78.8 ±1.5 and 78.4 ±1.3 %5mC for cord blood and 9 year old children, respectively. Alu methylation ranged from 22.9 to 27.0 and from 22.4 to 27.4 %5mC in newborns and 9-year olds, respectively. Means and standard deviations for Alu methylation levels were nearly identical at both ages: 25.3 ±0.7 %5mC. Methylation levels in umbilical cord blood were not correlated with methylation levels in 9-year old blood for either LINE-1 or Alu repeats as reported previously (26). Furthermore, levels of Alu and LINE-1 methylation were not correlated with each other at either time point.
DNA Methylation and Pubertal Development
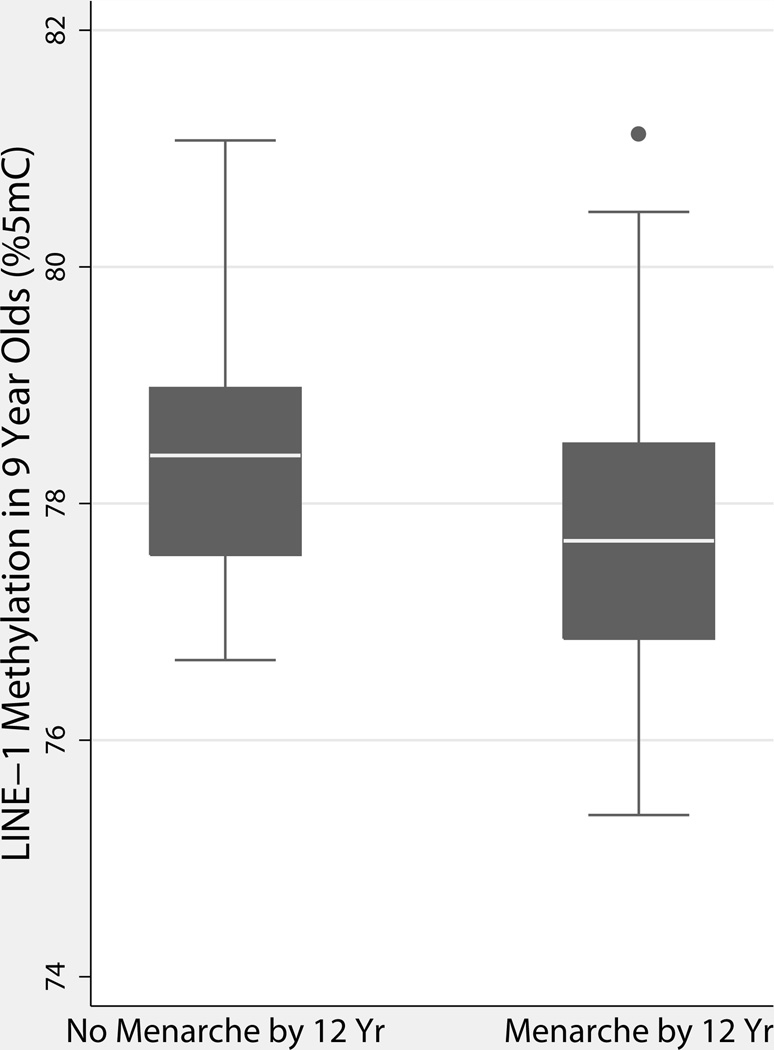
We did not find any significant associations between cord blood methylation and puberty status in girls or boys (Table 3). Among girls, LINE-1 methylation in age 9 blood was associated with decreased odds of having experienced menarche by age 12 (p=0.005; Figure 2; Table 4). LINE-1 and Alu methylation in 9-year-old boys were not significantly associated with odds of genital or pubic hair development (Table 4).
Table 3.
Associations of cord methylation with odds of pubertal development
| Alu | LINE-1 | ||||
|---|---|---|---|---|---|
| N | OR(95%CI) | p-value | OR(95%CI) | p- value |
|
| Girlsa | |||||
| Breast Development (9 yr) | |||||
| Methylation (%5mC) | 80 | 0.98(0.44,2.17) | 0.96 | 1.21(0.86,1.72) | 0.27 |
| BMI Z Score (9 yr) | 80 | 2.41(1.45,4.01) | 0.001 | 2.54(1.49,4.33) | 0.001 |
| Maternal Age of Menarche | 80 | 0.69(0.48,1) | 0.05 | 0.7(0.5,1) | 0.05 |
| Pubic Hair Development (10.5 yr) | |||||
| Methylation (%5mC) | 79 | 1.17(0.52,2.62) | 0.70 | 1.21(0.86,1.69) | 0.28 |
| BMI Z Score (10.5 yr) | 79 | 2.1(1.28,3.44) | 0.003 | 2.19(1.32,3.65) | 0.003 |
| Maternal Age of Menarche | 79 | 0.79(0.56,1.13) | 0.19 | 0.78(0.54,1.11) | 0.16 |
| Menarche (12 yr) | |||||
| Methylation (%5mC) | 86 | 1.13(0.57,2.24) | 0.72 | 0.87(0.64,1.19) | 0.38 |
| BMI Z-Score (12 yr) | 86 | 1.74(1.16,2.62) | 0.01 | 1.72(1.14,2.6) | 0.01 |
| Maternal Age of Menarche | 86 | 0.96(0.71,1.3) | 0.79 | 0.95(0.71,1.29) | 0.76 |
| Boysb | |||||
| Genital Development (10.5 yr) | |||||
| Methylation (%5mC) | 68 | 1.23(0.56,2.68) | 0.61 | 0.91(0.65,1.29) | 0.60 |
| Pubic Hair Development (12 yr) | |||||
| Methylation (%5mC) | 61 | 0.98(0.44,2.19) | 0.97 | 0.69(0.48,1.01) | 0.06 |
Girls models were adjusted for BMI Z-score at the concurrent age of puberty outcome and age of maternal menarche.
Boys models were not adjusted for any confounders.
Figure 2.
Box plot of LINE-1 Methylation in 9 year old blood by menarche status at age 12. On average, girls who had reached menarche by age 12 years also had lower LINE-1 methylation at age 9 (p=0.005) after controlling for maternal age of menarche and child BMI Z-score at age 12.
Table 4.
Associations of 9 yr methylation with odds of pubertal development
| Alu | LINE-1 | |||||
|---|---|---|---|---|---|---|
| N | OR(95%CI) | p-value | N | OR(95%CI) | p- value |
|
| Girlsa | ||||||
| Breast Development (9 yr) | ||||||
| Methylation (%5mC) | 126 | 1.76(0.89,3.46) | 0.10 | 126 | 1.00(0.73,1.37) | 0.99 |
| BMI Z Score (9 yr) | 126 | 1.86(1.28,2.71) | 0.001 | 126 | 1.88(1.30,2.73) | 0.001 |
| Maternal Age of Menarche | 126 | 0.69(0.52,0.90) | 0.01 | 126 | 0.70(0.53,0.92) | 0.01 |
| Pubic Hair Development (10.5 yr) | ||||||
| Methylation (%5mC) | 125 | 1.01(0.54,1.91) | 0.96 | 125 | 1.32(0.96,1.82) | 0.09 |
| BMI Z Score (10.5 yr) | 125 | 1.78(1.25,2.53) | 0.001 | 125 | 1.87(1.30,2.69) | 0.001 |
| Maternal Age of Menarche | 125 | 0.79(0.61,1.02) | 0.07 | 125 | 0.79(0.60,1.03) | 0.08 |
| Menarche (12 yr) | ||||||
| Methylation (%5mC) | 131 | 1.08(0.58,2.03) | 0.80 | 131 | 0.63(0.46,0.87) | 0.005 |
| BMI Z-Score (12yr) | 131 | 1.74(1.24,2.43) | 0.001 | 131 | 1.76(1.25,2.49) | 0.001 |
| Maternal Age of Menarche | 131 | 0.87(0.68,1.12) | 0.28 | 131 | 0.85(0.66,1.10) | 0.23 |
| Boysb | ||||||
| Genital Development (10.5 yr) | ||||||
| Methylation (%5mC) | 113 | 0.91(0.58,1.44) | 0.70 | 112 | 0.84(0.62,1.14) | 0.27 |
| Pubic Hair Development (12 yr) | ||||||
| Methylation (%5mC) | 98 | 0.86(0.53,1.38) | 0.53 | 97 | 1(0.74,1.36) | 0.98 |
Girls models were adjusted for BMI Z-score at the concurrent age of puberty outcome and maternal age of menarche.
Boys models did not adjust for any confounders.
DNA Methylation and Hormone Levels
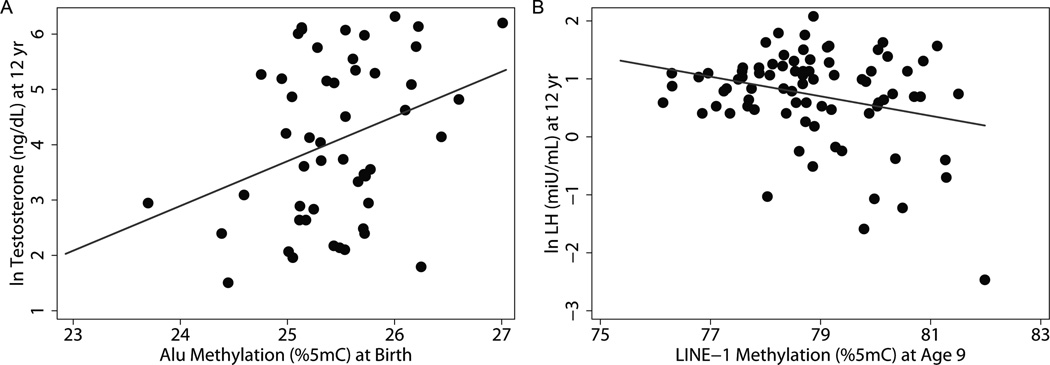
We also determined associations of Alu and LINE-1 methylation with hormone levels measured in serum collected from 12-year-old boys (Table 5). We found a marginally significant positive relationship between Alu methylation in umbilical cord blood and levels of T (β = 0.67; 95%CI = −0.05, 1.39). In contrast, we observed a consistent inverse relationship between Alu and LINE-1 methylation at age 9 with all 3 hormone levels. However, these relationships only reached statistical significance for LINE-1 and ln LH (p=0.05, Figure 3). When we removed outliers in this model (n=3) for sensitivity analyses, the association became more significant (β = −0.17; 95%CI: −0.32,−0.03; p = 0.02).
Table 5.
Associations of Alu and LINE-1 methylation at cord or 9 yr with hormone levels at 12 yr
| Alu | LINE-1 | |||||
|---|---|---|---|---|---|---|
| N | β(95%CI) | p-value | N | β(95%CI) | p-value | |
| Cord Blood Methylation | ||||||
| Testosterone (ng/dL)a | ||||||
| Methylation (%5mC) | 50 | 0.67(−0.05,1.39) | 0.07 | 50 | −0.14(−0.42,0.13) | 0.30 |
| BMI Z Score (12 yr) | 50 | −0.25(−0.67,0.17) | 0.24 | 50 | −0.41(−0.82,0) | 0.05 |
| LH (mIU/mL)a | ||||||
| Methylation (%5mC) | 50 | 0.18(−0.38,0.74) | 0.52 | 50 | 0.01(−0.2,0.22) | 0.92 |
| BMI Z Score (12 yr) | 50 | −0.32(−0.65,0.01) | 0.06 | 50 | −0.35(−0.67,−0.04) | 0.03 |
| FSH (mIU/mL)a | ||||||
| Methylation (%5mC) | 49 | 0.03(−0.22,0.28) | 0.81 | 49 | −0.05(−0.14,0.04) | 0.29 |
| BMI Z Score (12 yr) | 49 | −0.08(−0.22,0.06) | 0.25 | 49 | −0.1(−0.23,0.04) | 0.15 |
| 9 Yr Blood Methylation | ||||||
| Testosterone (ng/dL)a | ||||||
| Methylation (%5mC) | 85 | −0.21(−0.59,0.17) | 0.28 | 84 | −0.01(−0.25,0.23) | 0.93 |
| BMI Z score (12 yr) | 85 | −0.56(−0.9,−0.21) | 0.002 | 84 | −0.57(−0.92,−0.23) | 0.002 |
| LH (mIU/mL)a | ||||||
| Methylation (%5mC) | 85 | −0.03(−0.33,0.26) | 0.82 | 84 | −0.19(−0.37,0) | 0.05 |
| BMI Z score (9yr) | 85 | −0.38(−0.66,−0.11) | 0.01 | 84 | −0.4(−0.67,−0.14) | 0.003 |
| FSH (mIU/mL)a | ||||||
| Methylation (%5mC) | 85 | −0.05(−0.18,0.07) | 0.40 | 84 | −0.04(−0.12,0.04) | 0.36 |
| BMI Z score (9yr) | 85 | −0.1(−0.21,0.02) | 0.10 | 84 | −0.1(−0.21,0.02) | 0.09 |
Each regression model was constructed with ln Hormone (Testosterone, LH, or FSH) as the outcome and methylation (Alu or LINE-1) and BMI Z Score at age 12 as the independent variables in the model.
Figure 3.
Associations of (A) Alu Methylation in umbilical cord blood with ln Testosterone (n=50) and (B) LINE-1 methylation in 9 year old blood with ln LH in 12 year old boys (n=84). Alu methylations was positively associated (β(95%CI): 84.8(8.5,161.1)) with ln Testosterone at age 12 (p=0.07). Panel B shows the relationship of LINE-1 methylation with ln LH in boys after removing 3 outliers for sensitivity analyses. LINE-1 methylation in 9 year old boys was inversely associated with ln LH measured at age 12 (β(95%CI): −0.17(−0.30,−0.036)).
Discussion
In this study, we examined the association of repetitive element methylation with puberty status and hormone levels in CHAMACOS children. Although we did not observe significant associations of cord blood methylation with puberty status, we did find the DNA methylation in nine-year-old girls was inversely associated with odds of entering menarche by age 12. Testosterone levels in 12-year-olds boys were positively associated with cord blood Alu methylation, while all three hormone levels measured (T, LH, and FSH) at age 12 were inversely associated with Alu and LINE-1 methylation at child age 9.
To our knowledge, no other studies have examined associations of pubertal status and hormone levels with repetitive element methylation in adolescent children. One study in adults corroborates our findings related to menarcheal age (27). Demetriou and colleagues found that global methylation measured by Luminometric Methylation Assay (LUMA) was inversely associated with menarcheal age. While the LUMA assay covers methylation in CpG island promoters and repetitive elements, the study’s authors suggested that the relationship was likely driven by repetitive elements. Limited data also suggest that site specific methylation may also be linked to pubertal timing. For example, one human study has also reported an inverse association of early breast development with site-specific methylation of saliva DNA in CYP19A1, a gene involved in estrogen biosynthesis, among overweight girls (28).
The mechanistic link of methylation of repetitive elements with pubertal timing and hormonal levels is not yet clear. Differences in DNA methylation have previously been associated with a variety of exposures ranging from persistent organic pollutants to diet and adversity (29–31). It is possible that epigenetics is the mediator of environmental or other exposures that are resulting in earlier puberty as observed in girls over the last few decades. Interestingly, in our study, pubertal timing was associated with methylation in 9-year-old blood but not in cord blood, suggesting that physiological changes or exposures after birth and during childhood may be influencing pubertal timing. As with most population studies, the associations we identified indicate correlation, but not necessarily causation. Thus, it is important to note that the physiological changes occurring during pubertal development could also influence the methylation differences observed in CHAMACOS 9 year olds.
This study had some limitations. Methylation measurements were made in blood, which can be biased by cell heterogeneity. In sensitivity analyses, we ran models adjusting for white blood cell composition in the subset of samples with cell count data available as previously described(32). Results remained relatively unchanged suggesting that cell heterogeneity was not a significant source of bias in these models (data not shown). Our study was limited to Mexican-American children from low income families. In the future, it will be important to confirm findings in other cohorts. Finally, our analyses of hormone levels were limited only to boys (due to availability of funds), for whom we did observe a suggestive relationship of testosterone levels with DNA methylation.
There is a growing consensus that epigenetics may play a significant role in timing of pubertal development (23,33) yet few studies, particularly in humans, have examined these relationships. We found suggestive evidence of associations of DNA methylation of repetitive elements with pubertal timing and hormone levels in adolescent children. The strongest relationship observed was an inverse association between LINE-1 methylation and odds of experiencing menarche by age 12 in girls. These findings suggest a potential role for repetitive element methylation in relation to puberty initiation and development that should be confirmed in future studies.
Materials and Methods
Study subjects
CHAMACOS is a longitudinal birth cohort study of the effects of pesticides and other environmental exposures on pregnant Mexican-American women and their children living in the agricultural Salinas Valley, California. Eligibility criteria for enrollment of pregnant women included: ≥ 18 years of age, less than 20 weeks gestation, Spanish or English speaking, eligible for low-income health insurance, receiving prenatal care at one of the participating community clinics, and planning to deliver at the local public hospital. From 1999–2000, we enrolled 601 pregnant women, of whom 526 were still enrolled at delivery of a live, singleton newborn (34). CHAMACOS women were interviewed by bilingual, bicultural interviewers twice during pregnancy (~13 weeks and 26 weeks gestation) to obtain information on various characteristics including sociodemographics, mother’s reproductive and medical history, lifestyle exposures during pregnancy, diet, occupational and residential history, exposures to pesticides and other environmental chemicals, and housing quality. Assessments of the children’s growth and development were conducted at multiple time points through age 12 years. Clinical Tanner staging of pubertal development was conducted every 9 months between ages 9 and 12 years by trained examiners.
Alu and LINE-1 DNA methylation was measured in blood samples collected from children at delivery (umbilical cord blood representing fetal blood) and when they were 9 years old (mean=9.3 years, SD=0.3). Our study sample included a total of 266 children (140 girls and 126 boys) who had DNA samples available for methylation analysis at birth and/or age 9 and also had information on Tanner staging at ages 9, 10.5, and 12 years. Among the 266 children, 134 had methylation at both time points while 21 children had only cord blood measurements and 111 had only age 9 blood measurements. Sample sizes for methylation measurements were higher at age 9 because these children were more likely to have Tanner staging assessments at ages 9 through 12. Of the 126 boys included in analyses, 91 also had morning serum specimens collected at age 12 that were assessed for hormone levels. Study protocols were approved by the University of California, Berkeley Committee for Protection of Human Subjects. Written informed consent was obtained from all mothers and assent was provided by the children at the 9, 10.5, and 12-year assessments.
Cord blood collection and processing
Blood specimens from CHAMACOS children were collected from umbilical cords shortly after delivery and also by venipuncture when children were assessed at ages 9 and 12. Whole blood was collected in BD vacutainers (Becton, Dickinson and Company, Franklin Lakes, NJ) containing no anticoagulant. Vacutainers were centrifuged and separated components were subsequently divided into serum and clot aliquots that were stored at −80°C.
DNA methylation analyses
Isolation of DNA from clots was performed using a QIAamp Blood DNA Maxi kit (Qiagen, Hilden, Germany) as described previously (35). Bisulfite conversion of 500 ng of DNA was carried out using EpiTect Bisulfite Conversion Kits (Qiagen, Germantown, MD) and eluted into 20 µL Elution buffer. A non-CpG cytosine residue was used as an internal control to confirm bisulfite DNA conversion efficiency (99%). To analyze Alu and LINE-1 methylation status, we used a Pyromark Q96MD System (Qiagen) for pyrosequencing of PCR-amplified and bisulfite-treated DNA samples (17,36). The Alu and LINE-1 assays reports 4 CpG sites for all Alu and LINE-1 repeats across the genome. While the previously published method for Alu as described by Yang et al. (17) reported only 3 CpG sites, we were able to report methylation levels at the same 3 sites and also include one additional site because pyrosequencing read lengths have improved in the past few years. Repetitive element methylation (%5-mC) was calculated using Pyro Q-CpG Software (Qiagen). All samples were run in triplicate for each time-point/subject.
Quality assurance procedures included use of internal standards, repeats, and positive and negative controls for minimization of technical variability. All sample plates were run on the same day to ensure low batch variability. To further minimize experimental bias, all plates contained randomized encoded samples from different age groups. For Alu and LINE-1 triplicate measures, the coefficients of variation (CV) were 5 and 3 %, respectively. CV’s for intraplate replicates were in the same range.
Child anthropometric measurements
Children’s height and weight were measured at each visit. Height without shoes was measured using a stadiometer. Height measurements were performed in triplicate and then averaged. Child weight was determined using a calibrated Tanita electronic scale Model 1582(Tanita Corp, Tokyo, Japan). To ensure reliability, periodic refresher trainings were performed and measurements were compared among interviewers. BMI, calculated as weight divided by height2 (kg/m2) was compared to CDC reference data (37) to generate BMI Z-scores standardized by sex and age.
Tanner Staging
Pubertal progression was assessed by clinical examination of breast development in girls, genital development in boys, and pubic hair development in both as proposed by Marshall and Tanner (38). Development was classified into 5 stages of breast (B1–B5), genital (G1–G5), and pubic hair (PH1–PH5) with stage 1 defined as pre-pubertal and stage 5 representing adult development (39). Physical exams were conducted by staff trained by a pediatric endocrinologist. In girls, pubic hair development was assessed visually while breast development was determined by palpation of the breasts to distinguish adipose tissue from glandular breast tissue (40). Onset of puberty was indicated by the development of breast buds (B2). Menarche was determined by self-report. Specifically, at each visit beginning at age 9, the girls were asked if they had started menstruating. For boys, stages of genital and pubic hair development were conducted by visual assessment followed by a physical measurement of testicular volume using Prader orchidometer beads for comparison. Puberty onset is signaled by the initial enlargement of the scrotum and testes (G2), testicular volume of 3 ml or more, or androgenic stimulation of pubic hair development (PH2).
Pubertal hormone levels
Levels of luteinizing hormone (LH), follicle stimulating hormone (FSH), and testosterone (T) were measured in serum collected in the morning (before 9 am) from boys at age 12. LH and FSH were determined by electrochemiluminescent assay. Blanks, repeats, and internal standards were included for quality assurance. The intra- and inter-assay coefficients of variation (CVs) for both assays were less than 6% and 7%, respectively. T levels were analyzed using liquid chromatography with mass spectrometry detection after nonpolar solvent extraction. Samples were processed with inclusion of water blanks, 5 assay control pools, and a 200 pg standard to assess accuracy and precision of the assay. The intra-assay CV was <6% and the inter-assay CV was < 10%. All assays were performed by Esoterix Laboratory Services (Calabasas Hills, CA)
Statistical analyses
Pearson’s correlation coefficients were calculated to examine correlations between Alu and LINE-1 methylation at different time points, as well as between hormone levels at age 12.
We performed logistic regression models to determine the relationship of LINE-1 and Alu repeat methylation (cord and 9 year old blood) with odds of being at an advanced stage of puberty (Tanner stage >1) at a specific age point. In girls, separate models were constructed for breast development at age 9, pubic hair development at age 10.5, and menarche at age 12. For boys, similar models were constructed for genital development at age 10.5 and pubic hair development at age 12. These ages were chosen because they were the period at which close to half of the children had reached an advanced stage of puberty (e.g. Tanner stage 2 vs. stage 1). Since BMI Z-score at age of pubertal assessment and maternal age of menarche were associated with both methylation and Tanner staging among girls, we included these two variables as covariates in the girls’ models. We did not adjust for BMI Z-score in the boys’ models because it was not significantly associated with methylation measurements. Furthermore, no other covariates were included in the boys’ models. Linear regression modeling was performed to examine associations of Alu and LINE-1 methylation (cord and 9 year) with hormone levels (LH, FSH, and T) measured in boys at age 12. All hormone levels were natural log (ln) transformed to approximate a normal distribution. All models of hormone levels and methylation were adjusted for BMI Z-score at age 12 years. Sensitivity analyses included: a) rerunning regression models after removal of outliers (absolute values of residuals > 3) ; b) accounting for cell heterogeneity using differential cell count in a subset of cord blood samples and Houseman estimation by minfi in 9 year olds (32); and c) adjusting for prenatal concentrations of organophosphate metabolites (dialkyl phosphates) in urine (41) and persistent organic pollutants, dichlorodiphenyl trichloroethane (DDT) and polybrominated diphenyl ethers (comprised of congeners BDEs −47 −99 −100 and −153) measured in blood (26). Results did not change appreciably in sensitivity analyses adjusting for prenatal exposures to organophosphates, DDT, and PBDEs.
All statistical analyses were carried out using STATA software, version 12.0 (College Station, TX). P-values less than 0.05 were considered significant.
Acknowledgements
We are grateful to the laboratory and field staff and participants of the CHAMACOS study for their contributions
Statement of financial support: This publication was made possible by grants from the National Institute of Environmental Health Science (NIEHS) [PO1 ES009605, R01ES023067, R01ES017054, R01ES021369] and from the US Environmental Protection Agency (EPA) [RD83451301, R82670901]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIEHS or EPA.
Footnotes
Disclosure statement: We have no conflicts of interest to disclose.
References
- 1.Aksglaede L, Sorensen K, Petersen JH, Skakkebaek NE, Juul A. Recent decline in age at breast development: the Copenhagen Puberty Study. Pediatrics. 2009;123:e932–e939. doi: 10.1542/peds.2008-2491. [DOI] [PubMed] [Google Scholar]
- 2.Biro FM, Greenspan LC, Galvez MP, et al. Onset of breast development in a longitudinal cohort. Pediatrics. 2013;132:1019–1027. doi: 10.1542/peds.2012-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman-Giddens ME, Steffes J, Harris D, et al. Secondary sexual characteristics in boys: data from the Pediatric Research in Office Settings Network. Pediatrics. 2012;130:e1058–e1068. doi: 10.1542/peds.2011-3291. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Rhodes L, Demerath EW, Cousminer DL, et al. Association of adiposity genetic variants with menarche timing in 92,105 women of European descent. Am J Epidemiol. 2013;178:451–460. doi: 10.1093/aje/kws473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagen CP, Sorensen K, Aksglaede L, et al. Pubertal onset in girls is strongly influenced by genetic variation affecting FSH action. Sci Rep. 2014;4:6412. doi: 10.1038/srep06412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JM, Appugliese D, Kaciroti N, Corwyn RF, Bradley RH, Lumeng JC. Weight status in young girls and the onset of puberty. Pediatrics. 2007;119:e624–e630. doi: 10.1542/peds.2006-2188. [DOI] [PubMed] [Google Scholar]
- 7.Golub MS, Collman GW, Foster PM, et al. Public health implications of altered puberty timing. Pediatrics. 2008;121(Suppl 3):S218–S230. doi: 10.1542/peds.2007-1813G. [DOI] [PubMed] [Google Scholar]
- 8.Cousminer DL, Stergiakouli E, Berry DJ, et al. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum Mol Genet. 2014;23:4452–4464. doi: 10.1093/hmg/ddu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen K, Aksglaede L, Petersen JH, Juul A. Recent changes in pubertal timing in healthy Danish boys: associations with body mass index. J Clin Endocrinol Metab. 2010;95:263–270. doi: 10.1210/jc.2009-1478. [DOI] [PubMed] [Google Scholar]
- 10.Perry JR, Day F, Elks CE, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher MM, Eugster EA. What is in our environment that effects puberty? Reprod Toxicol. 2014;44:7–14. doi: 10.1016/j.reprotox.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 14.Thompson RF, Einstein FH. Epigenetic basis for fetal origins of age-related disease. J Womens Health (Larchmt) 2010;19:581–587. doi: 10.1089/jwh.2009.1408. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wang F, Guan J, et al. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am J Clin Nutr. 2010;91:1359–1367. doi: 10.3945/ajcn.2009.28858. [DOI] [PubMed] [Google Scholar]
- 16.Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su J, Shao X, Liu H, Liu S, Wu Q, Zhang Y. Genome-wide dynamic changes of DNA methylation of repetitive elements in human embryonic stem cells and fetal fibroblasts. Genomics. 2012;99:10–17. doi: 10.1016/j.ygeno.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Ayarpadikannan S, Kim HS. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014;12:98–104. doi: 10.5808/GI.2014.12.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA Methylation Changes after Exposure to Traffic Particles. Am J Respir Crit Care Med. 2009 doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byun HM, Motta V, Panni T, et al. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. 2013;10:28. doi: 10.1186/1743-8977-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price EM, Cotton AM, Penaherrera MS, McFadden DE, Kobor MS, Robinson W. Different measures of "genome-wide" DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics. 2012;7:652–663. doi: 10.4161/epi.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomniczi A, Wright H, Ojeda SR. Epigenetic regulation of female puberty. Front Neuroendocrinol. 2015;36:90–107. doi: 10.1016/j.yfrne.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Maarri O, Walier M, Behne F, et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS ONE. 2011;6:e16252. doi: 10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulrich CM, Toriola AT, Koepl LM, et al. Metabolic, hormonal and immunological associations with global DNA methylation among postmenopausal women. Epigenetics. 2012;7:1020–1028. doi: 10.4161/epi.21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huen K, Yousefi P, Bradman A, et al. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environ Mol Mutagen. 2014;55:209–222. doi: 10.1002/em.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demetriou CA, Chen J, Polidoro S, et al. Methylome analysis and epigenetic changes associated with menarcheal age. PLoS ONE. 2013;8:e79391. doi: 10.1371/journal.pone.0079391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stueve TR, Wolff MS, Pajak A, Teitelbaum SL, Chen J. CYP19A1 promoter methylation in saliva associated with milestones of pubertal timing in urban girls. BMC Pediatr. 2014;14:78. doi: 10.1186/1471-2431-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang FF, Morabia A, Carroll J, et al. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr. 2011;141:1165–1171. doi: 10.3945/jn.110.134536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Essex MJ, Boyce WT, Hertzman C, et al. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Dev. 2013;84:58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefi P, Huen K, Quach H, et al. Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environ Mol Mutagen. 2015;56:751–758. doi: 10.1002/em.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rzeczkowska PA, Hou H, Wilson MD, Palmert MR. Epigenetics: a new player in the regulation of mammalian puberty. Neuroendocrinology. 2014;99:139–155. doi: 10.1159/000362559. [DOI] [PubMed] [Google Scholar]
- 34.Eskenazi B, Bradman A, Gladstone E, Jaramillo S, Birch K, Holland N. CHAMACOS, a longitudinal birth cohort study: lessons from the fields. J Childrens Healt. 2003;1:3–27. [Google Scholar]
- 35.Holland N, Furlong C, Bastaki M, et al. Paraoxonase polymorphisms, haplotypes, and enzyme activity in latino mothers and newborns. Environ Health Perspect. 2006;114:985–991. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Royo JL, Hidalgo M, Ruiz A. Pyrosequencing protocol using a universal biotinylated primer for mutation detection and SNP genotyping. Nat Protoc. 2007;2:1734–1739. doi: 10.1038/nprot.2007.244. [DOI] [PubMed] [Google Scholar]
- 37.National Center for Health Statistics. CDC Growth Charts. United States; 2005. [Google Scholar]
- 38.Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child. 1970;45:13–23. doi: 10.1136/adc.45.239.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanner JM. Normal growth and techniques of growth assessment. Clin Endocrinol Metab. 1986;15:411–451. doi: 10.1016/s0300-595x(86)80005-6. [DOI] [PubMed] [Google Scholar]
- 40.Dorn LD, Dahl RE, Woodward HR, Biro F. Defining the boundaries of early adolescence: A user's guide to assessing pubertal status and pubertal timing in research with adolescents. Appl Dev Sci. 2006;10:30–56. [Google Scholar]
- 41.Bradman A, Eskenazi B, Barr DB, et al. Organophosphate urinary metabolite levels during pregnancy and after delivery in women living in an agricultural community. Environ Health Perspect. 2005;113:1802–1807. doi: 10.1289/ehp.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]