Abstract
Transcription of the reciprocally imprinted genes Insulin-like growth factor 2 (Igf2) and H19 is orchestrated by the 2.4-kb H19 Imprinting Control Region (H19ICR) located upstream of H19. Three known functions are associated with the H19ICR: (1) it is a germline differentially methylated region, (2) it is a transcriptional insulator, and (3) it is a transcriptional silencer. The molecular mechanisms of the DMR and insulator functions have been well characterized but the basis for the ICR’s silencer function is less well understood. In order to study the role the H19ICR intrinsically plays in gene silencing, we transferred the 2.4-kb H19ICR to a heterologous non-imprinted location on chromosome 5, upstream of the alpha fetoprotein (Afp) promoter. Independent of its orientation, the 2.4-kb H19ICR silences transcription from the paternal Afp promoter. Thus silencing is a function intrinsic to this DNA element. Further, ICR mediated silencing is a developmental process that, unexpectedly, does not occur through DNA methylation at the target promoter.
Keywords: Genomic Imprinting, gene silencing, H19
INTRODUCTION
Genomic imprinting is an epigenetic mechanism that regulates transcription of about 100 genes in mammals [1]. Imprinted genes display parent-of-origin specific gene expression and only one of the two parental alleles is expressed while the other becomes silenced. Imprinted genes are usually located in clusters scattered throughout the genome. One such imprinted cluster is the Igf2/H19 locus on mouse chromosome 7 (Fig. 1A). Loss of imprinting defects at this locus are associated with Beckwith Wiedemann and Russell-Silver syndromes and with many cancers [2].
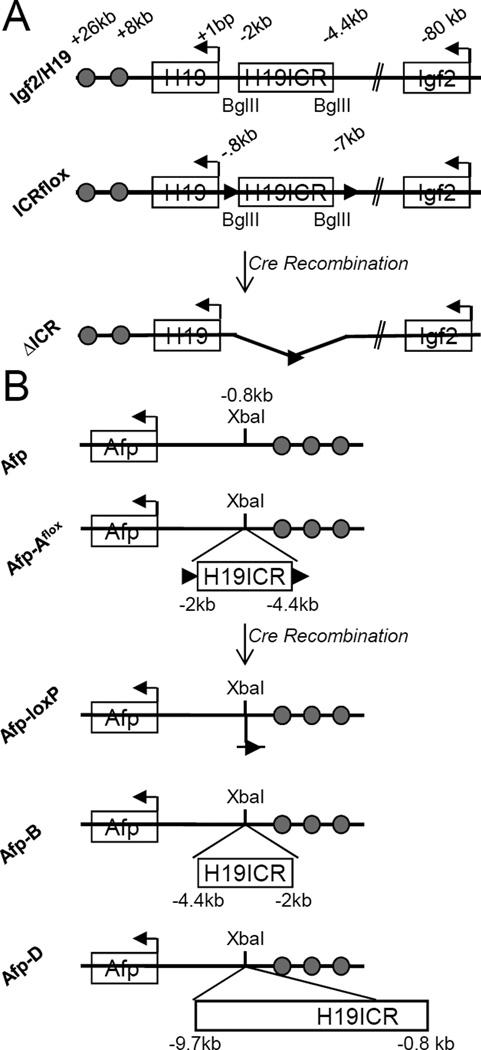
Figure 1. Mouse models used in this study.
(A) Igf2/H19 wild type and mutant alleles. The relative locations of the H19 and Igf2 genes and the H19ICR are depicted along with the shared endodermal and mesodermal enhancers (filled circles at +8 and +26 kb, respectively). All coordinates are relative to the H19 start site. The ICRflox allele [6] carries loxP insertions at −0.8 and −7-kb. Thus cre-mediated recombination results in deletion of the entire H19ICR but leaves the H19 promoter in tact. (B) Afp wild type and insertion alleles. The relative locations of the Afp gene and its three enhancer elements (located between −1 and −7.6-kb) are depicted along with the XbaI site where H19ICR sequences were inserted. These coordinates are relative to the Afp transcriptional start site. Afp-Aflox has the H19ICR carried on a 2.4-kb BglII fragment inserted at the XbaI site. Cre-mediated recombination of Afp-Aflox removes the H19ICR but a residual 194-bp including a single loxP site remains. Afp-B differs from Afp-A in the orientation of the inserted ICR. Afp-D carries the H19ICR inserted in the same orientation as Afp-B but has an additional 4.8-kb of sequences that separate the H19ICR from the Afp promoter. For Afp-Aflox, Afp-B, and Afp-D, the numbers below the ICR delineate the endogenous coordinates of the H19ICR fragment.
Igf2 and H19 are 80-kb apart and share enhancers that are located downstream of the H19 gene (Fig. 1A)[3, 4]. Transcription of both genes is regulated in cis by a 2.4-kb DNA element, the H19 imprinting control region (H19ICR)[5–7]. The ICR is located between the two genes, just upstream of the H19 promoter. Deletion and mutational studies at the endogenous locus have demonstrated three functions for the region. First, the ICR is the only germ line differentially methylated region (gDMR) at the locus and thus is responsible for establishing all the differences in DNA methylation that distinguish maternal and paternal chromosomes in later development [8–10]. Second, when non-methylated, as on the maternal chromosome, the ICR binds CTCF protein and forms a transcriptional insulator that blocks activation of the maternal Igf2 promoter by the shared downstream enhancers [11]. Third, a paternally inherited, methylated ICR is required for the developmentally regulated silencing of the adjacent paternal H19 promoter [6, 12].
To test whether these three functions are intrinsic to the 2.4-kb H19ICR we generated insertion mutation mouse models that carry the ICR at the non-imprinted alpha-fetoprotein (Afp) location on mouse chromosome 5 (Fig. 1B) [13]. The Afp gene is highly expressed in fetal liver but rapidly repressed after birth. Mice with one functioning copy of Afp are healthy and fertile.
We first looked at the DNA methylation patterns of maternally and paternally inherited ICR insertions and saw that the 65 CpGs within the H19ICR became methylated but only on paternal chromosomes. That is, there is no detectable methylation upon maternal inheritance but upon paternal inheritance, the ICR is fully protected from digestion by methylation sensitive enzymes. Thus the 2.4-kb H19ICR has intrinsic gDMR activity [13].
We next looked at transcriptional insulation. When inserted at the Afp location and maternally inherited, the ICR is not methylated, binds the protein CTCF, and therefore insulates the maternal Afp promoter from interacting with its enhancers so that maternal Afp expression is not detected [14]. In fact, multiple studies in transgenic mice and cell culture models support the idea that insulator function is entirely intrinsic to the H19ICR [11].
Our understanding of the methylated ICR as a transcriptional silencer is more limited. At the endogenous Igf2/H19 locus, silencing of the H19 promoter is a developmental process. Expression of the H19 gene is bi-allelic in early embryos. Repression of the paternal H19 allele in the epiblast correlates with the “spread” of DNA methylation from the ICR to the adjacent CpG-rich H19 promoter/exon 1 region [10, 15]. Sometime after the H19 promoter has become methylated, its silencing is permanent and independent of the continued presence of the paternal H19ICR [6, 9, 12]. For these reasons, it has been assumed that DNA methylation spreading is the mechanism that silences the paternal H19 gene.
In this study, we test whether gene silencing is an intrinsic property of the 2.4-kb H19ICR and whether the spread of DNA methylation is essential for the ICR’s function as a silencer. Unlike the H19 promoter, the Afp promoter does not harbor any CpG-rich motifs but only a few scattered CpG dinucleotides [16]. However, when upstream of the Afp gene, the H19ICR successfully silenced the paternal Afp allele without changing Afp promoter methylation. DNA methylation is therefore likely not the mechanism that is used by the ICR to silence nearby promoters. However, we present developmental analyses that are consistent with the idea that DNA methylation contributes to the stability of gene silencing.
MATERIALS AND METHODS
Mice
Animal research was approved by the NICHD Animal Care and Use Committee and done according to NIH guidelines. Key mouse lines are depicted in Figure 1. ICRflox and ΔICR are from Srivastava [6]. Afp-A, Afp-B and Afp-D are from Park [13]. Afp-Aflox was generated for this study and is similar to Afp-A except that the 2.4-kb BglII fragment carrying the H19ICR is flanked with loxP sites. To generate Afp-loxP animals, Afp-Aflox males were crossed to females transgenic for EIIa-Cre (Jackson Labs 003724). Albumin-cre [17] and Sox2-cre [18] mice were backcrossed into FVB to introduce a single nucleotide polymorphism that distinguishes FVB and SvJ129 alleles of Afp.
Parent-of-origin specific gene expression at Afp and H19
Total RNA was extracted from liver, converted into cDNA, and maternal and paternal specific transcription of Afp and H19 was quantitated using DNA melting curve analyses [19]. Data were analyzed using the unpaired t-test.
Bisulfite sequencing
Methods to quantitate DNA methylation are detailed in the legend to Supplemental Figure 1.
RESULTS
The 2.4-kb H19ICR silences the exogenous Afp promoter
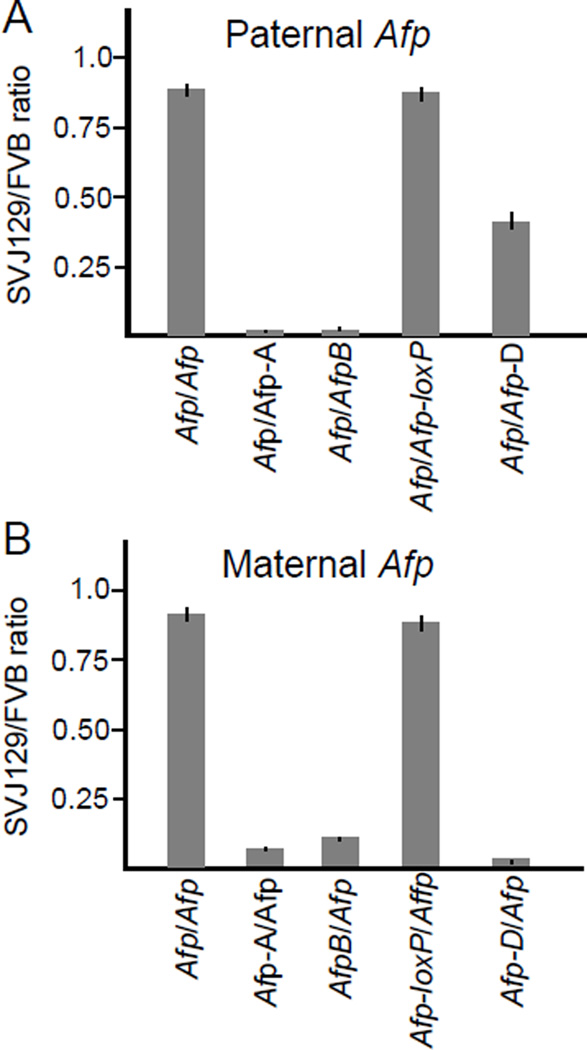
When paternally inherited, ICR insertions at the Afp locus are methylated at CpG dyads [13]. To test the ability of these methylated H19ICR elements to silence Afp expression we quantitated allele-specific expression of Afp upon paternal inheritance of each of the chromosomes described in Fig. 1B. In each experimental animal, the maternal Afp chromosome is wild type and served as an internal reference to normalize expression of the paternal Afp chromosome. Thus a value of 1 would indicate equal levels of expression from maternal and paternal chromosomes while lower values would demonstrate reduced expression of the paternal chromosomes.
In wild type animals, the paternal and maternal chromosomes are comparably expressed (Afp/Afp = 0.91±0.04, n=3) consistent with the fact that Afp is not an imprinted gene (Fig. 2A). However, insertion of the 2.4-kb H19ICR in either orientation results in >40-fold reduction in paternal Afp expression (Afp/Afp-A = 0.02±0.01, n=6; Afp/Afp-B = 0.03±0.01, n=5). We do not think that this loss in paternal Afp expression is due to an insertion mutation that disrupts key Afp regulatory elements because the paternal Afp-loxP chromosome expresses wild type levels of Afp (0.88±0.05, n=3).
Figure 2. Effect of H19ICR insertional mutations on Afp expression.
(A) The 2.4-kb H19ICR functions as a transcriptional silencer upon paternal inheritance. Crosses were set up to generate pups so that maternal chromosome 5 was always FVB and wild type in origin while paternal chromosome 5 was always SvJ129 in origin and was either wild type or mutant as described on the X-axis. RNAs were isolated from neonatal livers and assayed for allele specific expression of Afp. Results are depicted as the ratio of SVJ129 to FVB Afp RNAs. (B) The 2.4-kb H19ICR functions as a transcriptional insulator upon maternal inheritance. Crosses were set up similarly to those described in panel A except that the paternal chromosome 5 was always FVB and wild type in origin while maternal Afp was always SvJ129 and was either wild type or mutant as indicated on the X-axis. Allele specific expression is reported as the ratio of 129 to FVB Afp RNAs. Bar graphs depict mean ± SEM with N values indicated in the text.
It is interesting to see that the Afp-D insertion has only a mild phenotype (0.42±0.05, n=3; see also [14]). The Afp-D chromosome has the 2.4-kb H19ICR in the same orientation as Afp-B but carries an additional 4.8-kb that separate the inserted ICR from the Afp promoter (Fig. 1B). The mild Afp-D phenotype suggests a role for proximity of the ICR to its target promoter.
As additional controls we quantitated expression upon maternal inheritance of each of the alleles described in Fig. 1B. From previous studies we know that maternally inherited H19ICR elements are not methylated and therefore bind CTCF. CTCF binding organizes the maternal chromosome into structures that prevent physical interactions of the Afp enhancer and promoter [14]. In Fig. 2B, we see effects on expression that are consistent with these biochemical analyses. Insertions of the H19ICR in either orientation prevent activation of the maternal promoter (Afp-A/Afp = 0.07±0.02, n=10; Afp-B/Afp = 0.14±0.02, n=6) compared to wild type controls (Afp/Afp = 0.93±0.03, n=5). Most importantly for this study, the Afp-D insertion is highly effective in insulating the maternal Afp promoter from enhancer activation (Afp-D/Afp = 0.02±0.00, n=3). This contrast with the phenotype associated with paternal Afp-D inheritance emphasizes that repression of Afp by insulation (unmethylated maternal ICR) and by silencing (methylated paternal ICR) are distinct processes.
The Afp promoter becomes silenced without DNA methylation
The H19 promoter and the 5’ end of H19 exon 1 are embedded within a 5-kb very CpG-rich region. Silencing of the paternal H19 promoter at gastrulation correlates with the spread of DNA methylation from the H19ICR through the promoter [15]. We chose the Afp gene as the model locus for this study because it is depleted of CpG di-nucleotides. Even using non stringent criteria the nearest CpG-rich regions are 32-kb downstream of the Afp transcriptional start site and thus far outside the locations of known Afp regulatory sequences [20]. Given the prevailing models for silencing at the H19 locus and the overall dearth of CpG residues in the Afp promoter region, we were surprised to see that the ectopic H19ICR was able to completely abrogate transcription from the paternal Afp promoter.
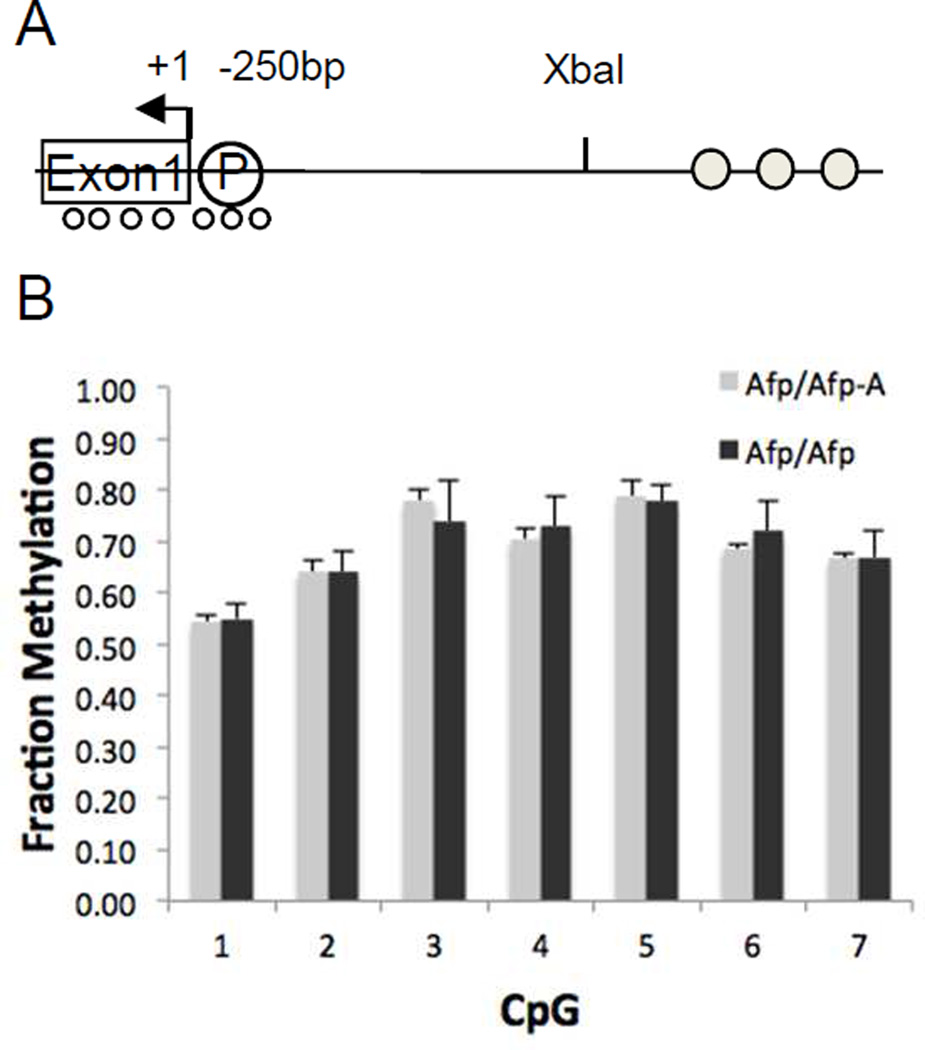
We therefore looked more carefully to see if there were any changes in DNA methylation of Afp gene sequences that were associated with paternal inheritance of the H19ICR insertion and that might explain Afp transcriptional silencing. Transgenic analyses have identified 150-bp upstream of the Afp transcriptional start site that are necessary and sufficient for Afp promoter function [20]. We concentrated on the three CpGs within this region and on the four CpGs at the 5’ end of Afp exon 1 (Fig. 3A). Using bisulfite sequencing, we determined their methylation patterns in E13.5 embryonic livers collected from wild type progeny or from embryos carrying an ICR inserted on the paternal chromosome (Afp/Afp-A). There were no differences between the degrees or the patterns of methylation in these samples (Fig. 3B, SFig.1 and STable 1).
Figure 3. Afp promoter CpG methylation does not change in response to the H19ICR insertion.
(A) Schematic depiction of the Afp locus showing the four CpGs within exon 1 and the three CpGs within the 250-bp Afp promoter that were analyzed. (B) Methylation frequencies for DNA isolated from livers of late gestation wild type mice (Afp/Afp), mice in which the inserted 2.4-kb H19ICR was paternally inherited (Afp/Afp-Aflox), and mice in which the 2.4-kb H19ICR was maternally inherited (Afp-Aflox/Afp). Methylation fractions were calculated from at least 50,000 reads from each of 10 samples as detailed in Sup Fig 1.
Transcriptional silencing depends on developmental processes
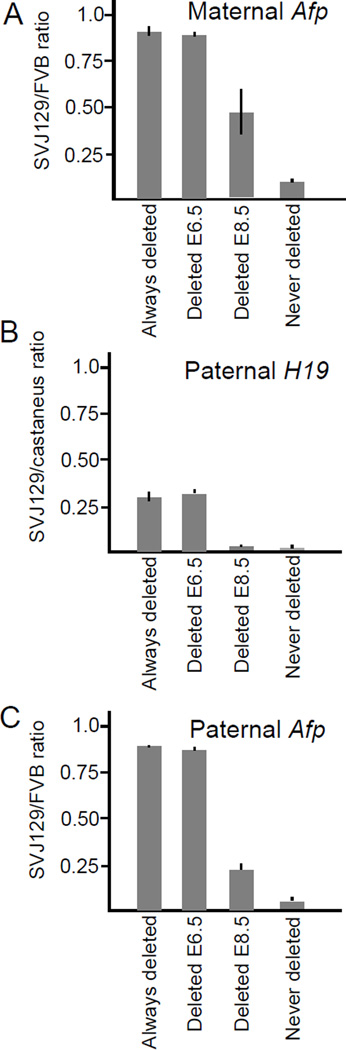
To better understand the mechanism for H19ICR mediated silencing, we wanted to test whether the ICR was required only to establish gene silencing or if its continued presence was necessary to prevent expression of the paternal Afp promoter. Therefore, we deleted the H19ICR at different points in development, using Sox2-cre [18] and Albumin-cre (Alb-cre) [17] to achieve recombination by E6.5 and E8.5, respectively. In each case, we measured gene activity in livers isolated from E16.5 embryos and compared expression profiles to littermates which did not carry the cre transgene (“Never Deleted” in Fig. 4) and also to age-matched animals where the ICR had been deleted in the germ line (“Always Deleted” in Fig. 4).
Figure 4.
A. Stability of transcriptional insulation after removal of the H19ICR from the maternal chromosome. Expression of the maternal Afp allele was quantitated as described in Fig. 2B in Afp-loxP/Afp pups (“Always deleted”), in Afp-Aflox/Afp pups carrying the Sox2-cre transgene (“Deleted E6.5”), in Afp-Aflox/Afp pups carrying the Alb-cre transgene (“Deleted E8.5”), and Afp-Aflox/Afp pups that did not carry a cre transgene (“Never deleted”). B. Stability of transcriptional silencing at the H19 locus. Expression of the paternal H19 allele (SvJ129) was quantitated relative to the expression of the maternal H19 allele that was always wild type and Mus castaneus in origin. “Always deleted” = H19+/H19ΔICR; “Deleted E6.5 = H19+/H19ICRflox + Sox2-cre transgene; “Deleted E8.5 = H19+/H19ICRflox + Alb-cre transgene; “Never deleted” = H19+/H19+. C. Stability of transcriptional silencing at the Afp locus. Crosses were set up as described for Fig. 2A. Expression of paternal Afp (SvJ129 allele) was quantitated relative to the expression of wild type maternal FVB Afp in Afp/Afp-loxP pups (“Always deleted”), in Afp/Afp-Aflox pups carrying the Sox2-cre transgene (“Deleted E6.5”), in Afp/Afp-Aflox pups carrying the Alb-cre transgene (“Deleted E8.5”), and in Afp/Afp-Aflox pups that did not carry a cre transgene (“Never deleted”).
To assure the efficacy of our model system, we first studied the effect of deletion on Afp expression when the H19ICR was maternally inherited (Fig. 4A). The maternal chromosomes serve as good controls because it is well established that transcriptional insulation is absolutely dependent upon the continual presence of the CTCF-bound ICR to prevent promoter-enhancer interactions. Here we show that cre-mediated deletion of the H19ICR from chromosome 5 at E6.5 or at E8.5 results in high levels of maternal Afp expression (Sox2-cre = 0.91±0.02, n=13; Alb-cre = 0.43±0.07; n=3) compared with control animals that did not carry the cre transgene (0.10±0.06; n=5). Similarly, we saw high levels of maternal Igf2 expression when the H19ICR was deleted with Sox2-cre or with Alb-cre using the ICRflox chromosome depicted in Fig. 1A (data not shown). Together, these results demonstrate the relative effectiveness of the two cre transgenes in removing the H19ICR at both the endogenous H19 and the exogenous Afp loci. Note that at both Igf2 and at Afp, the Alb-cre is only partially effective in recombination.
We next studied the effect of cre recombination on silencing of the endogenous H19 gene (Fig. 4B). Previous studies have shown that silencing of the paternal H19 promoter is a developmental process: the methylated ICR is needed to establish silencing but once silencing is established the ICR becomes superfluous and can be removed without re-activation of the paternal promoter [6, 12]. Consistent with these earlier findings, silencing of the paternal H19 is fully maintained when the ICR is removed by Alb-cre (Never deleted = 0.02±0.02, n=2; Deleted E8.5 = 0.02±0.01, n=8). However, deletion at E6.5 via Sox2-cre results in a phenotype that is indistinguishable from that seen in mice where the ICR was already removed in the germ line (germ line or “always deleted” = 0.30±0.04, n=5; Deleted E6.5 = 0.32±0.02, n=8). That is, having the ICR present only until E6.5 is no better than never having it there at all. These data show that the epigenetically stable silent state of the paternal H19 promoter is established between E6.5 and E8.5.
Our main interest was in the stability of silencing at the Afp locus. These results are depicted in Fig. 4C. Deletion of the H19ICR at E6.5 results in complete loss of silencing just as it does at the endogenous H19 locus (germ line or “always deleted” = 0.91±0.03, n=21; Deleted E6.5 = 0.89±0.02, n=6). However, deletion via Alb-cre at E8.5 has an intermediate phenotype (0.22±0.03, n=4) with the promoter showing increased activity relative to the “Never Deleted” littermates (0.05±0.01, n=3, p<0.01) but also showing repression relative to the “Always Deleted” control animals (p<0.001).
DISCUSSION
The H19ICR is essential for imprinting of the Igf2/H19 locus [5–7] and for organizing maternal and paternal chromosome domains into distinct 3D structures that promote high levels of monoallelic expression of H19 and Igf2, respectively [14, 21–23]. Deletion of the endogenous H19ICR results in maternal and paternal chromosomes that are functionally equivalent and that lack the long-range interactions characteristic of wild type chromosomes.
We tested the intrinsic properties and functionality of the ICR by transferring it to ectopic locations and assaying its ability to keep track of its own parental origin and to regulate the expression of its new neighboring genes. Others and we have shown that the 2.4-kb BglII fragment is sufficient to act as a DMR in whatever genomic context it has been tested [13, 24, 25]. There is one interesting caveat: the exact timing for acquisition of paternal specific methylation varies dependent upon the site of integration. At the endogenous locus, the H19ICR is fully methylated during spermatogenesis while at ectopic locations, DNA methylation is not established until after fertilization.
We have already shown that maternal (non-methylated) H19ICR insertions at the Afp locus bind CTCF and prevent physical interactions between the Afp promoter and its distal enhancers through CTCF-mediated processes just as the endogenous H19ICR prevents physical interactions between the maternal Igf2 promoters and their distal enhancers [14]. These results are consistent with numerous transgene studies in mice and in tissue culture that show that the H19ICR’s function as a transcriptional insulator is universal and dependent only on its location between any enhancer and its target promoters [11].
In this study we focus on the paternally inherited ICR and its ability to repress transcription of the adjacent Afp promoter. H19ICR mediated repression of paternal Afp is highly efficient and reduces promoter activity by 40-fold. Thus silencing activity of the H19ICR is intrinsic to the ICR and not dependent upon special features of the H19 promoter, its normal target. Upon paternal inheritance, the H19ICR inserted at Afp is methylated and cannot bind CTCF. Thus mechanisms for repression seen in Figure 2A (paternal inheritance) and Figure 2B (maternal inheritance) must be distinct. In this study, we refer to the repression of Afp mediated by the paternally inherited ICR as silencing, consistent with terminology used to described repression of the paternal H19 promoter.
Results in Figure 2 provide clear genetic evidence that two distinct mechanisms are used on the two parental chromosomes. Most importantly, the Afp-D insertion completely insulates Afp expression upon maternal inheritance but has only a minimal effect upon paternal inheritance. In fact, a comparison of expression from Afp-B and Afp-D chromosomes suggests that proximity of the ICR to its target promoter is necessary for effective gene silencing on the paternal chromosome but is not important for insulation on the maternal chromosome.
Altogether we see that each of the H19ICR’s key functions and properties are intrinsic to the 2.4-kb BglII fragment. The ability of the H19ICR to organize chromosomal structures and to establish gene expression patterns can be predicted on its own intrinsic properties and its activities are not dependent on unique features of the Igf2/H19 locus. Thus while many research labs, including our own, have identified and emphasized the critical significance of specific long-range interactions mediated by the H19ICR at the Igf2/H19 locus, ([11, 26], it is also important to keep in mind that the ICR is promiscuous in its ability to find suitable partners in regulating gene activity.
We note that we were actually surprised to see H19ICR-mediated silencing of the paternal Afp promoter given that the Afp gene does not have any CpG-rich regions and in fact, is overall quite deficient in CpG dyads. In fact, silencing by the H19ICR was not accompanied by any measurable changes in CpG methylation within the Afp sequences yet Afp repression is as efficient as that noticed for paternal H19.
The spread of DNA methylation from the ICR to the H19 promoter has seemed like an excellent explanation for H19 silencing. DNA methylation has been long associated with gene repression and especially CpG island (CGI) methylation is associated with robust repression [27]. Paternal H19 promoter methylation occurs in epiblasts and correlates with loss of paternal H19 expression. Moreover, genetic studies have clearly demonstrated that Dnmt1 methyltransferase activity is essential for normal H19 imprinting [28]. In contrast, our analyses of repression of paternal Afp suggest that DNA methylation is not essential for transcriptional silencing per se and that a paternally inherited H19ICR has the ability to repress transcription from an adjacent promoter without changing methylation frequency or patterns. We suggest that the developmental studies in Fig. 4B might address this paradox. By E8.5, silencing of H19 is epigenetically stable and continues even after removal of the H19ICR. In contrast, silencing at Afp is unstable. Altogether these results are consistent with the idea that other changes such as chromatin are the initial signals that direct promoter repression while CpG methylation functions as a stable cell memory for transcriptional state [27, 29].
Supplementary Material
Highlights.
The H19ICR can silence a heterologous promoter
Silencing by the H19ICR does not depend on promoter methylation.
Silencing by the H19ICR is a developmental process.
Acknowledgments
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Division of Intramural Research (ZIA HD 001804). Steve Coon and Tianwei Li generated the deep sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kelsey G, Bartolomei M. Imprinted Genes…and the Number is? PloS Genet. 2012;8:e1002601. doi: 10.1371/journal.pgen.1002601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman D, Halje M, Nordin M, Engstrom W. Insulin-like growth factor 2 in development and disease: a mini-review. Gerontology. 2013;59:240–249. doi: 10.1159/000343995. [DOI] [PubMed] [Google Scholar]
- 3.Kaffer C, Grinberg A, Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol Cell Biol. 2001;21:8189–8196. doi: 10.1128/MCB.21.23.8189-8196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 5.Kaffer CR, Srivastava M, Park K, Ives E, Hsieh S, Batlle J, Grinberg A, Huang SP, Pfeifer K. A transcriptional insulator at the imprinted H19/Igf2 Locus. Genes Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava M, Hsieh S, Grinberg A, Williams-Simon L, Huang S-P, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting element. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 7.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei MS, Walter J, Reik W. Epigenetic modification in an imprinting cluster are controlled by a heiracrchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava M, Frolova E, Rottinghaus B, Boe S, Grinberg A, Lee E, Lover P, Pfeifer K. Imprint control element mediated secondary methylation imprints at the Igf2/H19 locus. JBC. 2003;278:5977–5983. doi: 10.1074/jbc.M208437200. [DOI] [PubMed] [Google Scholar]
- 10.Tremblay KD, Duran KL, Bartolomei MS. A 5' 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace J, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–417. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorvaldsen J, Fedoriw A, Nguyen S, Bartolomei M. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol Cell Biol. 2006;26:1245–1258. doi: 10.1128/MCB.26.4.1245-1258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park K, Sellars E, Grinber A, Huang S, Pfeifer K. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol Cell Biol. 2004;24:3588–3595. doi: 10.1128/MCB.24.9.3588-3595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon Y, Jeong S, Rong Q, Park K-Y, JH C, K P. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 16.Pachnis V, Brannan C, Tilghman S. The structure and funcgtion of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7:673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postic C, Magnuson M. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi S, Lewis P, Pevny L, McMahon A. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 19.Jeong S, Hahn Y, Rong Q, Pfeifer K. Accurate quantitation of allele-specific expression patterns by analysis of DNA melting. Genome Research. 2007;17:1093–1100. doi: 10.1101/gr.6028507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spear B. Alpha-fetoprotein gene regulation: lessons from transgenic mice. Semin Cancer Biol. 1999;9:109–116. doi: 10.1006/scbi.1998.0087. [DOI] [PubMed] [Google Scholar]
- 21.Eun B, Sampley M, Good A, Gebert C, Pfeifer K. Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus. Nucleic Acids Res. 2013;42:817–826. doi: 10.1093/nar/gks1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Ju J-F, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu T, Hoffman A. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 24.Gebert C, Kunkel D, Grinberg A, Pfeifer K. H19 imprinting control region methylation requires an imprinted environment only in the male germ line. Mol Cell Biol. 2010;30:1108–1115. doi: 10.1128/MCB.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzaki H, Okamura E, Shimotsurm A, Fukamizu A, Tanimoto K. A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independent of its establishment in germ cells. Mol Cell Biol. 2009;29:4595–4603. doi: 10.1128/MCB.00275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanli I, Feil R. Chromatin mechanisms in the developmental control of imprinted gene expression. Int J Biochem Cell Biol. 2015;67:139–147. doi: 10.1016/j.biocel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Schuebeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 28.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 29.Jones P. Functions of DNA methylation: islands,start sites, gene bodies, and beyond. Nature Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.