Abstract
Notch signaling, involved in development and tissue homeostasis, is activated at the cell-cell interface through ligand-receptor interactions. Previous studies have implicated mechanical forces in the activation of Notch receptor upon binding to its ligand. Here we aimed to determine the single molecular force required for Notch activation by developing a novel low tension gauge tether (LTGT). LTGT utilizes the low unbinding force between single-stranded DNA (ssDNA) and E. coli ssDNA binding protein (SSB) (~4 pN dissociation force at 500 nm/s pulling rate). The ssDNA wraps around SSB and, upon application of force, unspools from SSB, much like the unspooling of a yoyo. One end of this nano yoyo is attached to the surface though SSB while the other end presents a ligand. A Notch receptor, upon binding to its ligand, is believed to undergo force-induced conformational changes required for activating downstream signaling. If the required force for such activation is larger than 4 pN, ssDNA will unspool from SSB and downstream signaling will not be activated. Using these LTGTs, in combination with the previously reported TGTs that rupture double stranded DNA at defined forces, we demonstrate that Notch activation requires forces between 4–12 pN, assuming an in vivo loading rate of 60 pN/s. Taken together, our study provides a direct link between single-molecular forces and Notch activation.
Keywords: Notch signaling, single-molecular forces, low tension gauge tether (LTGT), nano yoyo
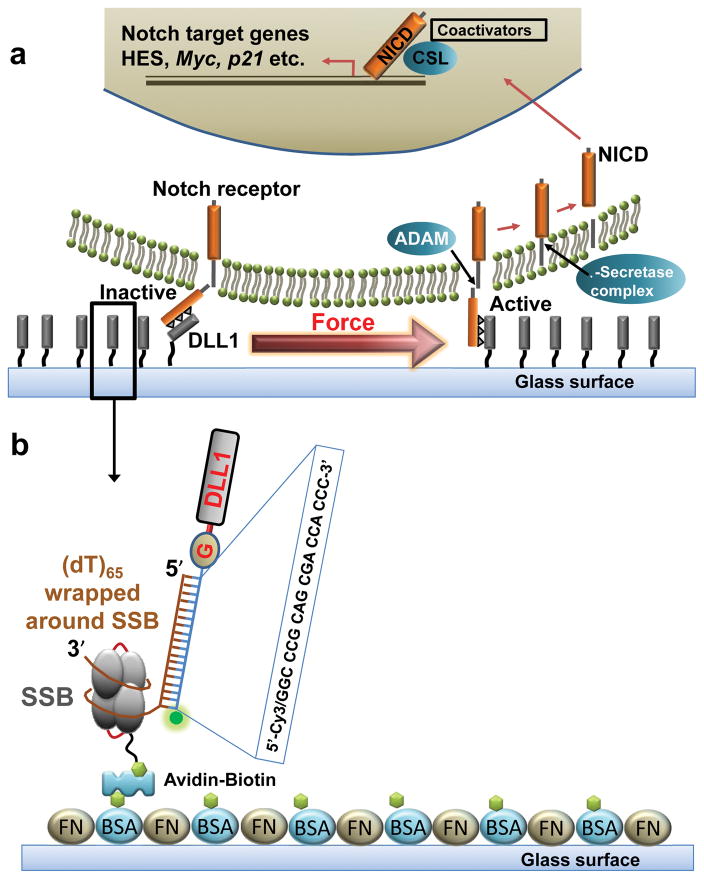
Notch signaling between neighboring cells regulates a host of critical cellular functions ranging from neurogenesis1, cell fate determination2, 3, immune responses4, and maintenance of adult tissue homeostasis5. Aberrant regulation of Notch signaling is associated with disease states such as cancer6–8. The transmembrane protein Notch receptor is a hetero-oligomer consisting of an extracellular domain, a single transmembrane region, and an intracellular domain. Upon binding to a delta ligand presented by a neighboring cell membrane, the Notch receptor undergoes conformational changes, exposing a cleavage site on its extracellular domain to initiate Notch signaling (Figure 1a). Previous studies have implicated mechanical forces in the initiation of Notch activation9–11. Recently, Gordon et al. showed that about 5 pN of force is necessary to expose the cleavage site in vitro12 but in a cell-based assay 1.5 pN of force was sufficient to activate Notch signaling. In addition, in most of the previous studies that evaluated the force requirement of Notch activation, the force was applied by the human practitioner rather than the cell. In order to evaluate the force requirement of Notch signal activation, we recently developed an array of Tension Gauge Tethers (TGTs), with molecular tension tolerance in the range of 10–60 pico-Newton (pN)13. We determined that Notch signaling can be activated even when the Notch ligands are presented through the TGTs of the lowest tension tolerance (~12 pN)13, leading to our proposal that Notch activation is either force independent or requires less than 12 pN of force. Here we developed a novel class of Low Tension Gauge Tether (LTGT) with tension tolerance below 12 pN so that we can better define the force requirement for Notch activation and other mechanical signaling processes.
Figure 1.
Design and working principle of DLL1-LTGT. (a) Force induced activation of Notch receptors upon binding to ligand DLL1 is shown. (b) Ligand DLL1 is conjugated to a double stranded DNA with an overhang of ssDNA (dT65) wrapped around a homotetrameric single-tailed SSB. DLL1-LTGT was immobilized to the passivated glass surface via biotin-neutravidin interactions. Glass surfaces were also coated with fibronectin to promote cell adhesion. A Cy3 fluorophore is conjugated to DLL1-LTGT so that we can monitor fluorescence signal loss in real time when a cell pull away the construct.
We reasoned that in order to engineer a tether that breaks at lower forces than 12 pN but is stable in the absence of force, we would need to utilize multivalent interactions. If two molecules are bound to each other through multiple weak bonds, it is highly unlikely that they would fully dissociate spontaneously. But, upon application of weak forces, the bonds can break one by one gradually. In fact, double stranded (ds) DNA rupture via application of force in the unzipping direction is one such example that gave us an approximately 12 pN of rupture force which is lower than the force required to rupture dsDNA when the force is applied in a shear configuration13.
E. coli single stranded DNA binding protein (SSB) forms a stable tetramer, that can bind ~65 nucleotides (nt) of single stranded (ss) DNA with very high affinity under moderately high salt conditions14 by wrapping the ssDNA around the tetramer15–20. We hypothesized that pulling on the ssDNA end may allow us to peel the ssDNA off the SSB protein surface gradually even at low forces. We previously showed that when the two ends of the ssDNA wrapped around a single SSB tetramer are under tension, the SSB protein is ejected from the ssDNA at approximately 9 pN of force21, 22. Here, we examined the SSB dissociation force under a different configuration where the force is applied between the SSB tetramer and the ssDNA (Figure 1). Due to the pulling configuration, as the ssDNA is unspooled from the SSB protein, the protein will rotate around the biotin attachment point, mimicking the unspooling of a yoyo. In order to attach the SSB to the surface, we added an in vivo biotinylation tag to the single C-terminal end of a previously described tandemly fused SSB tetramer23. Creation, purification, and characterization of tandemly fused SSB tetramer are described in the supporting information (Materials and Methods; Supporting Information Figure S1). Biotinylation was confirmed using the HABA assay24. If the rupture force between the ssDNA and the biotinylated tandem SSB tetramer (btSSB) is significantly below 12 pN, the lowest rupture force for double stranded DNA tethers, such a btSSB-ssDNA complex can serve as an LTGT.
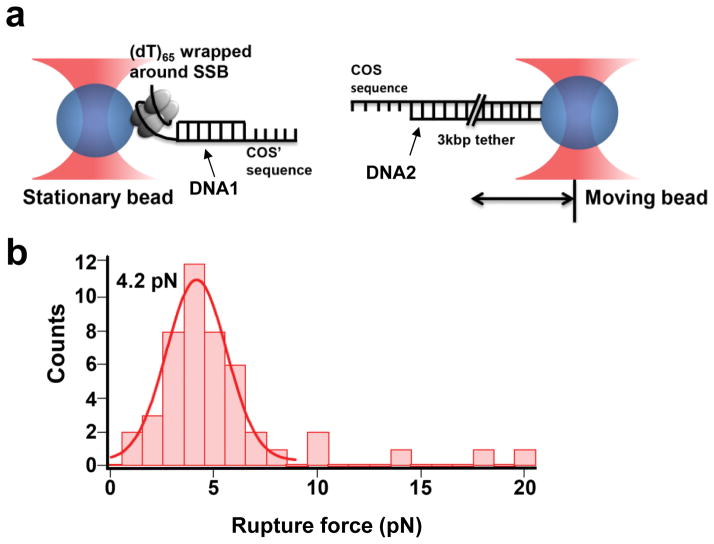
We determined the magnitude of force required to dissociate single ssDNA, (dT)65, from a single SSB tetramer using a dual-trap high-resolution optical tweezers (Figure 2a). In the dual-trap optical tweezers instrument, the stationary bead (Figure 2, left bead) carries the protein-ssDNA complex termed btSSB-DNA1 and the moving bead (Figure 2, right bead) carries a DNA construct termed DNA2. The DNA1 construct was prepared such that a 3′ (dT)65 and a 5′ COS′ sequence are separated by an 18 bp dsDNA (Fig. 2a, Supporting Information Figure S2). The 12 nucleotides (nt) COS′ sequence is not long enough to wrap around SSB or bind stably to SSB. As such, only the (dT)65 portion of DNA1 wraps around the btSSB. The btSSB-DNA1 complex was then immobilized on a stationary streptavidin-coated bead. DNA2 consists of a 3k base-pair (bp) double stranded (ds) DNA handle with a 5′ 12 nt ssDNA overhang with a COS sequence (complementary to the COS′ sequence in the stationary bead) at one end and a 5′ digoxigenin at the other. DNA2 was immobilized on the moving bead through the anti-digoxigenin antibodies coating it.
Figure 2.
Force calibration of btSSB: ssDNA LTGT. (a) High-resolution optical tweezers were used to determine required force for dissociation of ssDNA from btSSB. (b) A histogram of dissociation force between a single ssDNA and a single SSB is shown here (n=47). A Gaussian fit to the distribution gives a mean dissociation force of 4.1±0.1pN and a FWHM of 3.2±0.3 pN.
The moving bead was moved repeatedly close to and away from the stationary bead carrying DNA1. If the two DNA molecules anneal via their COS/COS′ sites, the force on the stationary bead increases as the moving bead is moved away until the connection between the two beads ruptures. Representative force vs. distance traces are shown in Supporting Information Figure S3 for a pulling speed of 500 nm/s. All optical tweezers experiments were performed at room temperature in a buffer containing 50 mM Na+ and 5 mM Mg+2 in which 65 nt of ssDNA wraps around SSB25. The histogram of force at the moment of rupture (n=47) (Figure 2b) shows a peak at 4.2 pN. We attribute the observed rupture events to the dissociation of DNA1 from btSSB for the following reasons. Control experiments without SSB or the COS sequence on DNA2 did not show any rupture events. Therefore, rupture events observed are due to either btSSB-ssDNA dissociation or shear opening of the 12 bp of COS/COS′ duplex. However, it is well known that the rupture force of DNA in the shearing geometry is in the range of 20–60 pN, depending on the number of base pairs under shear force26. In addition, we previously observed that the COS/COS′ duplex withstand forces that are needed to fully stretch short ssDNA (30–35 pN)27 or to unravel DNA from the nucleosome (up to about 20 pN)28. Therefore, the observed rupture force of a few pN must be due to disruption of the ssDNA-SSB interaction. Thus we achieved our goal of developing an LTGT that ruptures at forces significantly below 12 pN. When we repeated the measurement at a much higher pulling rate of 2000 nm/s, the peak of the rupture force histogram shifted to 8 pN (Supporting Information Figure S4), which is higher than the 4.2 pN obtained at 500 nm/s but is still below the 12 pN of the lowest TGT reported previously. Potential caveats concerning the accuracy of the rupture force caused by differences in ionic condition and temperature between the optical tweezers experiments and cell-based experiments are discussed in Supplementary Notes.
In order to determine the forces required for Notch signaling activation in living cells, we employed a Notch ligand, DLL1 (Delta-like protein 1), tethered to a surface through ssDNA wrapped around the btSSB (Figure 1b). In DLL1-LTGT, we first conjugated Protein G to 18 nt ssDNA (Supporting Information Figure S5)29 which is then hybridized to a complementary sequence connected to (dT)65 ssDNA to form an 18 bp dsDNA with a 3′overhang of (dT)65. The DLL1 ligand9 fused with the Fc domain was bound to Protein G conjugated to the DNA, followed by incubation with btSSB. The overhang of (dT)65 wraps around the btSSB to make the final product of btSSB: ssDNA: ProG: DLL1. This DLL1-LTGT was immobilized on a glass surface through a neutravidin-biotin linker (Figure 1). The surface was also coated with fibronectin to promote cell adhesion. If the force through a single Notch-DLL1 bond required to activate Notch signaling is larger than the force required to dissociate the ssDNA from the btSSB, the ssDNA will dissociate and Notch signaling will not be activated. On the other hand, if the required force is smaller than the dissociation force, the LTGT will endure, resulting in Notch signaling activation. To ensure the interaction of btSSB: ssDNA complexes is close to 1:1, we ran an electrophoretic mobility shift assay on (dT)65 constructs where (dT)65 was mixed with btSSB at a 1:1 molar ratio (Supporting Information Figure S6). Only a single band was observed confirming the formation of stable btSSB: ssDNA complexes.
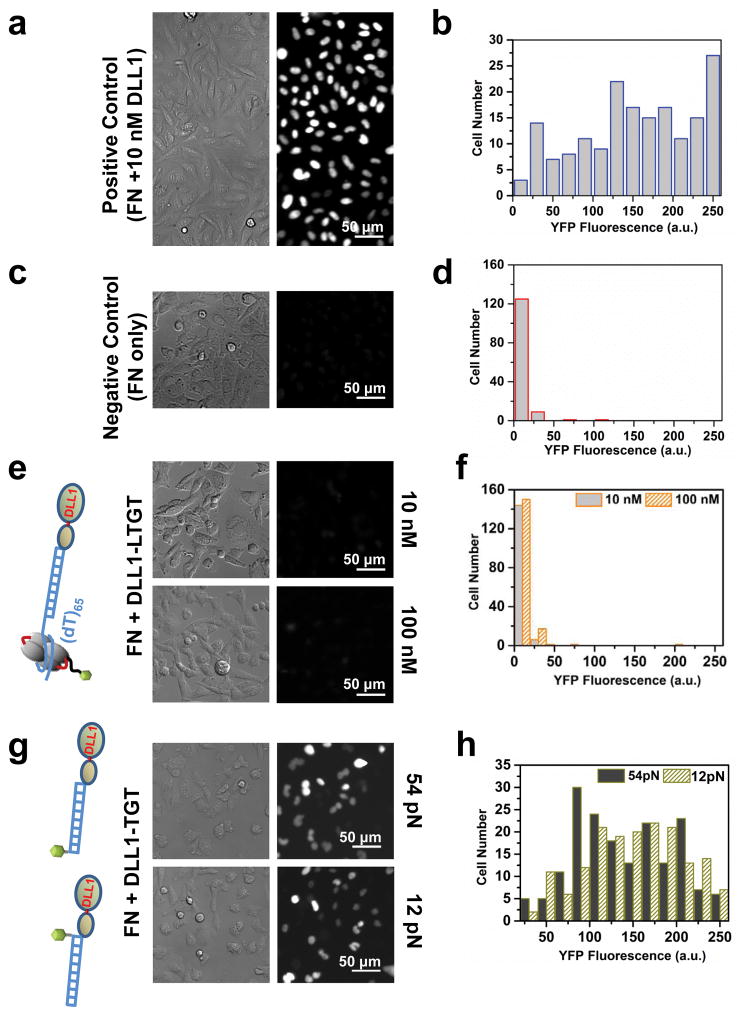
To read out Notch activation at the single cell level, we used CHO-K1 cells stably expressing human NOTCH1, whose intracellular domain is replaced by the transcription activator Gal4. This cell line is also transfected with Gal4-controlled H2B-YFP as a reporter so that after notch activation, H2B-YFP fluorescence is detected in the nucleus, reaching an optimal level after two days30. As a positive control, we seeded Notch reporter CHO cells30 on a glass surface coated with both fibronectin and 10 nM DLL1-Fc to see if activation of Notch signaling was represented by H2B-YFP fluorescence in the cell nucleus. We observed a high nucleus fluorescence signal reaching an optimal level after 48 hr from the positive control (Figure 3a, b). When we treated cells with DAPT31, 32, which prevents proteolytic cleavage mediated by γ-secretase, thus preventing Notch activation, we observed a reduction in nuclear fluorescence in a dose dependent manner (Supporting Information Figure S7). As a negative control, we omitted DLL1-Fc ligand and seeded cells on a fibronectin only surface. We observed only a faint fluorescence signal confirming that fibronectin alone cannot activate Notch signaling (Fig. 3c, d).
Figure 3.
Notch signaling in transgenic CHO-K1 cells is not activated in DLL1-LTGT assay as represented by low H2B-YFP expression. (a–b) High levels of YFP signal indicate Notch signaling is activated when ligand DLL1-Fc (10 nM) is directly immobilized on the surface. (c–d) When ligand DLL1 is excluded, Notch signaling is not activated. (e–f) Notch signaling is also not activated on the DLL1-LTGT surface even with ten-fold elevated concentration (100 nM). (g–h) Cells on 12 pN and 54 pN TGT surfaces show activation of Notch signaling. When btSSB: ssDNA part is excluded from the construct, one may design TGT in both 12 pN and 54 pN orientation simply changing the biotin position. It is to be noted that cells can activate Notch signaling even on 12 pN TGT engineered surfaces. This suggests Notch activation requires force between 4–12 pN.
To test our newly developed LTGT in Notch signaling, we prepared a surface coated with fibronectin and neutravidin (via biotinylated Bovine serum albumin) and then incubated 10 nM of DLL1-LTGT. Even 48 hr after plating the cells, we did not observe any fluorescence signal suggesting that Notch is not activated on the DLL1-LTGT (Figure 3e, f). Next, we increased the surface-incubation concentration of DLL1-LTGTs to 100 nM which increased the surface density of LTGTs according to fluorescence imaging of Cy3 fluorophores conjugated to them (Supporting Information Figure S8). Even with the increased DLL1-LTGT surface density, we only observed a faint nucleus fluorescence signal (Figure 3e, f).
Although the data presented so far suggest that Notch activation requires a force that is higher than the force needed to disrupt the ssDNA from SSB, it was possible that the bond between Protein G and the Fc-domain fused to DLL1 may be the weak link that ruptures in our experiment. To test this possibility, we prepared two DLL1-TGTs with the calculated rupture forces of 12 pN and 54 pN where DLL1-Fc is bound to Protein G conjugated to a double stranded DNA that is in turn immobilized to the surface through a biotin on the DNA. In the 12 pN TGT, the biotin is attached to the same duplex end as Protein G so that the cellular force applied through the Notch receptor-DLL1 bond applies an unzipping force to the DNA. In the 54 pN TGT, the biotin is moved to the opposite duplex end so that the cellular force is applied to the DNA in the shear configuration (Figure 3g, h). We found that for the same 10 nM concentration of DLL1-TGT incubation, Notch signaling is activated for both 12 pN and 54 pN TGTs, showing that the Fc-Protein G bond is not the weak link in our experiments. Lack of Notch activation on the DLL1-LTGT surface therefore is not due to the rupture of Fc-Protein G bond. Otherwise, we would have observed no Notch activation with 12 pN or 54 pN DLL1-TGTs. Instead, lack of Notch activation with DLL1-LTGT must be due to the rupture of btSSB: ssDNA, and we can deduce that the force through the single Notch-ligands bonds that is necessary for Notch activation is larger than the rupture force of ssDNA wrapped around btSSB, which is about 4 pN at 500 nm/s pulling rate.
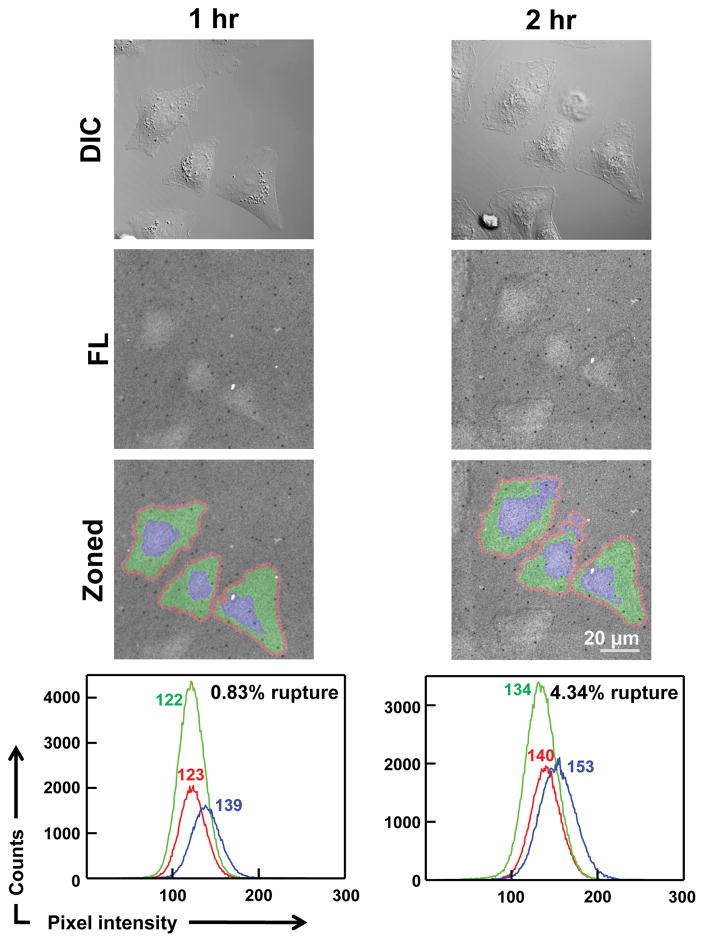
Next, in order to test if indeed the DLL1-LTGT is being ruptured by the cells, we monitored the fluorescence signal from Cy3 fluorophores conjugated to DLL1-LTGTs underneath the cells (Figure 4). Only a minimal amount of fluorescence loss (~0.8 %) was observed from the peripheral regions of the after 1 hour of cell plating but ~ 5 fold greater fluorescence loss (~4.3 %) was observed after 2 hours. We also observed that the fluorescence signal increases in the central region of the cell, likely due to migration of ruptured LTGTs toward the cell center. This indicates that Notch Receptors pulling on the DLL1-LTGT were able to remove the fluorescently tagged DNA from the surface immobilized btSSB, and therefore the cell must be applying forces larger than ssDNA-btSSB rupture force through the single Notch-DLL1 bonds. In contrast, when we monitored fluorescence loss on the 12 pN TGT surface, we did not observe any significant fluorescence loss in the cell periphery or increase in fluorescence in the central regions (Supporting Information Figure S9). Therefore, the force exerted by Notch receptor is below 12 pN.
Figure 4.
Time dependent rupture of DLL1-LTGT from the surface. DIC, surface fluorescence, and analyzed images show very little rupture after 1 hour of cell plating. However, there is a significant difference observed in terms of LTGT rupture after 2 hours suggesting that Notch receptors can dissociate ssDNA tethers from surface immobilized btSSB. Red, green and blue regions indicate background, rupture region, and cell nuclei respectively. The baseline value was obtained from non-fluorescent images and were corrected from both ruptured and background regions. A histogram of each region was plotted and the fit to the histogram was used to calculate rupture percentage. Values next to each peak indicate mean intensity of each region.
In a recent study, Narui et al. showed that restricting spatial movement of Notch ligands in supported lipid bilayers using a patterned surface can activate Notch signaling, indirectly implicating mechanical force33. If the Notch receptor is being pulled in a certain direction by the cell, the associated ligand on the lipid bilayers would move together, and only when the movement is resisted by a barrier, a sufficient force is developed to activate signaling. Such resisting force would develop if the ligands are immobilized directly on a surface but not for soluble ligands or mobile ligands on the membrane. However, in the study by Narui et al., the magnitude of the force required could not be determined. Recent advances of single molecule platforms including our own TGT technique can be leveraged to investigate such relevant tension forces in different mechanotransduction pathways13, 34–36. In our previous Notch study13, we found that Notch signaling can be activated even on the lowest 12 pN tension tolerance tethers. To conclusively pin down the role of mechanical forces in Notch activation, we designed a new low tension gauge tethers with a rupture force of 4 pN. Using our newly developed LTGT platform, we thus demonstrated that mechanical forces are required for Notch activation and the required force is between 4 pN and 12 pN, assuming that the rate of force increase used in our LTGT rupture experiments mimics the cellular rate (see discussion below). Recently, Gordon et al. performed single molecule mechanical perturbations to show that Notch receptor’s extracellular domain cleavage required for activation occurs at about 5 pN of force and externally applied constant force of about 1.5 pN can activate Notch signaling in the cell. These force values should be considered in agreement with our estimate of 4 pN-12 pN given that we do not know the manner in which the force is applied to the Notch-ligand bond in the cell. All of these forces are lower than 19 pN rupture force between Notch receptor and DLL1 ligand estimated using optical tweezers experiments37 so that the receptor-ligand bond would not rupture during mechanical changes to the Notch receptor. We note that the optical tweezers study did not specify the rate of force increase used.
Evidence has been presented that ligand endocytosis by the ligand-presenting cell is necessary for Notch activation and may be the primary source of force11, 12, 38, 39. However, our data indicate that such force can be internally generated from the receptor-presenting cells as long as the ligands are immobile (Fig. 4). We observed in live cell imaging using fluorescently labeled DLL1-LTGTs that fluorescence signaling is reduced at the cell periphery and increases at the cell center. Actin filaments can undergo retrograde flow during cell spreading40–44, and if Notch receptors are linked to the actin cytoskeleton, such retrograde flow toward the cell center may apply forces large enough to rupture LTGT and the ruptured LTGTs then accumulate near the cell center. We suggest that the internally generated acto-myosin forces, exerted at the cell-ECM interface, is also a potent source of mechanical force that can initiate Notch activation. The speed of actin retrograde flow has been estimated previously 41, 45, 46. However, the rate of force increase (also called the loading rate) cannot be determined from the actin flow rate alone because the local intracellular compliance remains unknown.
The measured LTGT rupture force values were dependent on the pulling speed (4 pN at 500 nm/s and 8 pN at 2000 nm/s). Because the 3 kb DNA tether between LTGT and the trapped bead has nonlinear elasticity, the force increases nonlinearly as the moving bead is moved at a constant speed. We calculated the loading rate at the corresponding rupture force and obtained ~58 pN/s at 4 pN (500 nm/s pulling speed) and ~630 pN/s at 8 pN (2000 nm/s pulling speed). Therefore, if the true loading rate in the cell is lower than ~58 pN/s, the force required for Notch activation can be lower than 4 pN. Regardless of the exact force value, our data clearly show that mechanical force is required for Notch activation because premature LTGT rupture induced by cellular pulling force prevents notch activation.
In summary, our live cell data demonstrate that Notch activation does require a mechanical force greater than 4 pN although the exact threshold force will depend on the cellular loading rate. Our newly developed LTGT platform of “nano yoyo” made of a spool of ssDNA around SSB protein can also be extended to explore force requirements in other signaling receptors, cell adhesion proteins and endocytosis.
Supplementary Material
Acknowledgments
We thank Irwin D. Bernstein for the generous gift of DLL1 and Michael B. Elowitz for providing genetically modified CHO cell-line to study Notch activation. Funding was provided by US National Science Foundation (PHY 1430124 to T.H. and Y.R.C.) and by the US National Institutes of Health grants (GM065367 to T.H., GM030498 to T.M.L.). F.C. acknowledges Carl R. Woese Institute for Genomic Biology Fellowship from the University of Illinois at Urbana-Champaign. T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions
T.H. and F.C. conceived the project and wrote the manuscript. F.C., I.L., T.N., T.H. designed experiments. I.L., T.N., B.L., X.W., B.C.K performed experiments and/or analyzed data. T.M.L and E.W conceived and designed the btSSB construct, JES purified and characterized the btSSB protein. All authors discussed the results.
The authors declare no competing financial interest.
This document includes Materials and Methods, supporting figures and information.
References
- 1.Louvi A, Artavanis-Tsakonas S. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand MD, Lake RJ. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Conboy IM, Rando TA. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 4.Radtke F, Fasnacht N, Macdonald HR. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Ables JL, Breunig JJ, Eisch AJ, Rakic P. Nat Rev Neurosci. 2011;12:269–83. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranganathan P, Weaver KL, Capobianco AJ. Nat Rev Cancer. 2011;11:338–51. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 7.Allenspach EJ, Maillard I, Aster JC, Pear WS. Cancer Biol Ther. 2002;1:466–76. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 8.Lobry C, Oh P, Aifantis I. J Exp Med. 2011;208:1931–5. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. J Cell Sci. 2000;113(Pt 23):4313–8. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 10.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 11.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Development. 2000;127:1373–85. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 12.Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, Blacklow SC. Dev Cell. 2015;33:729–36. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Ha T. Science. 2013;340:991–4. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlov AG, Lohman TM. J Mol Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 15.Lohman TM, Ferrari ME. Annu Rev Biochem. 1994;63:527–70. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 16.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Nat Struct Biol. 2000;7:648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 17.Bujalowski W, Lohman TM. Biochemistry. 1986;25:7799–802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 18.Lohman TM, Overman LB. J Biol Chem. 1985;260:3594–603. [PubMed] [Google Scholar]
- 19.Meyer RR, Laine PS. Microbiol Rev. 1990;54:342–80. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R, Kozlov AG, Roy R, Zhang J, Korolev S, Lohman TM, Ha T. Cell. 2011;146:222–32. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suksombat S, Khafizov R, Kozlov AG, Lohman TM, Chemla YR. Elife. 2015;4 doi: 10.7554/eLife.08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antony E, Weiland E, Yuan Q, Manhart CM, Nguyen B, Kozlov AG, McHenry CS, Lohman TM. J Mol Biol. 2013;425:4802–19. doi: 10.1016/j.jmb.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green NM. Biochem J. 1965;94:23C–24C. doi: 10.1042/bj0940023c. [DOI] [PubMed] [Google Scholar]
- 25.Roy R, Kozlov AG, Lohman TM, Ha T. J Mol Biol. 2007;369:1244–57. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatch K, Danilowicz C, Coljee V, Prentiss M. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78:011920. doi: 10.1103/PhysRevE.78.011920. [DOI] [PubMed] [Google Scholar]
- 27.Maffeo C, Ngo TT, Ha T, Aksimentiev A. J Chem Theory Comput. 2014;10:2891–2896. doi: 10.1021/ct500193u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo TT, Zhang Q, Zhou R, Yodh JG, Ha T. Cell. 2015;160:1135–44. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Rahil Z, Li IT, Chowdhury F, Leckband DE, Chemla YR, Ha T. Sci Rep. 2016;6:21584. doi: 10.1038/srep21584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S, Yin CT, Schenk DB, May PC, Altstiel LD, Bender MH, Boggs LN, Britton TC, Clemens JC, Czilli DL, Dieckman-McGinty DK, Droste JJ, Fuson KS, Gitter BD, Hyslop PA, Johnstone EM, Li WY, Little SP, Mabry TE, Miller FD, Audia JE. J Neurochem. 2001;76:173–81. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 32.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. EMBO Rep. 2002;3:688–94. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narui Y, Salaita K. Biophys J. 2013;105:2655–65. doi: 10.1016/j.bpj.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Nat Methods. 2012;9:64–7. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Ge C, Zhu C, Salaita K. Nat Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, Chen CS. Nat Methods. 2014;11:1229–32. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shergill B, Meloty-Kapella L, Musse AA, Weinmaster G, Botvinick E. Dev Cell. 2012;22:1313–20. doi: 10.1016/j.devcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Dev Cell. 2012;22:1299–312. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musse AA, Meloty-Kapella L, Weinmaster G. Semin Cell Dev Biol. 2012;23:429–36. doi: 10.1016/j.semcdb.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Curr Biol. 1998;8:1227–30. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 41.Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, Sheetz MP. Biophys J. 2006;91:3907–20. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Nature. 2003;424:334–7. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 43.Chan CE, Odde DJ. Science. 2008;322:1687–91. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 44.Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Cell. 2004;116:431–43. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 45.Burnette DT, Manley S, Sengupta P, Sougrat R, Davidson MW, Kachar B, Lippincott-Schwartz J. Nat Cell Biol. 2011;13:371–81. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.